1. Introduction
Astroviruses (AstVs) are non-enveloped (~28–30 nm in diameter), single-stranded, positive-sense RNA (6.2–7.8 kb) viruses belonging to family
Astroviridae [
1]. This family includes two genera,
Mamastrovirus and
Avastrovirus. The genus
Mamstrovirus infecting mammalian hosts includes 19 genotype species according to the 2010 classification of the International Committee on Taxonomy of Viruses (ICTV). The genus
Avastrovirus infects avian hosts. It has three genotype species. Fourteen newly identified AstV strains have since been proposed as genotype species of
Mamastrovirus [
2]. However, most newly discovered strains remain unclassified, including porcine astroviruses (PAstVs) [
3].
PAstVs belong to the genus
Mamastrovirus [
1]. They are enteric pathogens [
4]. PAstVs are divided into five distinct genotypes (PAstV 1–5) based on the open reading frame 2 (ORF2) region, which encodes the capsid protein [
4,
5,
6]. AstVs belonging to
Mamastrovirus are known to be gastroenteric viruses that cause diarrhea. However, since 2010, novel astroviral pathogens that infect the central nervous system (CNS) have been reported in humans and several animal species (mink, cattle, pigs, sheep, muskox, and alpaca) [
7]. Recently, some evidence supports the association of PAstV with cases of extraintestinal infections, including the CNS of pigs [
8,
9,
10,
11,
12]. In 2017, neuroinvasive PAstV (Ni-PAstV) 3 was detected in the CNS of pigs with encephalomyelitis in Hungary and the USA. Infected pigs exhibited clinical signs of astasia and knuckling, with a fatality rate of 75–100% [
13] and encephalomyelitis [
13,
14].
In South Korea, the prevalence of PAstV in domestic pigs was reported to be 20.1% and 9.2% in wild boars [
15,
16]. PAstV 4 was dominant (94.6%) in swine, with a few PAstV 2 present (5.4%) [
15,
16,
17]. However, previous studies in Korea have only analyzed fecal samples without considering disease symptoms. In this study, the presence of Ni-PAstV in South Korea was investigated using samples collected from domestic pigs showing neurological symptoms.
2. Materials and Methods
2.1. Sample collection, RNA extraction, and cDNA synthesis
Carcasses from five pigs with neurological symptoms from three farms were submitted to Jeonbuk National University Veterinary Diagnostic Center from August to September 2020 (case no. KOR/0983/2020, KOR/1096/2020, KOR/1295/2020, KOR/1006-1/2020 and KOR/1006-2/2020). Brain and intestinal tissue samples were collected from each pig. KOR/0983/2020, KOR/1096/2020 and the KOR/1295/2020 pigs were from farms in Jeongeup, Jeollabuk-do, Korea. KOR/1006-1/2020 and KOR/1006-2/2020 pigs were from a farm located in Hamyang, Gyeongsangnam-do, Korea. These tissues were stored at 80°C until further use.
For RNA extraction, tissue samples were washed twice with 5 mL of phosphate-buffered saline and homogenized in 1 mL of phosphate-buffered saline using TissueLyser II (Qiagen, Germany). Homogenized samples were then centrifuged at 13,000 rpm for 10 min. Total RNA was extracted from 200 µL of supernatant using a WizPrep™ Viral DNA/RNA Mini Kit (V2) (Wizbiosolutions, Seongnam, South Korea) according to the manufacturer’s instructions. Then 50 µL of cDNA was synthesized using a cDNA synthesis kit (Wizbiosolutions, South Korea) following the manufacturer’s instructions (42°C for 1 hr and 70°C for 10 min).
2.2. PCR screening and Sequencing
For detecting PAstV, partial RNA-dependent RNA polymerase (RdRp) gene of AstV was amplified using primer sets [
18,
19] with hemi-nested PCR. For the first-round PCR, AstV Pol F1 (5’-GARTTYGATTGGRCKCGKTAYGA-3’), AstV Pol F2 (5’-GARTTYGATTGGRCKAGGTAYGA-3’), and AstV Pol R1 (5’-GGYTTKACCCACATNCCRAA-3’) were used. For the second-round PCR, AstV Pol F3 (5’-CGKTAYGATGGKACKATHCC-3’), AstV Pol F4 (5’-AGGTAYGATGGKACKATHCC-3’), and AstV Pol R1 were used. PCR was performed under the following conditions: 94°C for 1 min, 40 cycles of 94°C for 30 sec, 50°C for 30 sec, and 68°C for 30 sec, and a final extension at 68°C for 5 min. PCR-amplified RdRp gene was cloned using a RBCT&A cloning kit (Real Biotech Corporation, Banqiao,Taiwan) and sequenced on a 3730xl DNA Analyzer platform (Applied Biosystems, Waltham, MA, USA). Additionally, multiplex RT-PCR was performed for PAstV typing [
20].
To amplify the complete capsid protein encoding ORF2 gene, RNA was extracted using TRIzol Reagent (Ambion, Austin, TX, USA) with a RNeasy Mini Kit (Qiagen, Hilden, Germany) and cDNA was amplified using AccuPower RocketScript RT PreMix. ORF2 genes were amplified using AstV ORF2 F (5’-CTSRATGGGAAACTCCT-3’) for forward primer, and the conserved stem–loop II-like motif (s2m) at the 3' end of PAstVs 1, 3, and 5 [
4]; AstV s2m R (5’-CCCTCGATCCTACTCGG-3’) for reverse primer, while PAstVs 2 and 4 were amplified using the 3' RACE method. PCR products were sequenced using barcode-tagged sequencing (Celemics, Seoul, South Korea).
2.3. Sequence analysis
Phylogenetic analyses were performed based on partial RdRp nucleotide sequences and capsid protein amino acid sequences. These sequences were edited using BioEdit v. 7.2.5 [
21] and aligned using Clustal Omega [
22]. Phylogenetic trees were constructed using the neighbor-joining method and the p-distance model with 1000 bootstrap replicates using the MEGA X program [
23]. Nucleotide and amino acid sequence identities were calculated using BioEdit with the sequence identity matrix method (p-distance).
2.4. Histopathology and in situ hybridization
To prepare formalin-fixed, paraffin-embedded (FFPE) blocks, brain tissues were fixed in 10% neutral-buffered formaldehyde. FFPE blocks were sectioned to 4 μm in thickness and used for hematoxylin and eosin (H&E) staining and in situ hybridization (ISH). H&E staining was conducted using a standard protocol [
24]. Histopathological examination was conducted at the Veterinary Diagnostic Center of Jeonbuk National University. For ISH, purified first PCR products of brain tissues were labeled using a PCR DIG Probe Synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s protocol. The second primer set (AstV Pol F3, AstV Pol F4, and AstV Pol R1) and conditions of the hemi-nested PCR targeting the partial RdRp gene were used in amplification steps for labeling probes. FFPE tissue slides were used for ISH, which was performed using a fully automated system (NexES IHC instrument; Ventana Medical Systems, Inc., Tucson, AZ, USA) and 3, 3′-diaminobenzidine (DAB) detection system (OptiView DAB IHC Detection kit, Ventana Medical Systems). They were then counterstained with hematoxylin.
2.5. Other laboratory diagnostics
Samples were tested for the presence of nine viral pathogens (Aujeszky’s disease virus (ADV), classical swine fever virus (CSFV), porcine circovirus type 2 virus (PCV2), porcine epidemic diarrhea virus (PEDV), porcine reproductive and respiratory syndrome virus (PRRSV), Japanese encephalitis virus (JEV), swine influenza virus (SIV), tick-borne encephalitis virus (TBEV) and transmissible gastroenteritis virus (TGEV)) and six bacterial pathogens (Brachyspira hyodysenteriae (swine dysentery), Escherichia coli (E. coli), Haemophilus parasuis (Glasser’s disease), Lawsonia intracellularis, Listeria spp., and Salmonella spp.), including those causing neurological diseases, using diagnostic manuals of the Jeonbuk National University Veterinary Diagnostic Center. Bacterial cultivation was conducted to identify additional infectious pathogens.
3. Results
3.1. Detection of porcine astorovirus
The 354–392 bp fragments of the partial RdRp genes of PAstV were successfully amplified using a heminested PCR from all ten tissue samples of each pig. Subsequent type-specific RT-PCR analysis revealed the presence of PAstV genotypes 1, 2, 4, and 5 with multiple genotypes coexisting within a single pig (
Table 1). Cloned partial RdRp sequences from single colony were sequenced and 131 sequences were obtained with all 5 genotypes. Additionally, 15 complete capsid protein encoded ORF2 gene sequences, ranging from 2231 to 2586 nucleotides of all 5 genotypes, were obtained.
3.2. Phylogenetic tree analysis
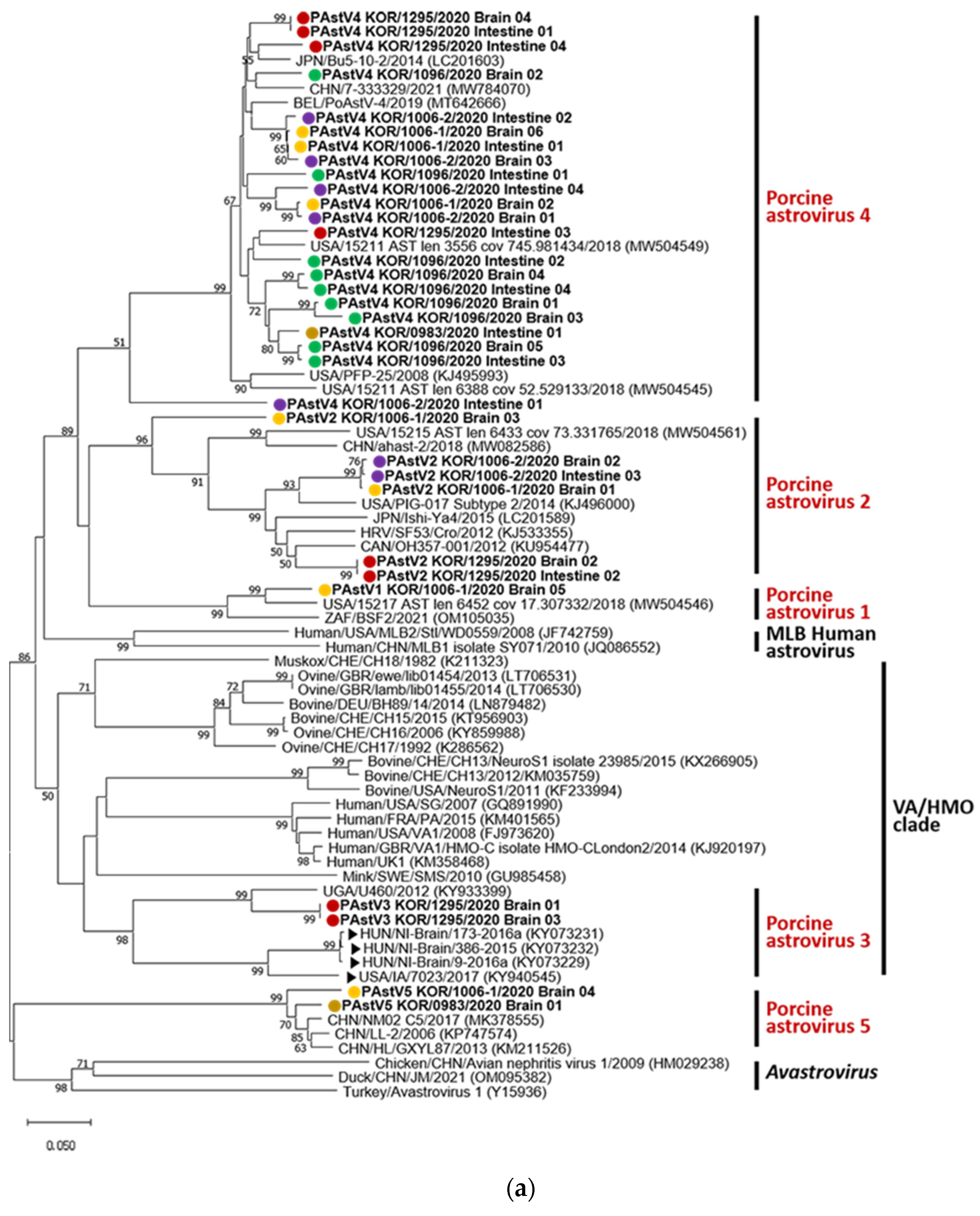
A phylogenetic tree (
Figure 1a) was constructed with PAstV genotypes 1–5,
Avastroviruses 1–3, and reported Ni-PAstV and AstV sequences using partial RdRp gene sequences. Using phylogenetic tree analysis and comparison of nucleotide sequence identity, 98 identical sequences were excluded and 33 sequences were used for phylogenetic tree analysis. The phylogenetic tree was divided into seven clades: PAstV genotypes 1, 2, 4, and 5, Melbourne (MLB) human astrovirus, Virginia and human-mink-ovine-like (VA/HMO) clade with PAstV 3, and
Avastrovirus. All PAstV genotypes (1–5) were detected in brain samples of Korean pigs. Multiple genotype PAstV infection was observed. Korean PAstVs sequences showed similarities ranging from 76.3% to 99.4% among six sequences in PAstV 2, from 74.0% to 99.4% among 22 sequences in PAstV 4, and 92.3% between 2 sequences in PAstV 5. PAstV3 exhibited 67.9%–69.2% sequence identities with Ni-PAstV3 reported in the USA (USA/IA/7023/2017) and Hungary (Ni-Brain/HUN) in 2017 based on BLASTn.
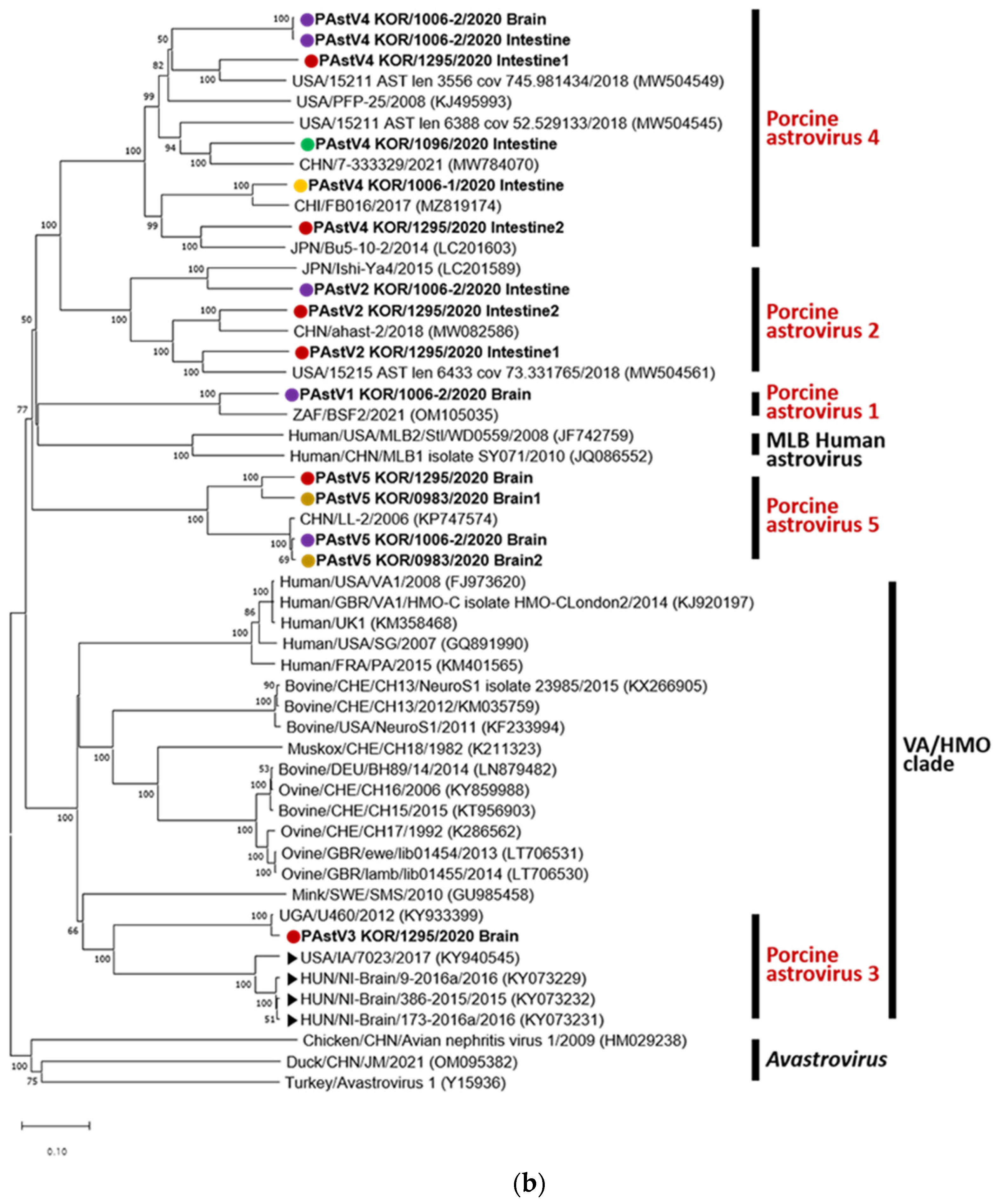
Similarly, all PAstV genotypes and multiple genotypes in a single Korean pig were identified in the phylogenetic tree of capsid protein amino acid sequences (
Figure 1b). PAstVs 1, 4, and 5 and PAstVs 3 and 5 were detected in brain samples of KOR/1006-2/2020 and KOR/1295/2020, respectively. The highest amino acid similarity in each sequence of BLASTp was observed as follows: 80.87% (OM105035.1; South Africa) in PAstV 1 KOR/1006-2/2020 Brain, 77.63% to 82.67% in PAstV 2 (OP413948.1 China with PAstV 2 KOR/1295/2020 Intestine1 and OP413947.1 China with PAstV 2 KOR/1006-2/2020 Intestine), 98.18% (KY933399.1; Uganda) in PAstV 3 KOR/1295/2020 brain, 72.01% to 93.68% in PAstV 4 (KX060809.1 China with PAstV4 KOR/1006-2/2020/Brain and OP413951.1 China with PAstV 4 KOR/1006-1/2020 Intestine), and 89.20% to 98.91% in PAstV 5 (MW504545.1; USA with PAstV 5 KOR/1295/2020 Brain and KP747574.1 China with PAstV 5 KOR/0983/2020 Brain2). PAstV 3 KOR/1295/2020 brain sequence showed 58.7%–59.2% and 59.1%–60.4% amino acid identities with Ni-PAstV 3 reported in the USA and Hungary, respectively.
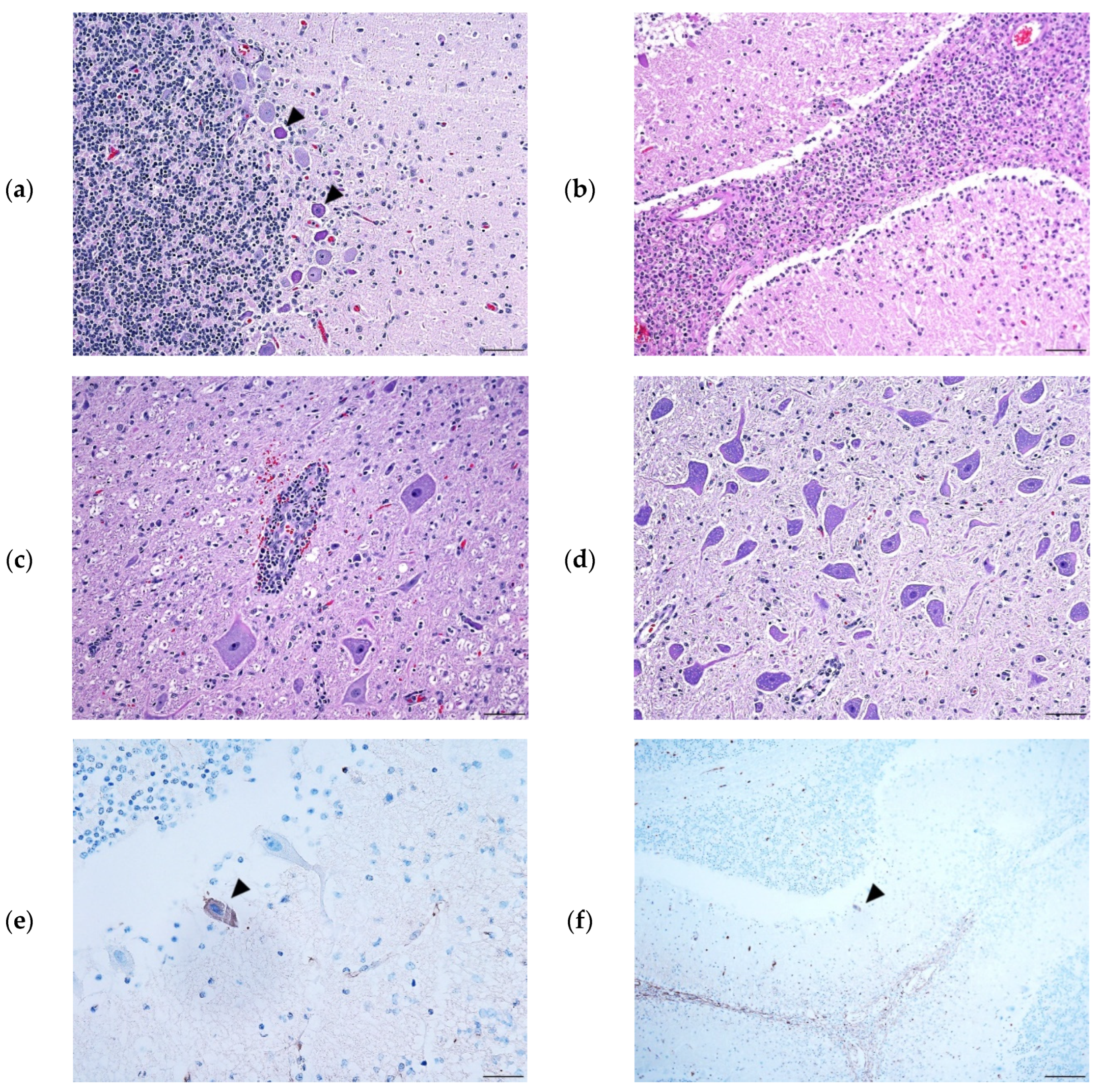
3.3. Histopathology and in situ hybridization
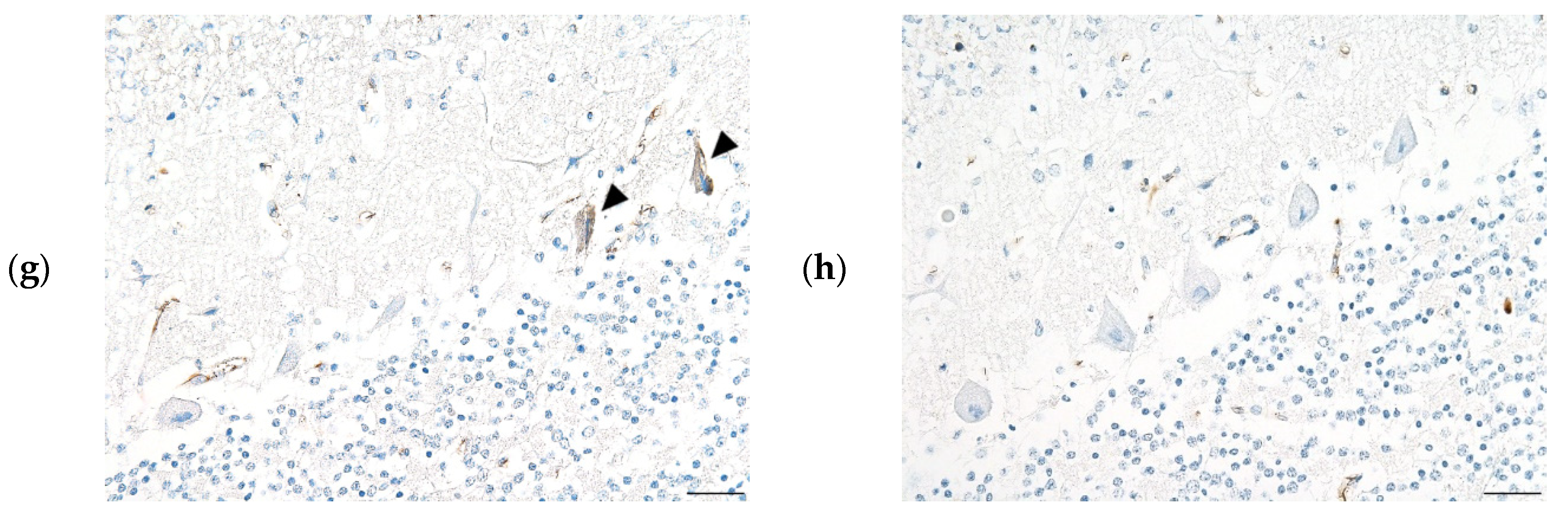
CNS histopathology revealed meningitis in both the brain and spinal cord (
Figure 2). Perivascular cuffing and neuronal necrosis (in the brain, Purkinje cells) with degeneration and myelitis were detected. The KOR/1096/2020 cerebrum showed mild meningitis, edema, and perivascular inflammation. ISH was performed and PAstV-infected Purkinje cells in the cerebellar cortex were stained dark brown (
Figure 2C–E).
3.4. Other laboratory diagnostics
The Veterinary Diagnostic Center of Jeonbuk National University performed PCR for viral and bacterial pathogens and bacterial cultivation. Lung sample from the KOR/0983/2020 pig was PCR-positive for PRRSV. Streptococcus suis was isolated from brain tissue. An intestinal sample of the KOR/1006-1/2020 pig was PCR-positive for E. coli STB. The lung sample of KOR/1295/2020 pig was PCR-positive for PRRSV. Streptococcus suis and Clostridium perfringens type A were isolated from the brain and intestine, respectively. KOR/1006-2/2020 and KOR/1096/2020 pigs were negative for tested other virus and bacterial pathogens by PCR and cultivation.
4. Discussion
In summary, five PAstV-positive pigs with neurological symptoms were identified in this study. Type specific RT-PCR detected PAstV genotypes 1, 2, 4, and 5. Four pigs were positive for two or more PAstV genotypes. Phylogenetic analysis of the RdRp gene nucleotide sequences and ORF2 gene amino acid sequences revealed that all PAstV genotypes 1–5 were present in brain samples. Histopathological examination revealed meningitis, perivascular cuffing, Purkinje cell necrosis, and CNS degeneration. In addition, ISH results revealed that PAstV infected neurons in brain tissues.
Genotypes of each sample were not identical in three experimental results (
Table 1). However, coinfection of multiple genotypes was detected. When the amplified RdRp gene was directly sequenced using the Sanger sequencing method, the analysis was unsuccessful except for two samples. The possibility of co-infection was confirmed through type-specific PCR results. Subsequently, TA cloning with RdRp gene was performed. Analysis of single colonies revealed the presence of various types of PAstVs. Due to high genetic diversity of PAstV, some type-specific primers might have not functioned properly. Additionally, in the case of RdRp gene sequencing, the limited number of sequencing attempts during colony sequence might have resulted in an incomplete collection of all sequences in amplicon. For the ORF2 gene sequence, PAstVs 1, 3, and 5 were sequenced using the s2m motif at the 3' end, while PAstVd 2 and 4 were amplified using the 3' RACE method, suggesting that results were obtained primarily from intestine samples that presumed to have relatively high viral concentrations.
In molecular analysis, obtained sequences showed high genetic diversity. In the partial RdRp gene, sequences showed a minimum nucleotide sequence identity of 74.0% within a single genotype. PAstV3 exhibited 67.9%–69.2% identities with Ni-PAstV3 reported in the USA and Hungary in 2017 based on BLASTn. The minimum amino acid sequences similarities of PAstVs 1, 2, 3, 4, and 5 among the highest amino acid similarity in BLASTp of each sequence were 80.87%, 77.63%, 98.18%, 72.01% and 89.20%, respectively. PAstV 3 KOR/1295/2020 brain sequence showed 58.7%–59.2% and 59.1%–60.4% amino acid identity with Ni-PAstV 3 reported in the USA and Hungary, respectively. These low sequence similarities could be interpreted as occurrence of various mutation events. However, the lack of analysis regarding the origin, routes of introduction into the country, and timing of introduction raises the possibility of an inaccurate interpretation. Additionally, limited research on PAstV with scarcity of sequence comparison groups might have contributed to these findings. Further detailed analysis should be conducted through subsequent studies employing methods such as evolutionary or recombination analysis.
The ISH examining the presence of PAstV RNA in neurons by hybridization of PAstV RNA and synthesized probes revealed PAstV infection in the CNS. However, there is a limitation in determining the genotype of infected PAstV confirmed through ISH due to its probe.
5. Conclusions
Recent studies have reported Ni-PAstV 3 causing encephalomyelitis in several pigs [
25]. In this study, all PAstV genotypes 1–5 were detected in brain samples. Coexistence of multiple genotypes was found. These results suggest the possibility of infection by multiple PAstV genotypes in the CNS. Additional analysis of full gene sequence and type-specific ISH and/or
in vivo experimental infection may help us further elucidate the pan-infection of PAstVs in the CNS.
Author Contributions
Conceptualization, J.S.P. and S.Y.L; Methodology, J.S.P.; Software, J.S.P.; Validation, J.S.P., C.G.J. and S.B.C.; Formal analysis, J.S.P.; Investigation, J.S.P.; Resources, J.K.O.; Data curation, J.S.P.; Writing—original draft preparation, J.S.P.; Writing—review and editing, J.S.P., C.G.J., S.B.C., S.Y.L. and J.K.O.; Visualization, J.S.P.; Supervision, J.K.O.; Project administration, C.G.J.; Funding acquisition, J.K.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant (2019R1A6A1A03033084) of the Basic Science Research Program of the National Research Foundation (NRF) funded by the Ministry of Education, Republic of Korea. This research was also funded by a grant (grant number Z-1543069-2023-25-01) of the Animal and Plant Quarantine Agency, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in NCBI database by accession no. OP643769.1 to OP643782.1.
Acknowledgments
We thank the Jeonbuk National University Animal Diagnostic Center for help with diagnostics and the Department of Animal and Plant Health Research of the Animal and Plant Quarantine Agency for technical assistance with ISH. We also thank Myun-sik Yang and Byungkwan Oh for their assistance with histopathological analysis.
Conflicts of Interest
The authors have no conflicts of interest relevant to this study to disclose.
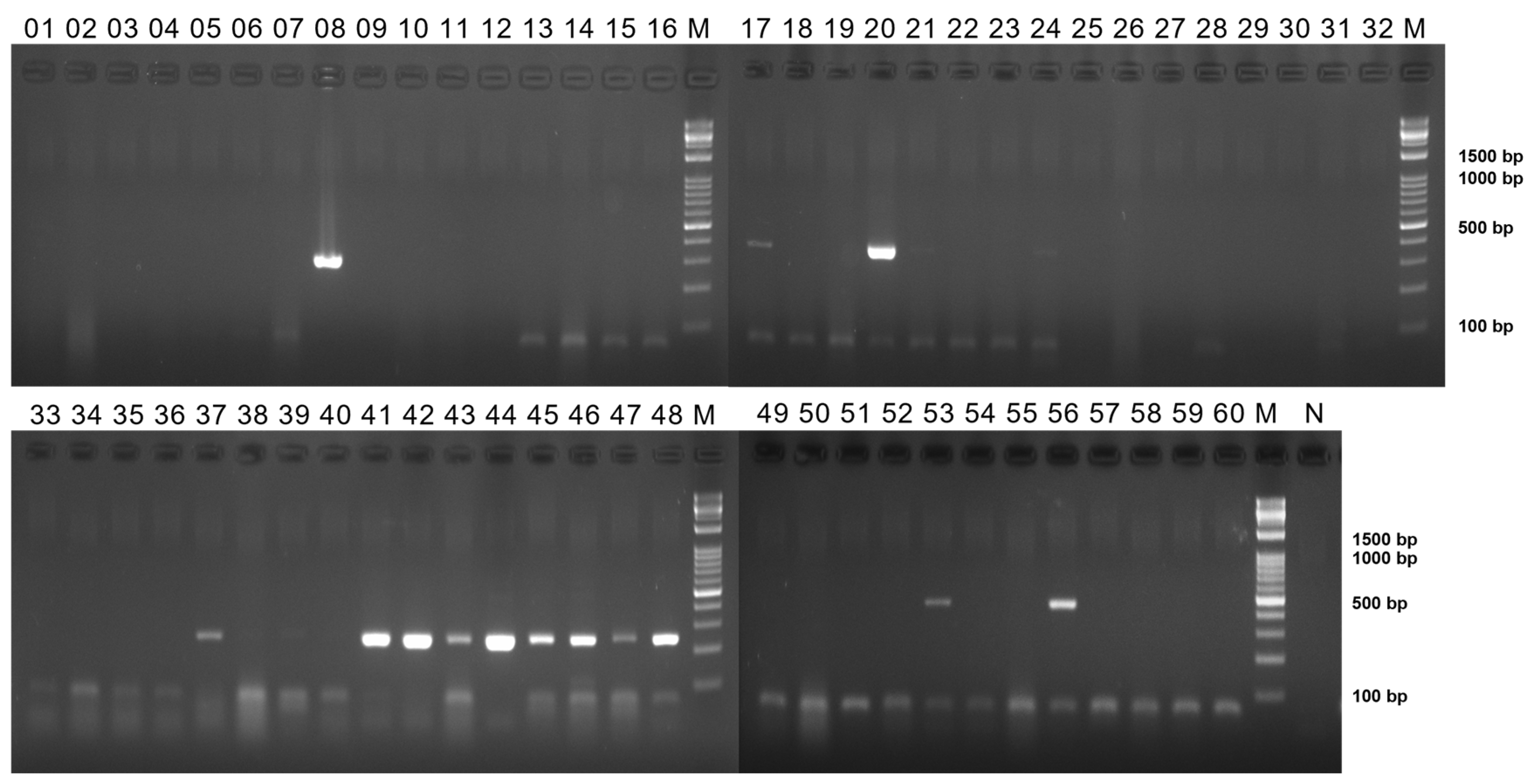
Appendix A
Figure A1.
13–24: PAstV 2, 25–36; PAstV 3, 37–48; PAstV 4, 49–60; PAstV 5. Lanes 1=KOR/0983/2020 brain1, 2=KOR/0983/2020 intestine1, 3= KOR/0983/2020 brain2, 4= KOR/0983/2020 intestine2, 5=KOR/1006-1/2020 brain, 6=KOR/1006-1/2020 intestine, 7=KOR/1006-2/2020 brain, 8=KOR/1006-2/2020 intestine, 9=KOR/1096/2020 brain, 10=KOR/1096/2020 intestine, 11=KOR/1295/2020 brain, 12=KOR/1295/2020 intestine; same combination with each genotype. Lanes M=marker, N=negative control.
Figure A1.
13–24: PAstV 2, 25–36; PAstV 3, 37–48; PAstV 4, 49–60; PAstV 5. Lanes 1=KOR/0983/2020 brain1, 2=KOR/0983/2020 intestine1, 3= KOR/0983/2020 brain2, 4= KOR/0983/2020 intestine2, 5=KOR/1006-1/2020 brain, 6=KOR/1006-1/2020 intestine, 7=KOR/1006-2/2020 brain, 8=KOR/1006-2/2020 intestine, 9=KOR/1096/2020 brain, 10=KOR/1096/2020 intestine, 11=KOR/1295/2020 brain, 12=KOR/1295/2020 intestine; same combination with each genotype. Lanes M=marker, N=negative control.
References
- Méndez, E.; Murillo, A.; Velázquez, R.; Burnham, A.; Arias, C.F. in Astrovirus Research 19-45 (Springer, 2012).
- Guix, S.; Bosch, A.; Pintó, R.M. in Astrovirus research 97-118 (Springer, 2012).
- Janowski, A.B. Beyond the Gastrointestinal Tract: The Emerging and Diverse Tissue Tropisms of Astroviruses. Viruses 2021, 5, 732. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; et al. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. Journal of General Virology 2013, 94, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Laurin, M.-A.; Dastor, M.; L’Homme, Y. Detection and genetic characterization of a novel pig astrovirus: relationship to other astroviruses. Archives of virology 2011, 156, 2095. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; et al. Multiple novel and prevalent astroviruses in pigs. Veterinary microbiology 2011, 149, 316–323. [Google Scholar] [CrossRef]
- Wildi, N.; Seuberlich, T. Neurotropic Astroviruses in Animals. Viruses 2021, 13, 1201. [Google Scholar] [CrossRef]
- Indik, S.; Valícek, L.; Smíd, B.; Dvoráková, H.; Rodák, L. Isolation and partial characterization of a novel porcine astrovirus. Vet Microbiol 2006, 117, 276–283. [Google Scholar] [CrossRef]
- Opriessnig, T.; Xiao, C.T.; Halbur, P.G. Porcine Astrovirus Type 5-Associated Enteritis in Pigs. J Comp Pathol 2020, 181, 38–46. [Google Scholar] [CrossRef]
- Padmanabhan, A.; Hause, B.M. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Archives of Virology 2016, 161, 2575–2579. [Google Scholar] [CrossRef]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Complete genome sequence of a newly identified porcine astrovirus genotype 3 strain US-MO123. J Virol 2012, 86, 13126. [Google Scholar] [CrossRef]
- Blomström, A.-L.; Ley, C.; Jacobson, M. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta veterinaria scandinavica 2014, 56, 1–6. [Google Scholar] [CrossRef]
- Bailey Arruda, P.A.; et al. Porcine astrovirus type 3 in central nervous system of swine with polioencephalomyelitis. Emerging infectious diseases 2017, 23, 2097. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; et al. Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, Hungary. Emerging infectious diseases 2017, 23, 1982. [Google Scholar] [CrossRef]
- Lee, M.H.; et al. Phylogenetic analysis of porcine astrovirus in domestic pigs and wild boars in South Korea. Virus Genes 2013, 46, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Choe, S.; Hyun, B.H.; An, D.J. Phylogenetic analysis of kobuviruses and astroviruses from Korean wild boars: 2016-2018. Arch Virol 2021, 166, 2591–2596. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jang, G.; Lee, C. Complete genome sequence of a porcine astrovirus from South Korea. Arch Virol 2015, 160, 1819–1821. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Poon, L.; Guan, Y.; Peiris, J. Novel astroviruses in insectivorous bats. Journal of virology 2008, 82, 9107–9114. [Google Scholar] [CrossRef]
- Lee, S.-Y.; et al. Genetic diversity and phylogenetic analysis of newly discovered bat astroviruses in Korea. Archives of virology 2018, 163, 3065–3072. [Google Scholar] [CrossRef]
- Zhang, Q.; et al. Multiplex gel-based PCR assay for the simultaneous detection of 5 genotypes of porcine astroviruses. Journal of Veterinary Diagnostic Investigation 2023, 35, 132–138. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: an important software for molecular biology. GERF Bull Biosci 2011, 2, 60–61. [Google Scholar]
- Sievers, F.; Higgins, D.G. Clustal omega. Current protocols in bioinformatics 2014, 48, 3–13. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Hematoxylin and eosin staining of tissue and cell sections. Cold spring harbor protocols 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, F.M.; et al. Experimental porcine astrovirus type 3-associated polioencephalomyelitis in swine. Veterinary Pathology 2021, 58, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).