1. Introduction
Breast cancer is one of the most prevalent cancers in females and one of the leading causes of cancer death worldwide (1, 2). Early detection and accurate characterization of breast malignancies are crucial factors in breast cancer management and positive treatment outcomes (3-12). Differentiation of benign from malignant breast lesions can aid clinicians in determining appropriate therapeutic plans. While histopathological examination of breast tissues extracted by biopsy is often required to confirm a suspicious lesion, a mammogram continues to be the gold standard for detection of breast cancer, but this approach has a high false positive rate (13). Multi-parametric MRI (mp-MRI), which includes dynamic contrast enhanced MRI (DCE-MRI), T2-weighted MRI and diffusion-weighted imaging (DWI) may allow differentiating benign and malignant breast lesions that present highly overlapping enhancement patterns. However, despite the potential to eliminate unnecessary biopsies and follow-up examinations of benign tumors, mp-MRI-based breast tumor differentiation still has increased false positive findings.
Cell density, organization, membrane integrity and cellular metabolism of breast tissues undergo changes in the presence of cancer. Magnetic resonance spectroscopic imaging (MRSI) is capable of detecting the changes in concentrations of various metabolites and lipids in the tissue that are altered due to cancer related changes in cellular metabolism (14-23). High cell density and altered tissue structure due to cancer also lead to restricted motion of water molecules in the tissue, which can be measured by the apparent diffusion coefficient (ADC) on DWI (12, 24-31). DCE-MRI, one of the most sensitive diagnostic techniques, highlights the areas of increased blood flow and blood volume in the breast tissues due to cancer with the help of a contrast agent (5-7, 12, 32-35).
Even though the sensitivity of mp-MRI methods can be affected by various factors like tumor size and aggressiveness, these methods are often reported to have relatively high sensitivity (in the range of 88–100% for DCE-MRI, 85-95% for DWI and 80% for MRSI) (9, 12, 36-39). Reported specificity on the other hand, is relatively low (69-74% for DCE-MRI, 75–82% for DWI, and 74% for MRSI), restricting the capability in classification of benign and malignant lesions (37-40). While single-voxel spectroscopy has reported 64–82% sensitivity and 85–91% specificity (41), the multi-voxel technique of MRSI can cover a larger area of the breast with a relatively higher spatial resolution. Advanced MRSI techniques like five-dimensional (5D) echo-planar correlated spectroscopic imaging (EP-COSI) can record two-dimensional (2D) correlated spectroscopy (COSY) from multiple regions in three-dimensional (3D) space (42). Achieving high specificity is challenging due to overlapping patterns of diagnostic measures between benign and malignant lesions.
One option to potentially improve the specificity while retaining the benefit of the non-invasive nature of these imaging modalities is to use machine learning (ML) models to identify subtle or complex differences in the multi-model data that differentiate benign and malignant lesions (43, 44). Development and validation of machine learning models have seen an impressive growth in the last decade due to their high accuracy and flexibility in handling a wide range of datatypes and features (45). While individual machine learning models may perform well, a meta-approach that combines individual models named ensemble learning could generate even more generalizable models that can reduce individual base learner’s variance or bias (46). In particular, advanced ensemble models like the gradient-boosted tree-based algorithm that combine multiple weak learners (decision trees) are shown to be capable of detecting key features of the multi-modal, multi-parametric imaging information for applications such as tissue/cancer grade classification (47-50).
Multiple studies have recently shown that the features extracted from DCE-MRI and DWI of breast tissues used in ML models are capable of predicting tumor grades, and classifying benign and malignant breast lesions (48-50). However, metabolite and lipid information from MRSI data has not been used in this context so far. Therefore, a major goal of this work was to evaluate the application of different machine learning models including ensemble learning techniques for the classification of benign and malignant breast lesions based on the 5D EP-COSI data with and without the corresponding ADC information from DWI data.
2. Materials and Methods
2.1. Subjects and Data Acquisition
The dataset consisted of 5D EP-COSI and DWI data from twenty-three subjects with malignant breast masses (mean age 52 [range:33–71] years and seventeen benign breast masses (mean age 37 [range:19–60] years). All scans were acquired on a Siemens 3T Skyra scanner (Siemens Healthineer, Erlangen, Germany). The 5D EP-COSI data was acquired using FOV = 160×160×120 mm3, matrix size = 16×16×8, TR/TE = 1500/35 ms, 64 t1 points and 512 t2 points with a spectral width of 1250 Hz and 1190 Hz along F1 and F2 respectively. A non-water-suppressed (NWS) 1D MRSI scan with one t1 point was acquired for eddy current phase correction and phase correction for combining signals from multiple-receive coils (51). The data was non-uniformly undersampled (NUS) along two spatial ky-kz and the spectral t1 dimensions with a total acceleration factor of 8, and was reconstructed using Group Sparsity (GS) - based compressed sensing technique (52, 53).
The DWI acquisition protocol included the following: 2D spin-echo echo-planar imaging (EPI) sequence (TR/TE of 3800/93ms; data matrix, 192 × 192; signal average, 3; slice thickness, 3 mm; distance factor, 20%) in the axial plane. Diffusion sensitizing gradients (DSG) in three orthogonal directions with b values of 50 and 800 s/mm2 were applied. The ADC maps were created automatically by the in-line scanner software using the trace-weighted images with b values of 50 and 800 s/mm2.
2.2. Pre-Processing
Tumor-containing slices in the DWI were selected and the boundaries of the lesion were marked by a radiologist. ADC values were then extracted from this delineated region of interest (ROI). The MRSI data were interpolated by a factor of 2 and the slices containing the tumor were identified similar to DWI. Spectroscopic voxels within the delineated region were extracted and the metabolite and lipid ratios were quantified in these voxels as described in (42). All variables were standardized with z-score normalization (zero mean and unit standard deviation) and voxels containing outlier measurements were removed. For the variables that followed a normal distribution, outliers were identified as three standard deviations away from the mean. For other variables, previously reported ranges of metabolite and lipid ratios were used as a guideline for outliers (42).
2.3. Feature Extraction
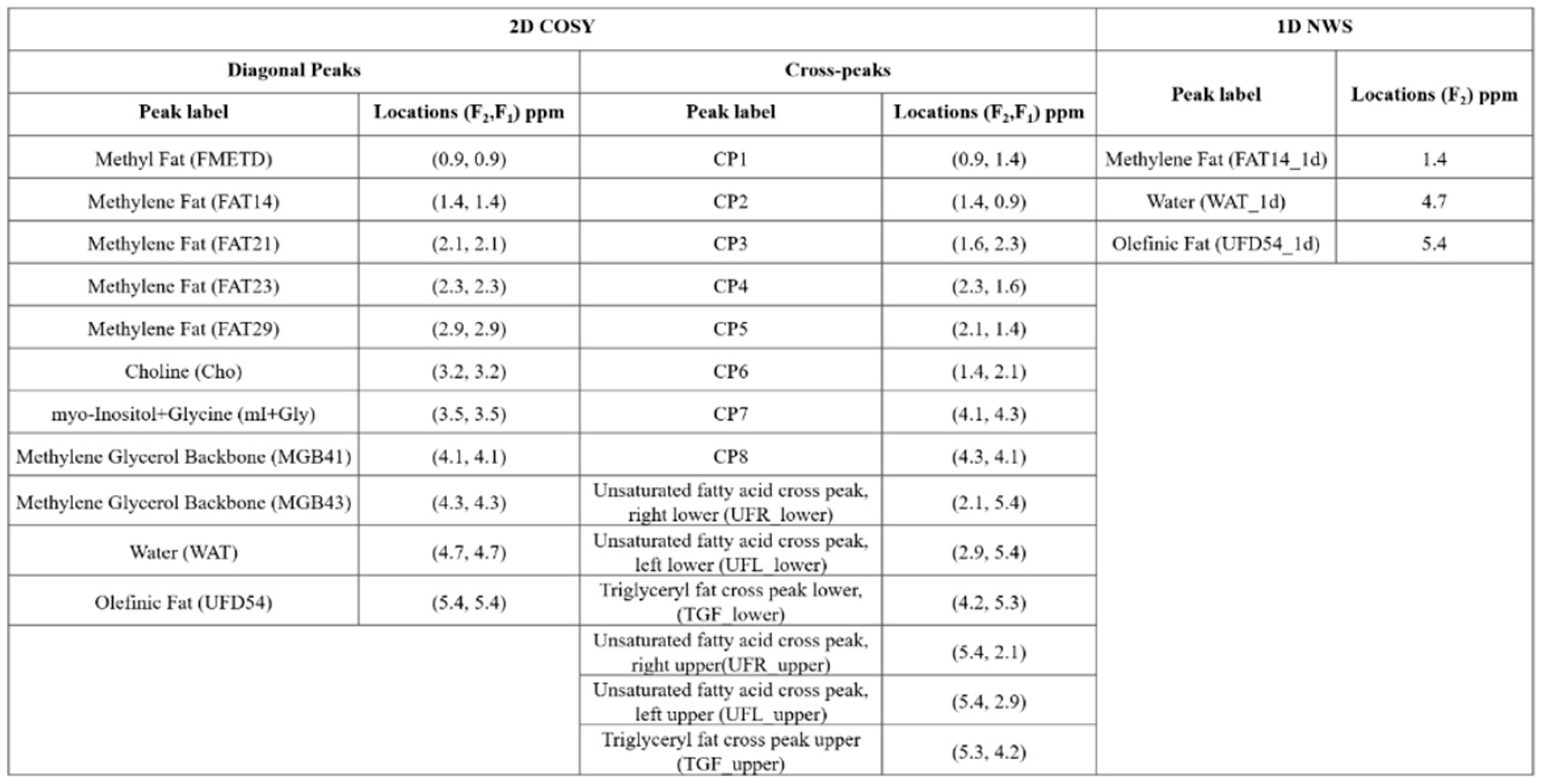
Ratios of 24 metabolites and lipids from from water-suppressed 2D spectra and 1D NWS spectra including choline (Cho), myo-Inositol+glycine (mI+Gly), Unsaturated fatty acid and Triglyceryl fat cross-peaks with respect to methylene fat, olefinic fat and water were estimated from the MRSI data. The full list of metabolites and lipids identified in the 2D correlated spectroscopy (COSY) and 1D NWS spectra are shown in
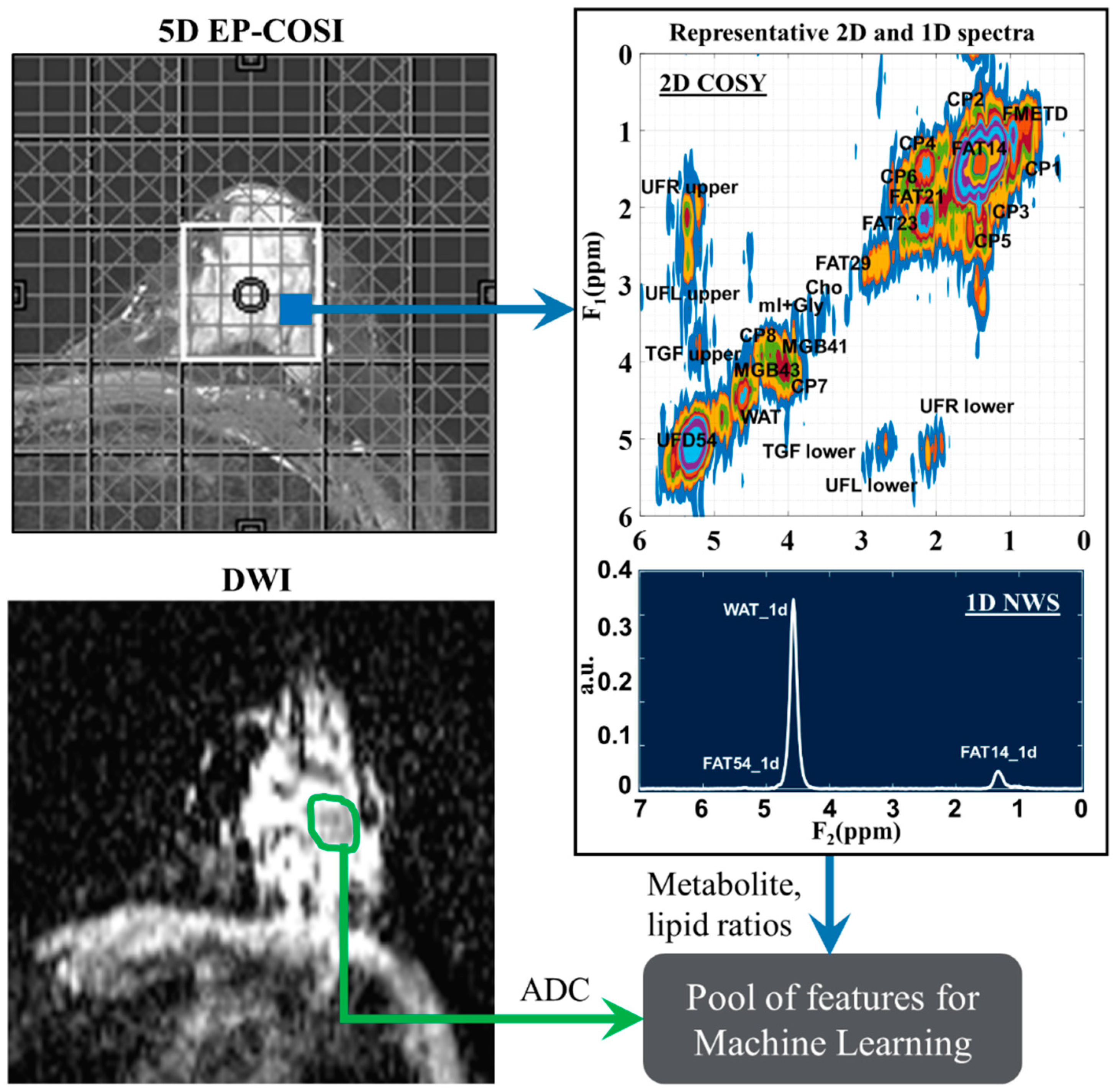
Table 1. The list of 99 quantitative features for ML analysis included metabolite and lipid ratios from 2D spectra, ADC values and water fraction, fat fraction and water-to-fat ratios from NWS spectra. A representative 2D COSY spectrum with labeled metabolite and lipid diagonal and cross peaks along with corresponding ADC map is shown in
Figure 1.
2.4. Feature Selection
The entire dataset was split into approximately 80% for training and 20% for testing in such a way that the metabolite and lipid ratios from different voxels in the same lesion were not present in both training and testing sets. This ensured that the samples in the training and testing set were independent, which in turn avoided overestimation of model performance due to data leakage so that the model will be generalizable to new data.
Recursive feature elimination (RFE) was used to determine the best combination of variables to maximize the model’s classification accuracy (54). A statistical significance test was used to narrow down the variable space before running the feature selection algorithms. Quantile-Quantile (Q-Q) plots of the data were used to check for normality and Levene's tests were used to check for the homogeneity of variances. Based on that, either a t-test or Mann-Whitney U (MWU) test for a statistical significance of p-value<0.05 was used. Correction for multiple comparisons was performed using Bonferroni correction methods. The base model for RFE was selected based on the model performance considering all the significant features identified in the statistical test.
2.5. Machine Learning Algorithms
The open-source machine learning library for Python, ‘scikit-learn’ was used for implementing different supervised learning algorithms for classification (55), which included support vector machine (SVM), Naive Bayes, and K-nearest neighbors (KNN) as well as ensemble learning techniques including Adaptive Boosting (AdaBoost), GradientBoost, Extreme Gradient Boost (XGBoost), Light Gradient Boost, Categorical Boost (CatBoost) RandomForest and Decision Tree based bagging classifiers (56-58). In bagging, the training data was divided into different parts by random sampling with replacement and multiple models were trained on these different subsets. It then combined the prediction of each of the models by averaging. Boosting, on the other hand, used multiple base learners like decision trees in a sequential manner where the successive learner corrected for the error in prediction by previous one.
2.6. Cross-Validation and Parameter Tuning
To ensure our classification model would generalize to unseen patient groups, the data was split into train and test sets in a way such that they were exclusive of each other - data from the same patient appears only in one set using the Shuffle Group Split. Likewise, the Grouped K-Folds cross validation method was used in both feature selection and hyperparameter tuning to return stratified folds with non-overlapping groups that are representative of the class distributions of the dataset. The train set was then standardized and the test set was standardized with the train set's statistics. The models were then optimized using the cross-validated Grid Search method, which exhaustively searched through the given hyperparameter space for the best combination of hyperparameters for the model and the training data.
2.7. Evaluation Metrics
The classification performance of the different machine learning models in the testing stage was compared based on the scores of (a) accuracy (ratio of correct predictions to total number of predictions), (b) F1 score (2*((precision*recall)/(precision+recall))) and (c) area under the receiver operating characteristic (ROC) curve.
2.8. Statistical Analysis
Statistical tests were performed to compare the performance of the machine learning models. One-way Analysis of Variance (ANOVA) test (in RStudio (version 4.1.1)) was used for this comparison based on the evaluation matrices for a statistical significance level of p-value<0.05. Tukey's HSD (honestly significant difference) post-hoc test was used for pair-wise analysis of these models with p-values adjusted for multiple comparisons using the Bonferroni method.
3. Results
3.1. Feature Selection and Comparison
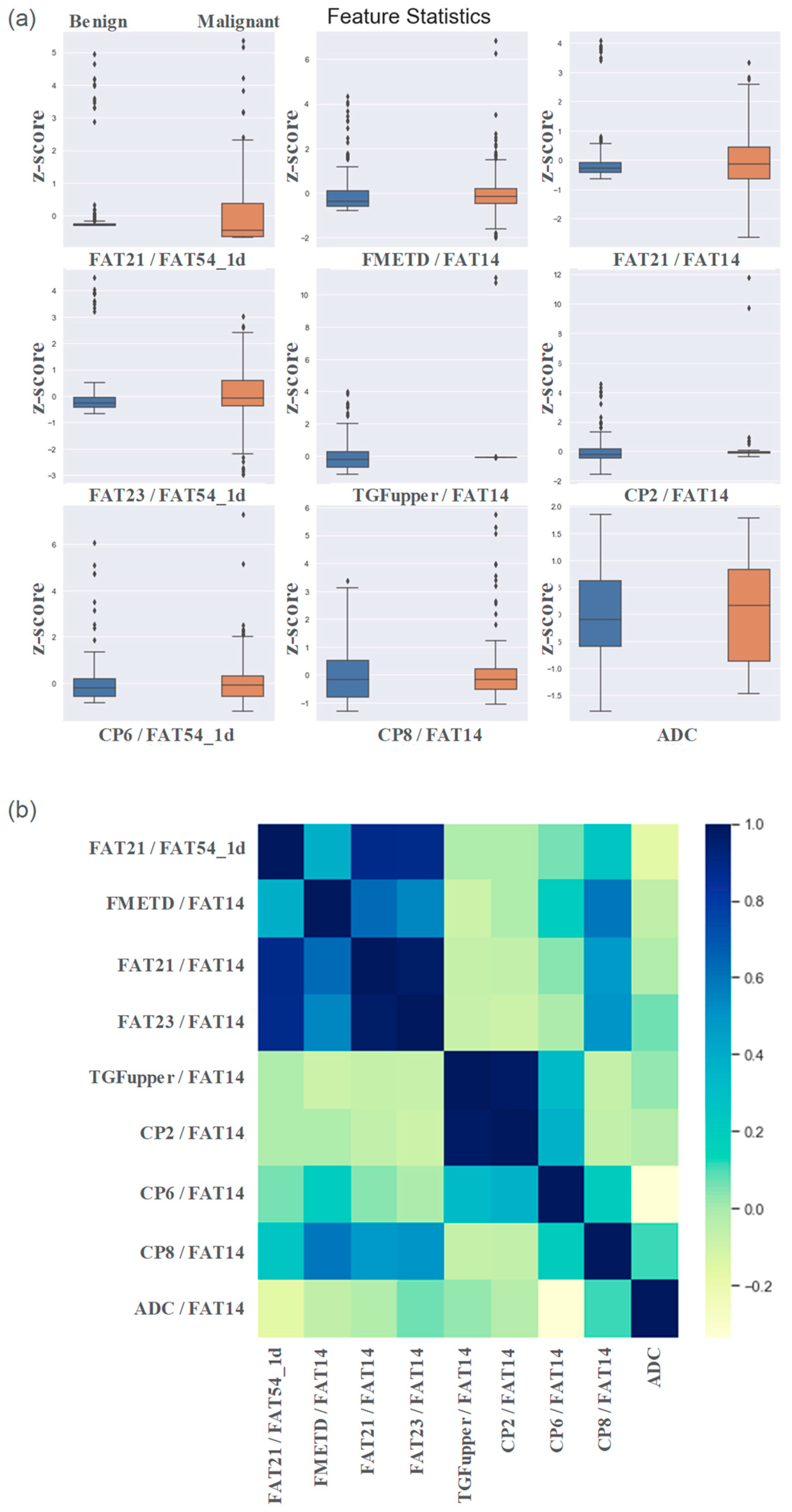
Based on the results of the Mann-Whitney U test comparing the benign and malignant classes, the feature set was narrowed down to 86 that were statistically significant at p ≤0.05. Nine out of these 86 features were identified as the most important by RFE, and included ADC, ratios of FAT21, FAT23, CP (1.4-0.9), CP (2.1-1.4), CP (4.3-4.1) and TGF upper with respect to FAT14, FMETD ratio with respect to WAT47 (1D) and the ratio of FAT21 with respect to FAT54 (1D). The boxplots of these most significant features are shown in
Figure 2a for both malignant and benign classes. The values were z-score normalized.
Figure 2b shows the correlation heatmap of these features.
3.2. Comparison of Models
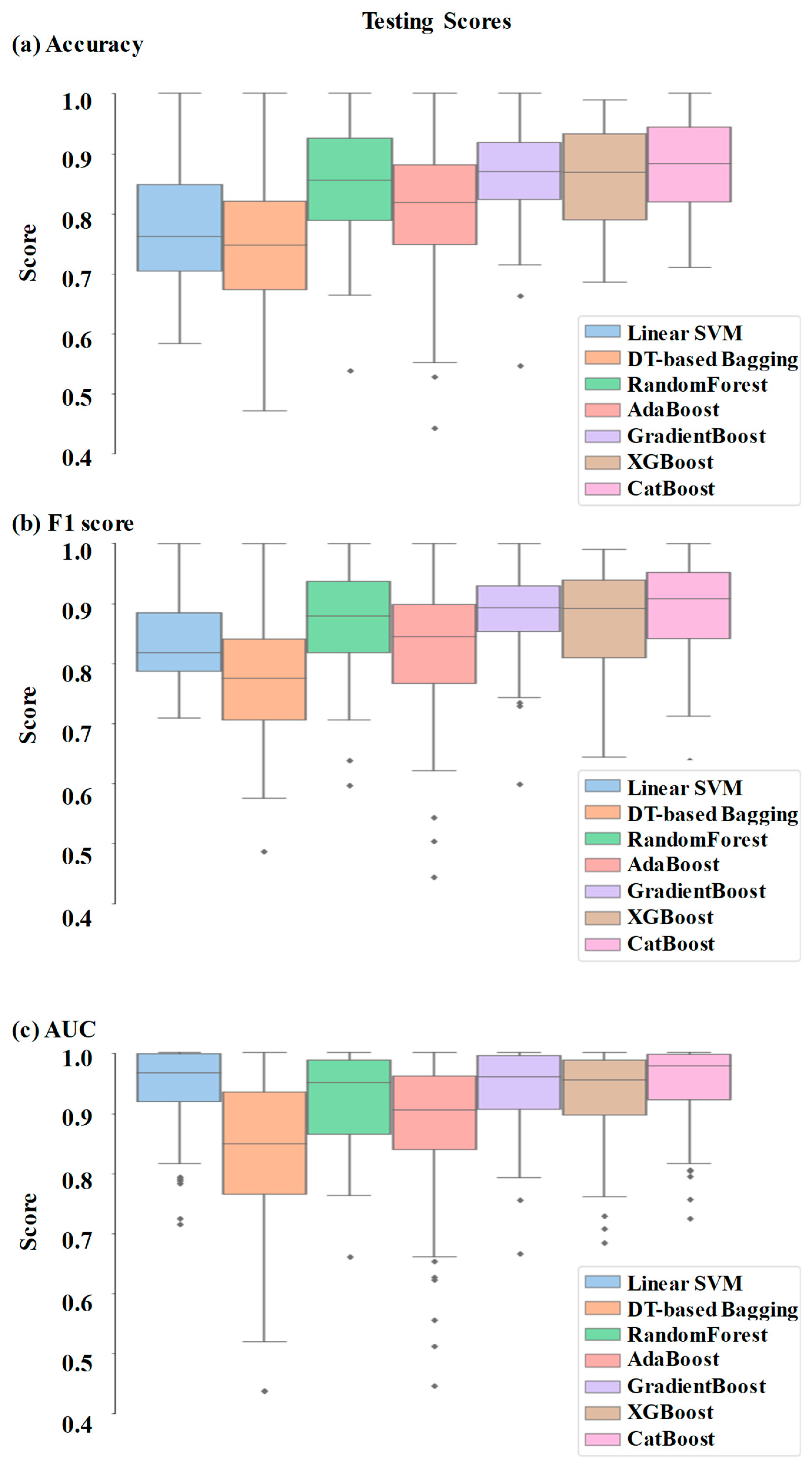
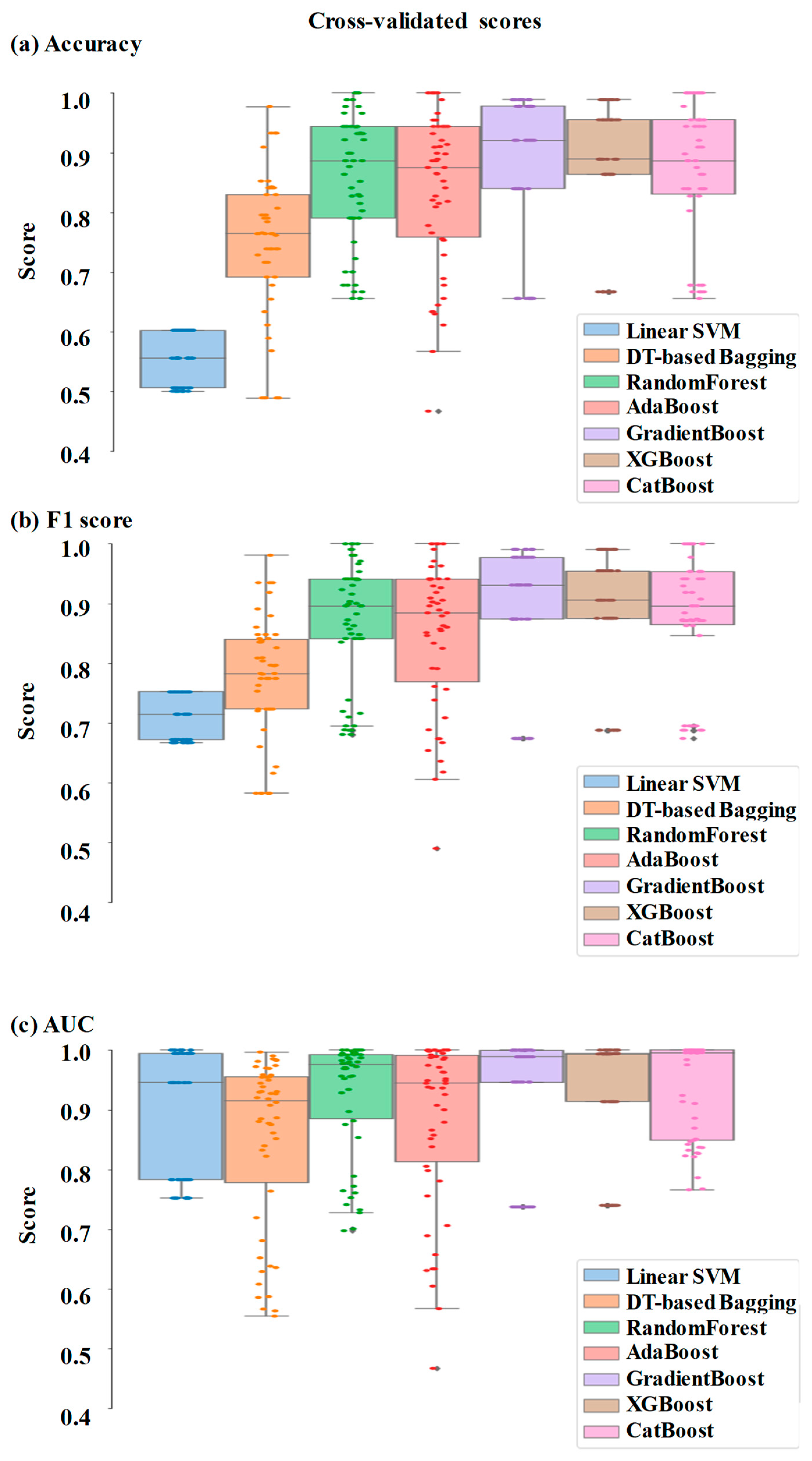
Comparative performance of linear SVM, DT-based bagging classifier, random forest, AdaBoost, GradientBoost, XGBoost and CatBoost are shown in
Figure 3,
Figure 4 and
Figure 5. These models were the best performing out of all the models considered in terms of performance metrics.
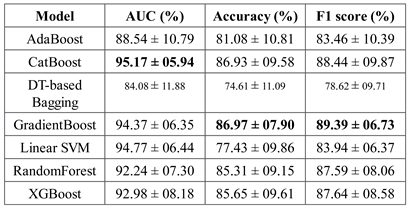
Figure 3 shows the AUC, F1 score, and accuracy of these seven classifiers in the testing stage repeated 100 times with randomized dataset split and model initializations, and
Figure 4 shows these scores in the cross-validation stage repeated 50 times. The respective box plots show the median and interquartile range (IQR) of these matrices, along with outliers. Their corresponding mean and standard deviation are listed in
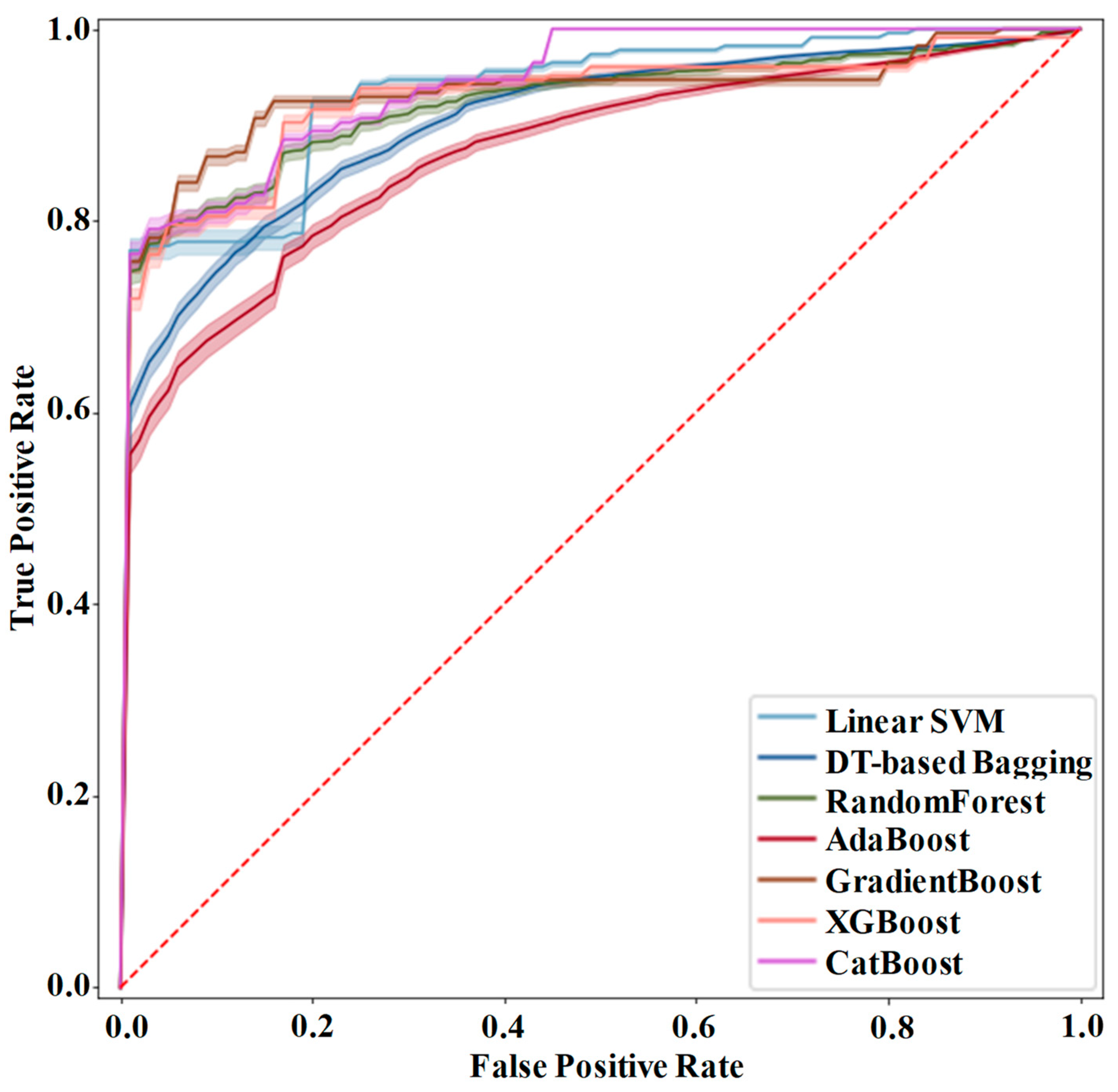
Table 2. ROC curves of these different models are shown in
Figure 5.
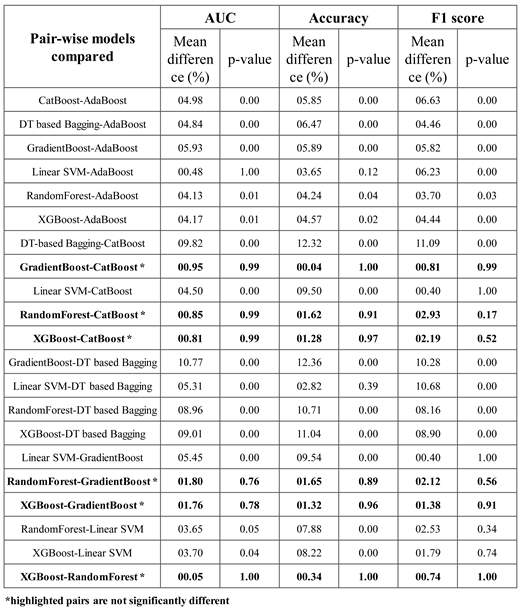
The results of Tukey's HSD post-hoc test following the ANOVA with p-values adjusted for multiple comparisons are shown in
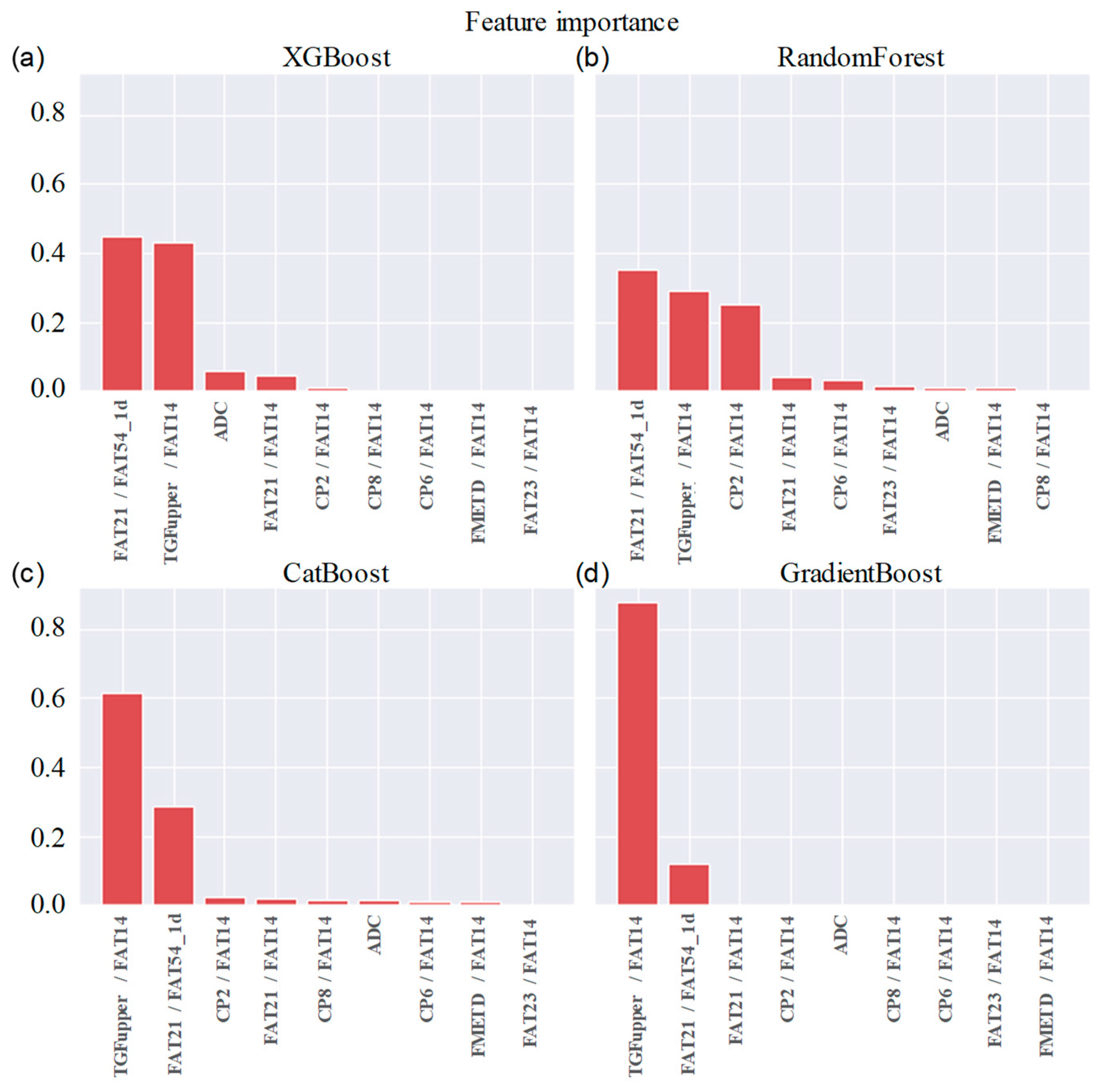
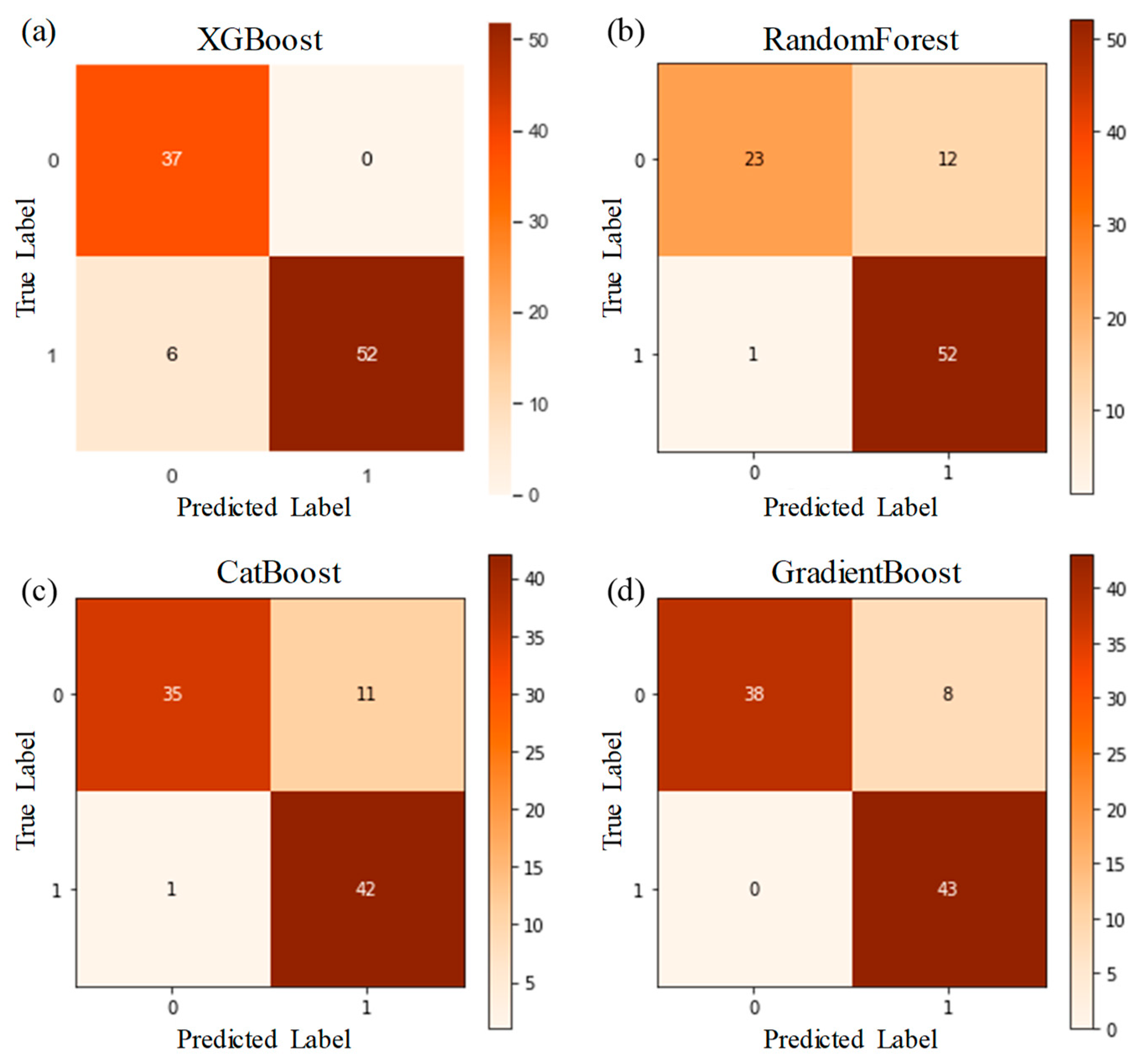
Table 3. It shows that the difference between the ensemble models XGboost, GradientBoost, CatBoost and RandonForest were not statistically significant in terms of Accuracy, AUC and F1 scores for the significance at p≤0.05. The feature importance and confusion matrix identified for these four classifiers are shown in
Figure 6 and
Figure 7, respectively.
3.3. Ablation Study
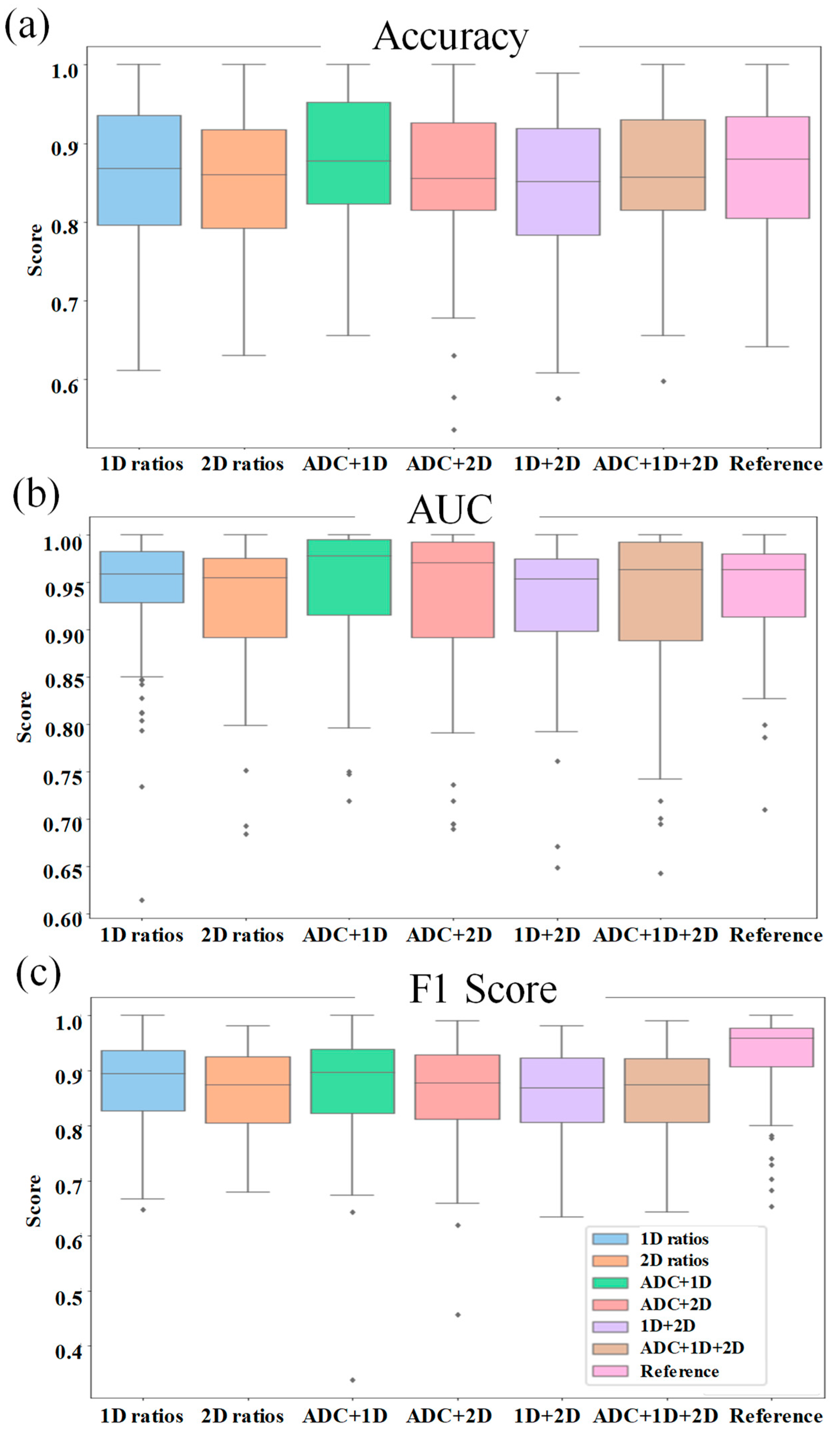
An ablation study was performed with choline, glycine + myo-Inositol, unsaturation index, and the features from non-water suppressed 1D spectra including water fraction, fat fraction and water-fat ratios. The model performance was also compared with and without ADC in the feature list. While the changes observed in the mean accuracy score over 100 repetitions were less than 1%, the change was positive in the presence of ADC (0.02%), while it was marginally negative in the presence of the additional metabolite ratios (<-0.009%). Boxplots comparing the performance matrices of the ablation study are shown in
Figure 8.
4. Discussion
This study showed the feasibility of using metabolite ratios from 5D EP-COSI and ADC values from the DWI data of breast cancer patients to train machine learning models for classifying benign and malignant lesions. While earlier studies have attempted to use lesion characterization using features extracted from the DWI and DCE-MRI data, these models did not use the quantitative measures of metabolite and lipid features which can be obtained with an MRSI examination (48-50). Although variations in water and fat levels can become ambiguous in glandular regions, especially in benign and healthy tissues, various lipid and metabolite ratios are reported to have statistically significant difference between the benign and malignant lesions (42). Building on this fact, our study pursued a detailed analysis of lesion characterization using 5D EP-COSI features in a machine learning framework.
The ensemble models were found to be performing better than the individual models. This is as expected, since they combine the strengths of multiple individual models (45). In fact, the ensemble models can use multiple base models to learn different aspects of the data and hence learn more complex relationships between the variables. They are also more robust to outliers and are also expected to reduce overfitting since they can compensate for the prediction errors of individual models. XGBoost, GradientBoost, RandomForest and CatBoost were found to be the best performing ensemble models in this study with 92% to 95% AUC, 85% to 87% accuracy and 87% to 90% F1 scores.
While the feature importance scores slightly varied among the top performing models, ADC was ranked among the top six. Four out of the top nine features were the ratios of cross-peaks, which are specific to the 2D COSY technique. The remaining four main features were the ratios of diagonal lipid peaks. It is interesting to note that the ratios of lipid cross peaks ranked higher than some of the conventional biomarkers like Cho and mI+Gly ratios for classifying benign and malignant lesions in the ML framework, despite both Cho and mI+Gly ratios being in the list of statistically significant variables in the MWU tests. The RFE process to select the significant features used XGBoost as the base model since it was one of the best-performing models based on all the variables from MWU tests. Therefore, it is to be noted that the top features were determined by their contribution in reducing loss function in the decision trees.
The number of datasets is one of the limitations in this study. Even though we have multiple voxels from the same dataset giving metabolite and lipid ratios, it is important to split the data based on the actual number of subjects rather than the voxels. It would be tempting to consider the individual voxels as separate data when splitting the training and testing sets. However, this approach could lead to severe data leakage, since multiple voxels from the same subject can have similar statistics, especially when interpolation is used to increase the number of voxels. Even otherwise, if the lesion spans multiple voxels in the spectroscopic data, the relatively low resolution and partial volume effects can potentially cause slightly overlapping information between the neighboring voxels. Therefore, if the train-test split is performed based on the voxels rather than individual subjects, it is reasonable to assume that during the training stage, the model would already see some of the statistics present in the testing data. This will artificially increase the score of test and validation performance matrices, but will not be generalizable to a new subject.
Even though these ML models should be generalizable to the MRSI/DWI data from different scanners and sites, it may be considered as another limitation of this study since there could be subtle/complex variations in the datasets from different scanners and sites so that the list of most important features could differ. A future study with a larger sample size ideally from different scanners and sites can further validate the results presented in this work.
Since the focus of this study was to analyze the performance of ML models with features from the 5D EP-COSI data, we have not considered some of the image-based features potentially available from DWI. For example, it has been recently shown that the features based on continuous-time random-walk (CTRW) and intravoxel incoherent motion (IVIM) models from DWI using multiple b-values can classify benign and malignant breast lesions using ensemble ML models (48). The ablation study shows that the presence of ADC values in the list of features improves the score of performance matrices. Therefore, more features from DWI as well as other modalities like DCE-MRI can be used in a future study to potentially further improve the model performance.
5. Conclusion
In this pilot validation of the multi-dimensional (5D EP-COSI) data for characterization of breast tissues, we have shown that ML based classification models can be trained using spectroscopic features in conjunction with ADC values from DWI to classify benign and malignant lesions. Multiple diagonal and cross-peaks from 2D COSY spectra were identified as important features, further asserting the advantage of 2D COSY spectra as compared to features derived from 1D spectra. GradientBoost, CatBoost, RandomForest and XGBoost were the best performing models with 92% - 96% AUC, 85% - 87% Accuracy and 87% - 90% F1-scores.
Author Contributions
A.J. reconstructed the raw data, performed machine learning analysis, and prepared original draft; M.L. did the machine learning analysis; A.S. performed the data acquisition; M.J. and S.L.F. assisted with lesion analysis and manuscript editing; P.M.M. and A.S. edited the manuscript; A.C., U.E., J.B. proof-read the final draft; M.A.T. conceived the experimental design, acquired funding and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a CDMRP grant from the US Army Breast Cancer Research Program:# W81XWH-16-1-0524.
Acknowledgments
Authors like to acknowledge the scientific support of Dr. Manoj Sarma, Dr Zohaib Iqbal, Dr Brian Burns, Dr Neil Wilson, Ms. Kavya Umachandran and Ms. Samantha Joseph. Also, authors would like to thank the support of Ms. Victoria Rueda with the recruitment of study subjects and the UCLA Radiology MRI technicians during data collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73(1):17-48. [CrossRef]
- Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020;70(4):313. [CrossRef]
- Al-Ajmi K, Lophatananon A, Yuille M, Ollier W, Muir KR. Review of non-clinical risk models to aid prevention of breast cancer. Cancer Causes Control. 2018;29(10):967-86. [CrossRef]
- Fu B, Liu P, Lin J, Deng L, Hu K, Zheng H. Predicting invasive disease-free survival for early stage breast cancer patients using follow-up clinical data. IEEE Trans. Biomed. Eng. 2018;66(7):2053-64. [CrossRef]
- Jagannathan N. Breast MR. NMR Biomed. 2009;22(1):1-2.
- Lehman CD, Isaacs C, Schnall MD, Pisano ED, Ascher SM, Weatherall PT, et al. Cancer yield of mammography, MR, and US in high-risk women: prospective multi-institution breast cancer screening study. Radiology. 2007;244(2):381-8. [CrossRef]
- Morris EA. Diagnostic breast MR imaging: current status and future directions. Radiol. Clin. North Am. 2007;45(5):863-80. [CrossRef]
- Pe M, Dorme L, Coens C, Basch E, Calvert M, Campbell A, et al. Statistical analysis of patient-reported outcome data in randomised controlled trials of locally advanced and metastatic breast cancer: a systematic review. The Lancet Oncology. 2018;19(9):e459-e69. [CrossRef]
- Pop CF, Stanciu-Pop C, Drisis S, Radermeker M, Vandemerckt C, Noterman D, et al. The impact of breast MRI workup on tumor size assessment and surgical planning in patients with early breast cancer. The breast journal. 2018;24(6):927-33. [CrossRef]
- Saslow D, Boetes C, Burke W, Harms S, Leach MO, Lehman CD, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J. Clin. 2007;57(2):75-89. [CrossRef]
- Weinreb JC, Newstead G. MR imaging of the breast. Radiology. 1995;196(3):593-610. [CrossRef]
- Zhang M, Horvat JV, Bernard-Davila B, Marino MA, Leithner D, Ochoa-Albiztegui RE, et al. Multiparametric MRI model with dynamic contrast-enhanced and diffusion-weighted imaging enables breast cancer diagnosis with high accuracy. J. Magn. Reson. Imaging. 2019;49(3):864-74. [CrossRef]
- Warner E. Breast-cancer screening. New England Journal of Medicine. 2011;365(11):1025-32. [CrossRef]
- Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59(1):80-4.
- Bolan PJ, Kim E, Herman BA, Newstead GM, Rosen MA, Schnall MD, et al. MR spectroscopy of breast cancer for assessing early treatment response: Results from the ACRIN 6657 MRS trial. J. Magn. Reson. Imaging. 2017;46(1):290-302. [CrossRef]
- Dorrius MD, Pijnappel RM, Jansen-van der Weide MC, Jansen L, Kappert P, Oudkerk M, et al. Determination of choline concentration in breast lesions: quantitative multivoxel proton MR spectroscopy as a promising noninvasive assessment tool to exclude benign lesions. New diagnostic developments to prevent unnecessary invasive procedures in breast cancer diagnostic work-up. 2011. [CrossRef]
- Gribbestad I, Sitter B, Lundgren S, Krane J, Axelson D. Metabolite composition in breast tumors examined by proton nuclear magnetic resonance spectroscopy. Anticancer Res. 1999;19(3A):1737-46.
- Haukaas TH, Euceda LR, Giskeødegård GF, Bathen TF. Metabolic portraits of breast cancer by HR MAS MR spectroscopy of intact tissue samples. Metabolites. 2017;7(2):18. [CrossRef]
- Jagannathan N, Seenu V, Kumar M. Potential of in vivo proton MR spectroscopy in the assessment of breast lesions without the use of contrast agent. Radiology. 2002;223(1):281-2. [CrossRef]
- Roebuck JR, Cecil KM, Schnall MD, Lenkinski RE. Human breast lesions: characterization with proton MR spectroscopy. Radiology. 1998;209(1):269-75. [CrossRef]
- Sharma U, Mehta A, Seenu V, Jagannathan N. Biochemical characterization of metastatic lymph nodes of breast cancer patients by in vitro 1H magnetic resonance spectroscopy: a pilot study. Magn. Reson. Imaging. 2004;22(5):697-706. [CrossRef]
- Thakur SB, Horvat JV, Hancu I, Sutton OM, Bernard-Davila B, Weber M, et al. Quantitative in vivo proton MR spectroscopic assessment of lipid metabolism: Value for breast cancer diagnosis and prognosis. J. Magn. Reson. Imaging. 2019;50(1):239-49. [CrossRef]
- Thomas MA, Binesh N, Yue K, DeBruhl N. Volume-localized two-dimensional correlated magnetic resonance spectroscopy of human breast cancer. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2001;14(2):181-6. [CrossRef]
- Amornsiripanitch N, Nguyen VT, Rahbar H, Hippe DS, Gadi VK, Rendi MH, et al. Diffusion-weighted MRI characteristics associated with prognostic pathological factors and recurrence risk in invasive ER+/HER2–breast cancers. J. Magn. Reson. Imaging. 2018;48(1):226-36. [CrossRef]
- Bammer R. Basic principles of diffusion-weighted imaging. Eur. J. Radiol. 2003;45(3):169-84. [CrossRef]
- Belli P, Costantini M, Bufi E, Magistrelli A, La Torre G, Bonomo L. Diffusion-weighted imaging in breast lesion evaluation. Radiol. Med. 2010;115(1):51-69. [CrossRef]
- Delbany M, Bustin A, Poujol J, Thomassin-Naggara I, Felblinger J, Vuissoz PA, et al. One-millimeter isotropic breast diffusion-weighted imaging: Evaluation of a superresolution strategy in terms of signal-to-noise ratio, sharpness and apparent diffusion coefficient. Magn. Reson. Med. 2019;81(4):2588-99. [CrossRef]
- deSouza NM. Diffusion-weighted MRI in multicenter trials of breast cancer: a useful measure of tumor response? : Radiological Society of North America; 2018. p. 628-9. [CrossRef]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401-7. [CrossRef]
- Newitt DC, Zhang Z, Gibbs JE, Partridge SC, Chenevert TL, Rosen MA, et al. Test–retest repeatability and reproducibility of ADC measures by breast DWI: Results from the ACRIN 6698 trial. J. Magn. Reson. Imaging. 2019;49(6):1617-28. [CrossRef]
- Sharma U, Danishad KKA, Seenu V, Jagannathan NR. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In vivo. 2009;22(1):104-13. [CrossRef]
- Furman-Haran E, Grobgeld D, Kelcz F, Degani H. Critical role of spatial resolution in dynamic contrast-enhanced breast MRI. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2001;13(6):862-7. [CrossRef]
- Hickman P, Moore N, Shepstone B. The indeterminate breast mass: assessment using contrast enhanced magnetic resonance imaging. The British Journal of Radiology. 1994;67(793):14-20. [CrossRef]
- Kvistad KA, Rydland J, Vainio J, Smethurst HB, Lundgren S, Fjøsne HE, et al. Breast lesions: evaluation with dynamic contrast-enhanced T1-weighted MR imaging and with T2*-weighted first-pass perfusion MR imaging. Radiology. 2000;216(2):545-53. [CrossRef]
- Liu P, Debatin J, Caduff R, Kacl G, Garzoli E, Krestin G. Improved diagnostic accuracy in dynamic contrast enhanced MRI of the breast by combined quantitative and qualitative analysis. The British journal of radiology. 1998;71(845):501-9. [CrossRef]
- Millet I, Pages E, Hoa D, Merigeaud S, Curros Doyon F, Prat X, et al. Pearls and pitfalls in breast MRI. The British journal of radiology. 2012;85(1011):197-207. [CrossRef]
- Bogner W, Gruber S, Pinker K, Grabner G, Stadlbauer A, Weber M, et al. Diffusion-weighted MR for differentiation of breast lesions at 3.0 T: how does selection of diffusion protocols affect diagnosis? Radiology. 2009;253(2):341-51. [CrossRef]
- Chen X, Li W-l, Zhang Y-l, Wu Q, Guo Y-m, Bai Z-l. Meta-analysis of quantitative diffusion-weighted MR imaging in the differential diagnosis of breast lesions. BMC Cancer. 2010;10(1):1-11. [CrossRef]
- Prvulovic Bunovic N, Sveljo O, Kozic D, Boban J. Is Elevated Choline on Magnetic Resonance Spectroscopy a Reliable Marker of Breast Lesion Malignancy? Front. Oncol. 2021;11:610354.
- Shahraki Z, Ghaffari M, Parooie F, Salarzaei M. Preoperative evaluation of breast cancer: Contrast-enhanced mammography versus contrast-enhanced magnetic resonance imaging: A systematic review and meta-analysis. Breast Dis. 2022;41(1):303-15. [CrossRef]
- Baltzer PA, Dietzel M. Breast lesions: diagnosis by using proton MR spectroscopy at 1.5 and 3.0 T—systematic review and meta-analysis. Radiology. 2013;267(3):735-46. [CrossRef]
- Joy A, Saucedo A, Joines M, Lee-Felker S, Kumar S, Sarma MK, et al. Correlated MR spectroscopic imaging of breast cancer to investigate metabolites and lipids: acceleration and compressed sensing reconstruction. BJR| Open. 2022;4:20220009. [CrossRef]
- Aamir S, Rahim A, Aamir Z, Abbasi SF, Khan MS, Alhaisoni M, et al. Predicting breast cancer leveraging supervised machine learning techniques. Comput. Math. Methods Med. 2022;2022. [CrossRef]
- Dou Y, Meng W. An Optimization Algorithm for Computer-Aided Diagnosis of Breast Cancer Based on Support Vector Machine. Frontiers in Bioengineering and Biotechnology. 2021;9:698390. [CrossRef]
- Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nature Reviews Molecular Cell Biology. 2022;23(1):40-55. [CrossRef]
- Zhou Z-H, Zhou Z-H. Ensemble learning: Springer; 2021.
- Qi C, Li Y, Fan X, Jiang Y, Wang R, Yang S, et al. A quantitative SVM approach potentially improves the accuracy of magnetic resonance spectroscopy in the preoperative evaluation of the grades of diffuse gliomas. NeuroImage: Clinical. 2019;23:101835. [CrossRef]
- Mehta R, Bu Y, Zhong Z, Dan G, Zhong P-S, Zhou C, et al. Characterization of breast lesions using multi-parametric diffusion MRI and machine learning. Phys. Med. Biol. 2023;68(8):085006. [CrossRef]
- Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ breast cancer. 2017;3(1):43. [CrossRef]
- Daimiel Naranjo I, Gibbs P, Reiner JS, Lo Gullo R, Sooknanan C, Thakur SB, et al. Radiomics and machine learning with multiparametric breast MRI for improved diagnostic accuracy in breast cancer diagnosis. Diagnostics. 2021;11(6):919. [CrossRef]
- Klose U. In vivo proton spectroscopy in presence of eddy currents. Magn. Reson. Med. 1990;14(1):26-30. [CrossRef]
- Burns BL, Wilson NE, Thomas MA. Group sparse reconstruction of multi-dimensional spectroscopic imaging in human brain in vivo. Algorithms. 2014;7(3):276-94. [CrossRef]
- Wilson NE, Burns BL, Iqbal Z, Thomas MA. Correlated spectroscopic imaging of calf muscle in three spatial dimensions using group sparse reconstruction of undersampled single and multichannel data. Magn. Reson. Med. 2015;74(5):1199-208. [CrossRef]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine learning. 2002;46:389-422. [CrossRef]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. the Journal of machine Learning research. 2011;12:2825-30.
- Buitinck L, Louppe G, Blondel M, Pedregosa F, Mueller A, Grisel O, et al. API design for machine learning software: experiences from the scikit-learn project. arXiv preprint arXiv:1309.0238. 2013. [CrossRef]
- Chen T, Guestrin C, editors. Xgboost: A scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; 2016. [CrossRef]
- Dorogush AV, Ershov V, Gulin A. CatBoost: gradient boosting with categorical features support. arXiv preprint arXiv:1810.11363. 2018. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).