Submitted:
17 April 2023
Posted:
18 April 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Results
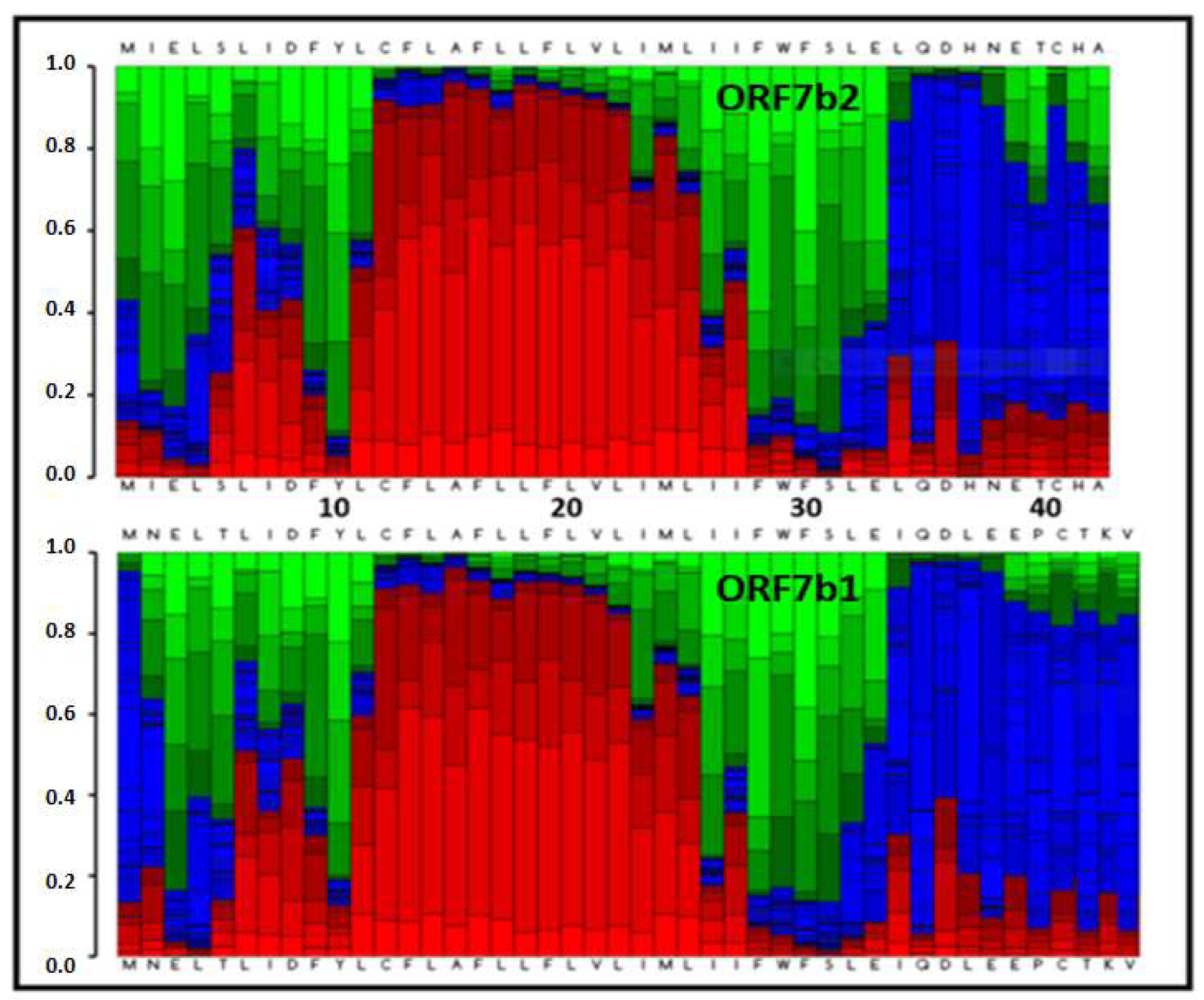
2.1. Sequence
2.2. Electrostatic properties
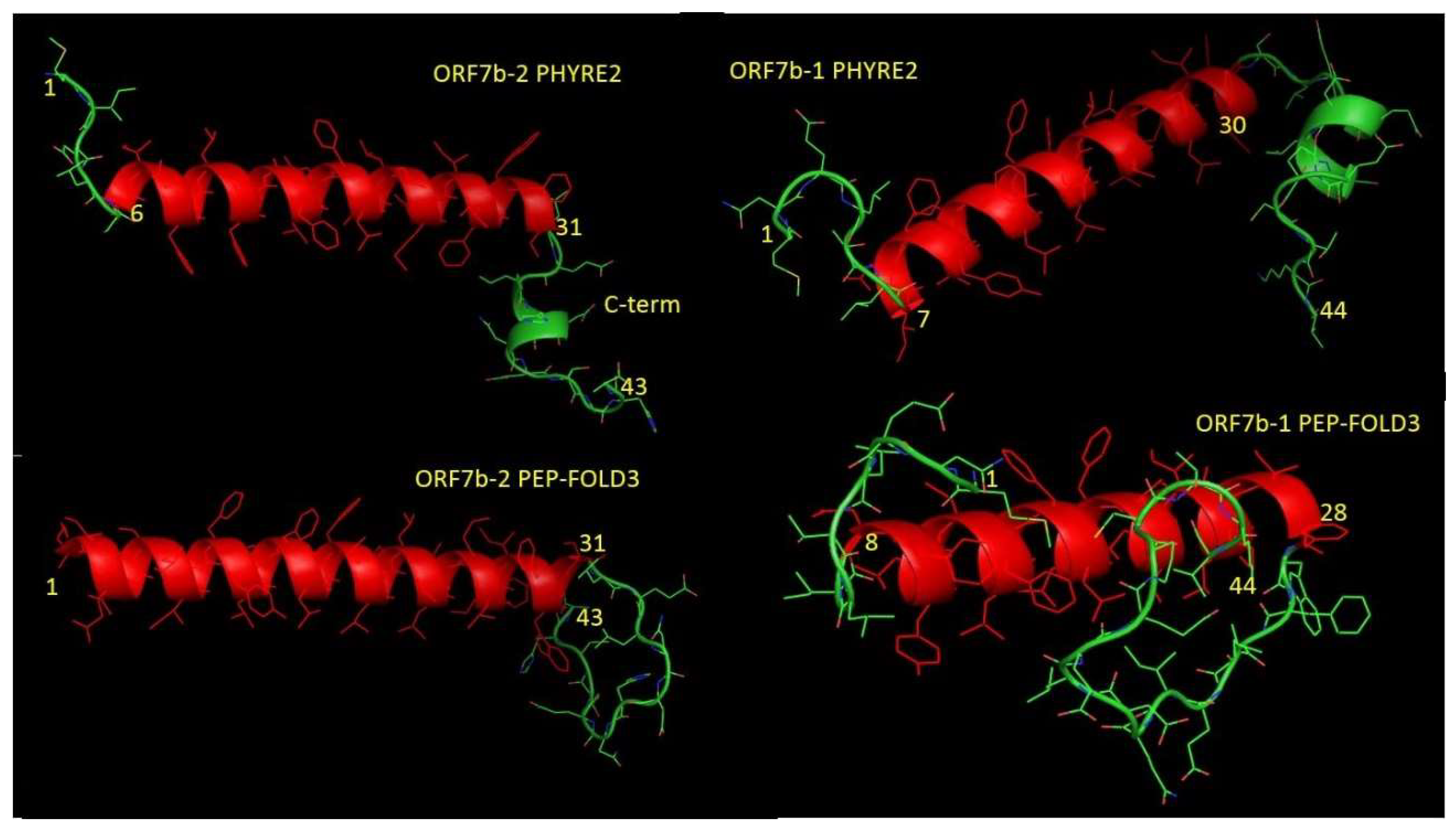
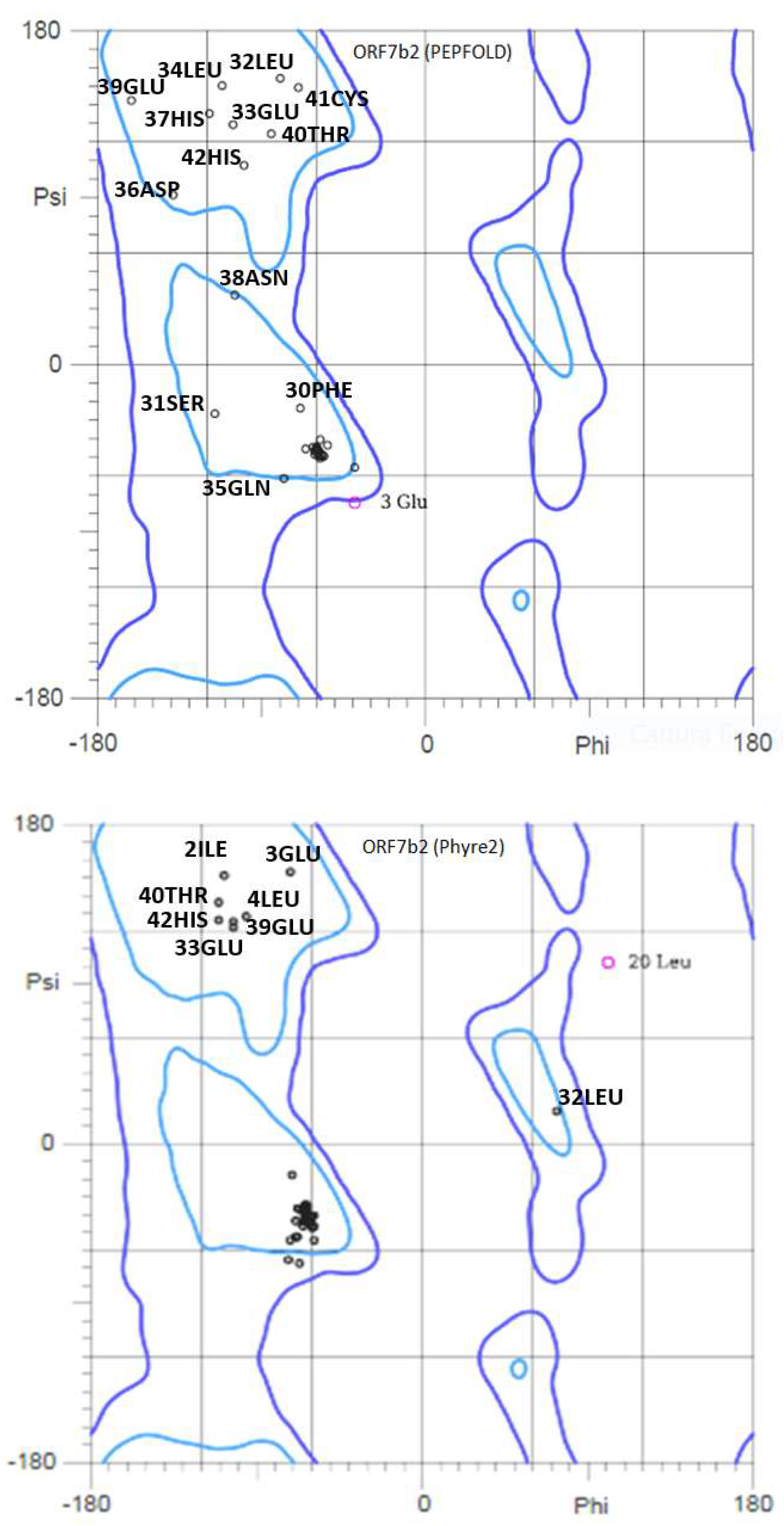
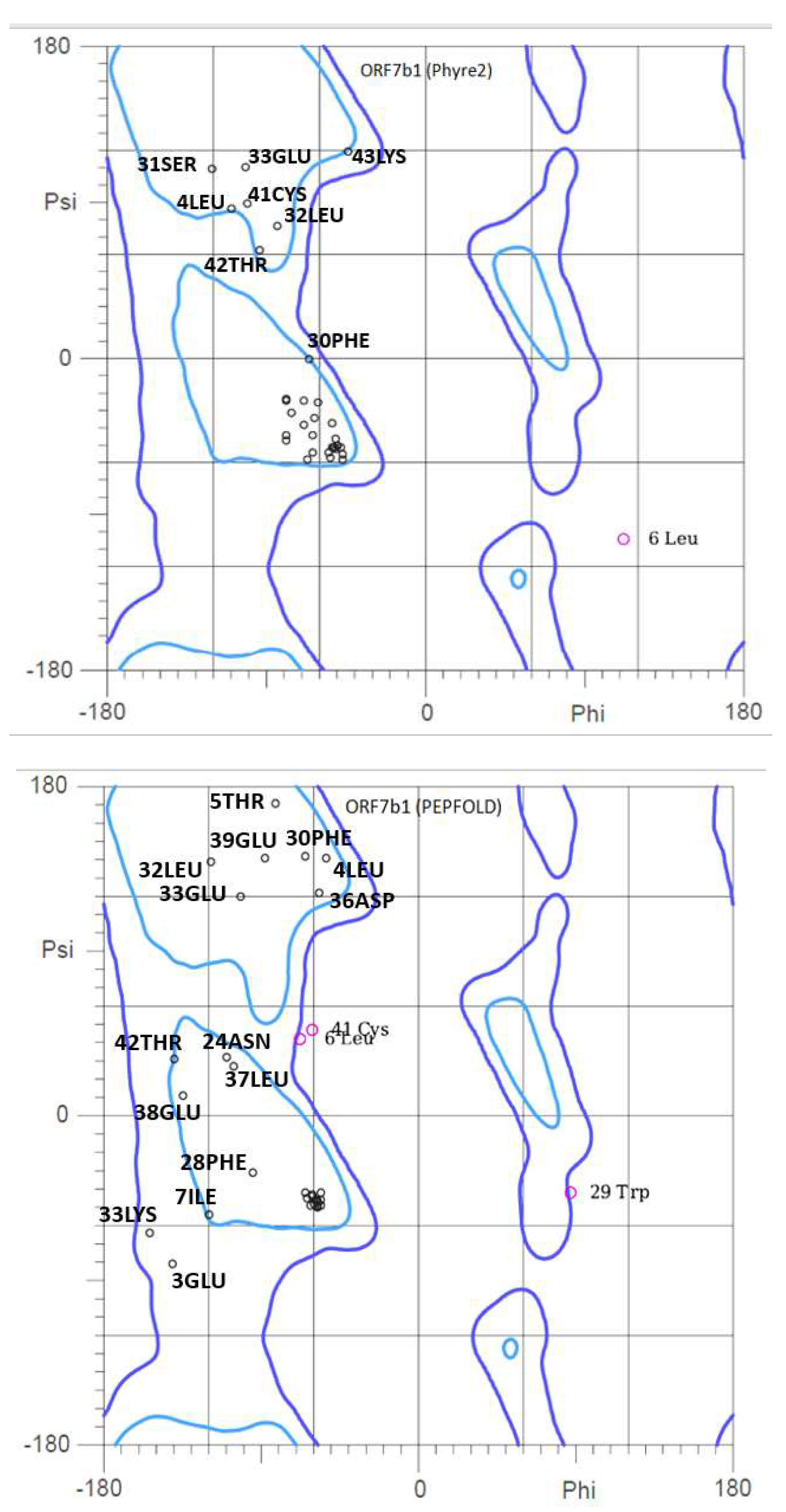
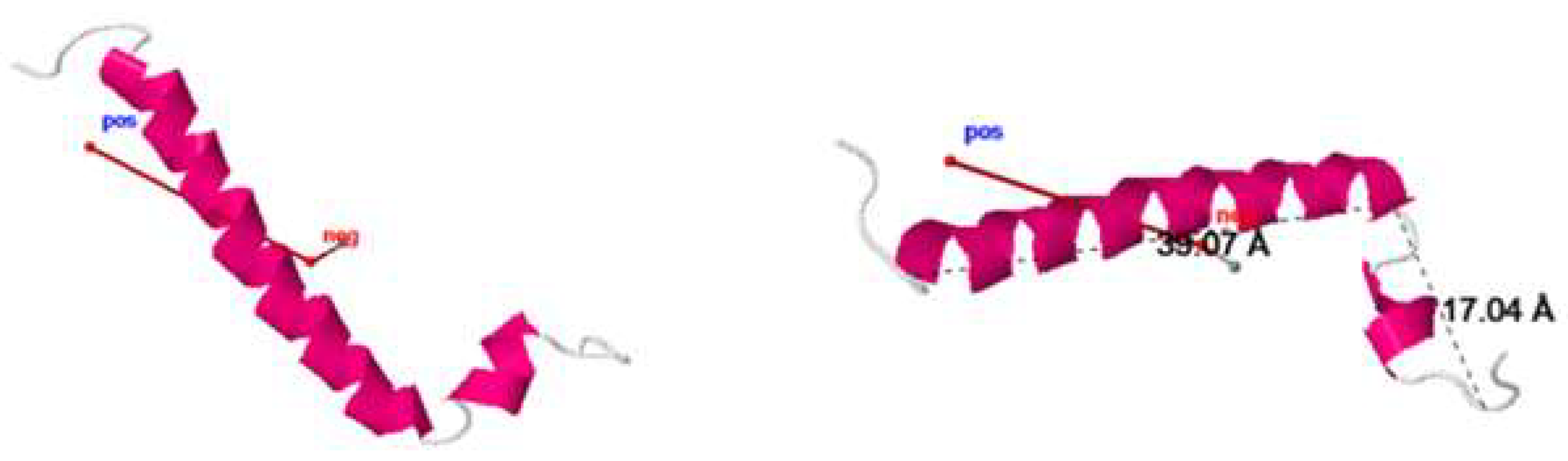
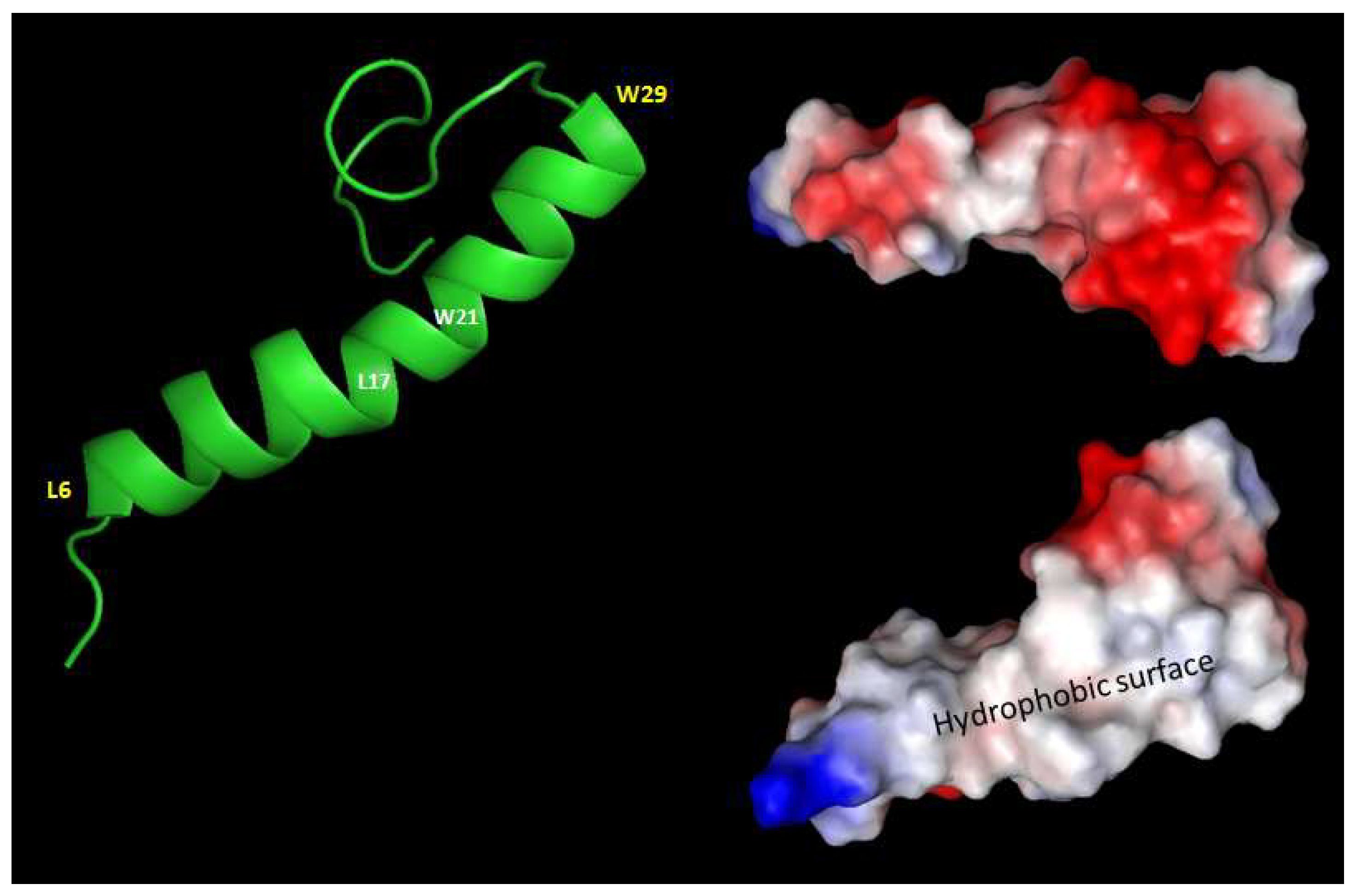
2.3. 3D structure
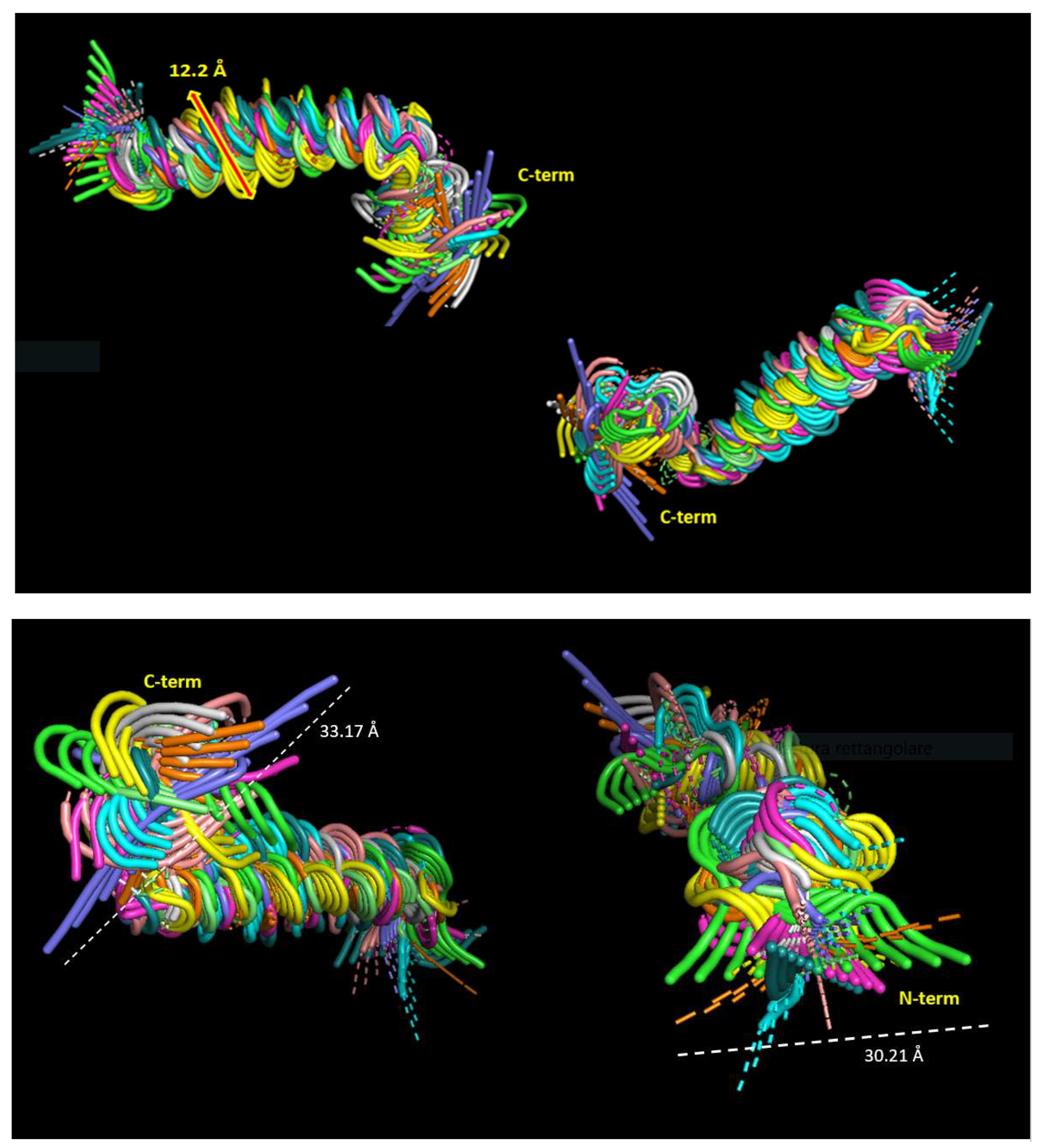
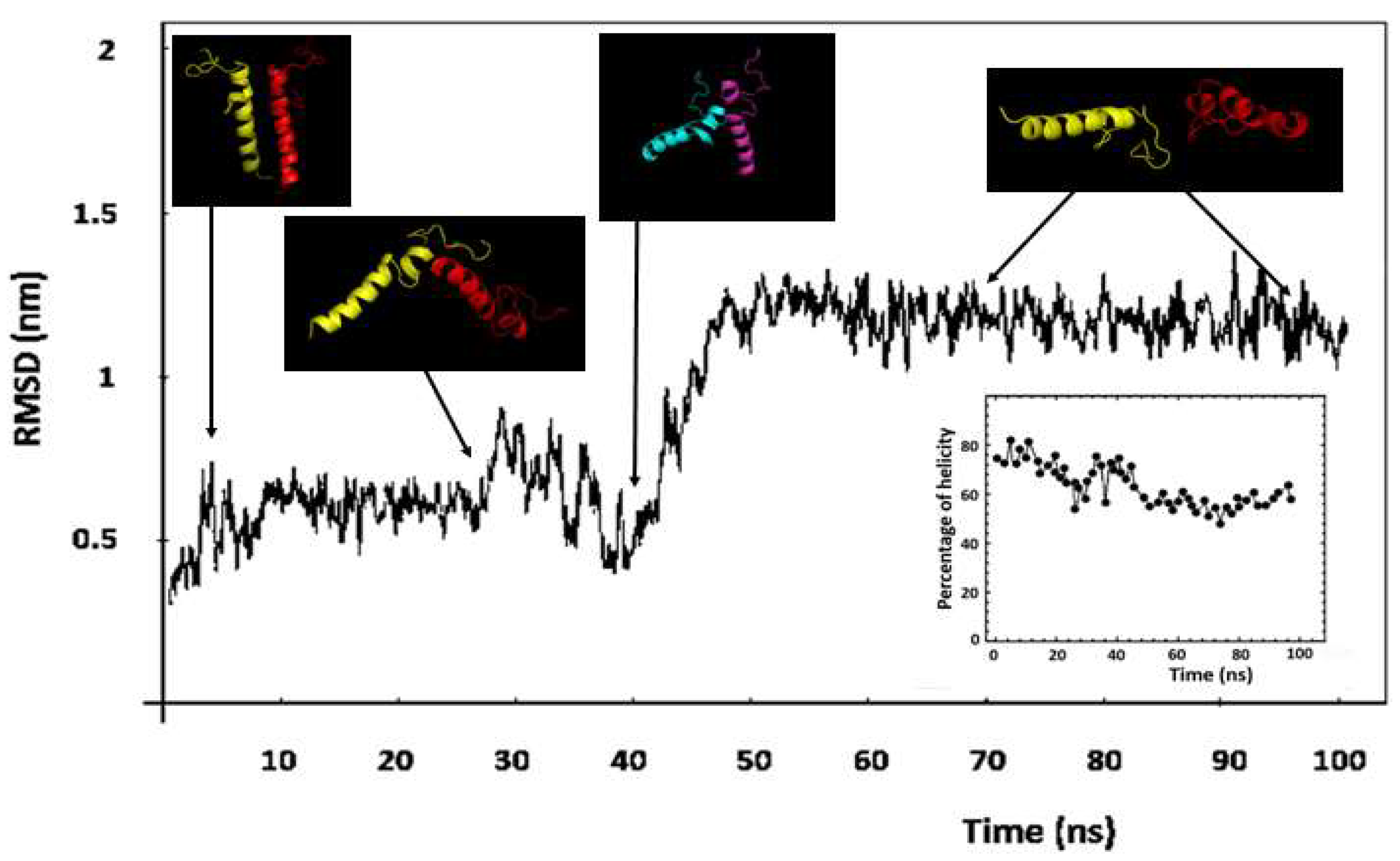
2.4. Dynamic properties of ORF7b-2
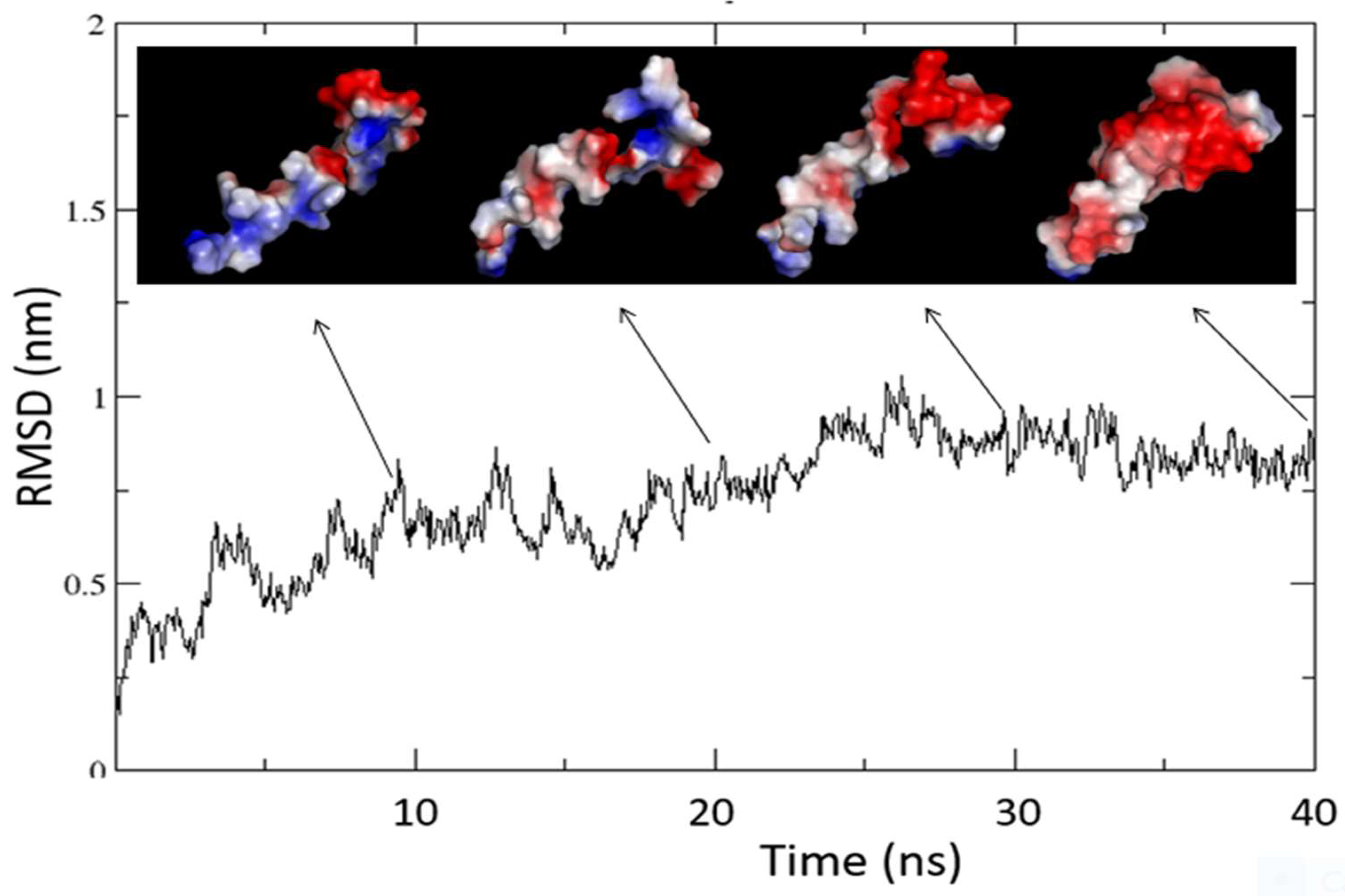
2.5. Molecular dynamics of ORF7b-2 in explicit water.
2.6. Molecular dynamics of ORF7b-2 in membrane.
3. Discussion
4. Materials and Methods
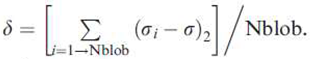
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Redondo N., Zaldivar-Lopez S., Garrido J.J., Montoya M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immun., [2021] VOL 12. ISSN 1664-3224. [CrossRef]
- Shang J, Han N, et al., Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Brief Bioinform. [2021] Mar 22;22[2]:1267-1278. PMID: 33126244; PMCID: PMC7665327. [CrossRef]
- Altincekic N., Korn S.M., et al., Large-Scale Recombinant Production of the SARS-CoV-2 Proteome for High-Thoughput and structural Biology Applications. Front. Mol. Biosci. [2021], Vol 8, Article 653148.
- Yang R, Zhao Q, et al., SARS-CoV-2 Accessory protein ORF7b Mediates Tumor Necrosis Factor-α-Induced Apoptosis in Cells. Front. Microbiol. [2021] 12:654709. [CrossRef]
- Ramasamy S, Subbian S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin. Microbiol. Rev. [2021]: 34:e00299-20. [CrossRef]
- Zhang, J., Cruz-cosme, R., et al.. A systemic and molecular study of subcellular localization of SARSCoV-2 proteins. Signal Transduct. Target. Ther. [2020] 5:269. [CrossRef]
- Schaecher SR, Diamond MS, Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J Virol. [2008] Oct;82[19]:9477-91. Epub 2008 Jul 16. PMID: 18632859; PMCID: PMC2546951. [CrossRef]
- Schaecher, S. R., J. M. Mackenzie, and A. Pekosz.. The ORF7b protein of severe acute respiratory syndrome coronavirus [SARS-CoV] is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. [2007] 81718-731. [CrossRef]
- Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory Proteins of SARS-CoV and Other Coronaviruses. Antiviral Res [2014] 109:97–109. [CrossRef]
- Debnath P, Khan U, Khan MS. Characterization and Structural Prediction of Proteins in SARS-CoV-2 Bangladeshi Variant Through Bioinformatics. Microbiol Insights. [2022] Aug 9; 15:11786361221115595. PMID: 35966939; PMCID: PMC9373114. [CrossRef]
- Mitch Leslie. A viral arsenal. SARS-CoV-2 wields versatile proteins to foil our immune system's counterattack. Science, [2022], vol 378, 6616, 128-131. [CrossRef]
- Marie-Laure Fogeron, Roland Montserret, et al., SARS-CoV-2 ORF7b: is a bat virus protein homologue a major cause of COVID-19 symptoms? - bioRxiv prep. [2021]. [CrossRef]
- Szente L, Singhal A, Domokos A, Song B. Cyclodextrins: Assessing the Impact of Cavity Size, Occupancy, and Substitutions on Cytotoxicity and Cholesterol Homeostasis. Molecules. [2018] May 20;23[5]:1228. PMID: 29783784; PMCID: PMC6100472. [CrossRef]
- Toft-Bertelsen TL, Jeppesen MG, et al., Amantadine has potential for the treatment of COVID-19 because it inhibits known and novel ion channels encoded by SARS-CoV-2. Commun Biol. [2021] Dec 1;4[1]:1347. Erratum in: Commun Biol. 2021 Dec 10;4[1]:1402. PMID: 34853399; PMCID: PMC8636635. [CrossRef]
- Scott R. Schaecher, Jason M. Mackenzie, Andrew Pekosz. The ORF7b Protein of Severe Acute Respiratory Syndrome Coronavirus [SARS-CoV] Is Expressed in Virus-Infected Cells and Incorporated into SARS-CoV Particles. J. Virol. [2007], Vol. 81, No. 2. [CrossRef]
- Campen, A., Williams, RM, Brown, C.J., Meng, J., Uversky, V.N., Dunker, A.K. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. [2008] Protein Pept Lett. 15[9] pp 956 – 963.
- Huyghues-Despointes BM, Scholtz JM, Baldwin RL. Effect of a single aspartate on helix stability at different positions in a neutral alanine-based peptide. Protein Sci. [1993] Oct;2[10]:1604-11. PMID: 8251935; PMCID: PMC2142265. [CrossRef]
- Guruprasad, K., Reddy, B.V.B. and Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. [1990] 4,155-161. [PubMed: 2075190].
- Deller MC, Kong L, Rupp B. Protein stability: a crystallographer's perspective. Acta Crystallogr F Struct Biol Commun. [2016] Feb;72[Pt 2]:72-95. Epub 2016 Jan 26. PMID: 26841758; PMCID: PMC4741188. [CrossRef]
- Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. PNAS [2013] Aug 13;110[33]:13392-7. Epub 2013 Jul 30. PMID: 23901099; PMCID: PMC3746876. [CrossRef]
- Lyle N, Das RK, Pappu RV. A quantitative measure for protein conformational heterogeneity. J Chem Phys. [2013] Sep 28;139[12]:121907. PMID: 24089719; PMCID: PMC3724800. [CrossRef]
- Holehouse AS, Das RK, Ahad JN, Richardson MO, Pappu RV. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys J. [2017] Jan 10;112[1]:16-21. PMID: 28076807; PMCID: PMC5232785. [CrossRef]
- Zeng X, Ruff KM, Pappu RV. Competing interactions give rise to two-state behavior and switch-like transitions in charge-rich intrinsically disordered proteins. Proc Natl Acad Sci U S A. [2022] May 10;119[19]:e2200559119. Epub 2022 May 5. PMID: 35512095; PMCID: PMC9171777. [CrossRef]
- Gurtovenko AA, Vattulainen I. Membrane potential and electrostatics of phospholipid bilayers with asymmetric transmembrane distribution of anionic lipids. J Phys Chem B. [2008] Apr 17;112[15]:4629-34. Epub 2008 Mar 26. PMID: 18363402. [CrossRef]
- Nordlund JR, Schmidt CF, Thompson TE. Transbilayer distribution in small unilamellar phosphatidylglycerol-phosphatidylcholine vesicles. Biochemistry. [1981] Oct 27;20[22]:6415-20. PMID: 7197988. [CrossRef]
- M.R.Moncelli, L.Becucci, R.Guidelli. The intrinsic pKa values for phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine in monolayers deposited on mercury electrodes. Biophys. J., [1994], Vol: 66, Issue: 6, Page: 1969-1980. ISSN: 0006-3495. PMCIDPMC1275922 PMID8075331. [CrossRef]
- Kelley LA et al. The Phyre2 web portal for protein modeling, prediction and analysis Nature Protocols [2015] 10, 845-858.
- Lamiable A, Thévenet P, et al., PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. [2016] Jul 8;44[W1]:W449-54.
- G.N. Ramachandran, C. Ramakrishnan & V. Sasisekharan: Stereochemistry of polypeptide chain configurations. J. Mol. Biol. [1963] vol. 7, p. 95-99. PMID 13990617.
- Camproux AC, Gautier R, Tuffery P. A hidden markov model derived structural alphabet for proteins. J Mol Biol. [2004] Jun 4;339[3]:591-605.
- Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J. Chem. Theor. Comput. [2014]; 10:4745-4758.
- William S. Young, Charles L. Brooks III, A Microscopic View of Helix Propagation: N and C-terminal Helix Growth in Alanine Helices, J Mol Biol, [1996] Volume 259, Issue 3, Pages 560-572, ISSN 0022-2836. [CrossRef]
- Wieczorek R, Dannenberg JJ. H-bonding cooperativity and energetics of alpha-helix formation of five 17-amino acid peptides. J Am Chem Soc. [2003] Jul 9;125[27]:8124-9. PMID: 12837081. [CrossRef]
- H. Frauenfelder, S.G. Sligar, P.G. Wolynes The energy landscapes and motions of proteins Science, [1991] 254, pp. 1598-1603.
- Bauer JA, Pavlovič J, Bauerová-Hlinková V. Normal Mode Analysis as a Routine Part of a Structural Investigation. Molecules. [2019] ;24:3293.
- Levy RM, Karplus M. Vibrational Approach to the Dynamics of an α-helix. Biopolymers. [1979] ;18:2465–2495.
- K. Suhre & Y.H. Sanejouand, ElNemo: a normal mode web-server for protein movement analysis and the generation of templates for molecular replacement. N Acid Res, [2004] 32, W610-W614.
- K. Suhre & Y.H. Sanejouand, On the potential of normal mode analysis for solving difficult molecular replacement problems. Acta Cryst. D [2004] vol.60, p796-799, International Union of Crystallography.
- Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: Automated Prediction of Hinges in Protein Structures. Proteins, [2008] 70[4]:1219-27.
- López-Blanco JR, Chacón P. New generation of elastic network models. Curr Opin Struct Biol. [2016]; 37:46–53.
- Atilgan, A. R., Durell, A. R., et al., Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J, [2001] 80, 505-515.
- Eldon G. Emberly, Ranjan Mukhopadhyay, Ned S. Wingreen, Chao Tang, Flexibility of alpha-Helices: Results of a Statistical Analysis of Database Protein Structures, JMB, [2003] Volume 327, Issue 1, Pages 229-237,ISSN 0022-2836. [CrossRef]
- T.E. Creighton, Proteins: Structures and Molecular Properties, WH Freeman and Co., New York [1993].
- W.G.J. Hol, P.T. van Duijen, H.J.C. Berendsen The α-helix dipole and the properties of proteins Nature, [1978] 273, pp.443-446.
- Clifford E. Felder, Jaime Prilusky, Israel Silman, and Joel L. Sussman, " A server and database for dipole moments of proteins", Nu Ac Res, [2007] 35, special Web Servers Issue. https://academic.oup.com/nar/article/35/suppl_2/W512/2922221.
- Baruah C, Mahanta S, Devi P, Sharma DK. In Silico Proteome Analysis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J Nanotechnol Nanomaterials. [2021]; 2[1]: 1-19.
- Sedla E., Fedunova D., et al., Polyanion Hydrophobicity and Protein Basicity Affect Protein Stability in Protein-Polyanion Complexes. Biomacromolecules (2009), 10, 2533–2538.
- Sedlák E., Antalı́k M, et al., Interaction of ferricytochrome c with polyanion Nafion, Biochimica et Biophysica Acta (BBA) - Bioenergetics, 1997, Volume 1319, Issues 2–3, Pages 258-266, ISSN 0005-2728. (https://www.sciencedirect.com/science/article/pii/S0005272896001703). [CrossRef]
- Erik Sedlák, Marian Antalík. Coulombic and noncoulombic effect of polyanions on cytochrome c structure. Biopolymers (1998) Vol46, No.3 Pag 145-154. [CrossRef]
- Antalı M., Bágelová J., et al., Effect of varying polyglutamate chain length on the structure and stability of ferricytochrome c. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, (2003) Vol 1646, Issues 1–2, Pages 11-20, ISSN 1570-9639. (https://www.sciencedirect.com/science/article/pii/S1570963902005435). [CrossRef]
- Gong J., Yao, P., et al., Structural Transformation of Cytochrome c and Apo Cytochrome c Induced by Sulfonated Polystyrene. Biomacromolecules (2003), Vol 4 Is 5, pg. 1293-1300. [CrossRef]
- Kokufuta, E., Shimizu, H., et al.,Salt linkage formation of poly(diallyldimethylammonium chloride) with acidic groups in the polyion complex between human carboxyhemoglobin and potassium poly(vinyl alcohol) sulfate. Macromolecules (1981), Vol 14, Is 5, 1178-1180 https://doi.org/10.1021/ma50006a008 ACS . [CrossRef]
- Tsuboi, A.; Izumi, T.; et al., Complexation of Proteins with a Strong Polyanion in an Aqueous Salt-free System, Langmuir 1996, 12, 6295–6303. ACS. [CrossRef]
- Ulmschneider MB, Sansom MS, Di Nola A. Properties of Integral Membrane Protein Structures: Derivation of an Implicit Membrane Potential. Proteins: Struct, Funct Genet. 2005; 59: 252–265. [CrossRef]
- Dong H, Sharma M, Zhou HX, Cross TA. Glycines: Role in Alpha-Helical Membrane Protein Structures and a Potential Indicator of Native Conformation. Biochemistry. 2012;51:4779–4789. ACS. [CrossRef]
- Slusky JS, Dunbrack RL., Jr, Charge Asymmetry in the Proteins of the Outer Membrane. Bioinformatics. 2013; 29: 2122–2128. [CrossRef]
- Monne M, Nilsson I, et al., Positively and Negatively Charged Residues Have Different Effects on the Position in the Membrane of a Model Transmembrane Helix. J Mol Biol. 1998; 284: 1177–1183. [CrossRef]
- Segrest JP, De Loof H, et al., Amphipathic Helix Motif: Classes and Properties. Proteins: Struct, Funct Genet. 1990;8:103–117. [CrossRef]
- Von Heijne G. Recent advances in the understanding of membrane protein assembly and structure. Quart Rev Biophys 2000; 32: 285–307.
- Hessa, T., Kim, H., Bihlmaier, K. et al. Recognition of transmembrane helices by the endoplasmic reticu-lum translocon. Nature 433, 377–381 (2005). [CrossRef]
- Hessa, T., Meindl-Beinker, N., Bernsel, A. et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007). [CrossRef]
- Liezel A. Lumangtad, Thomas W. Bell, The signal peptide as a new target for drug design, Bioorg&Med Chem Lett, [2020] Volume 30, Issue 10, 127115, ISSN 0960-894X. [CrossRef]
- M. Uhlén, L. Fagerberg, B.M. Hallström, et al. Tissue-based map of the human proteome. Science, [2015] 347, p. 1260419.
- J. Dudek, S. Pfeffer, P.-H. Lee, et al. Protein transport into the human endoplasmic reticulum J Mol Biol, [2015] 427, pp. 1159-1175.
- Simon SM, Blobel G. Signal peptides open protein-conducting channels in E. coli. Cell. [1992] May 15;69[4]:677-84. PMID: 1375130. [CrossRef]
- Fang J., Haasl R.J. Dong Y. Lushington G.H. Discover protein sequence signatures from protein-protein interaction data. BMC Bioinformatics. 2005; 6: 277-284.
- Naama Aviram and Maya Schuldiner, Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J Cell Sci [2017] 130, 4079-4085. [CrossRef]
- Harry F. Noller, Ribosomal RNA and Translation, Annual Review of Biochemistry (1991) Vol. 60: 191-227. [CrossRef]
- Miranda F. Mecha*, Rachel B. Hutchinson*, Jung Ho Lee, and Silvia Cavagnero Protein folding in vitro and in the cell: From a solitary journey to a team effort. Biophysical Chemistry, 2022, Volume 287,106821, ISSN 0301-4622. [CrossRef]
- Matthias P. Mayer Lila M. Gierasch. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones, JBC REVIEWS, (2019), Vol 294, Iss 6, Pgg 2085-2097. [CrossRef]
- Zahn M, Berthold N, Kieslich B, Knappe D, Hoffmann R, Sträter N. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol. 2013 Jul 24;425(14):2463-79. Epub 2013 Apr 2. PMID: 23562829. [CrossRef]
- Van Durme J, Maurer-Stroh S, Gallardo R, Wilkinson H, Rousseau F, Schymkowitz J. Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput Biol. 2009 Aug;5(8):e1000475. Epub 2009 Aug 21. PMID: 19696878; PMCID: PMC2717214. [CrossRef]
- Baeza-Delgado C, Marti-Renom MA, Mingarro I. Structure-based statistical analysis of transmembrane helices. Eur Biophys J. 2013;42(2–3):199–207. pmid:22588483.
- Killian J.A., von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000; 25: 429-434.
- Lundin C., Kim H., Nilsson I., White S.H., von Heijne G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc. Natl. Acad. Sci. USA. 2008; 105: 15702-15707.
- Wang S., Munro R., et al., Paramagnetic Relaxation Enhancement Reveals Oligomerization Interface of a Membrane Protein J. Am. Chem. Soc. 2012, 134, 41, 16995–16998. [CrossRef]
- Gupta, K., Donlan, J., Hopper, J. et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 (2017). [CrossRef]
- Keren Limor-Waisberg, Asaf Carmi, Avigdor Scherz, Yitzhak Pilpel, Itay Furman, Specialization versus adaptation: two strategies employed by cyanophages to enhance their translation efficiencies, Nucleic Acids Research, Volume 39, Issue 14, 1 August 2011, Pages 6016–6028. [CrossRef]
- Rampersad S, Tennant P. Replication and Expression Strategies of Viruses. Viruses. 2018:55–82. Epub 2018 Mar 30. PMCID: PMC7158166. [CrossRef]
- Stanley J Opella, Relating structure and function of viral membrane-spanning miniproteins, Current Opinion in Virology, Volume 12,2015, Pages 121-125, ISSN 1879-6257. [CrossRef]
- DiMaio D. Viral miniproteins. Annu Rev Microbiol. 2014;68:21-43. Epub 2014 Apr 10. PMID: 24742054; PMCID: PMC4430842. [CrossRef]
- Zhou HX, Pang X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem Rev. 2018 Feb 28;118(4):1691-1741. Epub 2018 Jan 10. PMID: 29319301; PMCID: PMC5831536. [CrossRef]
- Duarte, J. M., Biyani, N., Baskaran, K. & Capitani, G. An analysis of oligomerization interfaces in transmembrane proteins. BMC Struct. Biol. 13, 21 (2013).
- Stanley J Opella, Relating structure and function of viral membrane-spanning miniproteins, Current Opinion in Virology, Volume 12,2015, Pages 121-125, ISSN 1879-6257. [CrossRef]
- Chanel C. La, Lily E. Takeuchi, Srinivas Abbina, Sreeparna Vappala, Usama Abbasi, and Jayachandran N. Kizhakkedathu. Targeting Biological Polyanions in Blood: Strategies toward the Design of Therapeutics. Biomacromolecules 2020 21 (7), 2595-2621. [CrossRef]
- Alphonse DeLucia, Thomas W. Wakefield, et al., Efficacy and toxicity of differently charged polycationic protamine-like peptides for heparin anticoagulation reversal, Journal of Vascular Surgery, Volume 18, Issue 1, 1993, Pages 49-60, ISSN 0741-5214. (https://www.sciencedirect.com/science/article/pii/074152149370014P). [CrossRef]
- Armando S. Flores-Torres, Mario C. Salinas-Carmona, Eva Salinas, and Adrian G. Rosas-Taraco Eosinophils and Respiratory Viruses, Viral Immunology VOL. 32, NO. 5 Published Online: 4 Jun 2019. [CrossRef]
- Helene F. Rosenberg, Kimberly D. Dyer, Joseph B. Domachowske, Respiratory viruses and eosinophils: Exploring the connections, Antiviral Research, Volume 83, Issue 1, 2009, Pages 1-9, ISSN 0166-3542. (https://www.sciencedirect.com/science/article/pii/S0166354209002988). [CrossRef]
- Oughtred R, Rust J, et al., The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. [2020] Oct 18.
- Helisa H. Wippel, Juan D. Chavez, Xiaoting Tang, James E. Bruce, Quantitative interactome analysis with chemical cross-linking and mass spectrometry, CuOpChemBiol, [2022] Volume 66, 102076, ISSN 1367-5931. [CrossRef]
- Braun, P., Interactome mapping for analysis of complex phenotypes: Insights from benchmarking binary interaction assays. Proteomics, [2012] 12: 1499-1518. https://doi.org/10.1002/pmic.201100598. Hopp T.P., and Woods K.R. Amino acid scale: Hydrophilicity. [1981] Proc. Natl. Acad. Sci. U.S.A. 78:3824-3828.
- B. Hess, C. Kutzner, D. van der Spoel, E. Lindahl, J. Chem. Theory Comput. 2008, 4, 435.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. Shirts, J. Smith, P. Kasson, D. van der Spoel, B. Hess, E. Lindahl, Bioinformatics 2013, 29, 845.
- Raucci, R., Colonna, G., Castello, G. et al. Peptide Folding Problem: A Molecular Dynamics Study on Polyalanines Using Different Force Fields. Int J Pept Res Ther 19, 117–123 (2013). [CrossRef]
- Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. (2008) HingeProt: Automated Prediction of Hinges in Protein Structures. Proteins, 70(4):1219-27.
- Bahar, I., Atilgan A. R., Erman, B. (1997) Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Folding and Design, (1997), 2, 173-181.
- Atilgan, A. R., Durell, A. R., Jernigan, R. L., Demirel, M. C. , Keskin, O. , Bahar, I. (2001), Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophysical Journal, 80, 505-515.
- Gordon, D.E., Jang, G.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature [2020] 583, 459–468. [CrossRef]
- Lukasz P. Kozlowski, Proteome-pI: proteome isoelectric point database, in Nu Ac Res, [2017] vol. 45, D1, pp. D1112–D1116, The UniProt Consortium: a hub for protein information. [CrossRef]
- Kyte J., Doolittle R., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. [1982] 157, pp 105– 132.
- Simon SM, Blobel G. Signal peptides open protein-conducting channels in E. coli. Cell. [1992] May 15;69[4]:677-84. PMID: 1375130. [CrossRef]
- Bevacqua A, Bakshi S, Xia Y (2021) Principal component analysis of alpha-helix deformations in transmembrane proteins. PLOS ONE 16(9): e0257318. [CrossRef]
- Zagrovic B., Jayachandran G.,, et al., How Large is an Helix? Studies of the Radii of Gyration of Helical Peptides by Small-angle X-ray Scattering and Molecular Dynamics, J Mol Biol, [2005] Vol 353, Issue 2, Pags 232-241, ISSN 0022-2836. [CrossRef]
- Deller MC, Kong L, Rupp B. Protein stability: a crystallographer's perspective. Acta Crystallogr F Struct Biol Commun. [2016] Feb;72[Pt 2]:72-95. Epub 2016 Jan 26. PMID: 26841758; PMCID: PMC4741188. [CrossRef]
- S Costantini, G Colonna, AM Facchiano. Amino acid propensities for secondary structures are influenced by the protein structural class. Bioch Biophys Res Comm [2006] 342 [2], 441-451. [CrossRef]
- Yan Y, Tao H, He J, Huang S-Y.* The HDOCK server for integrated protein-protein docking. Nature Protocols, 2020; [CrossRef]
- Lomize AL, Todd SC, Pogozheva ID. (2022) Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. 31:209-220.
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995 May 26;268(5214):1144-9.
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res, 35, W522-5, 2007.
- Weller PF, Spencer LA. Functions of tissue-resident eosinophils. Nat Rev Immunol. 2017 Dec;17(12):746-760. Epub 2017 Sep 11. PMID: 28891557; PMCID: PMC5783317. [CrossRef]
- Sharma A, Colonna G., System-Wide Pollution of Biomedical Data: Consequence of the Search for Hub Genes of Hepatocellular Carcinoma Without Spatiotemporal Consideration. Mol Diagn Ther. (2021); 25(1): 9-27. Epub 2021 Jan 21.PMID: 33475988 Review. [CrossRef]
| ORF7b-2* | ORF7b-1** | ||||
|---|---|---|---|---|---|
| Amino acid | Number of residues | Percentage % |
Number of residues |
Percentage % |
|
| Ala [A] | 2 | 4.7 | 1 | 2.3 | |
| Asn [N] | 1 | 2.3 | 1 | 2.3 | |
| Asp [D] | 2 | 4.7 | 2 | 4.5 | |
| Cys [C] | 2 | 4.7 | 2 | 4.5 | |
| Gln [Q] | 1 | 2.3 | 1 | 2.3 | |
| Glu [E] | 3 | 7.0 | 4 | 9.1 | |
| His [H] | 2 | 4.7 | - | - | |
| Ile [I] | 5 | 11.6 | 5 | 11.4 | |
| Leu [L] | 11 | 25.6 | 11 | 25.0 | |
| Lys [K] | - | - | 1 | 2.3 | |
| Met [M] | 2 | 4.7 | 2 | 4.5 | |
| Phe [F] | 6 | 14.0 | 6 | 13.6 | |
| Pro [P] | - | - | 1 | 2.3 | |
| Ser [S] | 2 | 4.7 | 1 | 2.3 | |
| Thr [T] | 1 | 2.3 | 2 | 4.5 | |
| Trp [W] | 1 | 2.3 | 1 | 2.3 | |
| Tyr [Y] | 1 | 2.3 | 1 | 2.3 | |
| Val [V] | 1 | 2.3 | 2 | 4.5 | |
| Protein | Sequence 5 10 15 20 25 30 35 40 |
|---|---|
| ORF7b-2 | MIELSLIDFYLCFLAFLLFLVLIMLIIFWFSLELQDHNETCHA |
| ORF7b-1 | MNELTLIDFYLCFLAFLLFLVLIMLIIFWFSLEIQDLEEPCTKV |
| Physical-chemical parameters |
ORF7b-1 |
ORF7b-2 |
Notes |
|
|---|---|---|---|---|
| N [MW] | 44 [5301.51] | 43 [5179.31] | Number of residues and M.W. |
|
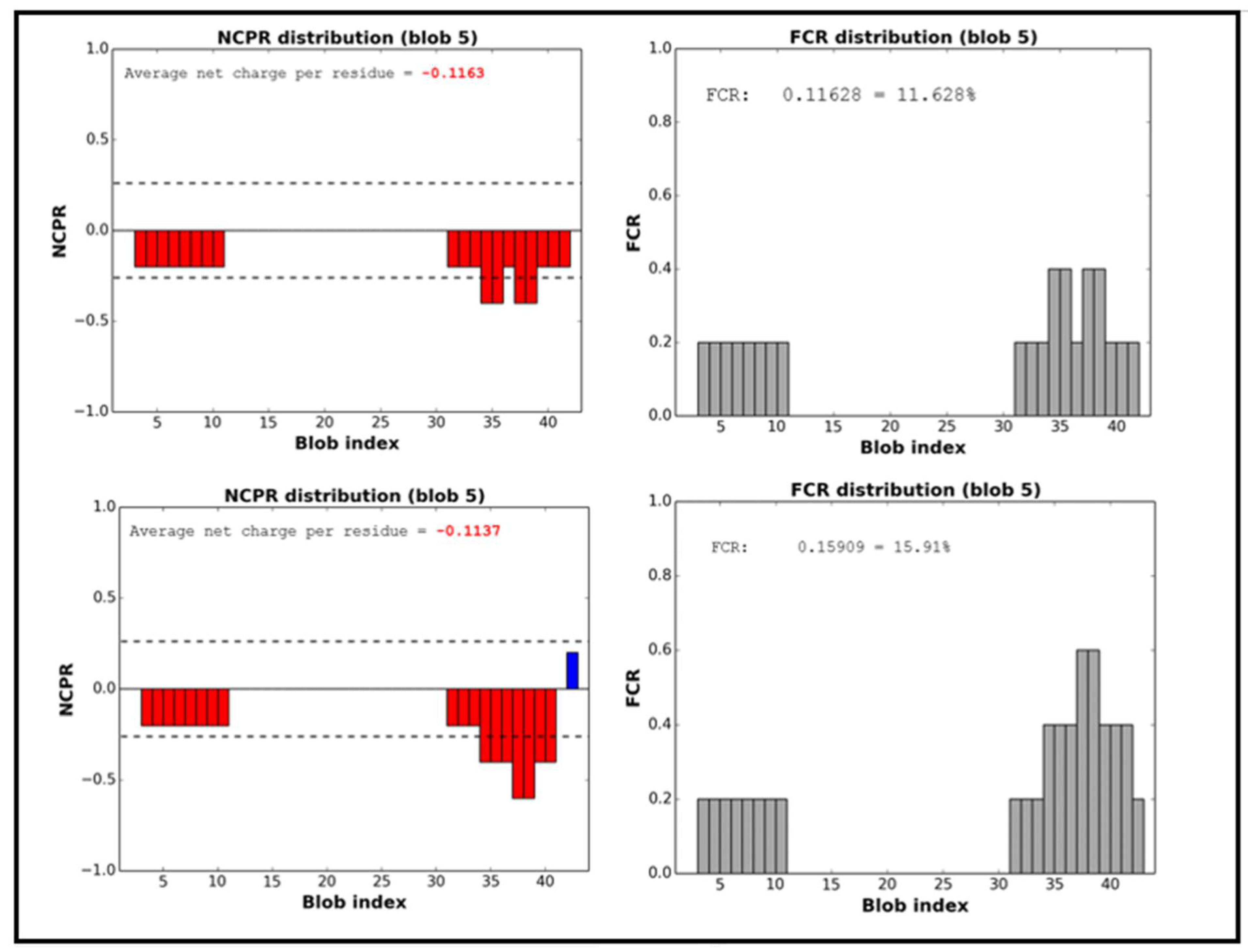
| f- | 0.13636 | 0.11628 | Fraction of negative residues | |
| f+ | 0.02273 | 0.00000 | Fraction of positive residues | |
| FCR | 0.15909 | 0.11628 | Fraction of charged residues | |
| NCPR | -0.11364 | -0.11628 | Net charge per residue | |
| Sigma | 0.08117 | 0.11628 | Charge asymmetry | |
| K | 0.35577 | 0.25372 | Charge patterning parameter | |
| Delta | 0.03182 | 0.01706 | ||
| Max Delta | 0.08945 | 0.06725 | ||
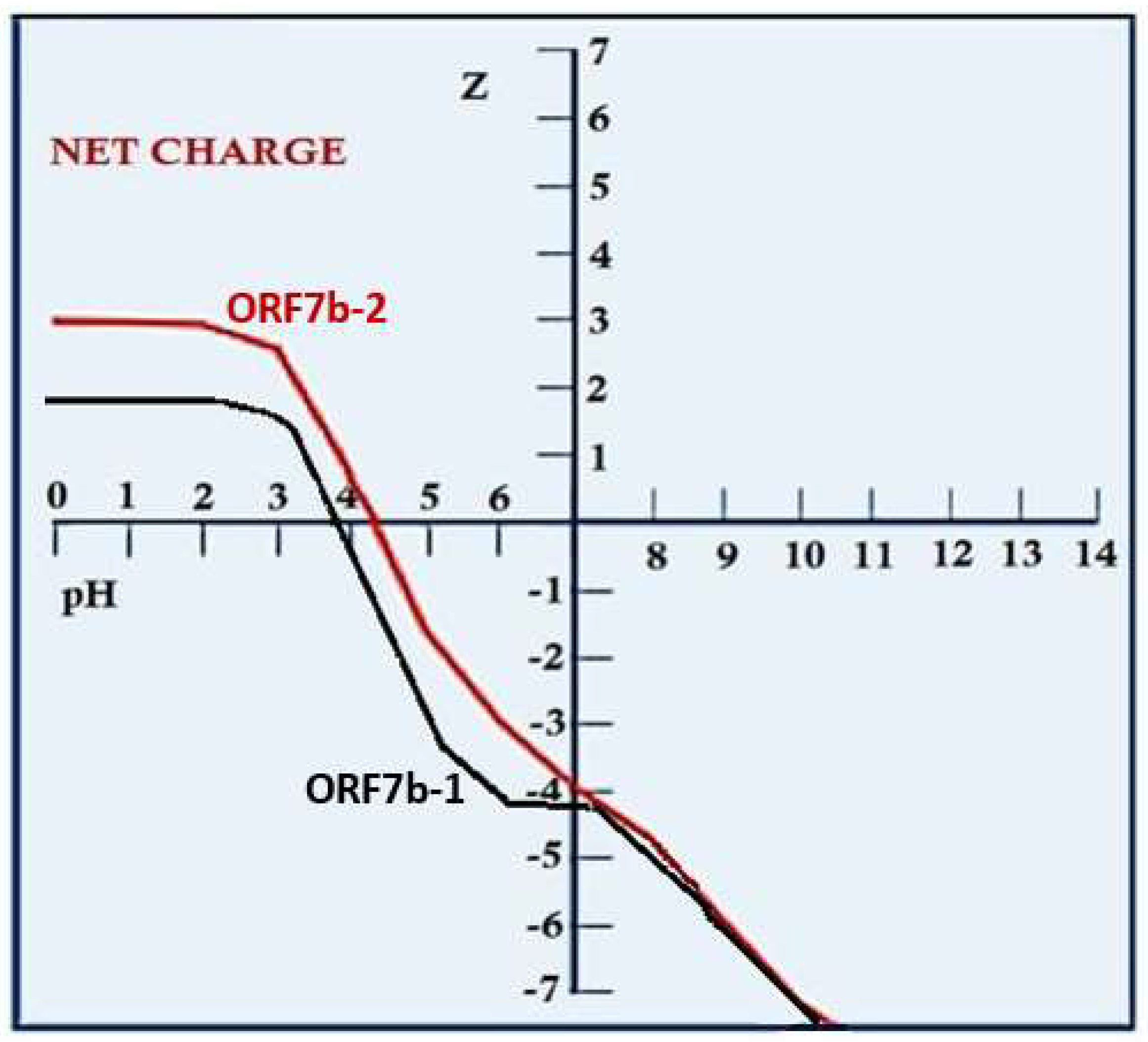
| pI | 3.72 | 4.32 | Isoelectric point | |
| AH | -0.83 | -0.98 | Average hydrophilicity | |
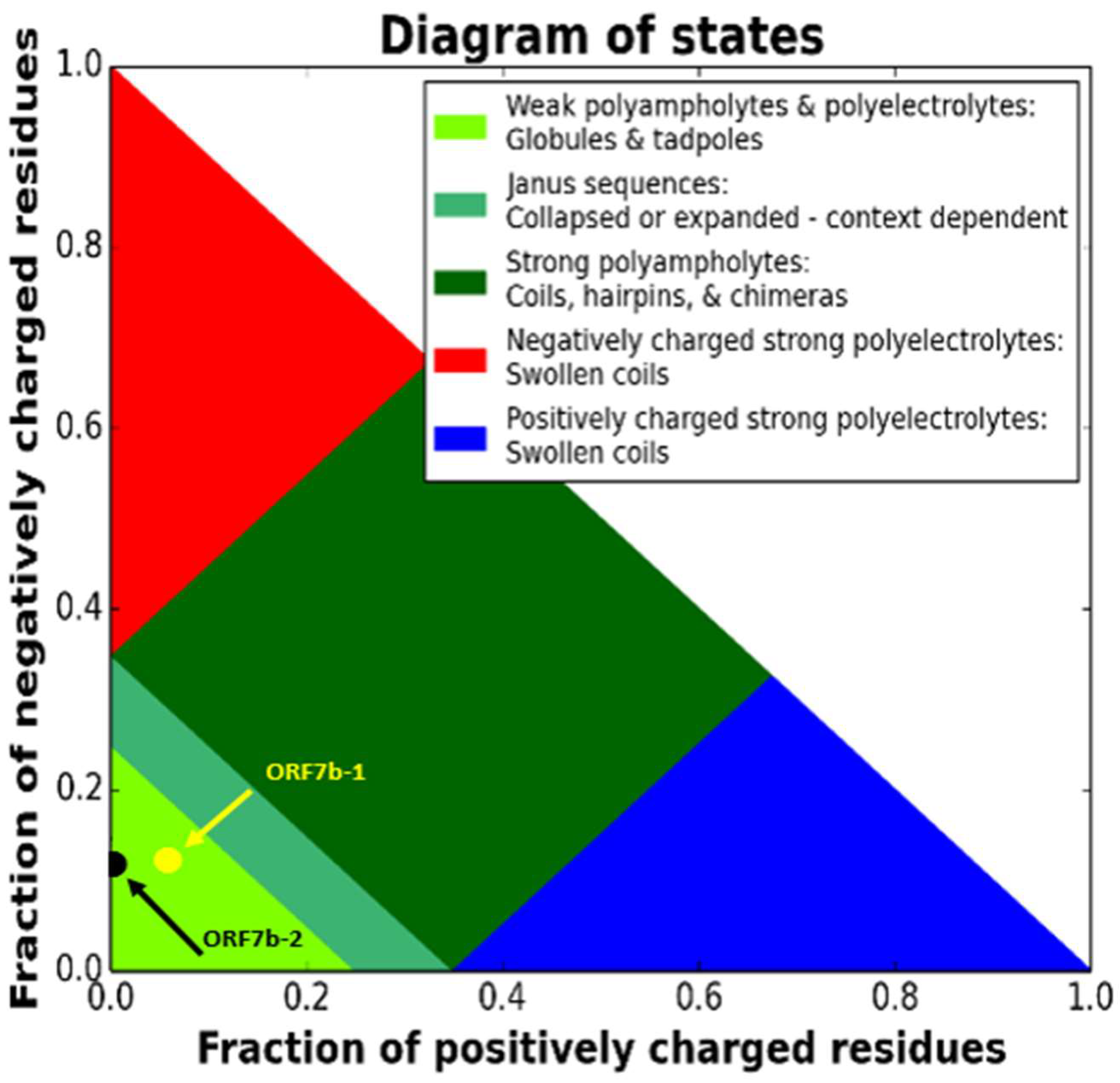
| Phase Plot Region | 1 | 1 | [see State diagram] | |
| Phase Plot Annotation | Globule/Tadpole | Globule/Tadpole | Prolate elongated structures | |
| Polymeric State | Weak polyampholyte | Polyanion (Weak negative polyelectrolyte) |
| Slowest mode 1 | |||
|---|---|---|---|
| Rigid Part No | Residues | Score | Hinge residues |
| 1 | 1-20 | 0.88 | 20 |
| 2 | 21-43 | 0.9 | 20 |
| Slowest mode 2 | |||
| Rigid Part No | |||
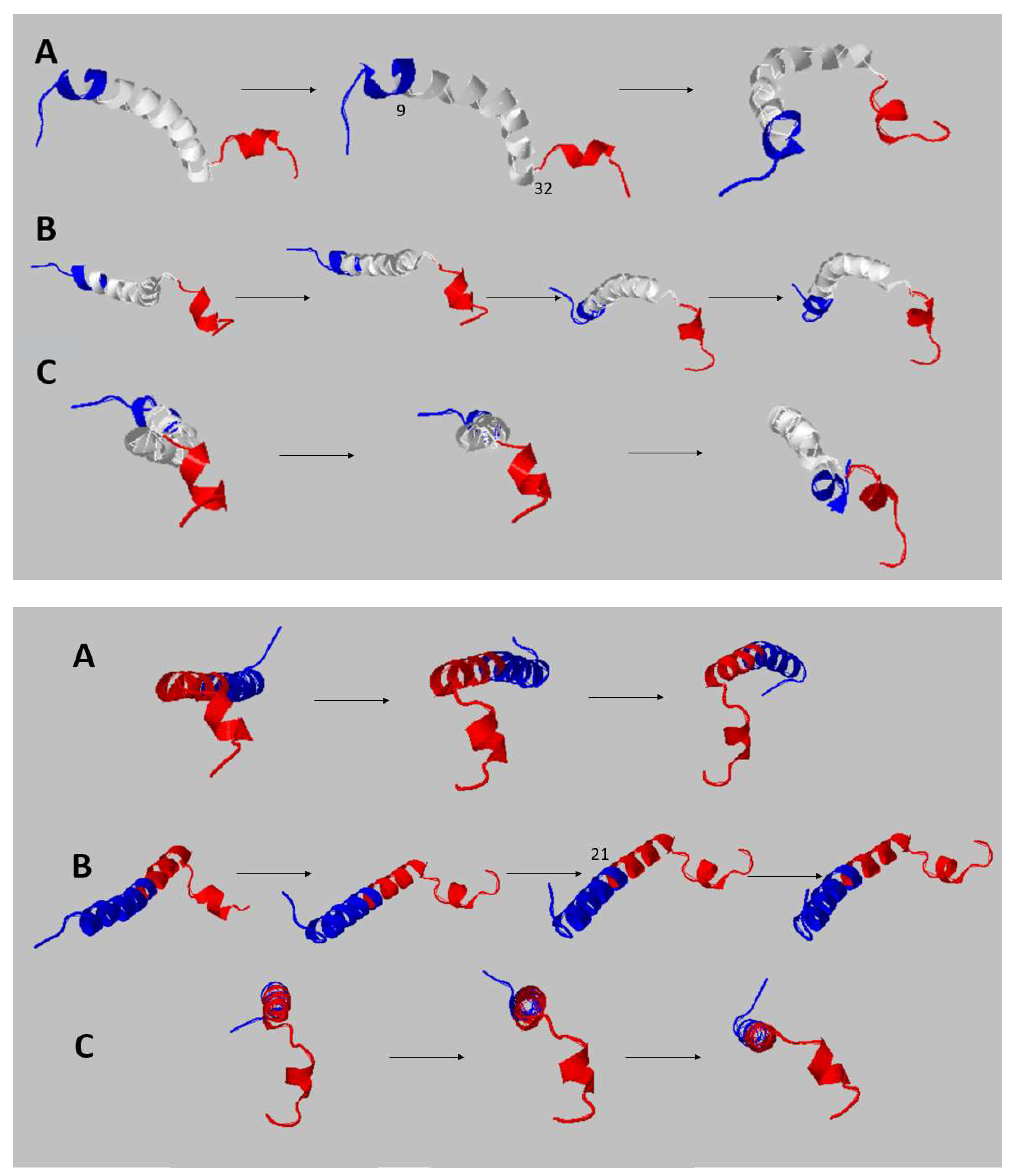
| 1 | 1-9 | 0.68 | 9 |
| 2 | 10-32 | 0.82 | 32 |
| 3 | 33-43 | 0.85 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




