Submitted:
30 May 2023
Posted:
01 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results and discussion
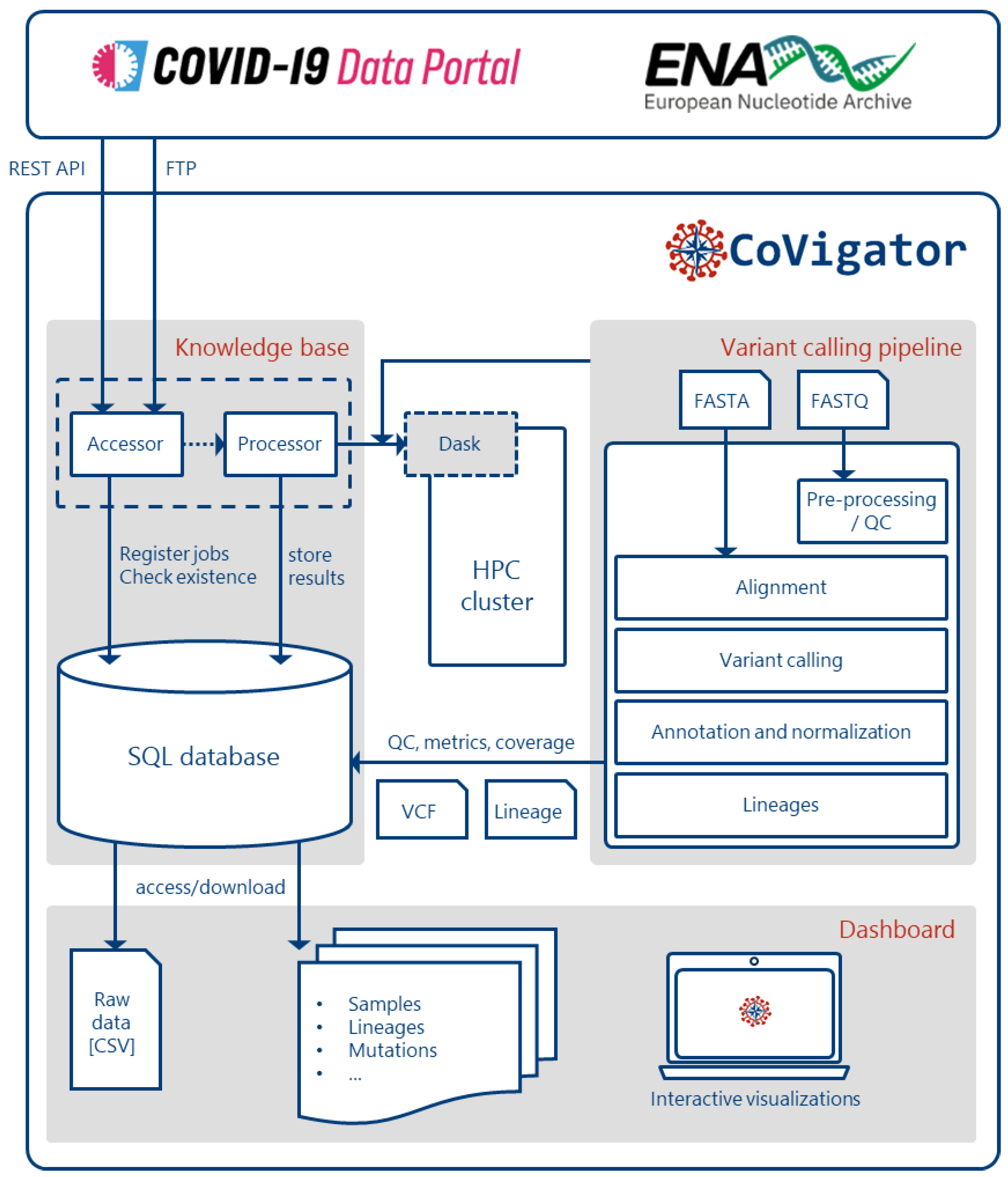
System description
Knowledge base
Analysis pipeline
Dashboard
Samples
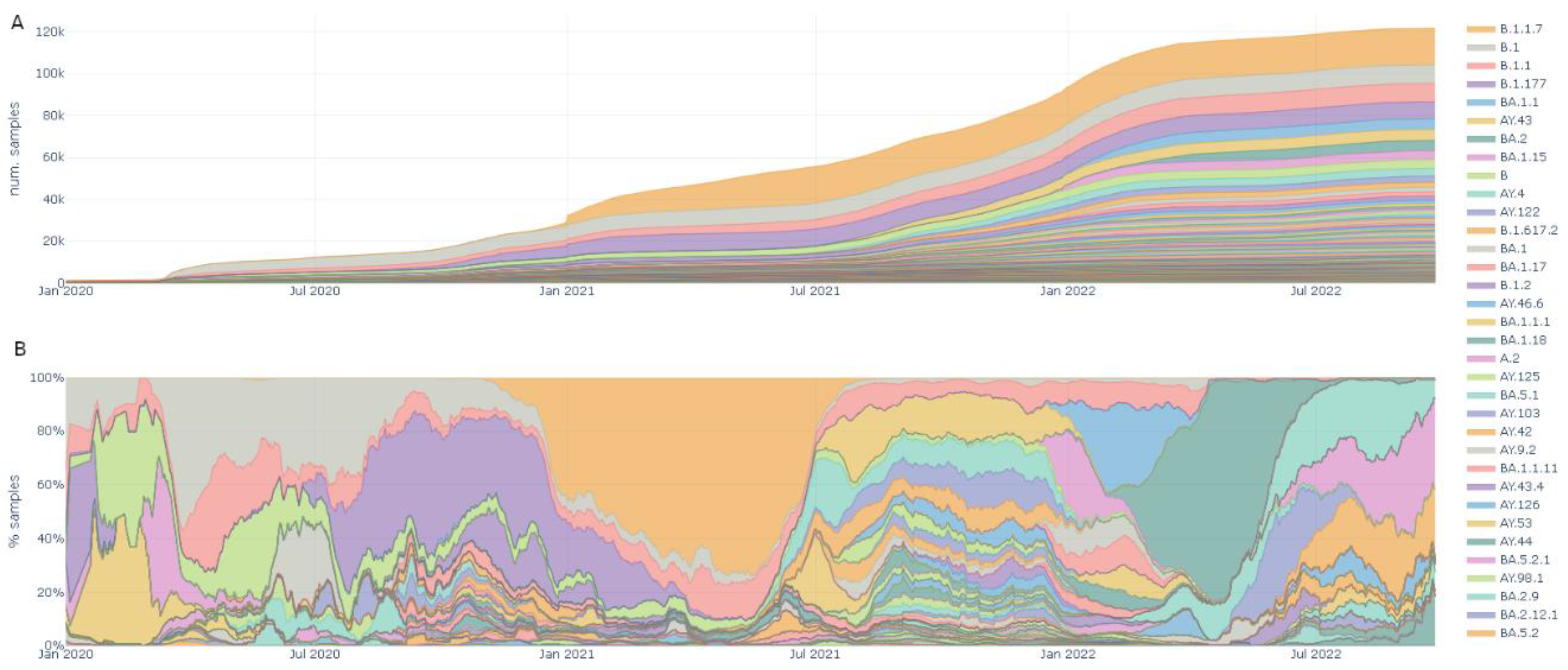
Lineages
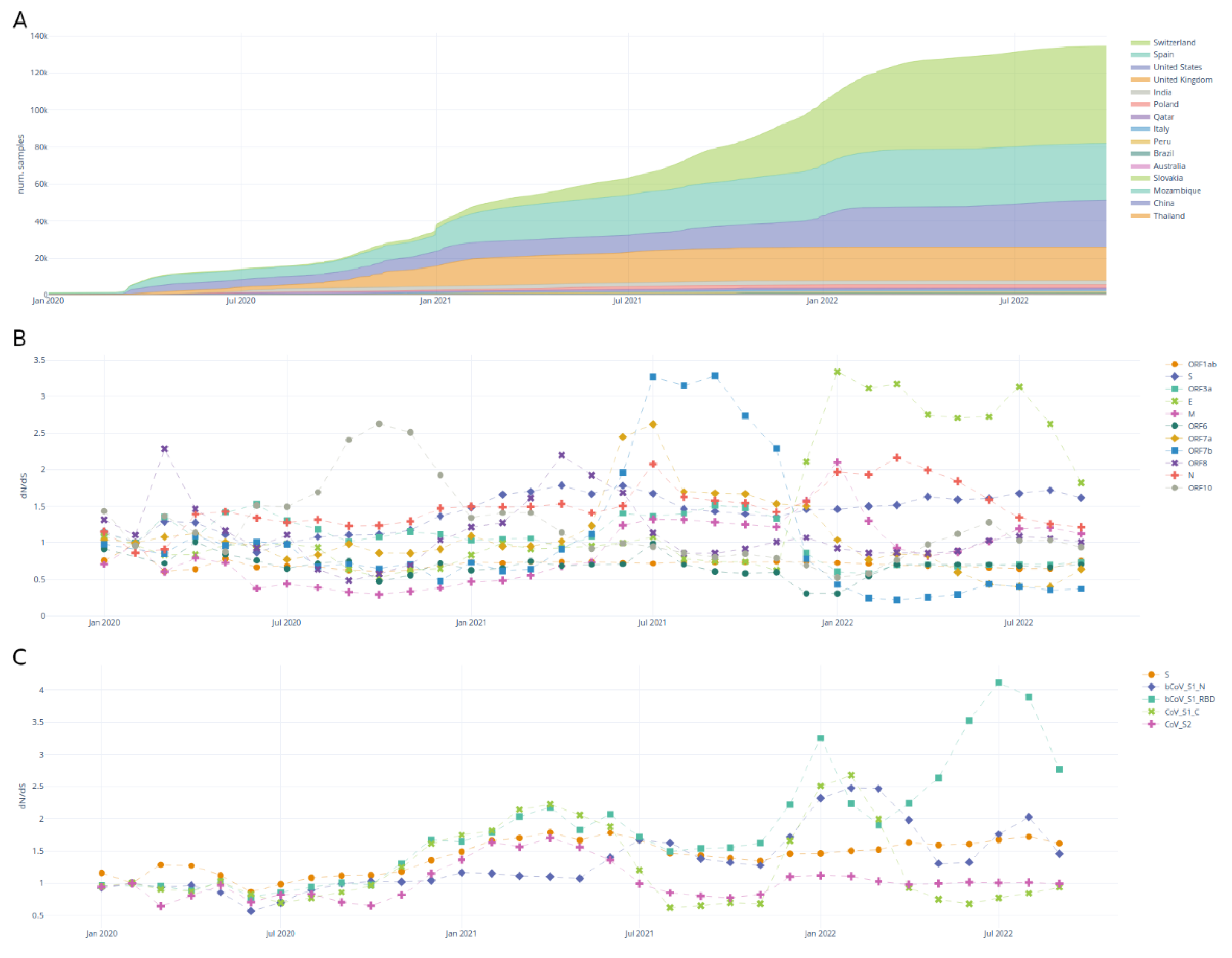
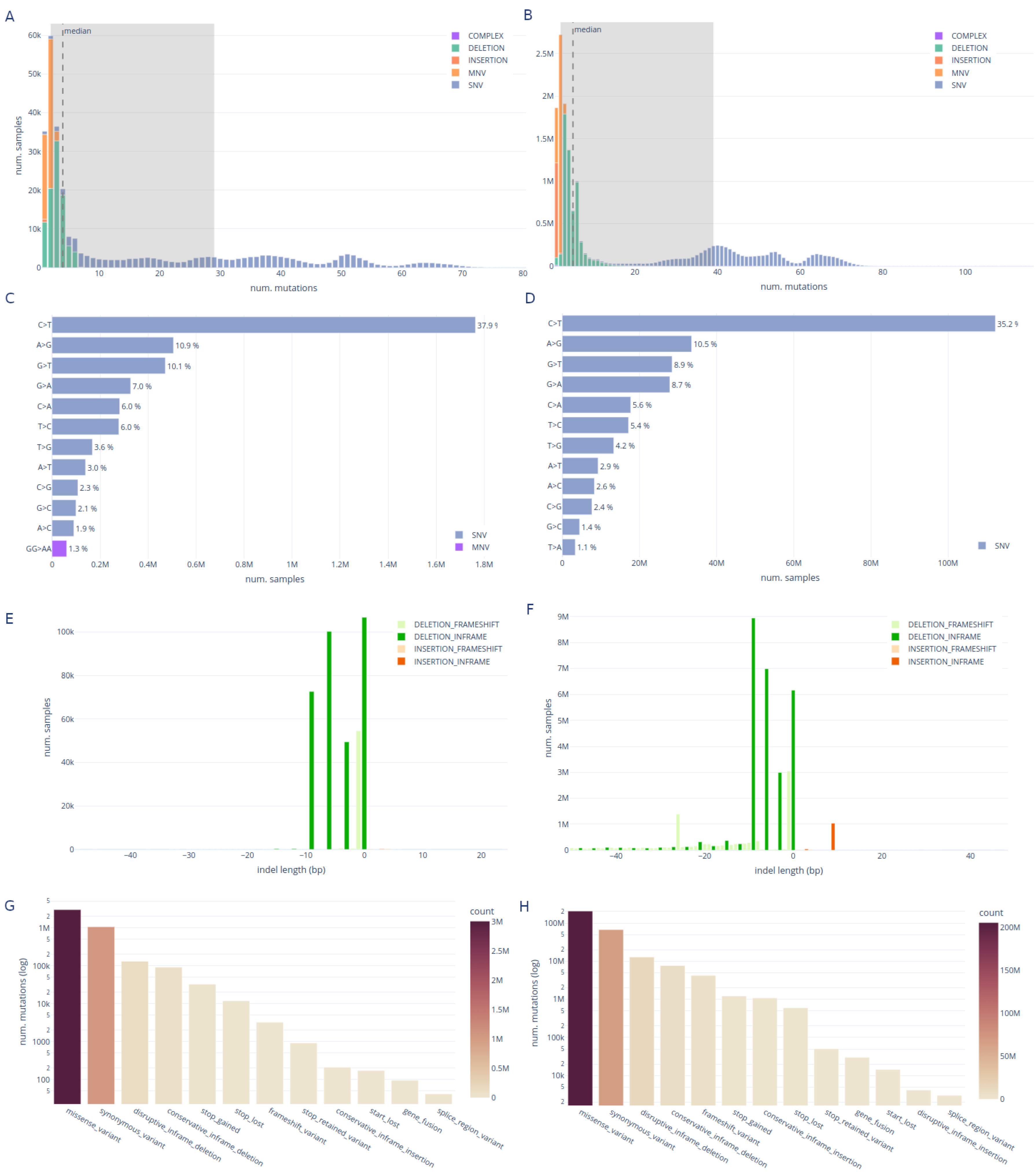
Mutation statistics
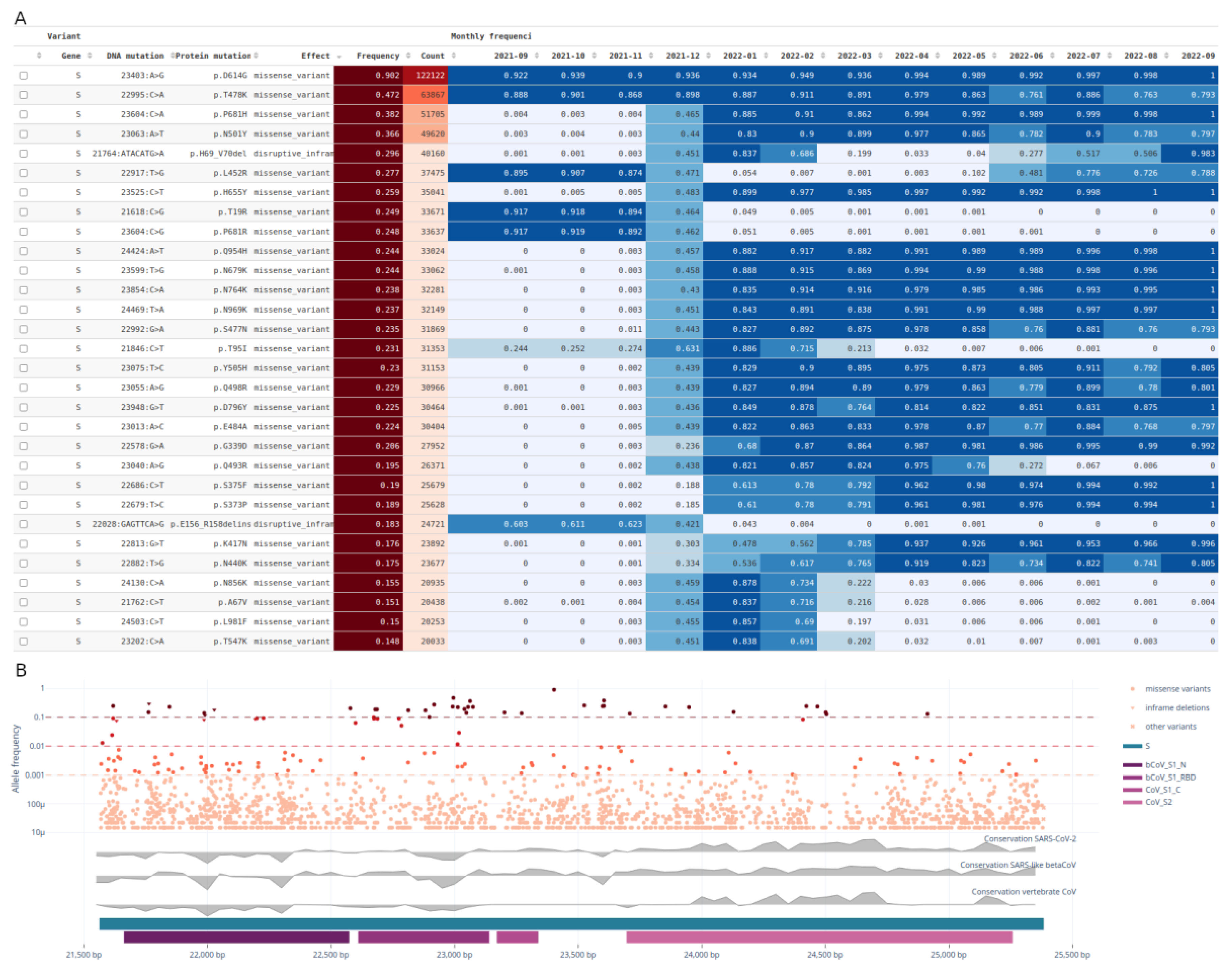
Recurrent mutations
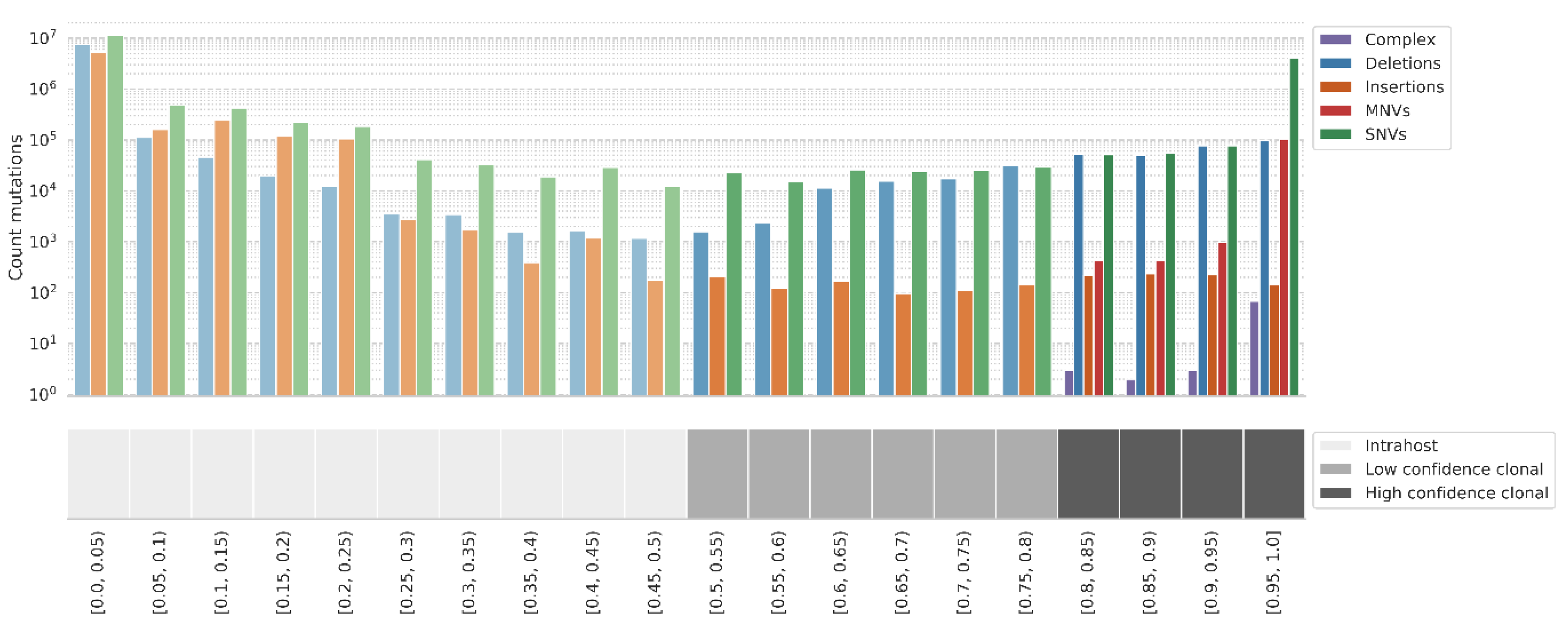
Clonal and intrahost mutations in the raw reads dataset
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflict of Interest
References
- Moorthy, V.S.; Karam, G.; Vannice, K.S.; Kieny, M.-P. Rationale for WHO's new position calling for prompt reporting and public disclosure of interventional clinical trial results. PLoS Med. 2015, 12, e1001819. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Ventura, C.V.; Maia, M.; Bravo-Filho, V.; Góis, A.L.; Belfort, R. Zika virus in Brazil and macular atrophy in a child with microcephaly. The Lancet 2016, 387, 228. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Shang, W.; Yang, Y.; Rao, Y.; Rao, X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines 2020, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID's Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Schrörs, B.; Riesgo-Ferreiro, P.; Sorn, P.; Gudimella, R.; Bukur, T.; Rösler, T.; Löwer, M.; Sahin, U. Large-scale analysis of SARS-CoV-2 spike-glycoprotein mutants demonstrates the need for continuous screening of virus isolates. PLoS One 2021, 16, e0249254. [Google Scholar] [CrossRef] [PubMed]
- Riesgo-Ferreiro, P. VAFator; Github: https://github.com/TRON-Bioinformatics/vafator.git: https://github.com/TRON-Bioinformatics/vafator.git, 2022.
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.; Gifford, R.; Cotten, M.; Robertson, D. CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation, 2020.
- Mercatelli, D.; Triboli, L.; Fornasari, E.; Ray, F.; Giorgi, F.M. Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations. J. Med. Virol. 2021, 93, 3238–3245. [Google Scholar] [CrossRef]
- Maier, W.; Bray, S.; van den Beek, M.; Bouvier, D.; Coraor, N.; Miladi, M.; Singh, B.; Argila, J.R. de; Baker, D.; Roach, N.; et al. Ready-to-use public infrastructure for global SARS-CoV-2 monitoring. Nat. Biotechnol. 2021, 39, 1178–1179. [Google Scholar] [CrossRef]
- Nicholls, S.M.; Poplawski, R.; Bull, M.J.; Underwood, A.; Chapman, M.; Abu-Dahab, K.; Taylor, B.; Colquhoun, R.M.; Rowe, W.P.M.; Jackson, B.; et al. CLIMB-COVID: Continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Genome Biol. 2021, 22, 196. [Google Scholar] [CrossRef] [PubMed]
- Wittig, A.; Miranda, F.; Hölzer, M.; Altenburg, T.; Bartoszewicz, J.M.; Beyvers, S.; Dieckmann, M.A.; Genske, U.; Giese, S.H.; Nowicka, M.; et al. CovRadar: Continuously tracking and filtering SARS-CoV-2 mutations for molecular surveillance, 2021.
- Harrison, P.W.; Lopez, R.; Rahman, N.; Allen, S.G.; Aslam, R.; Buso, N.; Cummins, C.; Fathy, Y.; Felix, E.; Glont, M.; et al. The COVID-19 Data Portal: Accelerating SARS-CoV-2 and COVID-19 research through rapid open access data sharing. Nucleic Acids Res. 2021, 49, W619–W623. [Google Scholar] [CrossRef] [PubMed]
- Cecret; GitHub: GitHub, Accessed 2021.
- Harshil, P.; Sarai, V.; Sara, M.; Jose, E.-C.; Michael, L.H.; Gisela, G.; nf-core bot; Phil, E.; Miguel, J.; Stephen, K.; et al. nf-core/viralrecon; zenodo, 2021.
- connor-lab. ncov2019-artic-nf; GitHub: GitHub, 2022.
- Al Khatib, H.A.; Benslimane, F.M.; Elbashir, I.E.; Coyle, P.V.; Al Maslamani, M.A.; Al-Khal, A.; Al Thani, A.A.; Yassine, H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front. Cell. Infect. Microbiol. 2020, 10, 575613. [Google Scholar] [CrossRef]
- Armero, A.; Berthet, N.; Avarre, J.-C. Intra-Host Diversity of SARS-Cov-2 Should Not Be Neglected: Case of the State of Victoria, Australia. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Karamitros, T.; Papadopoulou, G.; Bousali, M.; Mexias, A.; Tsiodras, S.; Mentis, A. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J. Clin. Virol. 2020, 131, 104585. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; Cesare, M. de; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372. [Google Scholar] [CrossRef] [PubMed]
- Moreno, G.; Katarina M. Braun; Peter J. Halfmann; Trent M. Prall, Kasen K. Riemersma, Amelia K. Haj, Joseph Lalli, Kelsey R. Florek, Yoshihiro Kawaoka, Thomas C. Friedrich, David H. O’Connor. Limited SARS-CoV-2 diversity within hosts and following passage in cell culture 2020. [CrossRef]
- Popa, A.; Genger, J.-W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaço, H.; et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.; Nolan, D.J.; Moot, S.; Feehan, A.; Cross, S.; Garcia-Diaz, J.; Lamers, S.L. Intra-host site-specific polymorphisms of SARS-CoV-2 is consistent across multiple samples and methodologies, 2020.
- Siqueira, J.D.; Goes, L.R.; Alves, B.M.; Carvalho, P.S. de; Cicala, C.; Arthos, J.; Viola, J.P.B.; Melo, A.C. de; Soares, M.A. SARS-CoV-2 genomic and quasispecies analyses in cancer patients reveal relaxed intrahost virus evolution. bioRxiv 2020. [CrossRef]
- Tonkin-Hill, G.; Martincorena, I.; Amato, R.; Lawson, A.R.J.; Gerstung, M.; Johnston, I.; Jackson, D.K.; Park, N.R.; Lensing, S.V.; Quail, M.A.; et al. Patterns of within-host genetic diversity in SARS-CoV-2 2020. [CrossRef]
- Sapoval, N.; Mahmoud, M.; Jochum, M.D.; Liu, Y.; Elworth, R.A.L.; Wang, Q.; Albin, D.; Ogilvie, H.A.; Lee, M.D.; Villapol, S.; et al. SARS-CoV-2 genomic diversity and the implications for qRT-PCR diagnostics and transmission. Genome Res. 2021, 31, 635–644. [Google Scholar] [CrossRef]
- Zhou, Z.-Y.; Liu, H.; Zhang, Y.-D.; Wu, Y.-Q.; Peng, Min-Sheng: Li, Aimin; Irwin, D.M.; Li, H.; Lu, J.; Bao, Y.; Lu, X.; et al. Worldwide tracing of mutations and the evolutionary dynamics of SARS-CoV-2. bioRxiv 2020. [CrossRef]
- James, S.E.; Ngcapu, S.; Kanzi, A.M.; Tegally, H.; Fonseca, V.; Giandhari, J.; Wilkinson, E.; Chimukangara, B.; Pillay, S.; Singh, L.; et al. High Resolution analysis of Transmission Dynamics of Sars-Cov-2 in Two Major Hospital Outbreaks in South Africa Leveraging Intrahost Diversity. medRxiv 2020. [CrossRef]
- Sashittal, P.; Luo, Y.; Peng, J.; El-Kebir, M. Characterization of SARS-CoV-2 viral diversity within and across hosts. bioRxiv 2020. [CrossRef]
- Shen, Z.; Xiao, Y.; Kang, L.; Ma, W.; Shi, L.; Zhang, L.; Zhou, Z.; Yang, J.; Zhong, J.; Yang, D.; et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 713–720. [Google Scholar] [CrossRef]
- Valesano, A.L.; Rumfelt, K.E.; Dimcheff, D.E.; Blair, C.N.; Fitzsimmons, W.J.; Petrie, J.G.; Martin, E.T.; Lauring, A.S. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog. 2021, 17, e1009499. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, L.; Sun, W.; Zhang, Z.; Chen, W.; Zhu, A.; Huang, Y.; Xiao, F.; Yao, J.; et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021, 13, 30. [Google Scholar] [CrossRef]
- Conda; Anaconda Software Distribution: https://docs.conda.io/: https://docs.conda.io/, 2022.
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), 2019; pp 314–324.
- van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Wilm, A.; Aw, P.P.K.; Bertrand, D.; Yeo, G.H.T.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; McCarthy, S.A. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics 2017, 33, 2037–2039. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; Jesus, J.G. de; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Le Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef]
- O'Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Kwon, S.B.; Ernst, J. Single-nucleotide conservation state annotation of the SARS-CoV-2 genome. Commun. Biol. 2021, 4, 698. [Google Scholar] [CrossRef]
- Ensembl annotations SARS-CoV-2; ftp://ftp.ensemblgenomes.org/pub/viruses/json/sars_cov_2/sars_cov_2.json: ftp://ftp.ensemblgenomes.org/pub/viruses/json/sars_cov_2/sars_cov_2.json, Accessed 2021.
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The population genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Spielman, S.J.; Wilke, C.O. The relationship between dN/dS and scaled selection coefficients. Mol. Biol. Evol. 2015, 32, 1097–1108. [Google Scholar] [CrossRef]
- Kistler, K.; Huddleston, J.; Bedford, T. Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2. bioRxiv 2022. [CrossRef]
- Rogozin, I.B.; Saura, A.; Bykova, A.; Brover, V.; Yurchenko, V. Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Garushyants, S.K.; Rogozin, I.B.; Koonin, E.V. Insertions in SARS-CoV-2 genome caused by template switch and duplications give rise to new variants that merit monitoring. bioRxiv 2021. [CrossRef]
- Montgomery, S.B.; Goode, D.L.; Kvikstad, E.; Albers, C.A.; Zhang, Z.D.; Mu, X.J.; Ananda, G.; Howie, B.; Karczewski, K.J.; Smith, K.S.; et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013, 23, 749–761. [Google Scholar] [CrossRef]
- DeMaio, N.; Conor Walker; Rui Borges; Lukas Weilguny; Greg Slodkowic; Nick Goldman. Issues with SARS-CoV-2 sequencing data. Available online: https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473.
| Tool | Purpose | Settings | References | Version | FASTQ | FASTA |
|---|---|---|---|---|---|---|
| fastp | Adapter trimming | [42] | 0.20.1 | X | ||
| BWA mem 2 | Alignment | Default | [43] | 2.2.1 | X | |
| GATK | Variant calling and alignments preprocessing | MQ>=20, BQ>=20, ploidy=1 | [44] | 4.2.0.0 | X | |
| sambamba | Read deduplication | MQ>=20, BQ>=20, ploidy=1 | [45] | 0.8.2 | X | |
| samtools | Coverage analysis | [46] | 1.12 | X | ||
| LoFreq | Variant calling | MQ>=20, BQ>=20 | [47] | 2.1.5 | X | |
| BCFtools | Variant calling, normalization and annotation | MQ>=20, BQ>=20 | [48] | 1.14 | X | X |
| iVar | Variant calling | MQ>=20, BQ>=20 | [49] | 1.3.1 | X | |
| Biopython | Custom variant calling on assemblies sequences based on Needleman-Wunsch global alignment | aligner.mode = 'global'aligner.match = 2aligner.mismatch = -1aligner.open_gap_score = -3aligner.extend_gap_score = -0.1aligner.target_end_gap_score = 0.0aligner.query_end_gap_score = 0.0 | [50] | 1.79 | X | |
| SnpEff | Functional annotations | [51] | 5.0 | X | X | |
| VAFator | Technical annotations | MQ>0, BQ>0 | [14] | 1.2.5 | X | |
| Pangolin | Lineage calling | [52] | 4.1.2 | X | X | |
| ConsHMM | Conservation annotations | [53] | NA | X | X | |
| Pfam | SARS-CoV-2 protein domains | [54] | NA | X | X |
| Approach | Sample filters | Variant filters |
|---|---|---|
| Valesano-like [39] | >= 50,000 mapped reads>= 29,000 bp horizontal coverage | VAF >= 2%, VAF < 50 %DP >= 100>= 10 supporting reads |
| Sapoval-like [34] | >= 20,000 mapped reads | VAF >= 2%, VAF < 50 %DP >= 10Mask extremes of genome + homoplasmic positions [63] |
| Tonkin-Hill-like [33] | Excessive number iSNVs (99.9th percentile)Outlier number of iSNVs with mid-VAFs, between 40% and 80 % | VAF >= 5%, VAF < 50 %DP >= 100>= 5 supporting reads |
| CoVigator approach | >= 50,000 mapped reads>= 29,000 bp horizontal coverageExcessive number iSNVs (99.9th percentile)Outlier number of iSNVs with mid-VAFs, between 40% and 80% | VAF >= 2%, VAF < 50 %DP >= 100>= 10 supporting readsMask extremes of genome + homoplasmic positions [63] from indels <= 10 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





