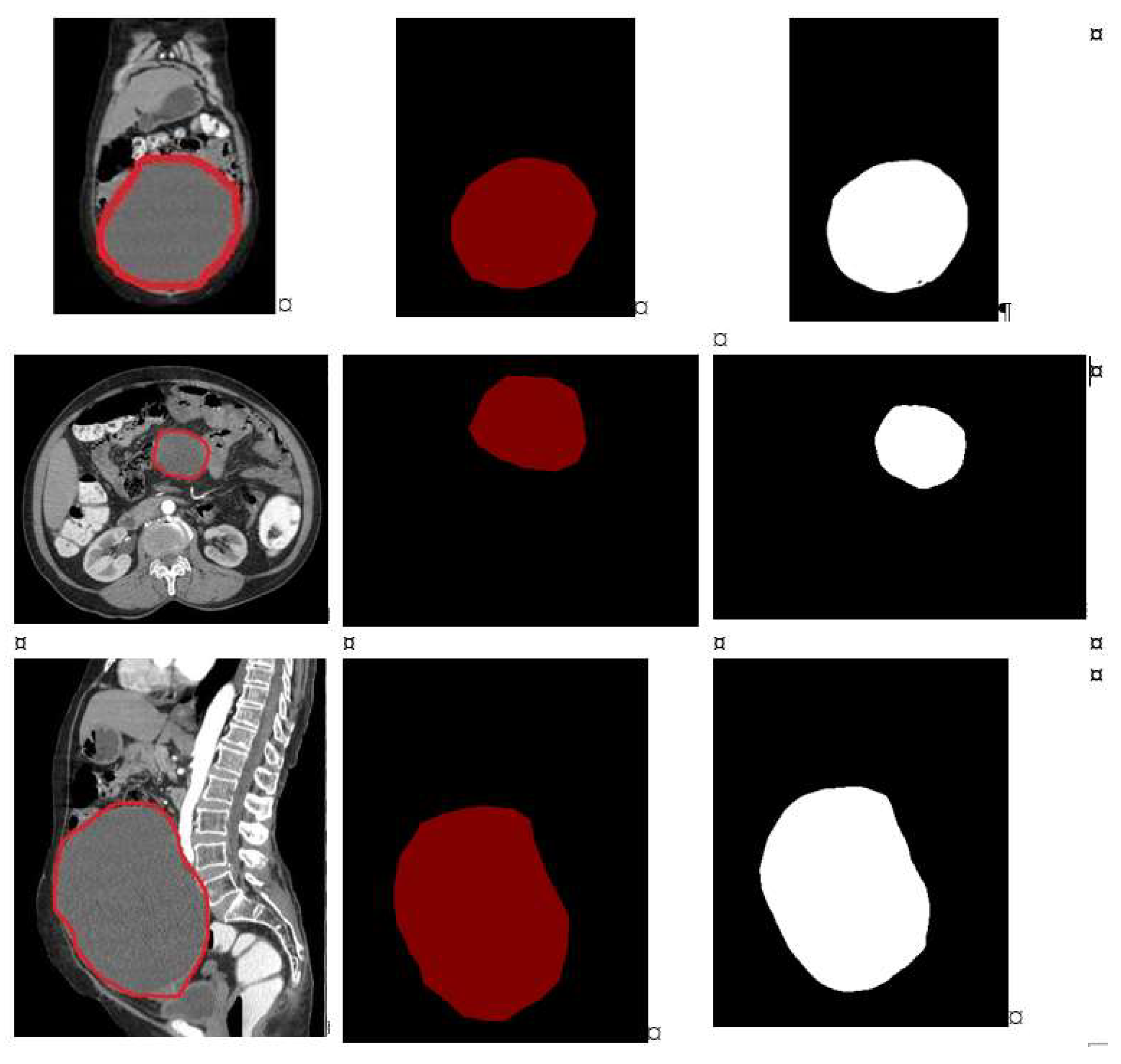
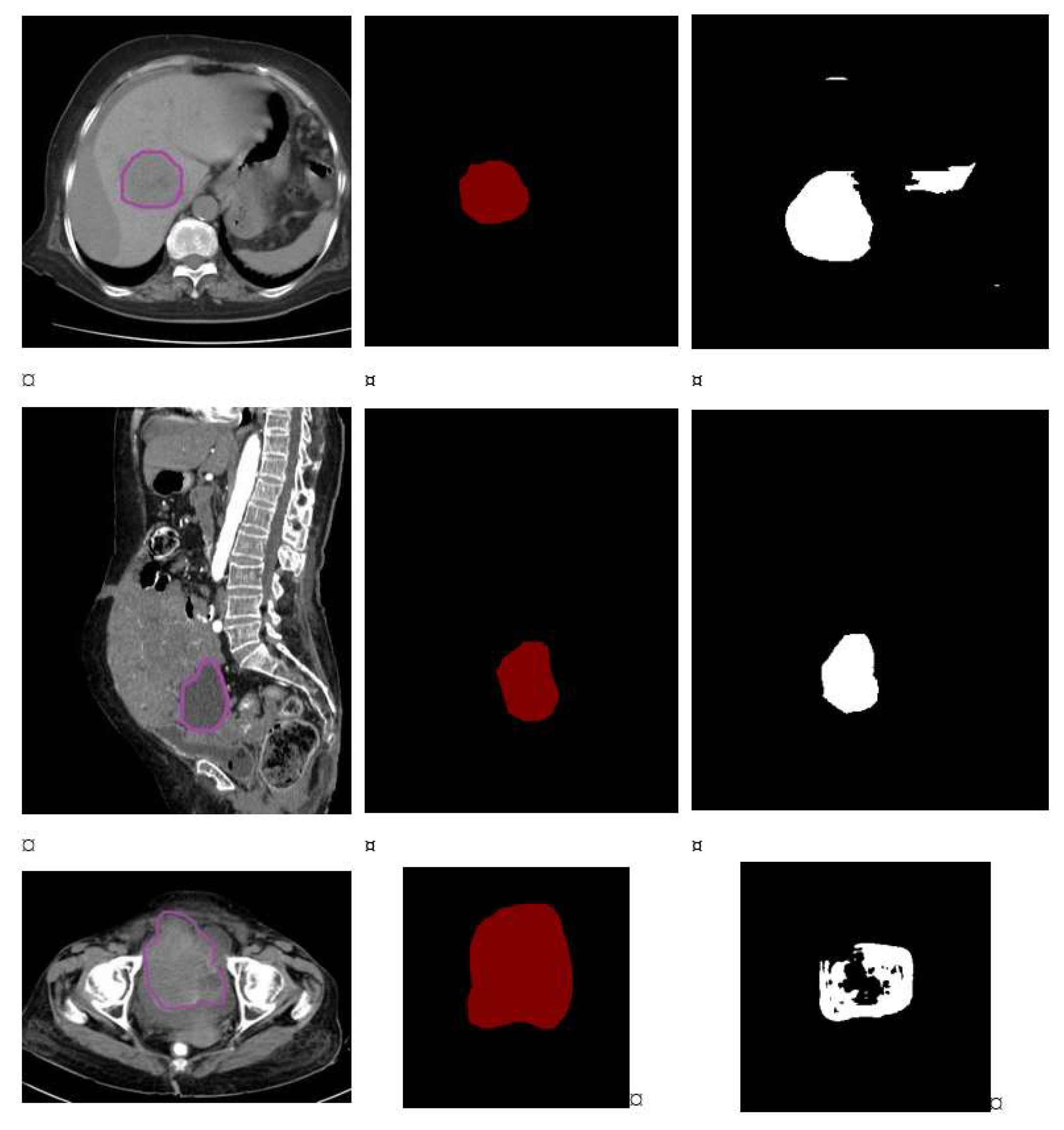
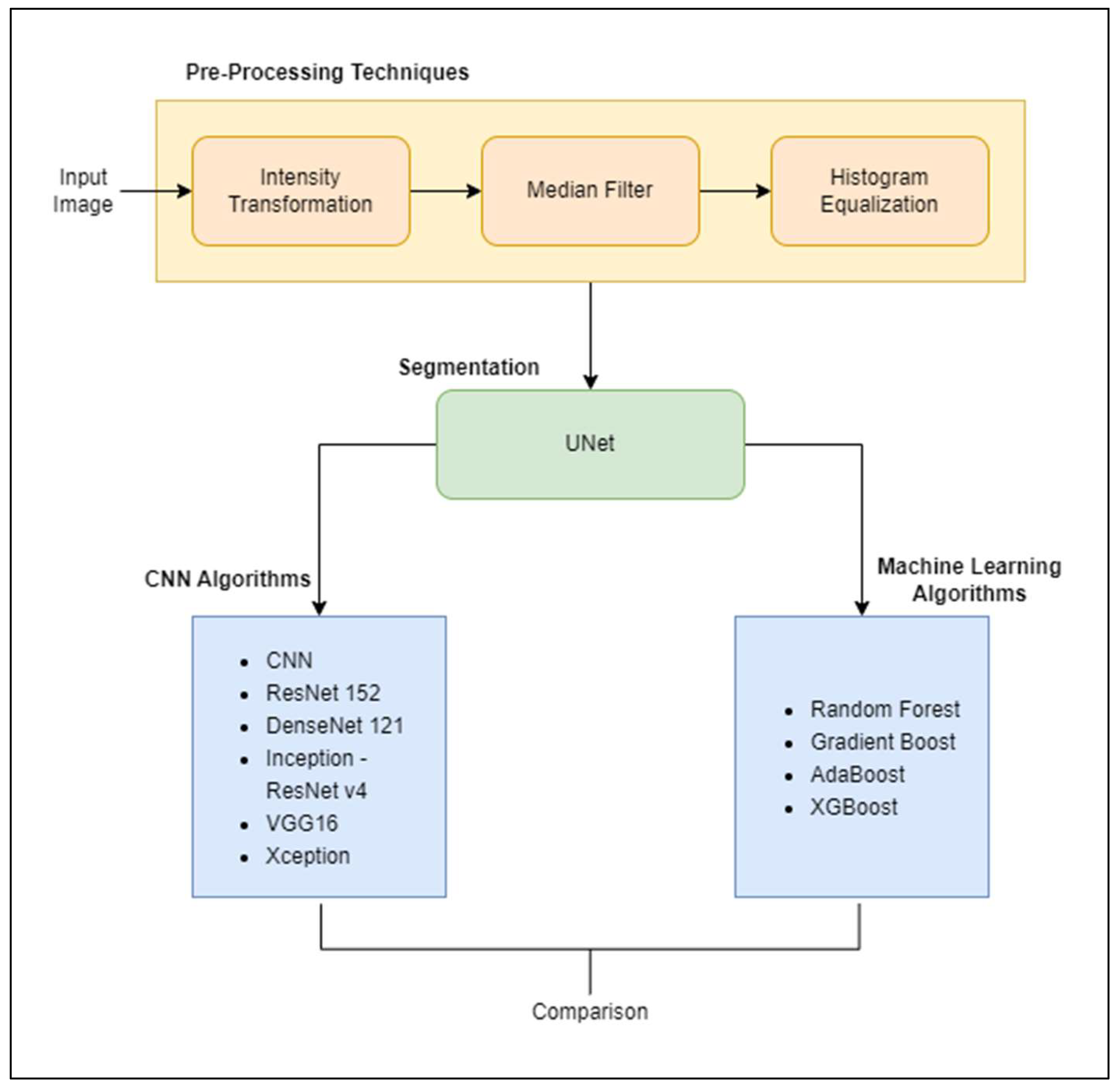
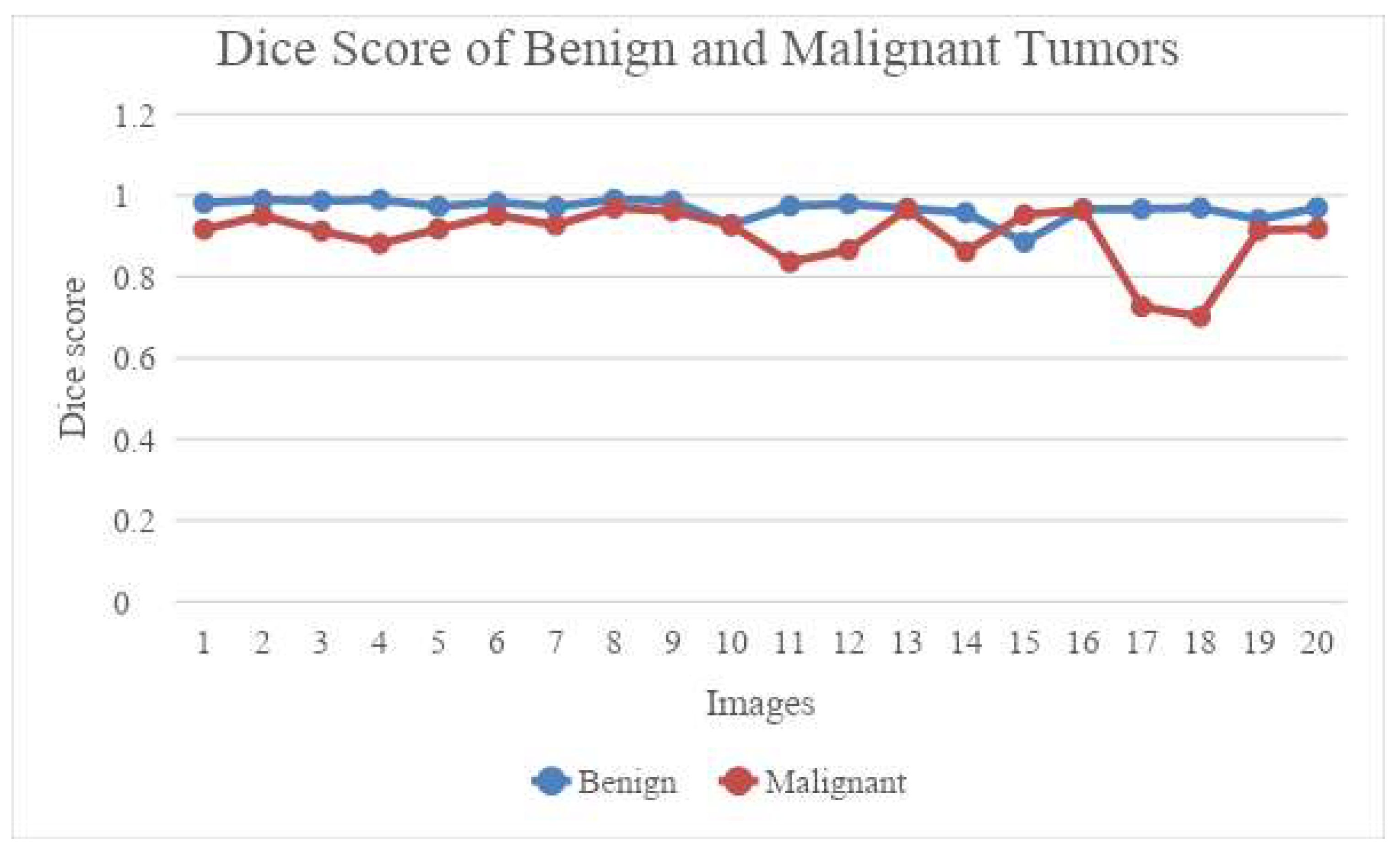
1. Introduction
Ovarian cancer stands out as a commonly diagnosed type of cancer worldwide. Considering the fact that it usually goes unrecognised until terminal stages, ovarian cancer is a leading reason for high mortality rates among women with gynaecological illnesses. Ranking fifth in cancer deaths among women, the risk of being diagnosed with Ovarian cancer peaks between the ages 55 and 64 on an average [
1]. Silent symptoms and undetermined causes act as major factors for late diagnosis and ineffective screening methods.
The American Cancer Society claims that around 19,710 women would be diagnosed with ovarian cancer, and that around 13,270 deaths would occur from ovarian cancer in 2023 in the United States [
2]. In the past few years, significant developments in the field of biomedical imaging have contributed to the domain of cancer detection. With interdisciplinary approaches being popularized to solve objectives, Medical Imaging can be combined with Machine Learning and Deep Learning disciplines to effectively detect and categorize tumours. Ultrasound and CT scan images contain large amounts of information making it an ideal use case for implementing Deep Learning algorithms.
This paper aims in providing a comparative study of the detection and classification of Ovarian tumors using Machine learning and Deep Learning algorithms, using CT scanned ovarian images. Multiple ML models and CNN variants are used for this purpose and are compared inter-categorically as well as intra-categorically. The Literature Survey section covers latest developments and ongoing research, not only in ovarian cancer detection but also in how state-of-the-art Deep Learning algorithms are used in other medical scenarios.
Section 2 reports the literature survey of the latest research performed in the area of biomedical imaging and of several Learning algorithms used.
Section 3 demonstrates the methodology along with the steps and the models used in the current work.
Section 4 presents the experimental results and the discussion.
Section 5 provides the conclusion thoughts.
2. Literature Review
Jung et al. [
3] use Ultrasound images of the lower body region of females to remove unwanted information in the frame and classify the ovaries into five classes – normal, cystadenoma, mature cystic teratoma, endometrioma, and malignant tumour. They use a texture-based analysis for tumour detection and train a convolutional autoencoder or CNN-CAE which is robust to the noise and removes irrelevant data such as callipers, annotations in the images or even names of the patient in order to maintain anonymity, which also makes it easier to detect the tumour. The images before and after the autoencoder are both fed into CNNs such as Inception, ResNet and different variants of DenseNets. Weighted class activation mapping or Grad-CAM has been used to visualise the result. It can be noted that the model classifies better when unnecessary data is removed using CNN-CAE. DenseNet121 and DenseNet161 are better performers amongst all the algorithms used when parameters like accuracy, sensitivity, specificity, and the area under the curve (AUC) are considered to be metrics of performance.
Wang et al. [
4], use pelvic CT scan images to detect and segment out Ovarian cancer tumours simultaneously i.e., create a multi task deep learning model. They propose a model called YOLO-OCv2 which is an enhancement to their previously proposed algorithm, YOLO-OC where the OC stands for Ovarian Cancer, a tumour detection model. They also state that Ultrasound images have lesser clarity when compared to CT scan images. The YOLO-OCv2 is based on YOLOv5 architecture rather than YOLOv3, as used in the prior work. Mosaic enhancement is also used here, in order to improve the background information of the object which contributes to the robustness of the model by making the calculations a little easier. A decoupling head is used for converting the result of the model into semantic parameters like category and confidence levels. One of the drawbacks that the authors mention is that the model is complex in nature and therefore is computationally expensive. However, the multitask model YOLO-OCv2, has outperformed other algorithms like Faster-RCNN, SSD and RetinaNet which were trained on the COCO dataset.
In this work, Mahmood et al. [
5] create a Nuclei segmentation model that can be used to segment out the nuclei in multiple locations of the body. Training a model dedicated to every site or organ in the body can get tedious and be difficult due to lack of data. Hence, the authors build a generalised model that can segment pixels in H&E stained histopathological images into two categories, namely Nuclear and Non-nuclear. Simple Digital Image processing approaches to segment out the nuclei are found to face difficulties such as meticulous selection of parameters or high computation levels. Deep Learning algorithms were more robust to these challenges. However, a critical challenge to these CNNs is to segment overlapped and clumped nuclei. To overcome this, they have used Conditional Generative Adversarial Networks or cGAN as they can control the GAN training output depending on a class. The model is trained on synthetically generated data along with real data in order to make sure that sufficient input is present. The model is trained with data from nine organs and is tested on four organs where it outperforms its peers such as FCN, U-Net and Mask R-CNN.
Guan et al. [
6] use mammographic images to detect breast cancer which is the second most common type of cancer in women, using CNN models. One of the main obstacles in building such models is the lack of data. To overcome this, data augmentation is a solution. This can be done in several ways but in the current work, the authors have focused on Affine transformations and synthetic data generation using GANs. The images are collected from Digital Database for Screening Mammography (DDSM) from which Region of Interests or ROIs are cropped, annotated and are saved in PNG format. This is called the Original ROIs or ORG ROIs. On these, affine transformations such as shifting, shearing, scaling etc. are performed at randomly chosen degrees to obtain Affine ROIs or AFF ROIs. The original ROIs are also fed as an input to the discriminator of the GAN which produces synthetically generated GAN ROIs which is also used as data to train the classification models. The images used for testing are classified into a healthy category called Normal and a tumour depicting category called Abnormal. Results show that when the neural network is trained with original data along with GAN generated data, it performs better than the other combinations. It is also important to note that the work includes mammography images in two different views known as CC view and MLO view.
In addition, the detailed literature review was made to understand the advancement of deep learning in the medical imaging segmentation.
According to Karimi et al. [
7], Convolution is an important process in segmentation which has contributed significantly to computer vision algorithms. However, as their weights are defined during training itself, it is not capable of altering its bias based on other images or their parts. Attention-based neural networks hold this capability and are designed to overcome this obstacle as the weights of its architecture are only partially fixed and partially are able to assign the weights based on the input data. Transformers are one of the most widely used Attention-based models. Transformers are known to avoid the vanishing gradient problem. They also allow parallel input processing thereby allowing lower training time. However, transformers require large amounts of data when compared to CNNs as the bias is changing constantly with every input provided unlike CNNs whose weights are decided and are fixed during training. Handling pixel level data could also be a challenge as there would be comparatively a very large amount of data in each image. To overcome this, Vision Transformers (ViT) is used to divide images into Image Patches. The proposed algorithm using transformers does not use any convolution operations to segment the Brain cortical plate and the Hippocampus in MRI images of the brain. The results are compared with FCN architectures like 3D UNet++, Attention UNet, SE-FCN and DSRNet. The proposed network performs segmentation accurately when compared to the other and with significantly a smaller number of labelled training images.
Xu et al. [
8] work on histopathological whole-slide images (WSIs) to detect ovarian cancer using CNNs trained on images of multiple resolution. The algorithms find it challenging to process whole slide images and are thus broken down into smaller patches and aggregated later to obtain results. Recently, algorithms like Vision Transformer (ViT), instance-based Vision Transformer (i-ViT) etc are used to combine the patch-level results. The authors propose a new algorithm by introducing efficient patch selection to extract and aggregate multi-resolution patches to form Multi-Resolution Vision Transformer or MR-ViT. The model is aimed to recognise long-range relations between several patches. A modified version of ResNet50 called Heatmap ResNet50 is used for CNN-based patch selection and ResNet18 along with MR-ViT is used for ViT based slide classification. These are applied onto the OVCARE dataset to classify the instances into normal and tumour images. The future scope of the work is said to be finding the optimal parameters to make sure the algorithms perform better.
Li et al. [
9] introduce a variation of UNet known as CR-UNet to simultaneously segment out Ovaries and Follicles from Transvaginal Ultrasound (TVUS) images. These types of images are usually prone to noise and acoustic shadows. Although algorithms like CNNs can be used for segmentation, using them to segment ovaries and follicles would be challenging as the size, shape, number and texture vary greatly for different instances. A simple UNet could be the solution but as UNet is prone to lose spatial relative information of neighbouring pixels, it might affect detecting the ovaries even when follicles might be detected. To overcome this issue, the CR-UNet is introduced which contains the structure of a UNet along with spatial Recurrent Neural Networks (RNN) added into each layer between the encoder-decoder. The Tanh activation function is replaced with ReLU in a plain RNN to form spatial RNN which speeds up the training process when compared to other types of RNNs. Since large input feature size might be an issue during this implementation, the spatial RNNs are removed in the first and last layer of the model to form CR-Unet-v2. To avoid vanishing gradient problems created by the RNNs, deep supervision is used to inject auxiliary losses at each layer in order to compensate. These algorithms are applied on the dataset collected and annotated by radiologists. It is then compared with models like DeepLabV3+, PSPNet-1, PSPNet-2 and U-Net to find that the proposed model outperforms them all.
In the proposed work by Goodfellow et al. [
10], an adversarial net framework is suggested that loosely resembles a minimax two-player game. A generative model G which captures the data distribution is pitted against a discriminative model D which gauges if the data is from training instead of G. G has to maximise the probability of D making a mistake and D has to improve recognising the instances more accurately. Both G and D are multilayer perceptrons. The paper also provides theoretical validations for the proposed models. From their experiments on MNIST, the Toronto Face Database, and CIFAR-10 datasets, they deduce that even if the framework is not better than existing models, it provides competitive results with better generative models. It concludes that although the method has disadvantages like there is no explicit representation of the distribution of the generator over data x and D and G must be accurately synchronised, there are many computational and statistical advantages like not using Markov chains, no inference needed during learning and using gradients and not data examples to update the generator network.
Nagarajan et al. [
11], in their research work, provide three approaches that are used to classify ovarian cancer types using CT images. The first approach uses a deep convolutional neural network (DCNN) based on AlexNet which does not provide satisfactory results. So, a combination of AlexNet, GoogleNet and VGG is used in the second approach by fusing the SoftMax values of the SoftMax layer of each network structure using a weighted sum to obtain the result. The dataset includes 350 training images and 147 test images with 50 and 21 images in each ovarian cancer type category respectively. Since there is not a huge dataset, the second approach has an overfitting problem. To overcome this, GAN is used in the third approach to augment the image samples along with the DCNN which provides the best results out of the three approaches in metrics such as precision, recall, f-measure and accuracy.
In the work of Zhao et al. [
12], in order to explore the representation capacity of multi-modality ultrasound ovarian tumor images, a Multi-Modality Ovarian Tumor Ultrasound (MMOTU) image dataset which has 1469 2d ultrasound images along with 170 contrast enhanced ultrasonography (CEUS) images is taken. These images are given pixel-wise and global-wise annotations. Four baseline architectures - CNN-based “Encoder-Decoder'', transformer-based “Encoder-Decoder”, U-shape networks and spatial-context based two-branch networks for semantic segmentation are provided. A Dual-Scheme Domain-Selected Network which has a feature alignment-based architecture using adversarial learning to assist in the domain shift of encoders so that both source and target images have better representation capacity is proposed to extract domain-distinct and domain-universal features. This method provides an insight to solve domain shift problems using feature decoupling techniques. The dataset does not include images for all categories and the effect of unbalanced sample size is yet to be observed which provides room for future improvement.
The research work of Saha et al. [
13] includes a novel 2d segmentation network called MU-net is proposed, which is a combination of MobileNetV2 and U-Net used to segment out follicles in ovarian ultrasound images. USOVA3D Training Set 1 dataset is used. For the encoder part of the U-net architecture, a pre-trained MobileNetV2 model is used to help the model converge faster. The decoder of the U-net model is retained. The semantic segmentation of U-nets classifies every pixel and provides high accuracy. The proposed model is evaluated against several other models from previous literature and is shown to be more accurate with an accuracy of 98.4%.
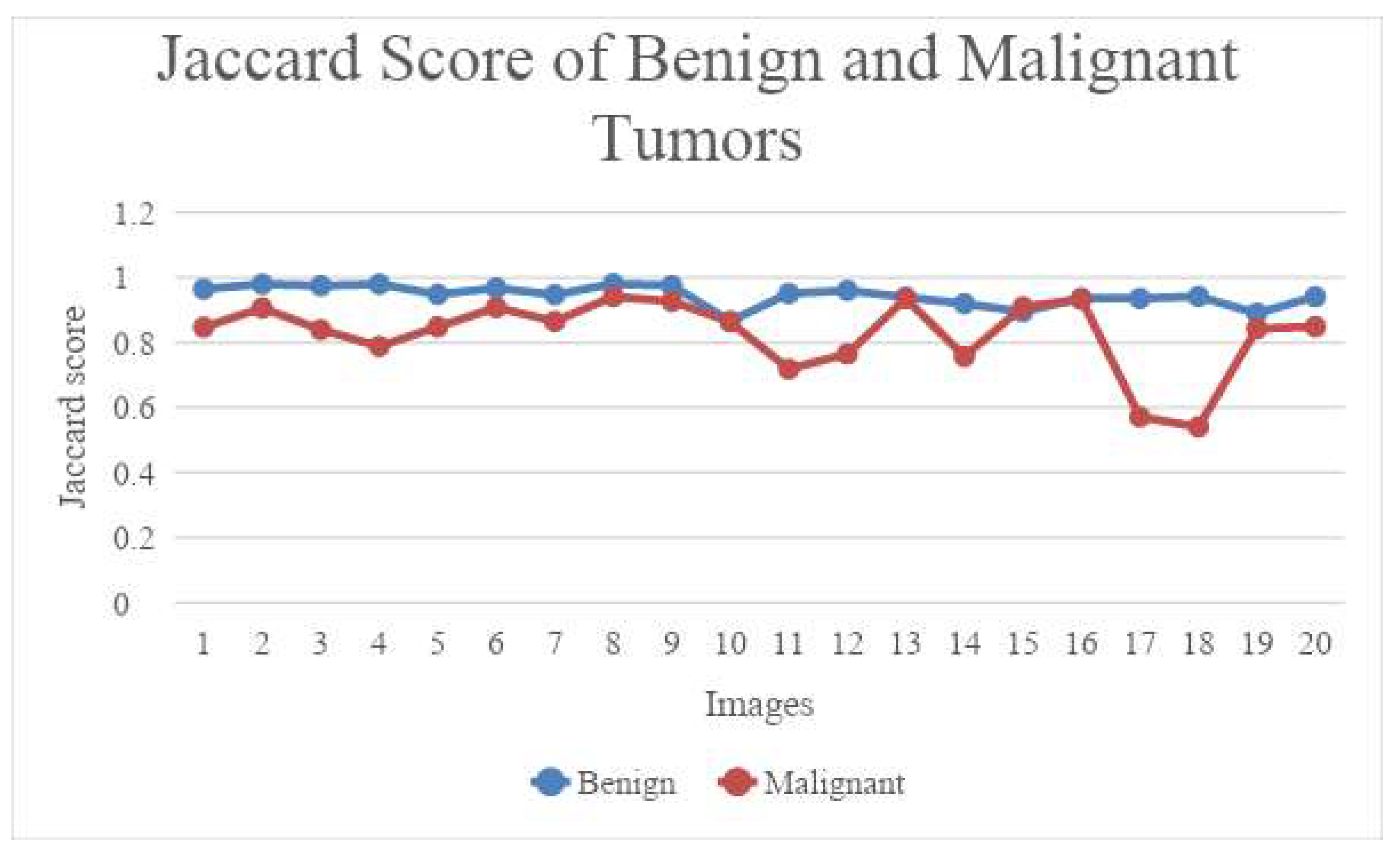
Jin, J et al. [
14], for their work, use a dataset containing 469 ultrasound images divided into 353 test images, 23 validation images and 93 test images are used, for which manual segmentation was delineated as ground truth. Four UNet models - U-net, U-net++, U-net with Resnet and CE-Net are used to perform automatic segmentations. First, Python and Pyradiomics 2.2.0 was used on the segmented target volume to extract radiomic features the reliability of which was evaluated with intraclass correlation coefficients (ICC) and Pearson correlation. 97 features were extracted from the delineated target volume. Jaccard similarity coefficient (JSC), dice similarity coefficient (DSC), and average surface distance (ASD) were used to evaluate the accuracy of automatic segmentation. U-net with Resnet and CE-Net outperformed the other models and it was concluded that CE-net gives the best radiomics reliability.
In this paper by Thangamma et al. [
15], k-means algorithm and fuzzy c-means algorithm are used on ultrasound images of ovaries. It is concluded that the fuzzy c-means algorithm provides a better result than the k-means algorithm as it is insensitive to noise and that the results are dependent on the number of iterations.
The work by Hema et al. [
16] involves FaRe-ConvNN which applies annotations on the image dataset where the images have three categories: epithelial, germ and stroma cells. In order to avoid overfitting and other issues due to small dataset size, image augmentation using image enhancement and transformation techniques like resizing, masking, segmentation, normalization, vertical or horizontal flips and rotation is done. Noise reduction and filtering is carried out on these images. FaRe-ConvNN is used to compensate for manual annotation. After the region-based training in FaRe-ConvNN, a combination of SVC and Gaussian NB classifiers is used to classify the images which resulted in impressive precision and recall values.
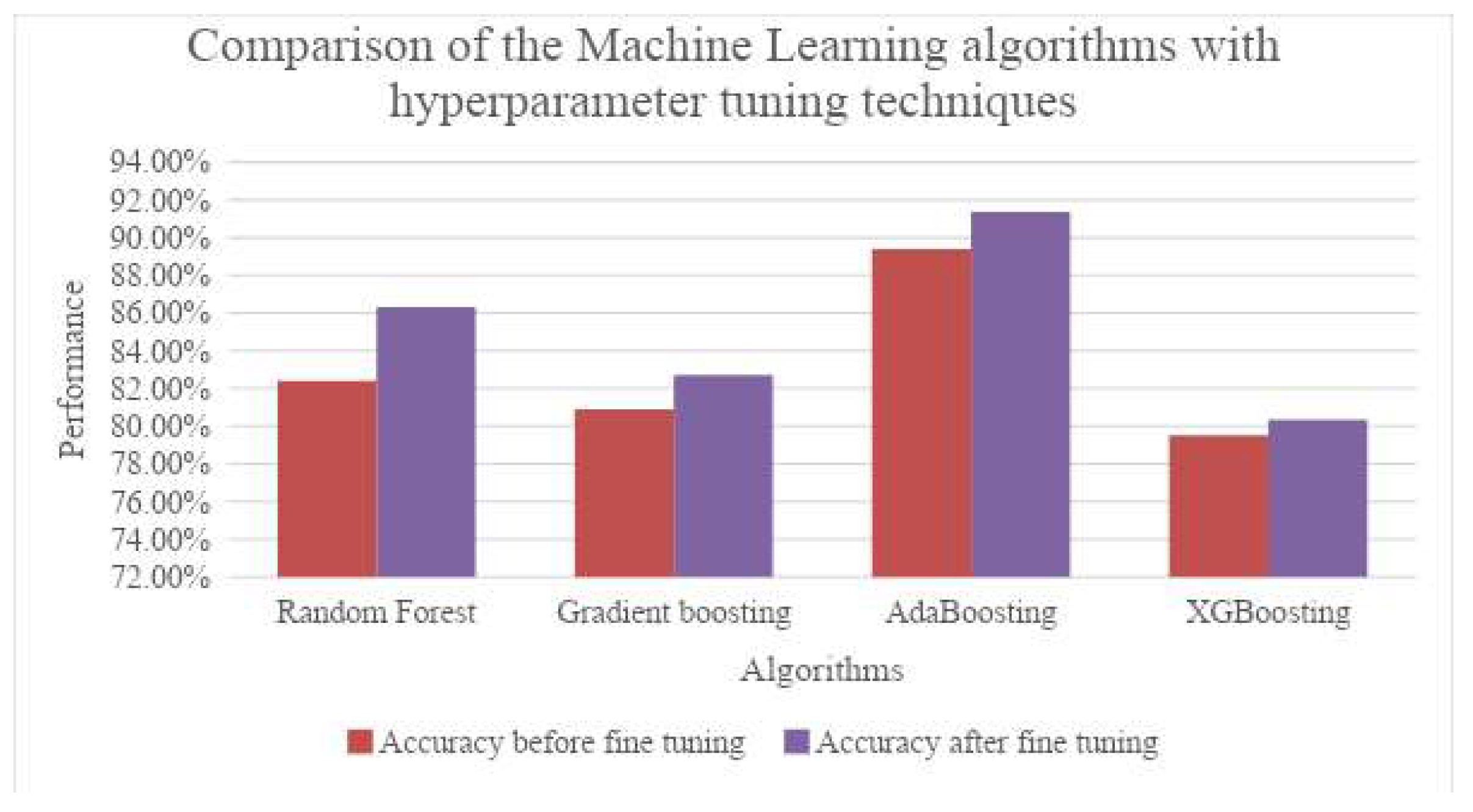
In the paper by Ahmad et al. [
17], 349 patient data that has three subgroups, blood routine test, general chemistry and tumor marker, is taken to identify significant blood biomarkers using statistical analysis like Student's t-test and Mann-Whitney U-test. Several machine learning models like Logistic Regression (LR), Light Gradient Boosting Machine (LGBM), Decision Tree (DT), Random Forest (RF), Support Vector Machine (SVM), Gradient Boosting Machine (GBM), and Extreme Gradient Boosting Machine (XGB) are used separately and metrics like accuracy, precision, recall, F1-score, Area Under Curve (AUC) and log-loss are used for evaluation. Along with identifying several significant blood biomarkers using statistical methods, it is deduced that GBM and LGBM have the highest accuracy, F1-score and AUC while RF displays highest recall score and DT shows the highest precision with least log-loss value given by LGBM for the blood samples dataset. RF showed the maximum accuracy and AUC and minimum log-loss with LGBM giving the highest precision and F1-score with SVM having the highest recall value for the general chemistry dataset. In the marker dataset, highest recall, accuracy, AUC and lowest log-loss were obtained by both XCBoost and RF with DT showing maximum precision and RF having maximum F1-score.
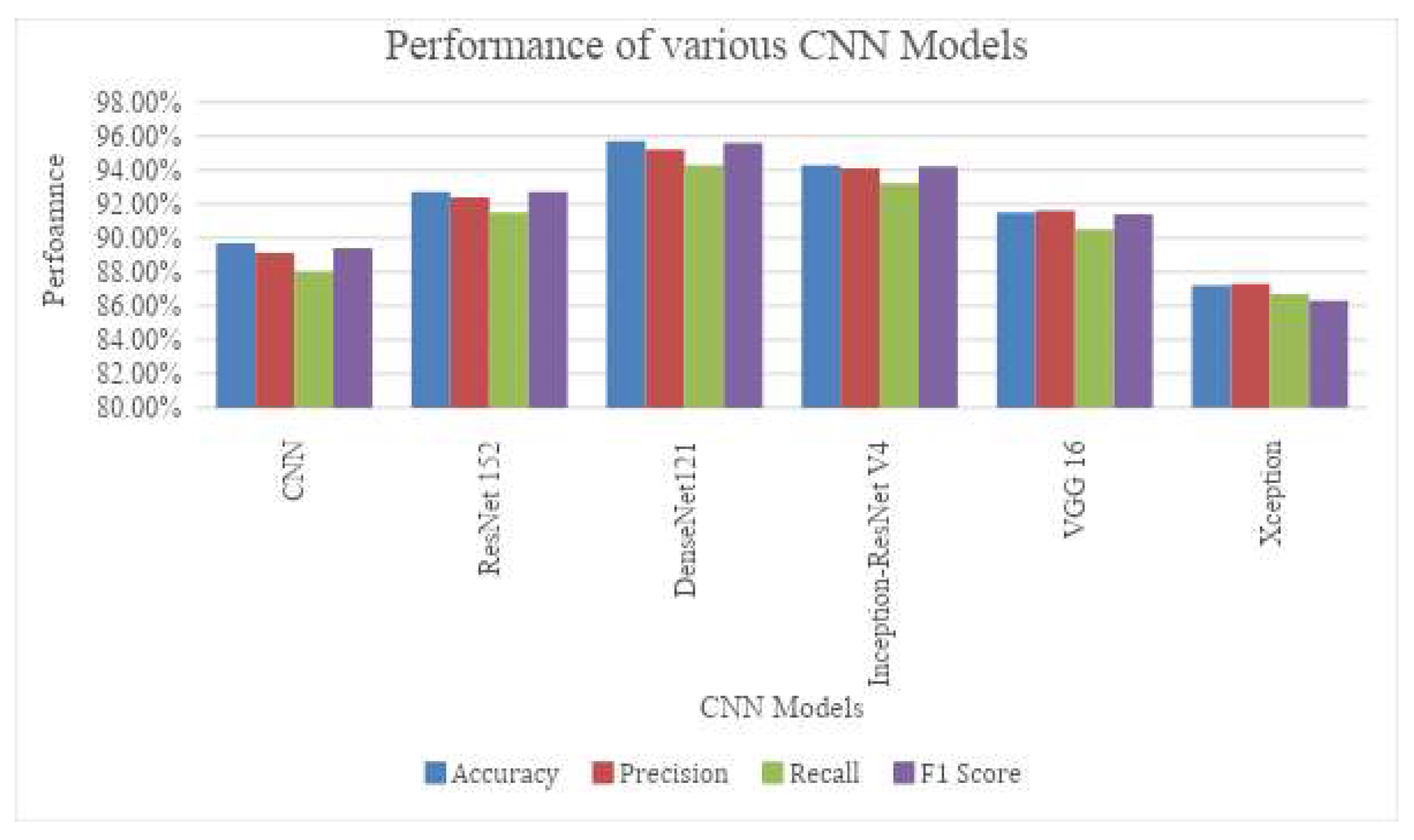
In the works carried out by Ashwini et al. [
18,
19,
20], the various deep learning models were used to segment the CT scanned images and classified using variants of CNN. In the work [
18,
19], the Otsu’s method was used to segment the tumor and obtained the dice score of 0.82 and Jaccard score of 0.8356. Further the performance of the segmentation was used cGAN [
20], in this study the segmentation and classification of tumors were carried out in the single pipeline and obtained the dice score of 0.91 and the Jaccard score of 0.89. Similarly, the works carried by Fernandes et al [
21,
22], according to the work [
21], the authors have proposed segmentation of brain MRI images using entropy based. As per [
22], the detection and classification of brain tumors by parallel processing using big data tools such as kafka and Pyspark.