Submitted:
01 June 2023
Posted:
02 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
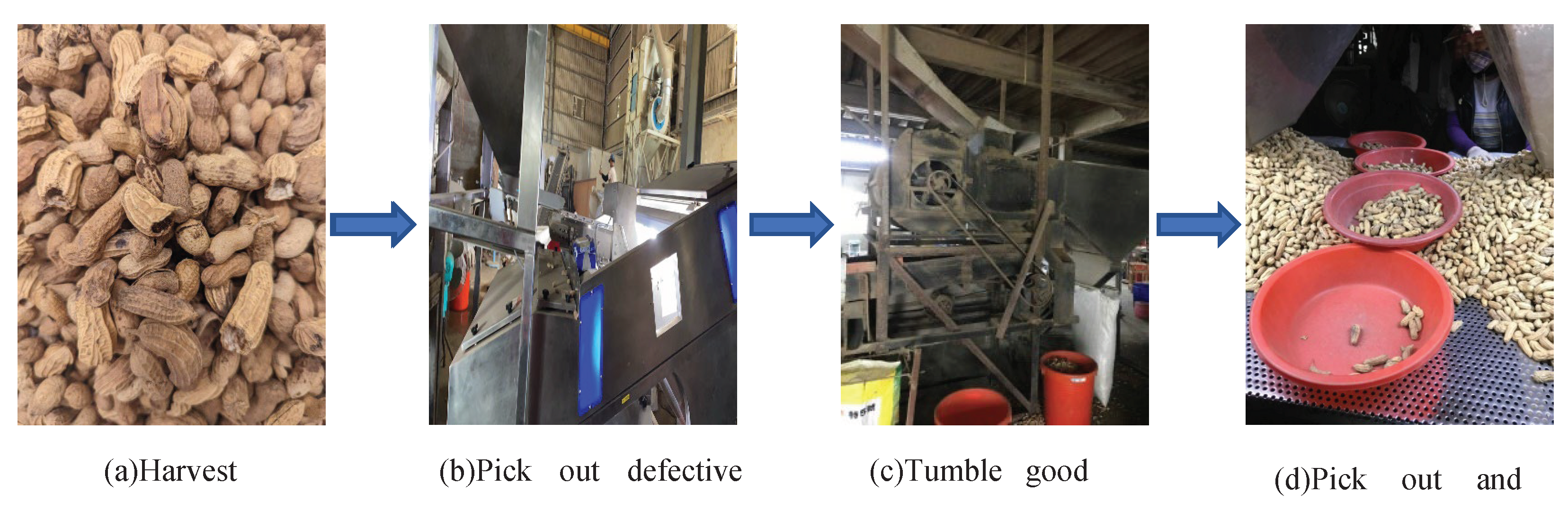
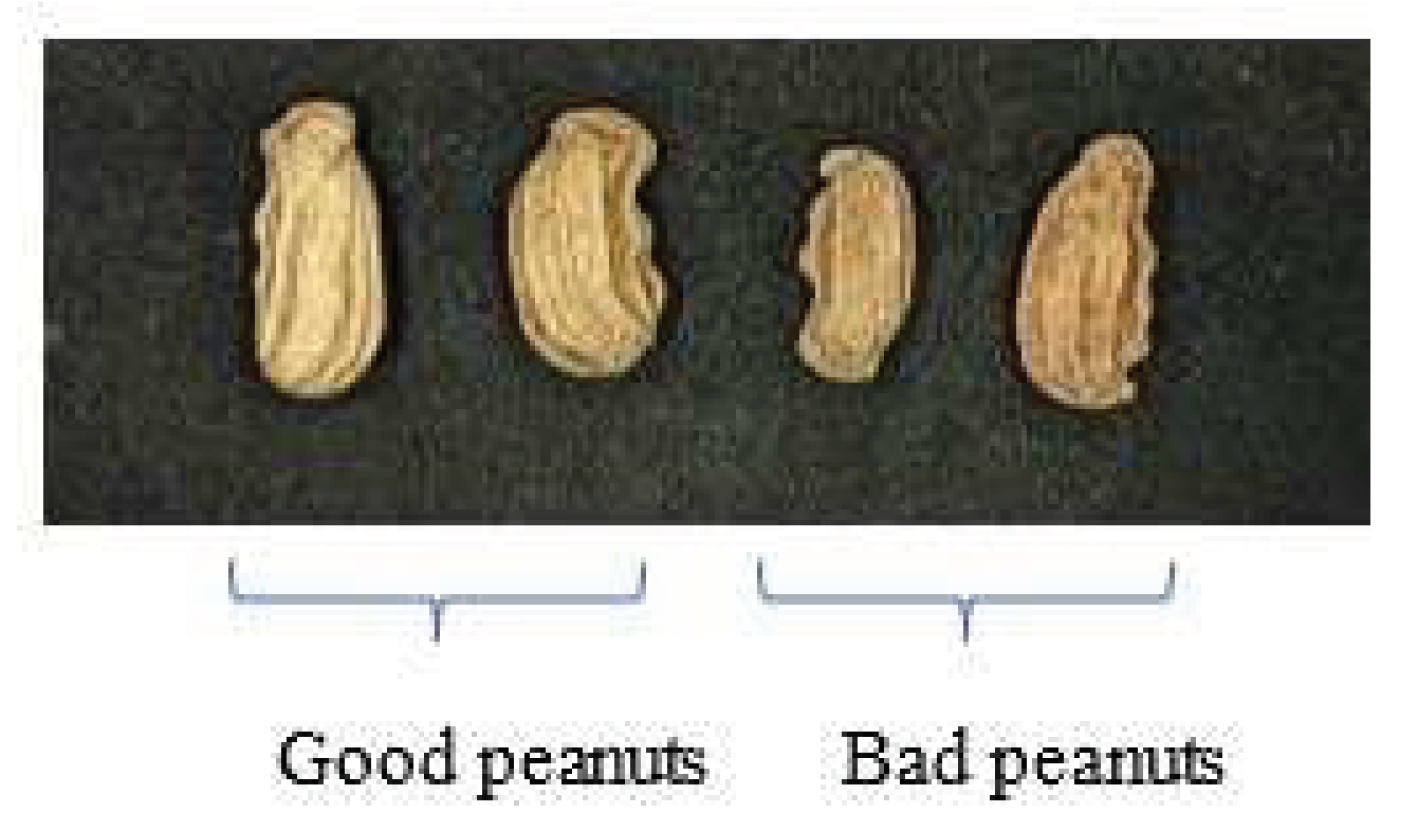
2.1. Peanut samples
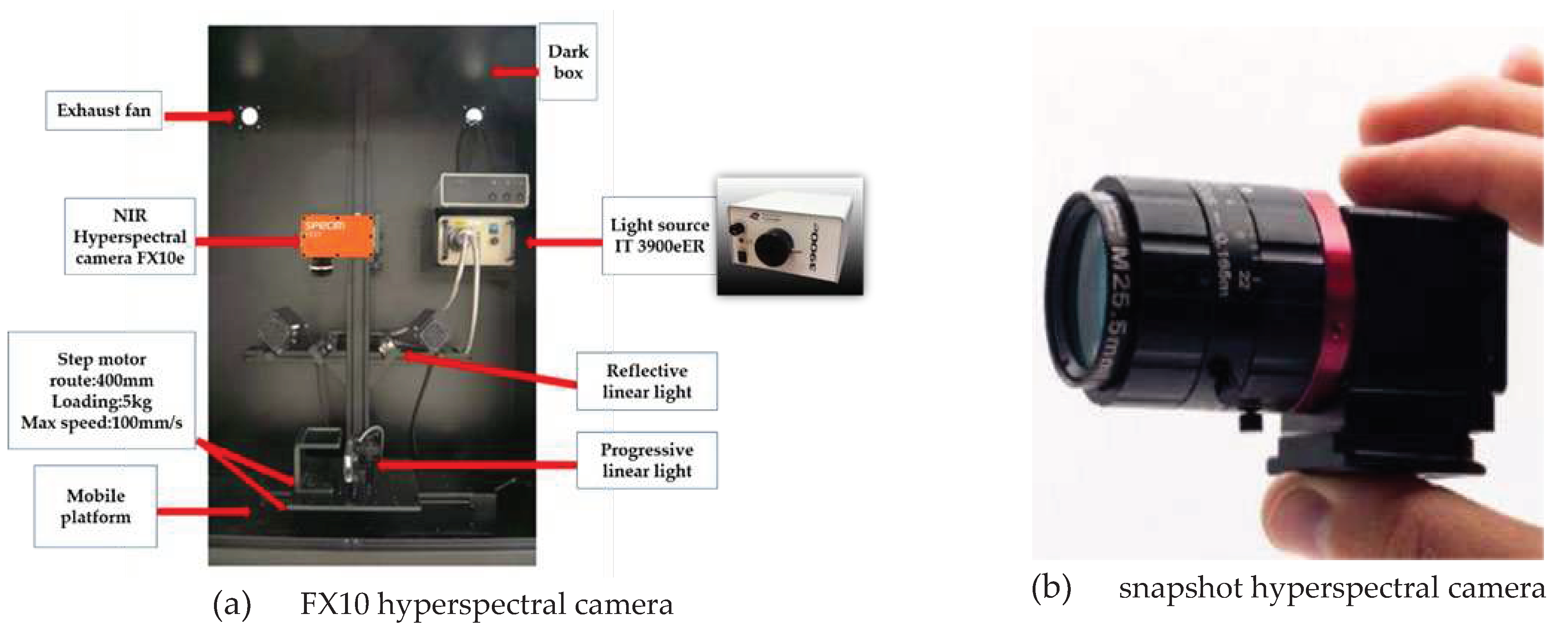
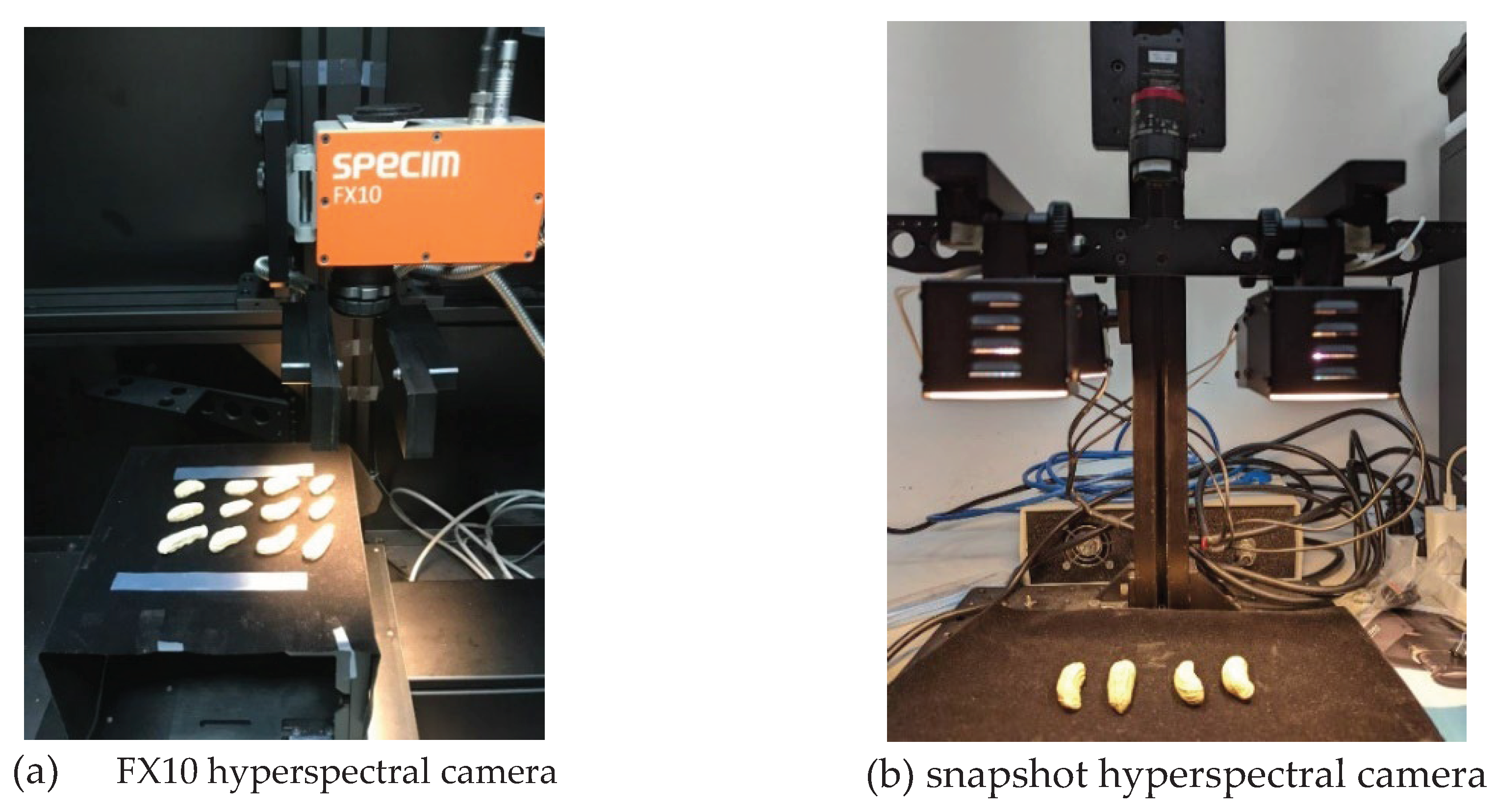
2.2. Hyperspectral imaging system
2.3. Principal Component Analysis (PCA)
2.4. Preprocessing
2.5. Convolutional Neural Network (CNN)
2.5.1. 1D-CNN
2.5.1.1. 1D-CNN model
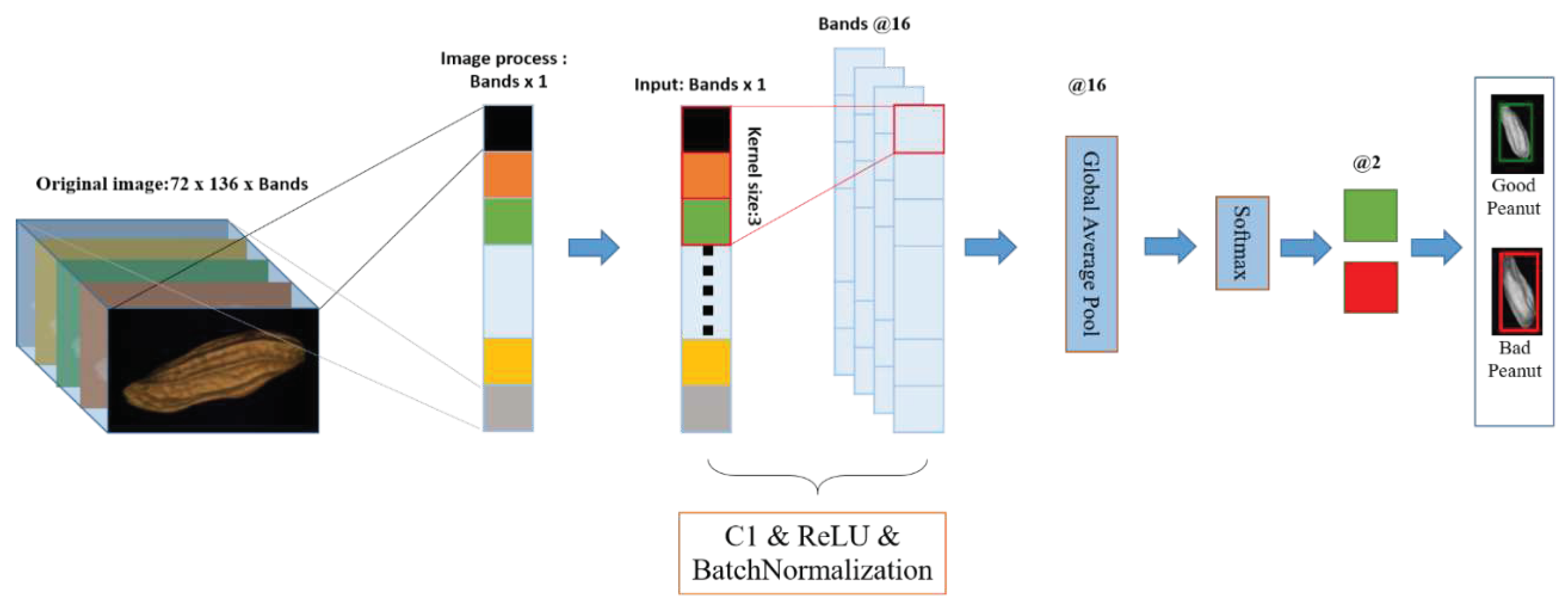
2.5.1.2. Realtime-1DCNN
2.5.2. 2D-CNN
2.5.2.1. 2D-CNN model
2.5.2.2. Realtime-2DCNN
2.5.3. 3D-CNN
2.5.3.1. 3D-CNN model
2.5.3.2. Realtime-3DCNN
2.6. Flow chart
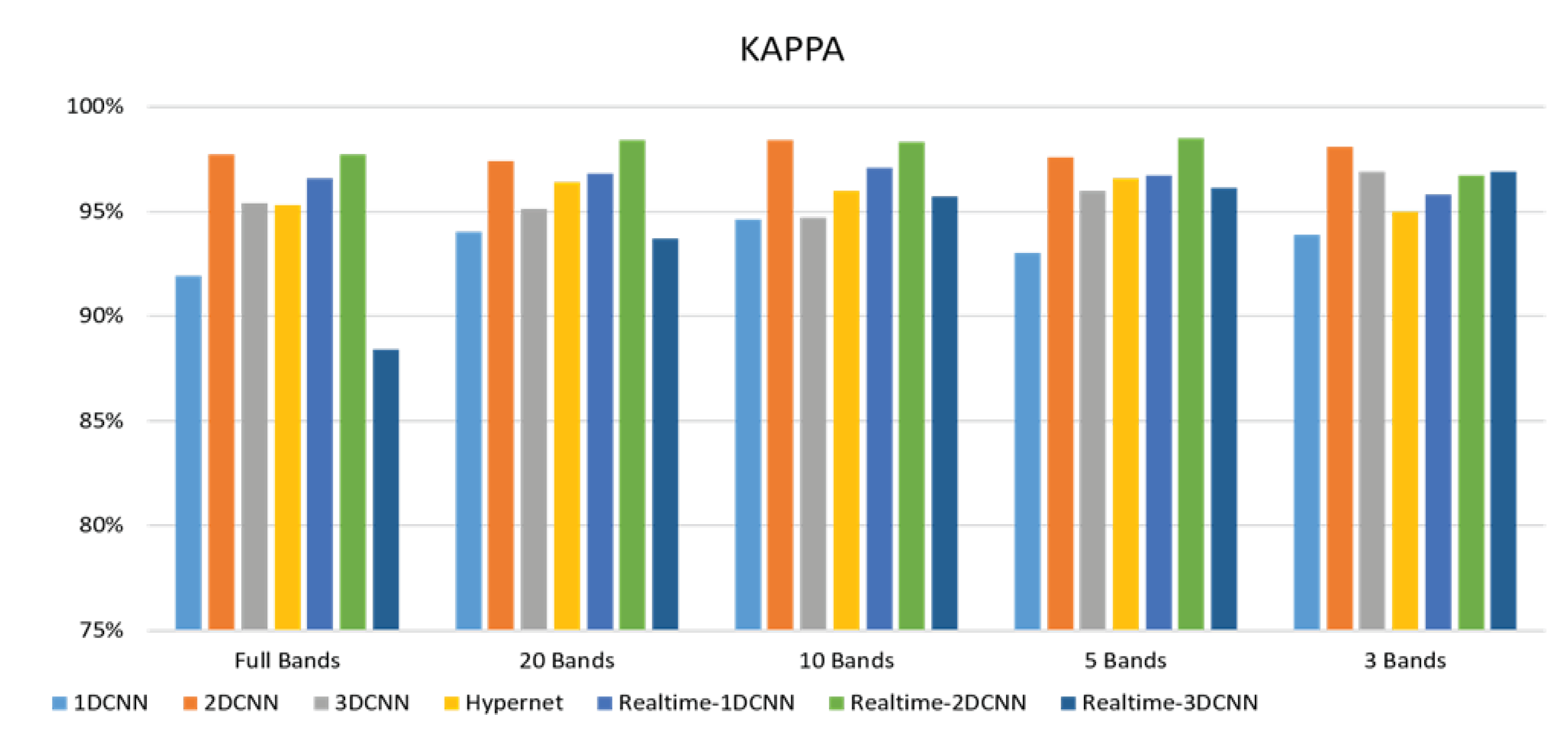
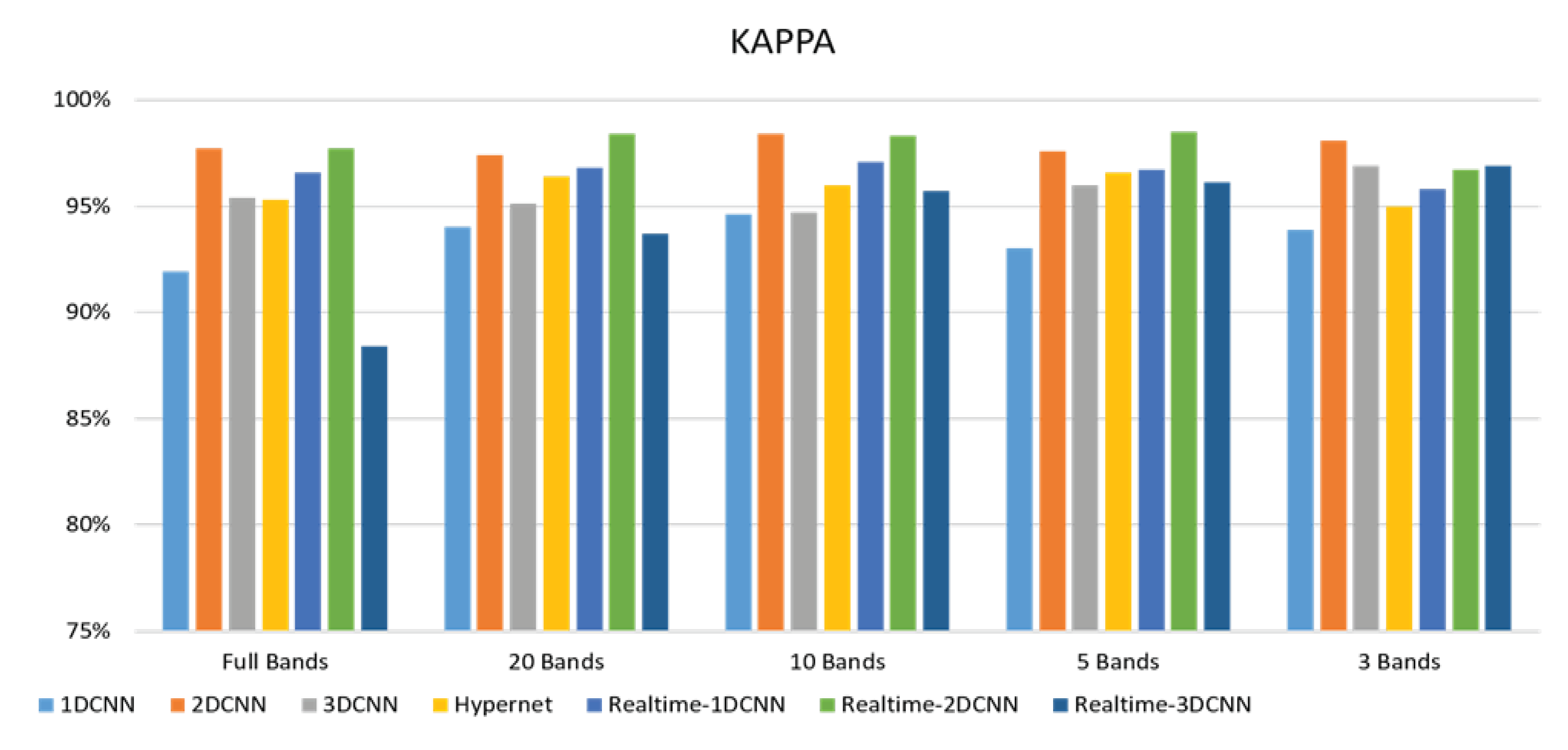
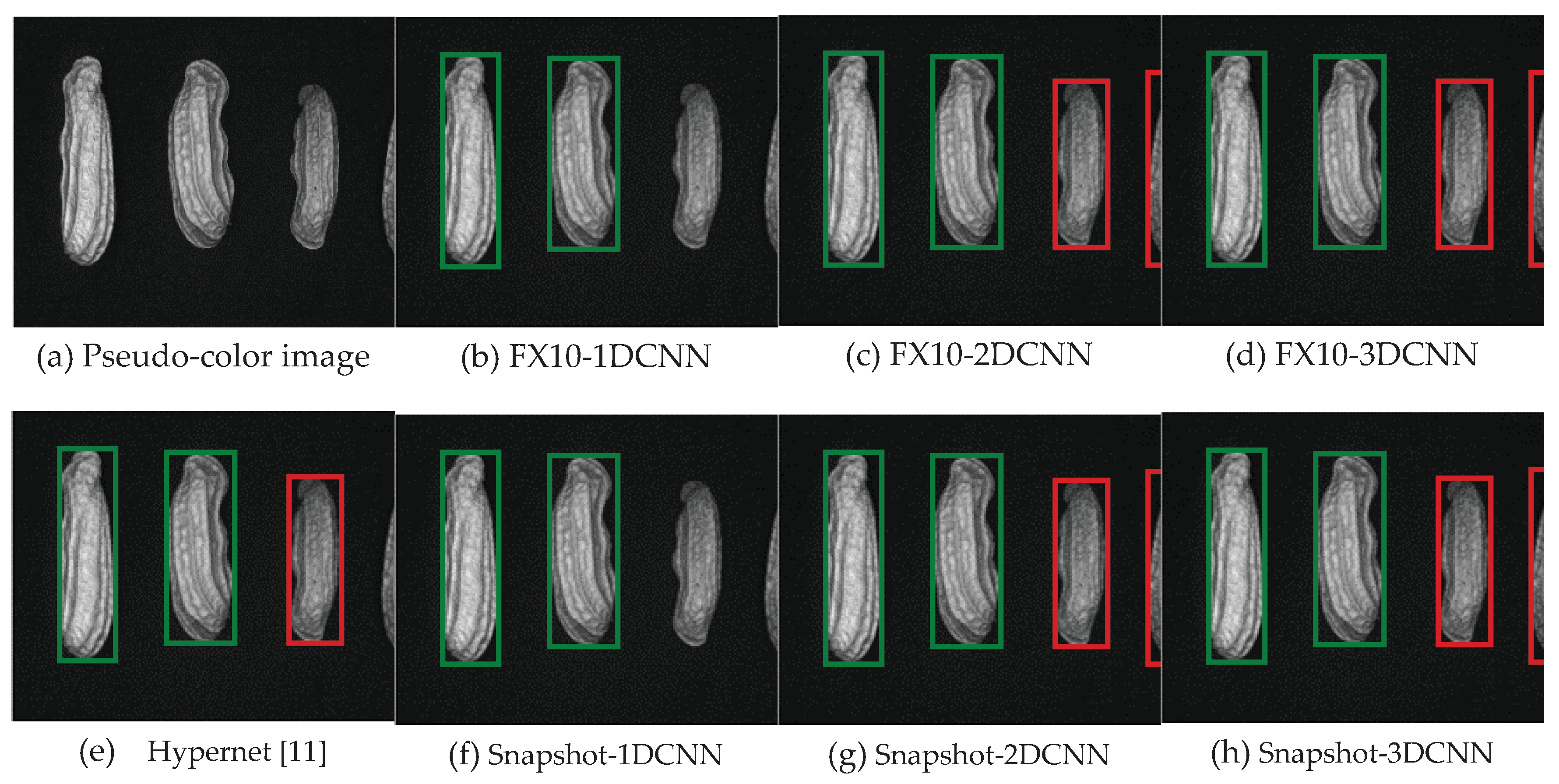
3. Results
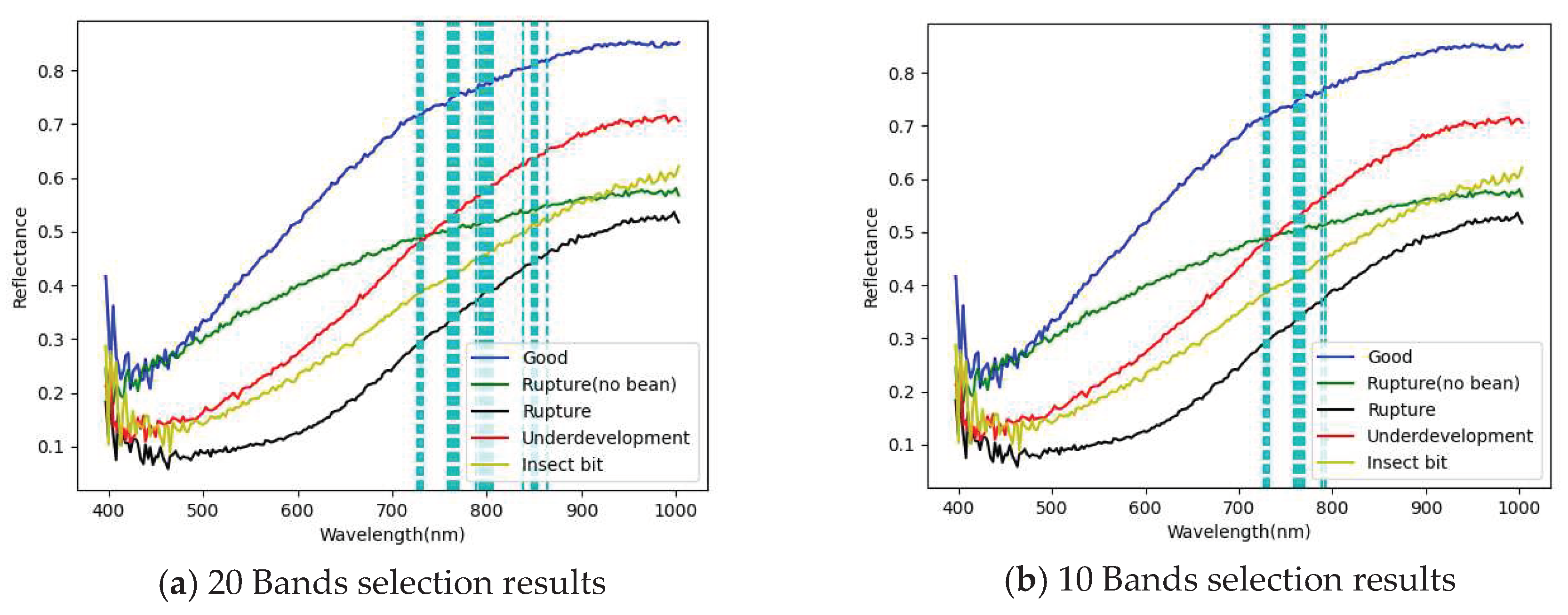
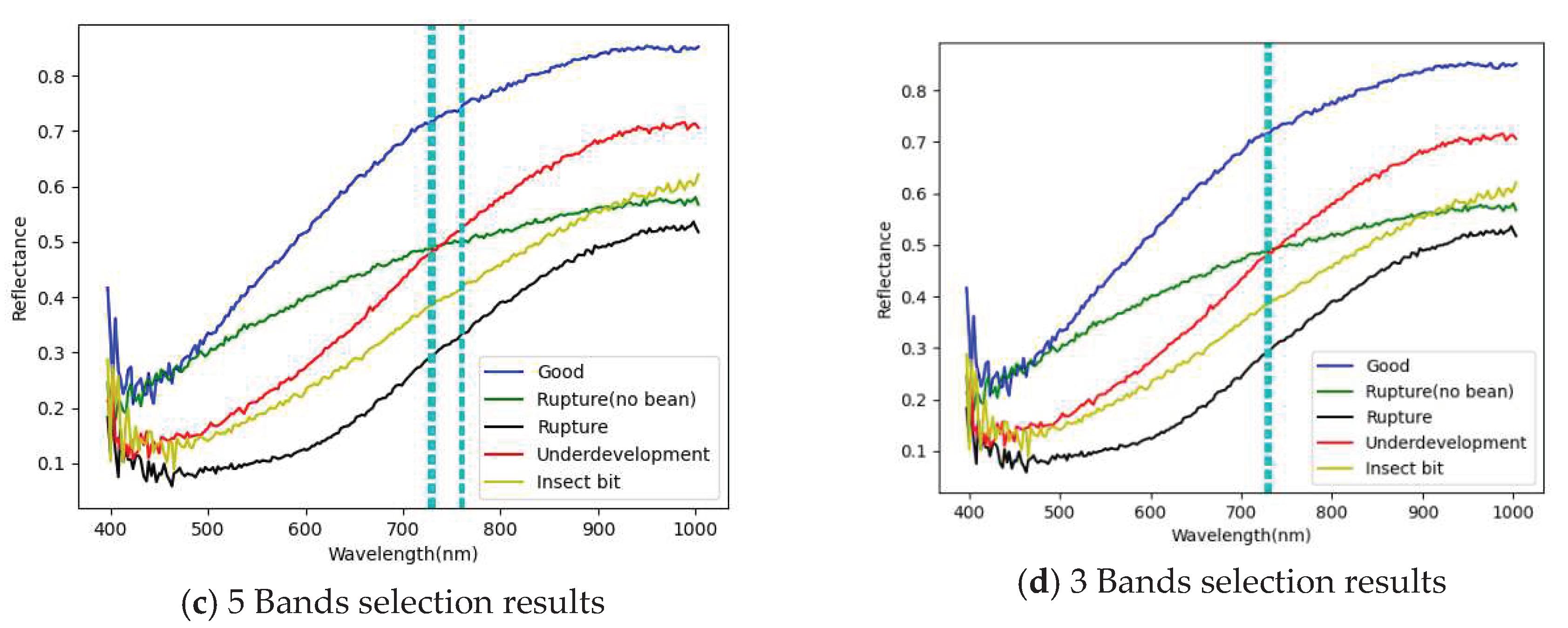
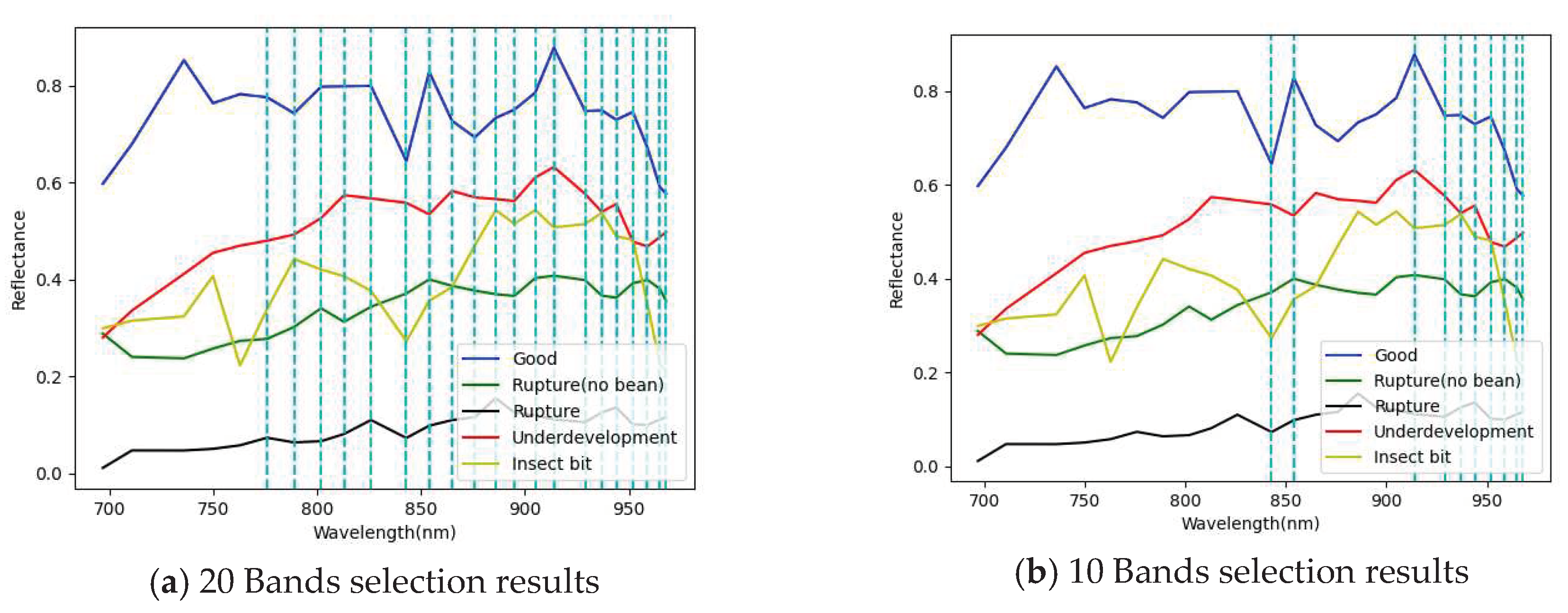
3.1. Band selection results
3.1.1. FX10 hyperspectral image band selection results
3.1.2. Snapshot band selection results
3.2. Classification results flow
3.2.1. FX10 data

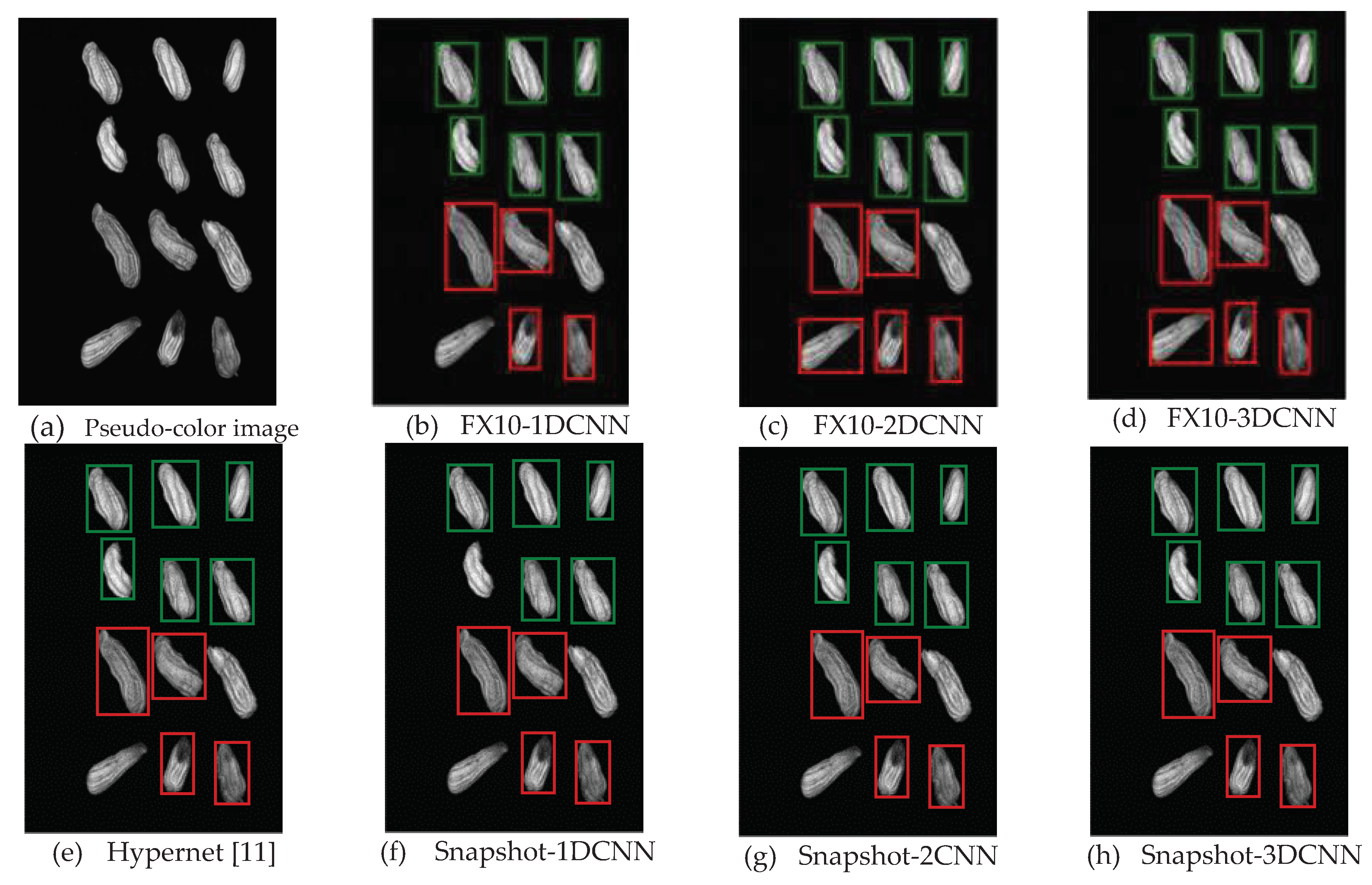
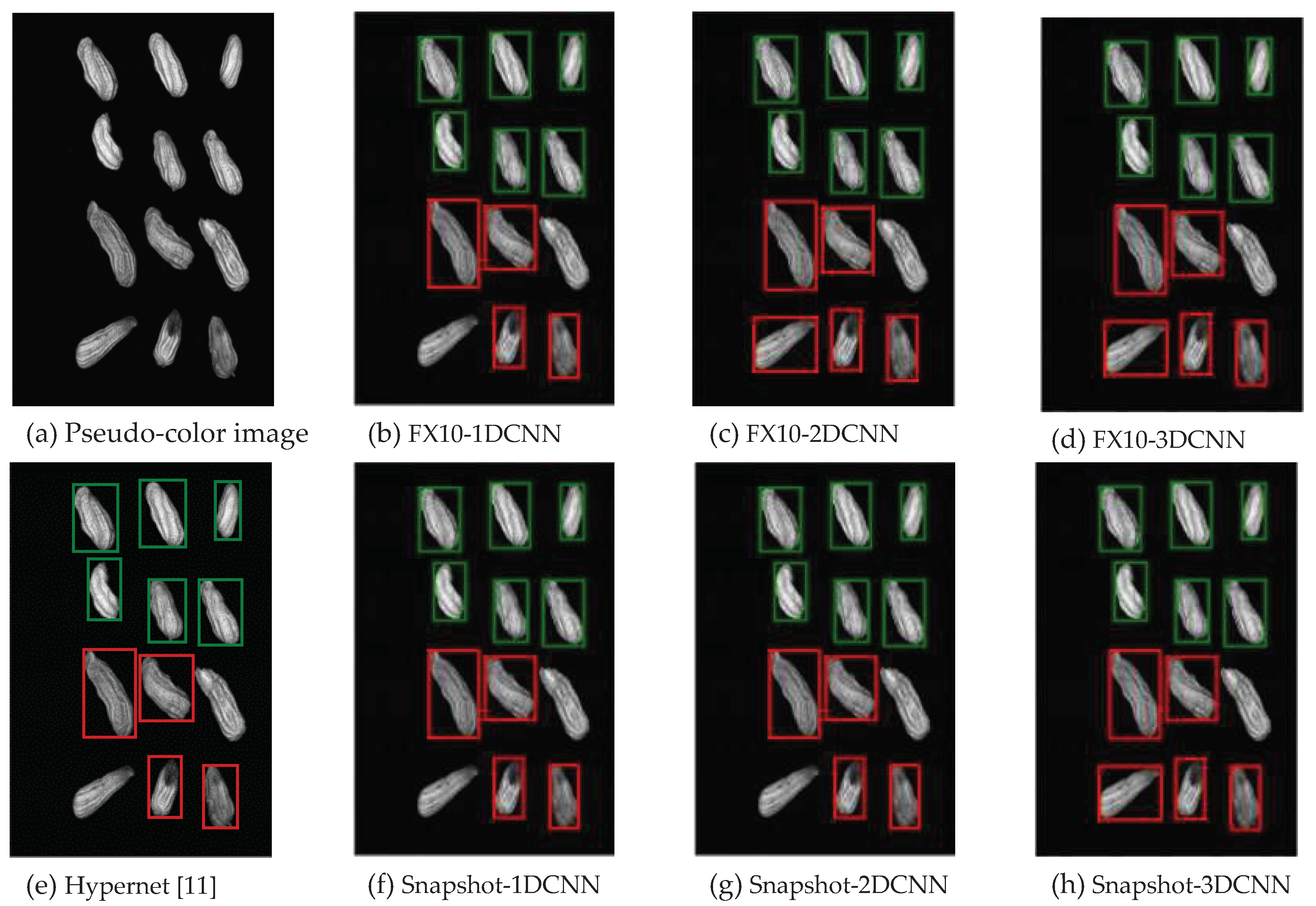
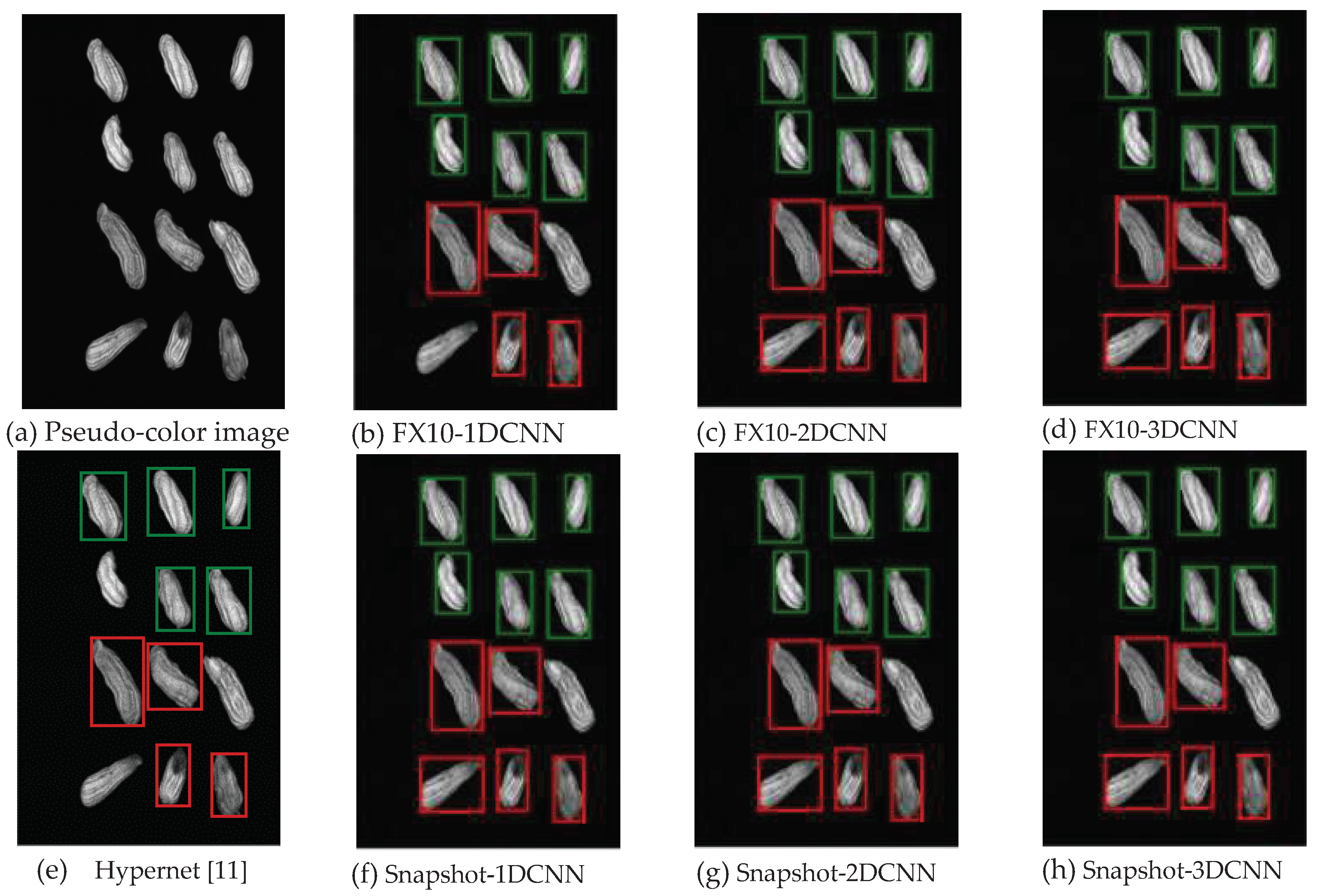
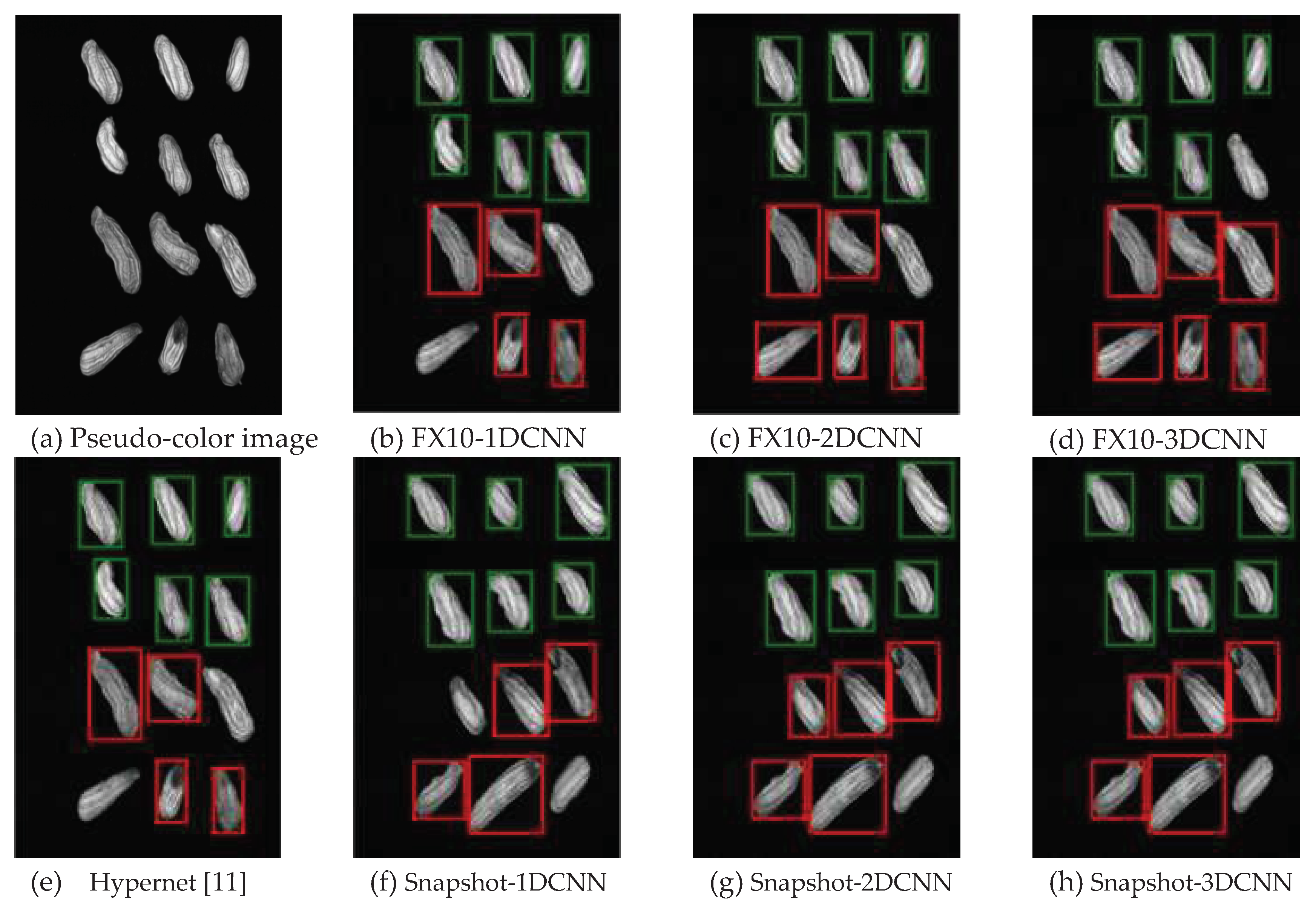
3.2.1.Snapshot data
| Model | Number of bands | TPR | FPR | Acc | Kappa | Runtime(ms) |
| 1D-CNN | Full band | 0.971 | 0.098 | 0.933 | 0.867 | 23.9 |
| 20 band | 0.919 | 0.086 | 0.917 | 0.833 | 23.5 | |
| 10 band | 0.915 | 0.126 | 0.893 | 0.787 | 23.4 | |
| 5 band | 0.933 | 0.145 | 0.890 | 0.780 | 23.3 | |
| 3 band | 0.931 | 0.222 | 0.837 | 0.673 | 23.3 | |
| 2D-CNN | Full band | 0.991 | 0.036 | 0.977 | 0.954 | 30.7 |
| 20 band | 0.997 | 0.047 | 0.974 | 0.949 | 29.4 | |
| 10 band | 0.994 | 0.025 | 0.982 | 0.969 | 27.8 | |
| 5 band | 0.980 | 0.028 | 0.976 | 0.951 | 26 | |
| 3 band | 0.988 | 0.025 | 0.981 | 0.963 | 25.5 | |
| 3D-CNN | Full band | 0.976 | 0.066 | 0.954 | 0.909 | 47.1 |
| 20 band | 0.968 | 0.064 | 0.951 | 0.903 | 33.1 | |
| 10 band | 0.962 | 0.067 | 0.947 | 0.894 | 31.3 | |
| 5 band | 0.971 | 0.050 | 0.960 | 0.920 | 29.6 | |
| 3 band | 0.980 | 0.042 | 0.969 | 0.937 | 28.7 | |
| Hypernet [11] | Full band | 0.930 | 0.021 | 0.953 | 0.906 | 33 |
| 20 band | 0.979 | 0.050 | 0.964 | 0.929 | 31.5 | |
| 10 band | 0.997 | 0.072 | 0.960 | 0.920 | 30.4 | |
| 5 band | 0.979 | 0.047 | 0.966 | 0.931 | 26.5 | |
| 3 band | 0.939 | 0.038 | 0.950 | 0.900 | 24.7 | |
| Realtime-1DCNN | Full band | 0.92 | 0.17 | 0.86 | 0.73 | 23.8 |
| 20 band | 0.907 | 0.269 | 0.793 | 0.586 | 23.4 | |
| 10 band | 0.871 | 0.169 | 0.850 | 0.700 | 23.2 | |
| 5 band | 0.98 | 0.08 | 0.96 | 0.91 | 23 | |
| 3 band | 0.880 | 0.090 | 0.930 | 0.870 | 22.8 | |
| Realtime-2DCNN | Full band | 0.990 | 0.037 | 0.977 | 0.954 | 29.1 |
| 20 band | 1.000 | 0.031 | 0.984 | 0.969 | 28.3 | |
| 10 band | 1.000 | 0.033 | 0.983 | 0.965 | 25.9 | |
| 5 band | 1.000 | 0.031 | 0.985 | 0.973 | 24.8 | |
| 3 band | 0.980 | 0.045 | 0.967 | 0.934 | 24.2 | |
| Realtime-3DCNN | Full band | 0.996 | 0.187 | 0.884 | 0.769 | 28.4 |
| 20 band | 0.948 | 0.073 | 0.937 | 0.874 | 27.9 | |
| 10 band | 0.997 | 0.077 | 0.957 | 0.914 | 26.9 | |
| 5 band | 0.985 | 0.060 | 0.961 | 0.922 | 26.3 | |
| 3 band | 0.988 | 0.050 | 0.969 | 0.937 | 24.4 |
4. Discussion
4.1. Data analysis
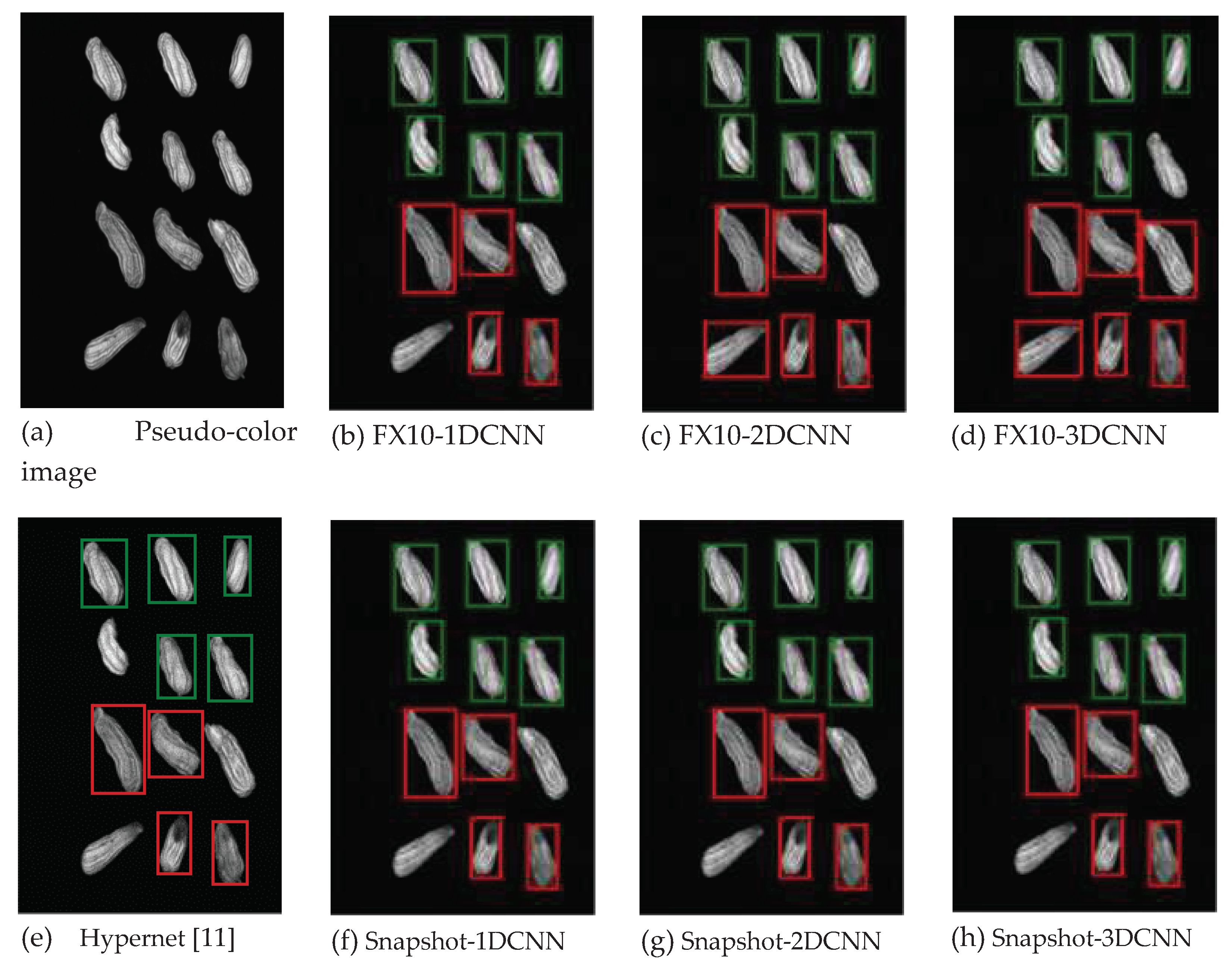
4.1.1. FX10 Data
4.1.2. FX10 Data
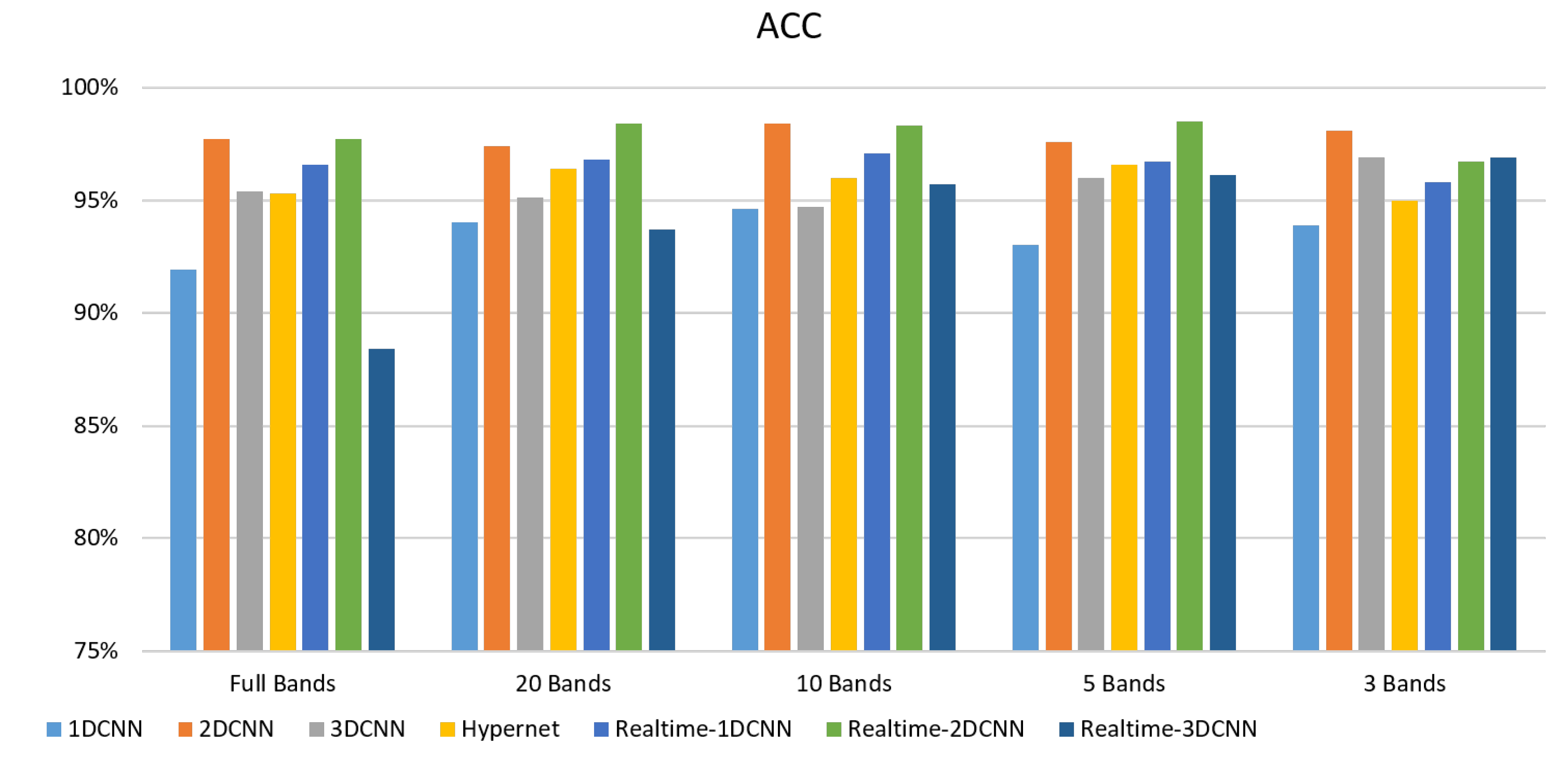

5. Conclusions
- (1)
- The FX10 had a lot of data/information. The ACC was 98% when the 3DCNN was used for analysis. On the contrary, the ACC of the snapshot was 98% when the 2DCNN was used. The number of convolution layers was reduced.
- (2)
- With the assistance of peanut farmers, more than 4,000 hyperspectral peanut images were collected. A database was created to get closer to the practical situation.
- (3)
- A 2DCNN model based on spatial information was developed. A good recognition rate could be achieved. The recognition speed could reach real-time, which was about 40 (FPS).
- (4)
- The PCA has been successfully used for selecting important and representative bands. The main characteristic bands were between 700 and 850 nm.
References
- Thenkabail, P. S., Smith, R. B., & De Pauw, E. (2000). Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote sensing of Environment, 71(2), 158-182.
- Dale, L. M., Thewis, A., Boudry, C., Rotar, I., Dardenne, P., Baeten, V., & Pierna, J. A. F. (2013). Hyperspectral imaging applications in agriculture and agro-food product quality and safety control: A review. Applied Spectroscopy Reviews, 48(2), 142-159.
- Adão, T., Hruška, J., Pádua, L., Bessa, J., Peres, E., Morais, R., & Sousa, J. J. (2017). Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sensing, 9(11), 1110.
- Council of Agriculture, Executive Yuan. (2001). Peanut. https://www.coa.gov.tw/ws.php?id=1012&print=Y.
- Huang, L., He, H., Chen, W., Ren, X., Chen, Y., Zhou, X., Xia, Y., Wang, X., Jiang, X., & Liao, B. (2015). Quantitative trait locus analysis of agronomic and quality-related traits in cultivated peanut (Arachis hypogaea L.). Theoretical and Applied Genetics, 128(6), 1103-1115.
- Bindlish, E., Abbott, A. L., & Balota, M. (2017). Assessment of peanut pod maturity. 2017 IEEE Winter Conference on Applications of Computer Vision (WACV).
- Jiang, J., Qiao, X., & He, R. (2016). Use of Near-Infrared hyperspectral images to identify moldy peanuts. Journal of Food Engineering, 169, 284-290.
- Qi, X., Jiang, J., Cui, X., & Yuan, D. (2019). Identification of fungi-contaminated peanuts using hyperspectral imaging technology and joint sparse representation model. Journal of food science and technology, 56(7), 3195-3204.
- Han, Z., & Gao, J. (2019). Pixel-level aflatoxin detecting based on deep learning and hyperspectral imaging. Computers and Electronics in Agriculture, 164, 104888.
- Xueming He, Chen Yan, Xuesong Jiang, Fei Shen, Jie You, Yong Fang (2021) Classification of aflatoxin B1 naturally contaminated peanut using visible and near-infrared hyperspectral imaging by integrating spectral and texture features.
- Liu, Z., Jiang, J., Qiao, X., Qi, X., Pan, Y., & Pan, X. (2020). Using convolution neural network and hyperspectral image to identify moldy peanut kernels. LWT, 132, 109815.
- Zou, S., Tseng, Y.-C., Zare, A., Rowland, D. L., Tillman, B. L., & Yoon, S.-C. (2019). Peanut maturity classification using hyperspectral imagery. biosystems engineering, 188, 165-177.
- Qi, H., Liang, Y., Ding, Q., & Zou, J. (2021). Automatic Identification of Peanut-Leaf Diseases Based on Stack Ensemble. Applied Sciences, 11(4), 1950.
- Dunteman, G. H. (1989). Principal components analysis. Sage.
- Martinez, A. M., & Kak, A. C. (2001). Pca versus lda. IEEE transactions on pattern analysis and machine intelligence, 23(2), 228-233.
- Chang, C.-I., Du, Q., Sun, T.-L., & Althouse, M. L. (1999). A joint band prioritization and band-decorrelation approach to band selection for hyperspectral image classification. IEEE Transactions on Geoscience and Remote Sensing, 37(6), 2631-2641.
- Al-Amri, S. S., & Kalyankar, N. V. (2010). Image segmentation by using threshold techniques. arXiv preprint arXiv:1005.4020.
- Wu, K., Otoo, E., & Shoshani, A. (2005). Optimizing connected component labeling algorithms. Medical Imaging 2005: Image Processing,.
- Suzuki, S. (1985). Topological structural analysis of digitized binary images by border following. Computer vision, graphics, and image processing, 30(1), 32-46.
- Wang, Ping, et al. “Deep Convolutional Neural Network for Coffee Bean Inspection.” Sensors and Materials 33.7 (2021): 2299-2310.
- Murphy, R. J., Monteiro, S. T., & Schneider, S. (2012). Evaluating classification techniques for mapping vertical geology using field-based hyperspectral sensors. IEEE Transactions on Geoscience and Remote Sensing, 50(8), 3066-3080.
- Singh, C. B., Jayas, D. S., Paliwal, J. N. D. G., & White, N. D. G. (2009). Detection of insect-damaged wheat kernels using near-infrared hyperspectral imaging. Journal of stored products research, 45(3), 151-158.
- Qin, J., Burks, T. F., Ritenour, M. A., & Bonn, W. G. (2009). Detection of citrus canker using hyperspectral reflectance imaging with spectral information divergence. Journal of food engineering, 93(2), 183-191.
- Hestir, E. L., Brando, V. E., Bresciani, M., Giardino, C., Matta, E., Villa, P., & Dekker, A. G. (2015). Measuring freshwater aquatic ecosystems: The need for a hyperspectral global mapping satellite mission. Remote Sensing of Environment, 167, 181-195.
- Chen, S. Y., Lin, C., Tai, C. H., & Chuang, S. J. (2018). Adaptive window-based constrained energy minimization for detection of newly grown tree leaves. Remote Sensing, 10(1), 96.
- Chen S-Y, Lin C, Chuang S-J, Kao Z-Y. Weighted Background Suppression Target Detection Using Sparse Image Enhancement Technique for Newly Grown Tree Leaves. Remote Sensing. 2019; 11(9):1081.
- Tiwari, K. C., Arora, M. K., & Singh, D. (2011). An assessment of independent component analysis for detection of military targets from hyperspectral images. International Journal of Applied Earth Observation and Geoinformation, 13(5), 730-740.
- Chen, S. Y., Cheng, Y. C., Yang, W. L., & Wang, M. Y. (2021). Surface defect detection of wet-blue leather using hyperspectral imaging. IEEE Access, 9, 127685-127702.
- Kelley, D. B., Goyal, A. K., Zhu, N., Wood, D. A., Myers, T. R., Kotidis, P & Müller, A. (2017, May). High-speed mid-infrared hyperspectral imaging using quantum cascade lasers. In Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XVIII (Vol. 10183, p. 1018304). International Society for Optics and Photonics.
- Dai, Q., Cheng, J. H., Sun, D. W., & Zeng, X. A. (2015). Advances in feature selection methods for hyperspectral image processing in food industry applications: A review. Critical reviews in food science and nutrition, 55(10), 1368-1382.
- Ariana, D. P., Lu, R., & Guyer, D. E. (2006). Near-infrared hyperspectral reflectance imaging for detection of bruises on pickling cucumbers. Computers and electronics in agriculture, 53(1), 60-70.
- Li, D., Zhang, J., Zhang, Q., & Wei, X. (2017, October). Classification of ECG signals based on 1D convolution neural network. In 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom) (pp. 1-6). IEEE.
- Hsieh, T.-H., & Kiang, J.-F. (2020). Comparison of CNN algorithms on hyperspectral image classification in agricultural lands. Sensors, 20(6), 1734.
- Pang, L., Men, S., Yan, L., & Xiao, J. (2020). Rapid vitality estimation and prediction of corn seeds based on spectra and images using deep learning and hyperspectral imaging techniques. IEEE Access, 8, 123026-123036.
- Yang, D., Hou, N., Lu, J., & Ji, D. (2022). Novel leakage detection by ensemble 1DCNN-VAPSO-SVM in oil and gas pipeline systems. Applied Soft Computing, 115, 108212.
- Laban, N., Abdellatif, B., Moushier, H., & Tolba, M. (2021). Enhanced pixel based urban area classification of satellite images using convolutional neural network. International Journal of Intelligent Computing and Information Sciences, 21(3), 13-28.
- Wang, C., Liu, B., Liu, L., Zhu, Y., Hou, J., Liu, P., & Li, X. (2021). A review of deep learning used in the hyperspectral image analysis for agriculture. Artificial Intelligence Review, 1-49.
- M Rustowicz, R., Cheong, R., Wang, L., Ermon, S., Burke, M., & Lobell, D. (2019). Semantic segmentation of crop type in Africa: A novel dataset and analysis of deep learning methods. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops,.
- Moreno-Revelo, M. Y., Guachi-Guachi, L., Gómez-Mendoza, J. B., Revelo-Fuelagán, J., & Peluffo-Ordóñez, D. H. (2021). Enhanced Convolutional-Neural-Network Architecture for Crop Classification. Applied Sciences, 11(9), 4292.
- Toraman, S. (2020). Preictal and Interictal Recognition for Epileptic Seizure Prediction Using Pre-trained 2DCNN Models. Traitement du Signal, 37(6).
- Ji, S., Xu, W., Yang, M., & Yu, K. (2012). 3D convolutional neural networks for human action recognition. IEEE transactions on pattern analysis and machine intelligence, 35(1), 221-231.
- Saveliev, A., Uzdiaev, M., & Dmitrii, M. (2019). Aggressive action recognition using 3d cnn architectures. 2019 12th International Conference on Developments in eSystems Engineering (DeSE).
- Ji, S., Zhang, C., Xu, A., Shi, Y., & Duan, Y. (2018). 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sensing, 10(1), 75.
- Ji, S., Zhang, Z., Zhang, C., Wei, S., Lu, M., & Duan, Y. (2020). Learning discriminative spatiotemporal features for precise crop classification from multi-temporal satellite images. International Journal of Remote Sensing, 41(8), 3162-3174.

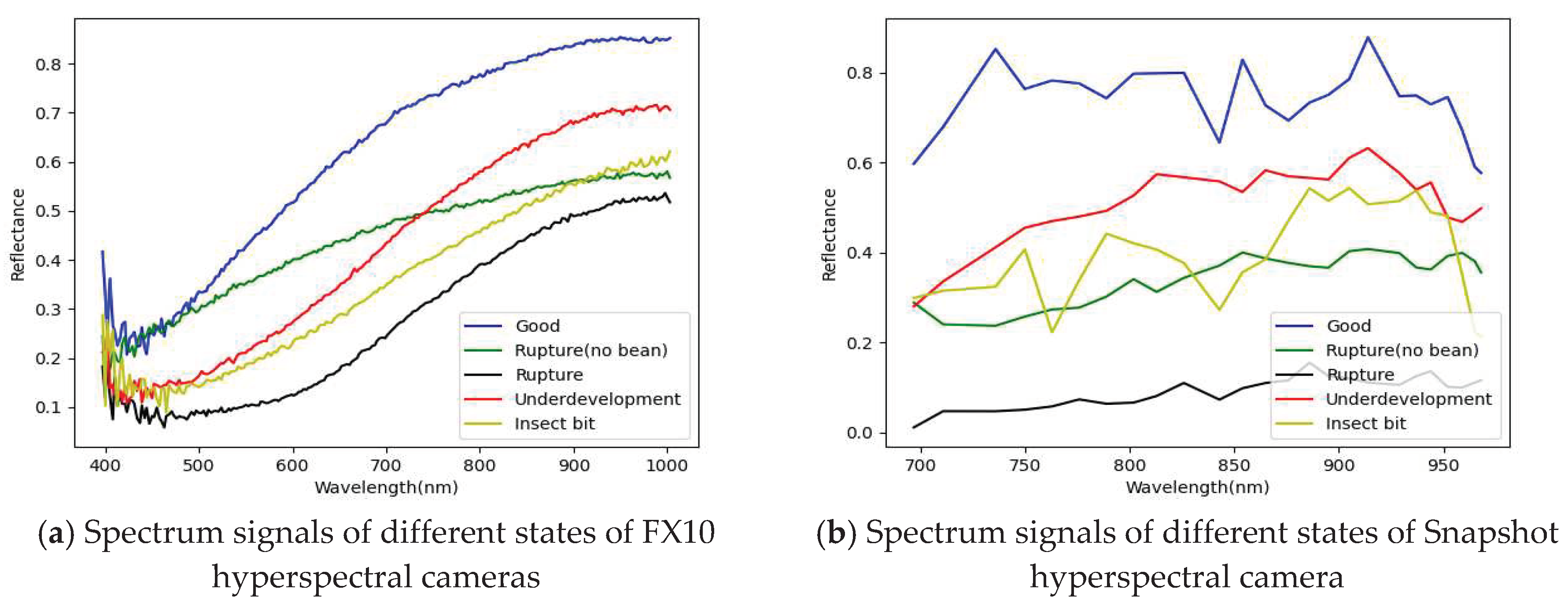
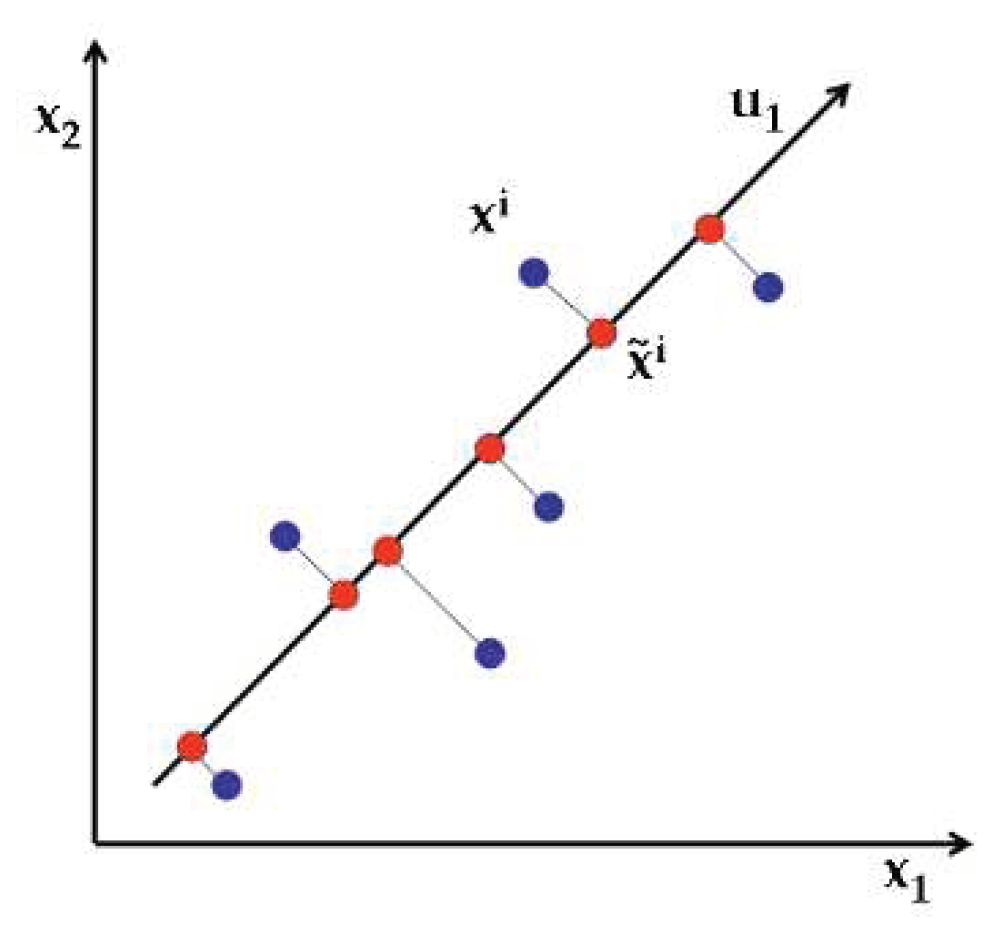
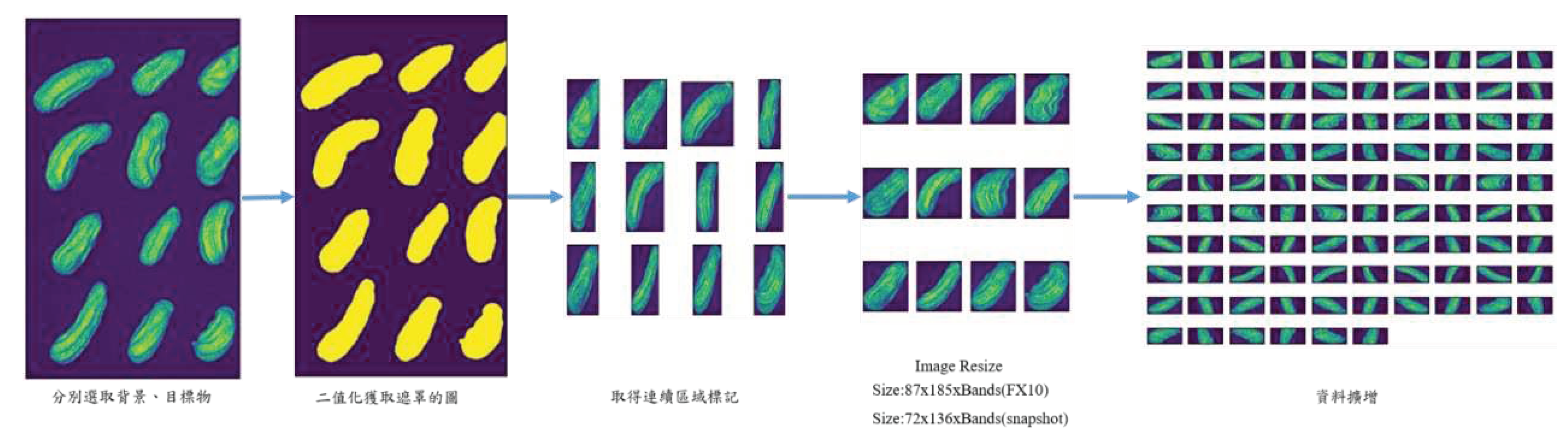
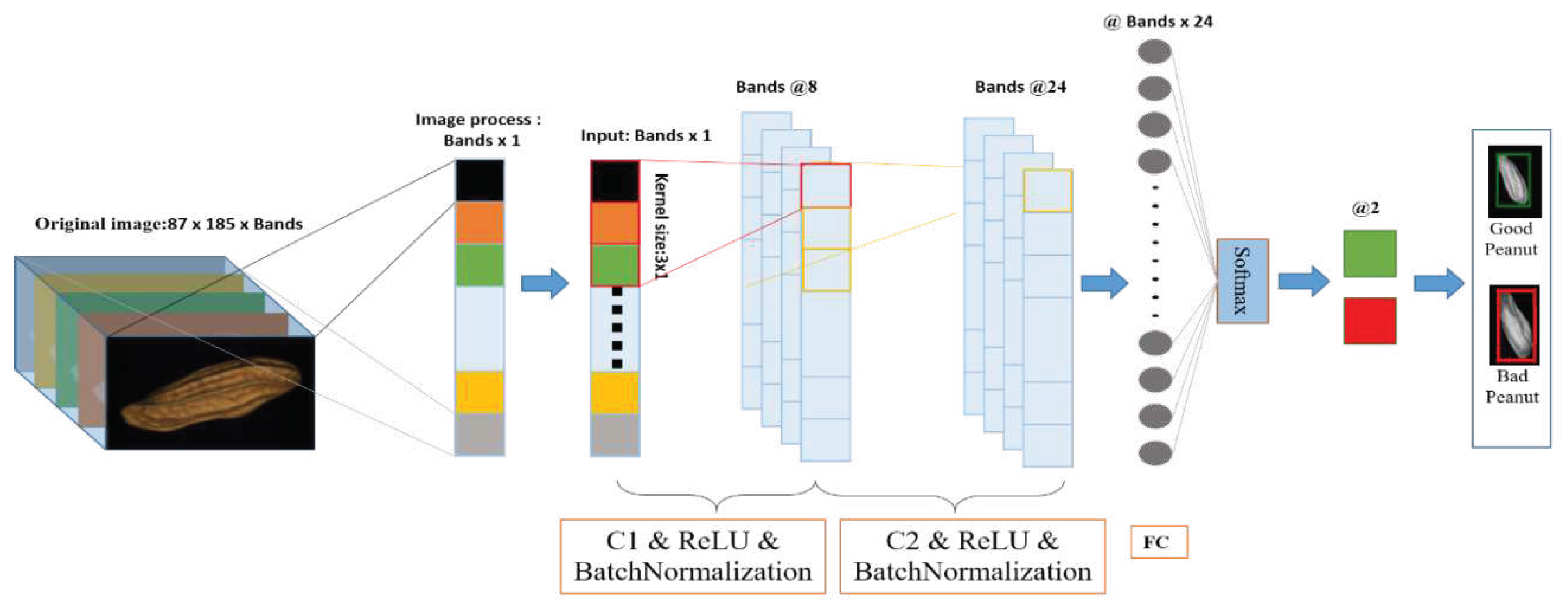
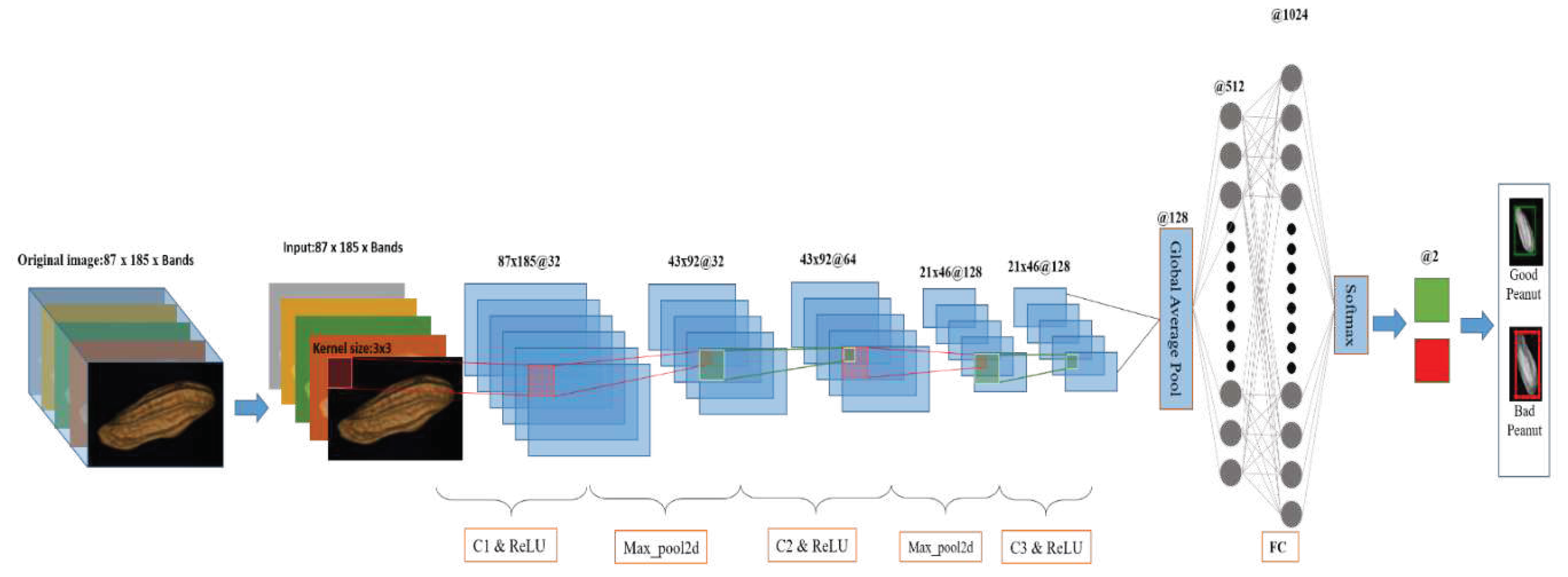
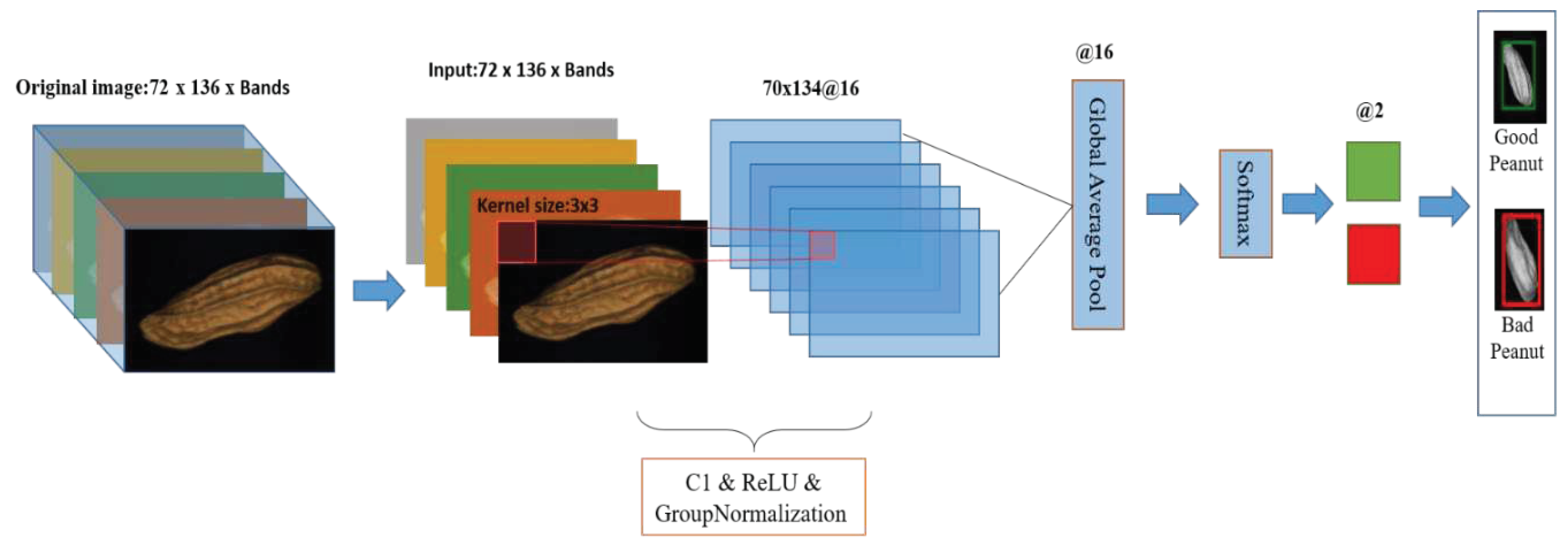
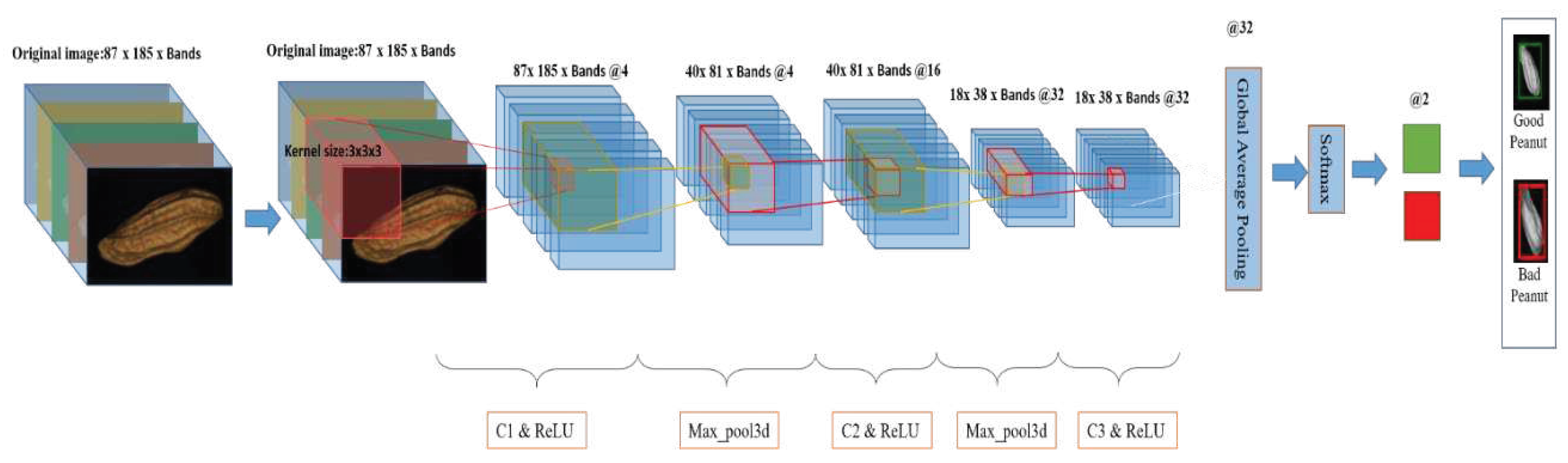

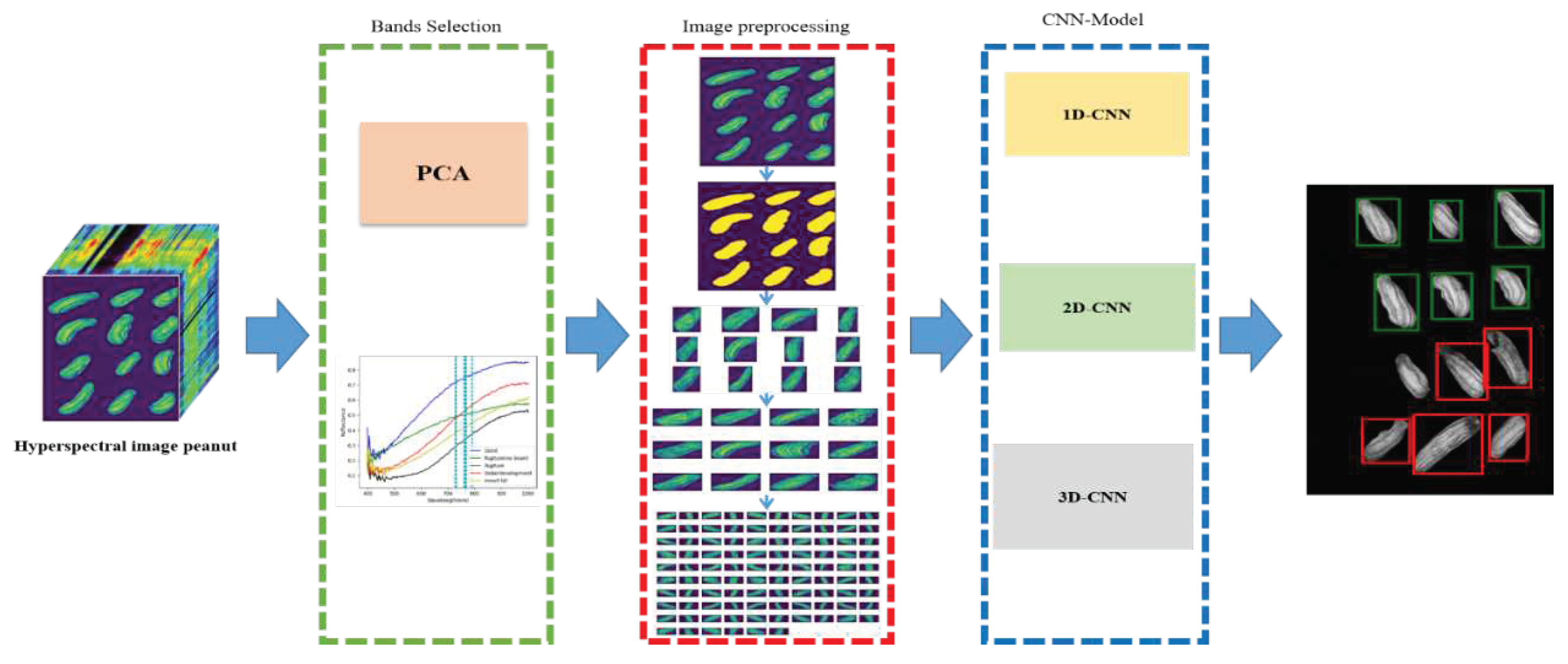


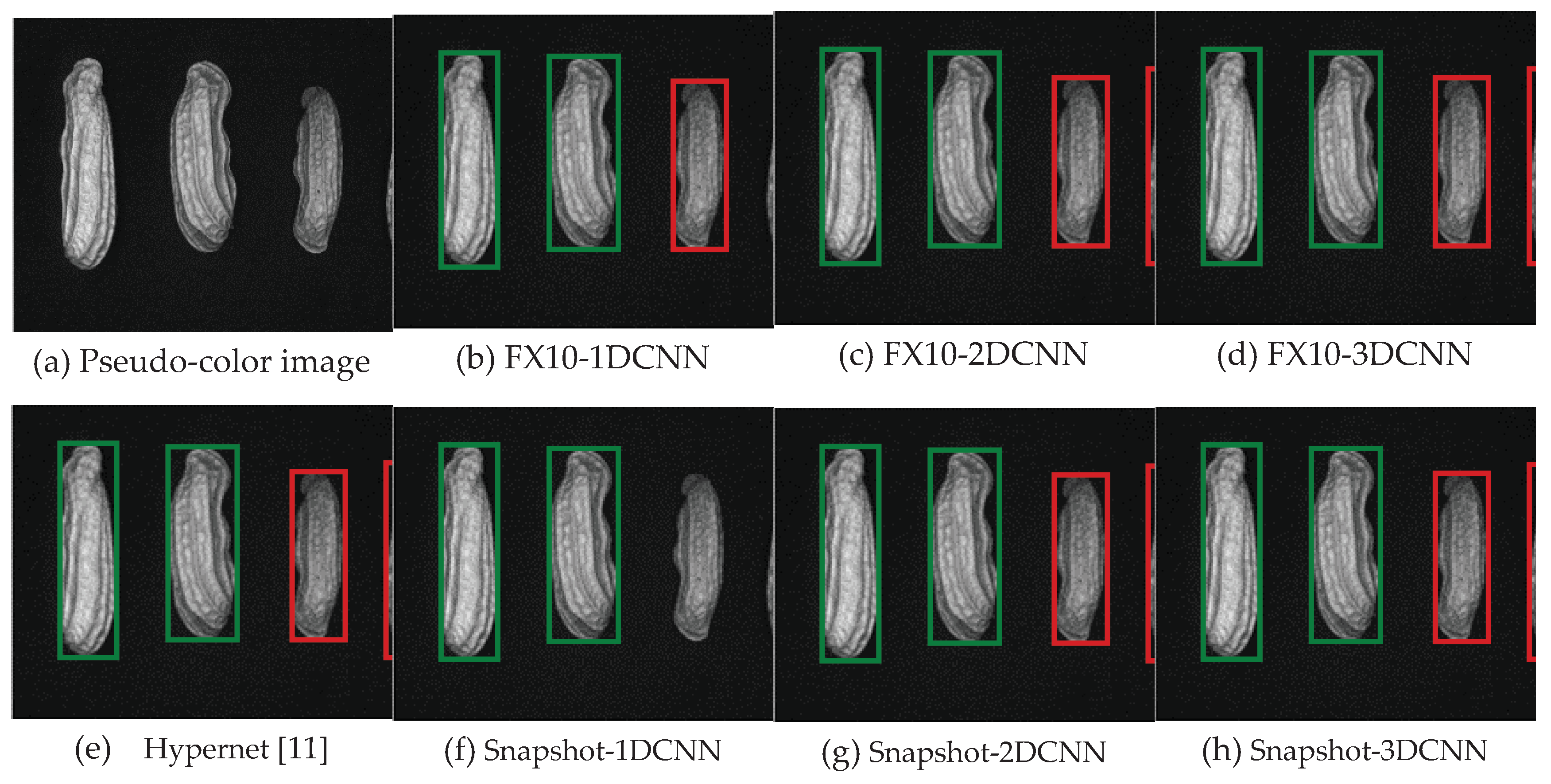
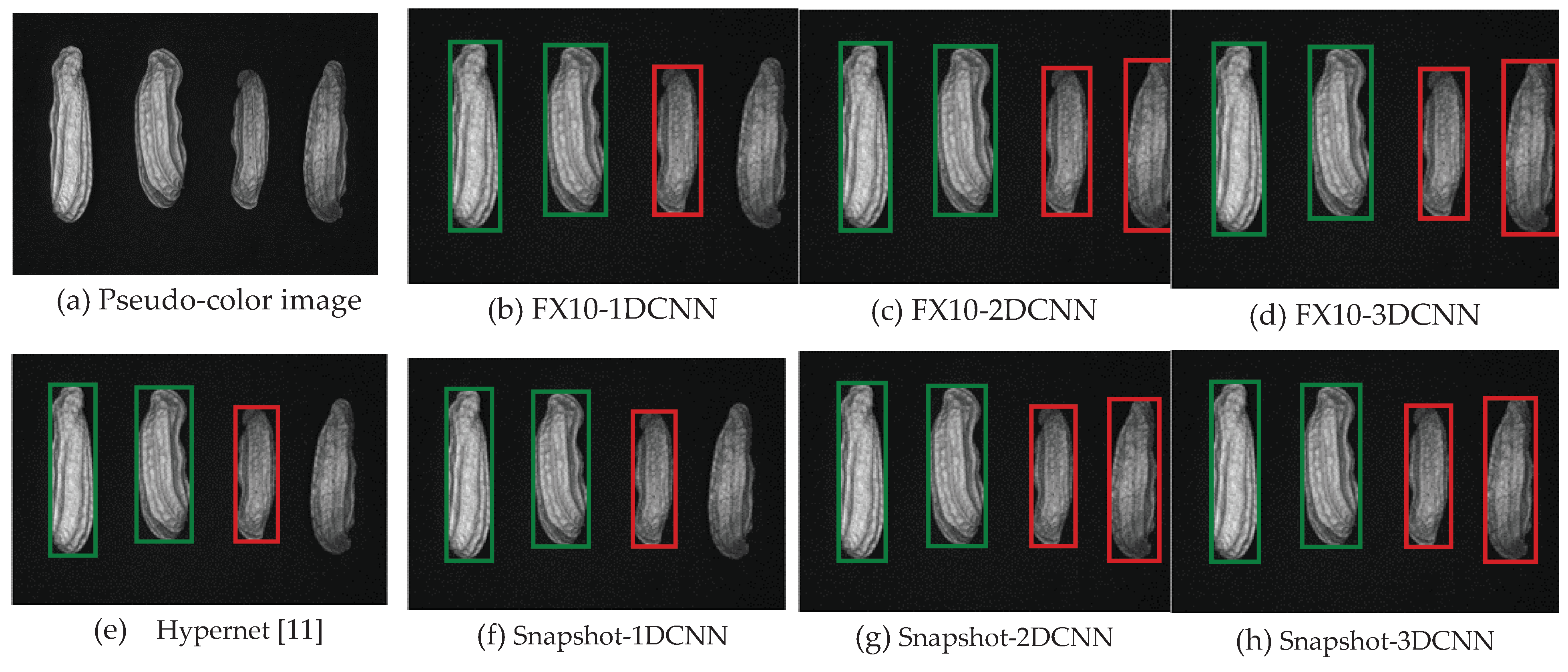
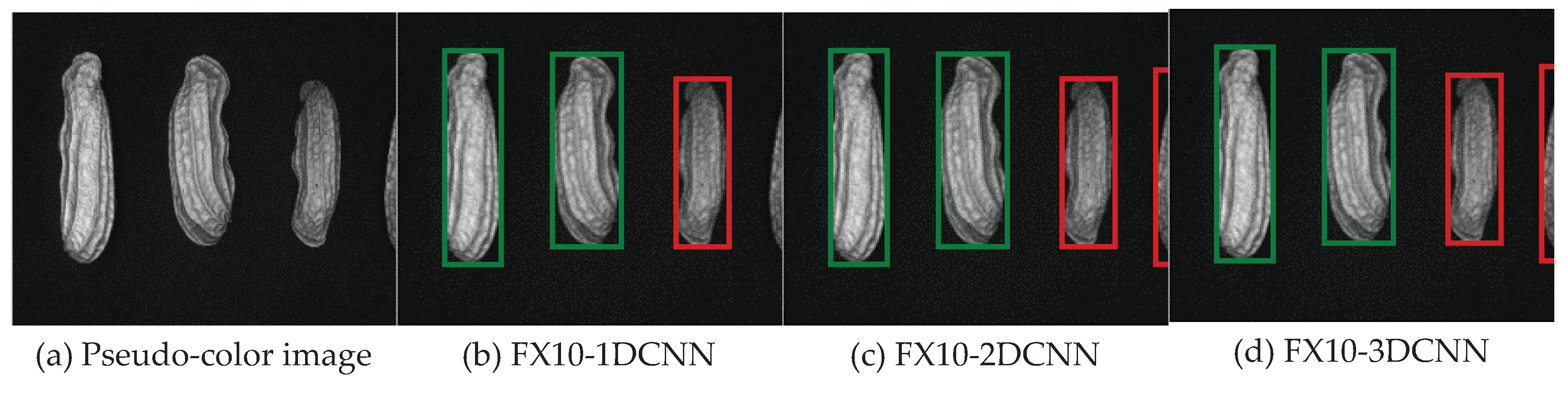
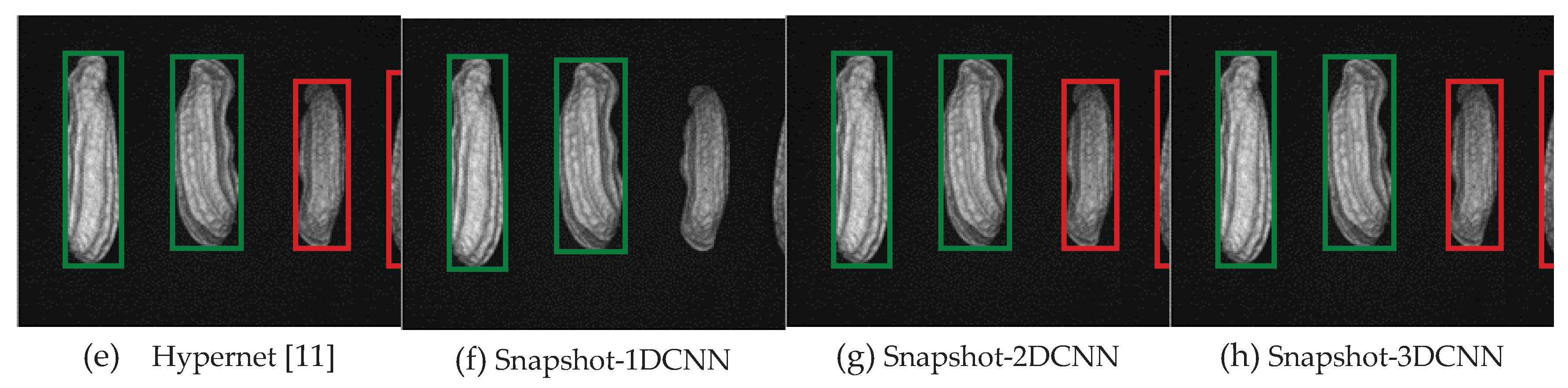
| References | Detection target | Spectral range (nm) | Data quantity | Data analysis | Overall accuracy |
|---|---|---|---|---|---|
| (Jiang et al., 2016) [7] | Aflatoxin | 970-2,570 nm | 149 pcs | PCA+ Threshold | 98.7 |
| (Qi et al., 2019) [8] | Aflatoxin | 967-2,499 nm | 2,312 pixels | JSRC+SVM | 98.4 |
| (Han & Gao, 2019) [9] | Aflatoxin | 292-865 nm | 146 pcs | CNN | 96 |
| (Zou et al., 2019) [12] | Peanut shell maturity | 400-1,000 nm | 540 pcs | LMM+RGB | 91.15 |
| (Liu et al., 2020) [11] | Aflatoxin | 400-1000 nm | 1,066 pcs | Unet, Hypernet | 92.07 |
| (Qi et al., 2021) [13] | Peanut leaves | RGB | 3,205 images | GoogleNet | 97.59 |
| (Xueming et al., 2021) [10] | Aflatoxin | 400-1,000 nm | 150 pcs | SGS+SNV | 94 |
| Classification | Normal Peanut | Defect Peanut | Total |
|---|---|---|---|
| Training amount | 237 | 240 | 477 |
| Testing amount | 540 | 540 | 1080 |
| Total | 777 | 780 | 1557 |
| Classification | Normal Peanut | Defect Peanut | Total |
|---|---|---|---|
| Training amount | 350 | 350 | 700 |
| Testing amount | 1000 | 1000 | 2000 |
| Total | 1350 | 1350 | 2700 |
| Analysis method | Important band |
|---|---|
| PCA 20 Band | 726.33,729.06,731.79,759.14,761.88,764.62,767.36,770.11, 789.33,792.08,794.84,797.59,800.34,803.1, 805.85,838.98,847.29,852.83, 850.06, 863.92 |
| PCA 10 Band | 726,729,731,759,761,764,767,770,789,792, |
| PCA 5 Band | 726,729,731,759,761 |
| PCA 3 Band | 726,729,731 |
| Analysis method | Important band |
|---|---|
| 20 Band | 854, 843, 929, 914, 959, 968, 965, 952, 944, 937, 905, 895, 886, 876, 865, 826, 813, 802, 789, 776 |
| 10 Band | 854, 843, 929, 914, 959, 968, 965, 952, 944, 937 |
| 5 Band | 854, 843, 929, 914, 959 |
| 3 Band | 854, 843, 929 |
| Model | Number of bands | TPR | FPR | Acc | Kappa | Runtime(ms) |
|---|---|---|---|---|---|---|
| 1D-CNN | Full band | 0.92 | 0.29 | 0.83 | 0.67 | 44.2 |
| 20 bands | 0.937 | 0.002 | 0.967 | 0.935 | 35.4 | |
| 10 bands | 0.924 | 0.024 | 0.962 | 0.908 | 32.8 | |
| 5 bands | 0.913 | 0.023 | 0.956 | 0.907 | 31.1 | |
| 3 bands | 0.91 | 0.03 | 0.954 | 0.904 | 30.7 | |
| 2D-CNN | Full band | 0.94 | 0.07 | 0.971 | 0.940 | 58 |
| 20 bands | 0.959 | 0.004 | 0.977 | 0.956 | 32.6 | |
| 10 bands | 0.956 | 0.062 | 0.977 | 0.933 | 28.6 | |
| 5 bands | 0.957 | 0.011 | 0.973 | 0.941 | 26.9 | |
| 3 bands | 0.94 | 0.07 | 0.96 | 0.93 | 26 | |
| 3D-CNN | Full band | 0.990 | 0.170 | 0.910 | 0.820 | 61 |
| 20 bands | 0.980 | 0.013 | 0.983 | 0.967 | 37 | |
| 10 bands | 0.967 | 0.035 | 0.981 | 0.961 | 32.8 | |
| 5 bands | 0.961 | 0.068 | 0.978 | 0.957 | 30.5 | |
| 3 bands | 1.0 | 0.05 | 0.97 | 0.94 | 29.3 | |
| Hypernet [11] | Full band | 0.14 | 0.93 | 0.85 | 54.5 | |
| 20 bands | 0.964 | 0.0736 | 0.9444 | 0.888 | 32.3 | |
| 10 bands | 0.9634 | 0.1020 | 0.9277 | 0.855 | 29.6 | |
| 5 bands | 0.98787 | 0.07692 | 0.9527 | 0.90546 | 25.7 | |
| 3 bands | 0.9875 | 0.1 | 0.938 | 0.87 | 25.4 | |
| Realtime-1DCNN | Full band | 0.92307 | 0.1764 | 0.86666 | 0.73333 | 51.2 |
| 20 bands | 1.0 | 0.150 | 0.9111 | 0.82222 | 23.4 | |
| 10 bands | 0.98 | 0.07 | 0.96 | 0.922 | 23.2 | |
| 5 bands | 0.976 | 0.077 | 0.958 | 0.91 | 23 | |
| 3 bands | 0.8823 | 0.09 | 0.9333 | 0.866666 | 22.8 | |
| Realtime-2DCNN | Full band | 0.96 | 0.1 | 0.94 | 0.85 | 51.8 |
| 20 bands | 0.98 | 0.04 | 0.93 | 0.87 | 31.4 | |
| 10 bands | 0.98 | 0.07 | 0.9505 | 0.90 | 28 | |
| 5 bands | 0.961 | 0.068 | 0.978 | 0.957 | 26 | |
| 3 bands | 1 | 0.06 | 0.96 | 0.92 | 25.2 | |
| Realtime-3DCNN | Full band | 0.99 | 0.17 | 0.91 | 0.82 | 72.1 |
| 20 bands | 0.99 | 0.05 | 0.966 | 0.933 | 34.4 | |
| 10 bands | 0.99 | 0.05 | 0.966 | 0.933 | 32.3 | |
| 5 bands | 0.961 | 0.068 | 0.978 | 0.957 | 29.3 | |
| 3 bands | 1 | 0.07 | 0.95 | 0.91 | 24.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





