Submitted:
15 March 2023
Posted:
17 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1 Datasets
- StarPepDB. It is a graph database embedded in the StarPep toolbox that consists of 45120 peptides with annotated activities retrieved from 40 bioactive databases and other sources [16]. A sub-dataset consisting of 2004 hemolytic peptides was extracted from this database to generate HSPNs, METNs and discover new hemolytic motifs. In addition, the complete StarPepDB was also used in the motif enrichment process to help find the most representative hemolytic motifs.
- HemoPI-1. It encompasses 552 experimentally validated highly hemolytic peptides (positive) and 552 random peptides extracted from Swiss-Prot (negative) [8]. This dataset was only used in motif enrichment analysis.
- Big-Hemo. It is a non-redundant combination of several datasets that contain either hemolytic or highly hemolytic peptides as positive samples and non-hemolytic or low hemolytic peptides as negative samples. The datasets used to generate the Big-Hemo dataset are HemoPI-2 Main and Validation [8], HemoPI-3 Main and Validation [8], HAPPENN [1], HLPred-Fuse Layer 2 Training and Independent datasets [4] and HemoNet [9]. To construct Big-Hemo, only positive samples labeled as “highly hemolytic” were retrieved from these datasets to handle the problem of lack of agreement and standardization at considering when a peptide is hemolytic or not, and the way of measuring this property, respectively [1,31]. Although HAPPENN dataset contains positive samples not labelled as highly hemolytic, its positive samples were also included in Big-Hemo in order to gain more diversity and a better representation of hemolytic peptides. Thus, this dataset was addressed to evaluate whether our novel motifs are enriched in highly hemolytic peptides, which are more concerning when designing therapeutic peptides. In addition to redundancy removal, peptides containing ‘X’ several times in a sequence and Nphe or Nleu in their sequences were also discarded. The resulting Big-Hemo dataset contains 2196 highly hemolytic peptides. Like HemoPI-1 dataset, Big-Hemo was also used for motif enrichment analysis.
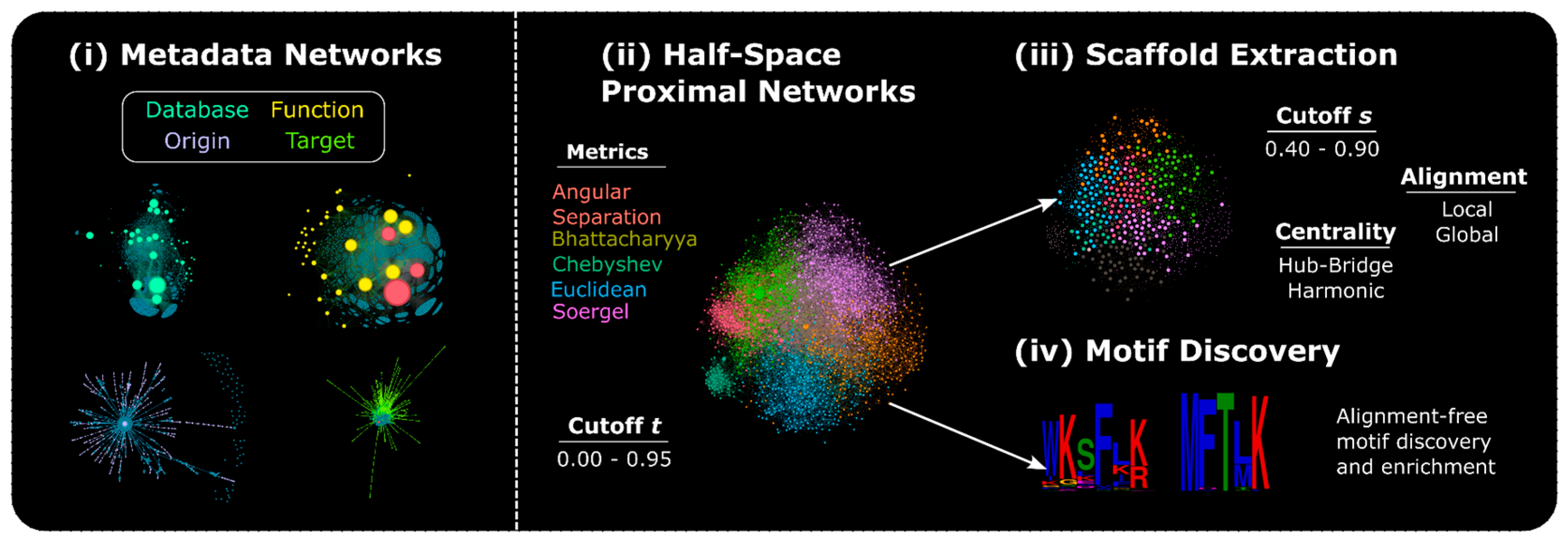
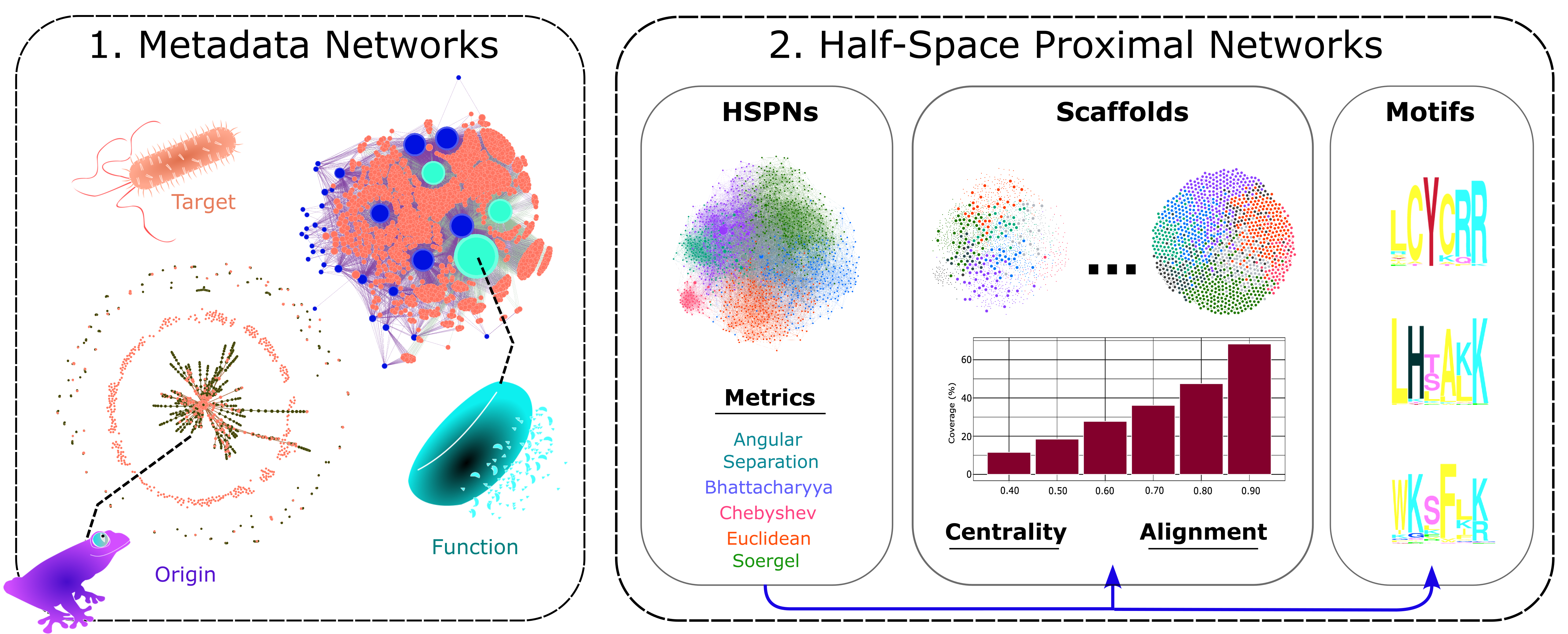
2.2 Network Generation and Analysis
2.2.1. Metadata Networks (METNs)
2.2.2. Half-Space Proximal Networks (HSPNs)
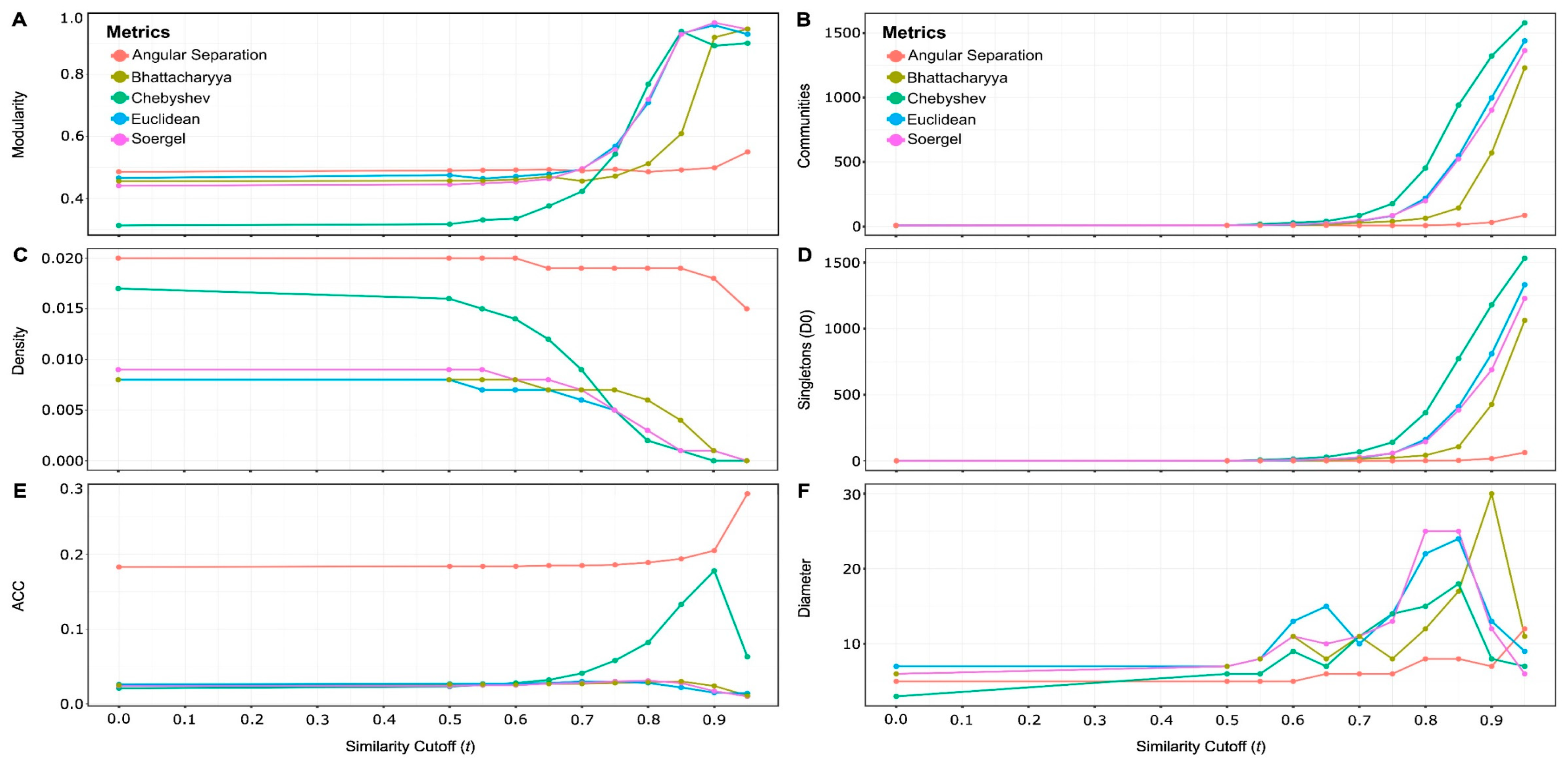
- A (dis)similarity measure is calculated for each pair of nodes using the vectors of peptide features. Then these values are normalized (min-max normalization). This forms a symmetric similarity matrix of size where represents the number of hemolytic peptides and represents the similarity score between the nodes and , being 1 the highest similarity value and 0 the lowest. Then a rule called Half-Space Proximal (HSP) test [26] is applied to construct the HSPN, which is a strongly connected but sparse network [25], that preserves the number of nodes while containing a relatively low number of edges compared to the counterparts, CSNs [25].
- Finally, a threshold or cutoff value can be applied to the weighted edges to further reduce the density of the graph by removing edges whose similarity values are lower than . This helps to study the topology of the resulting graphs and subsequently find the best representative network of the chemical space occupied by hemolytic peptides. It is worth mentioning that for the construction of HSPNs, using a value is not mandatory.
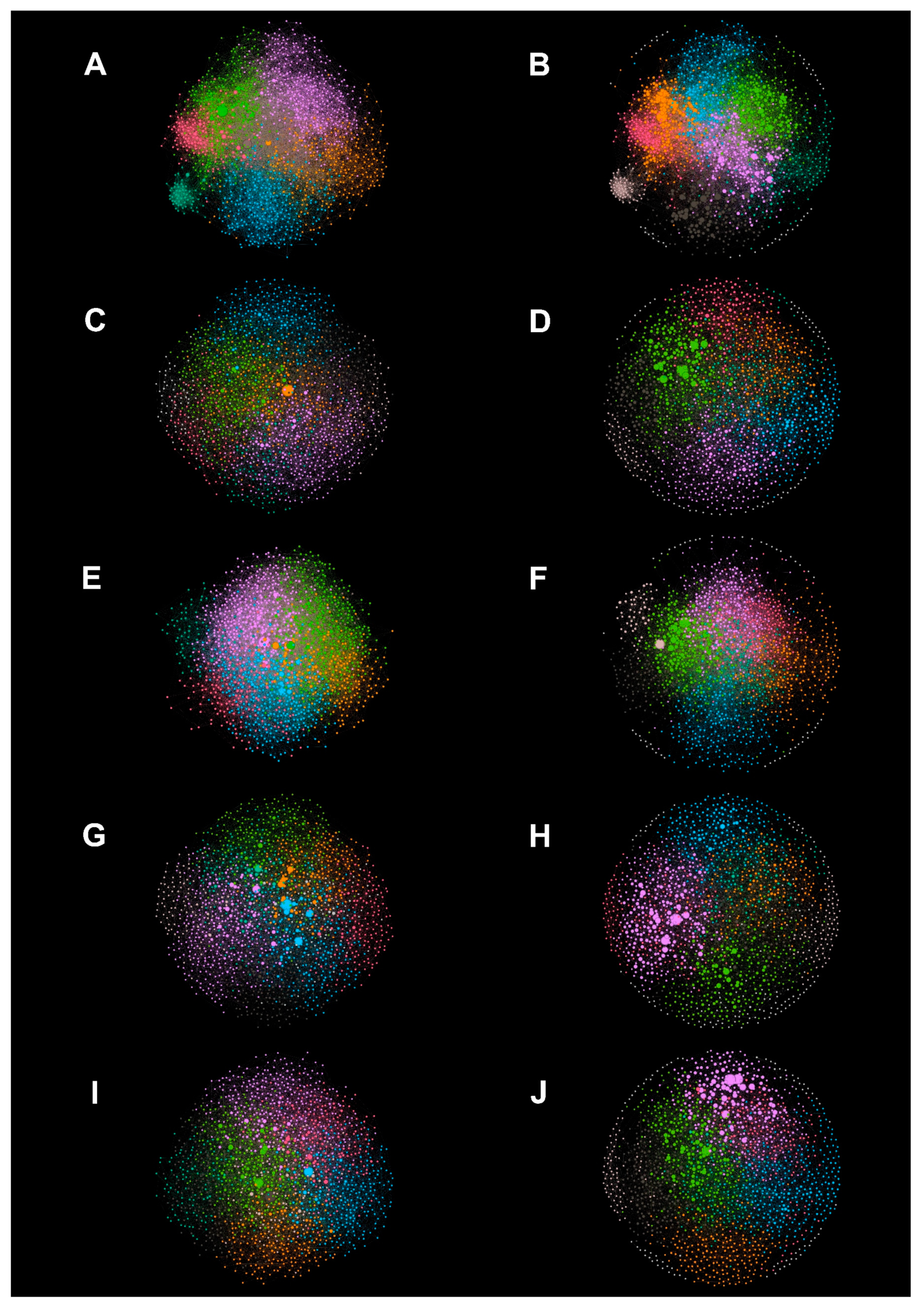
2.2.3. Network Visualization
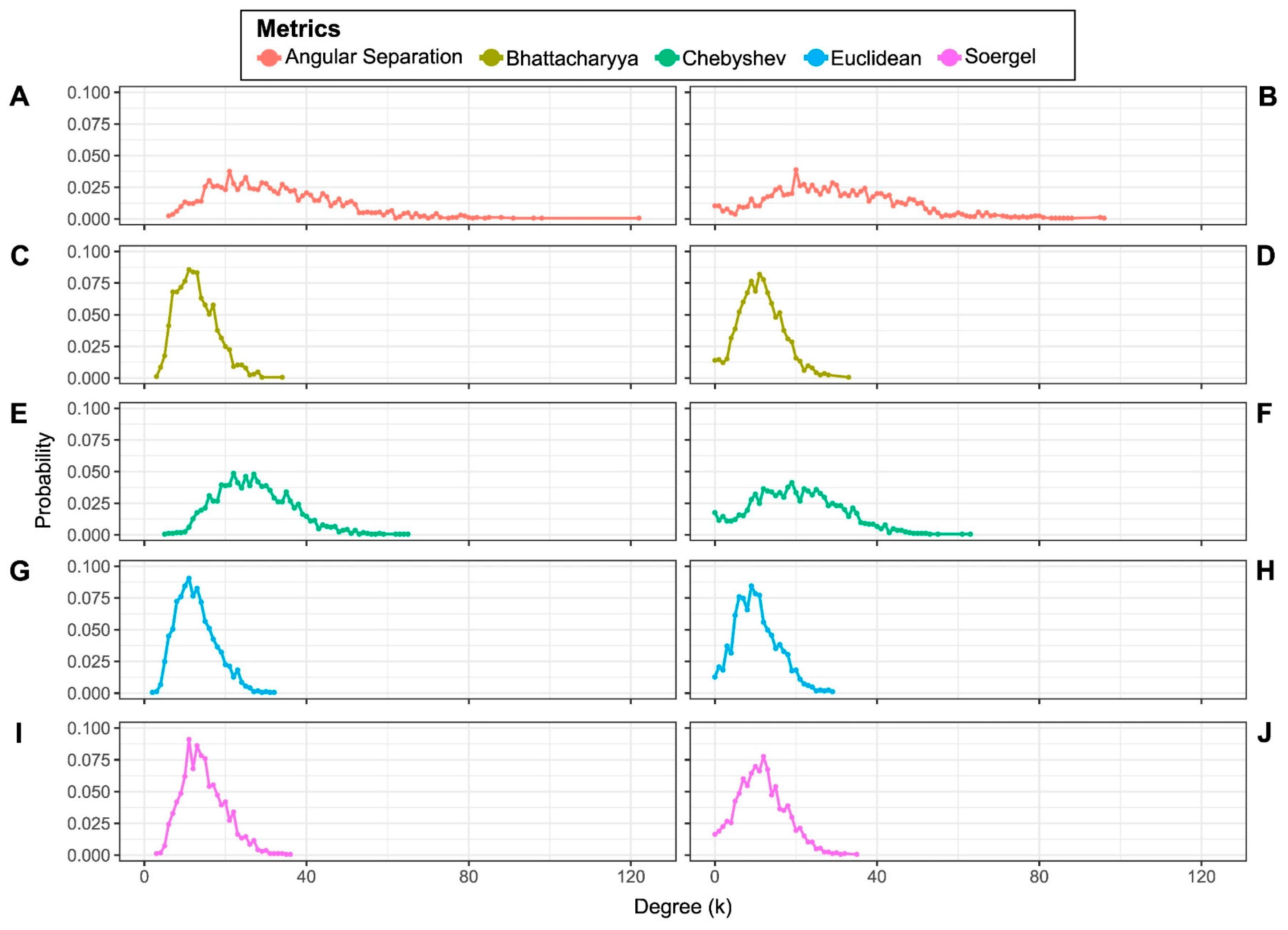
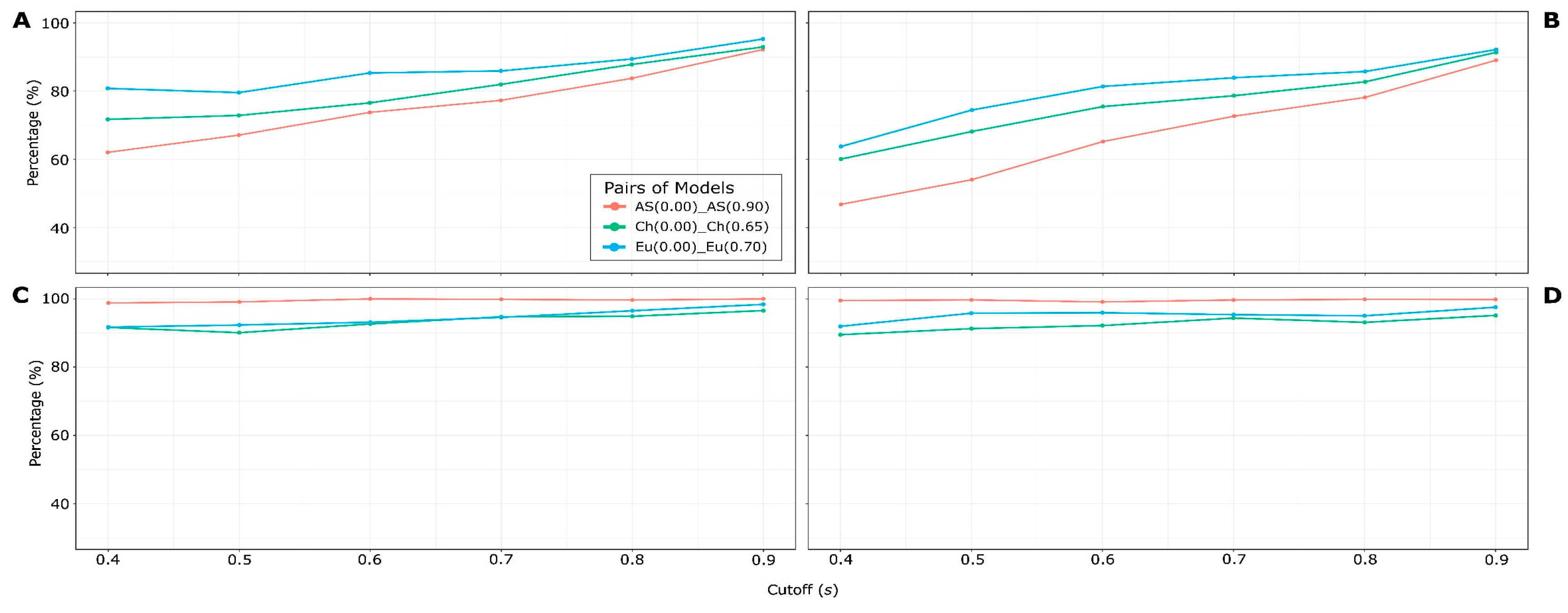
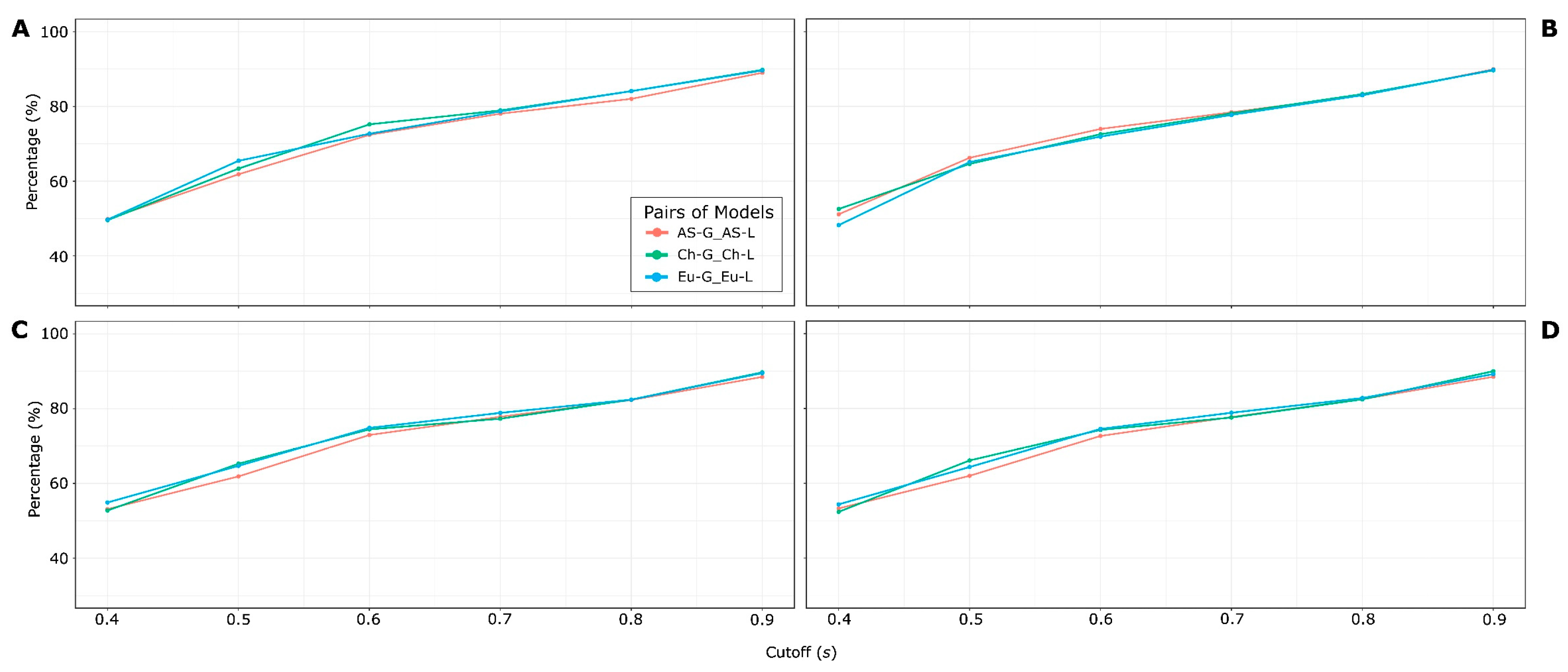
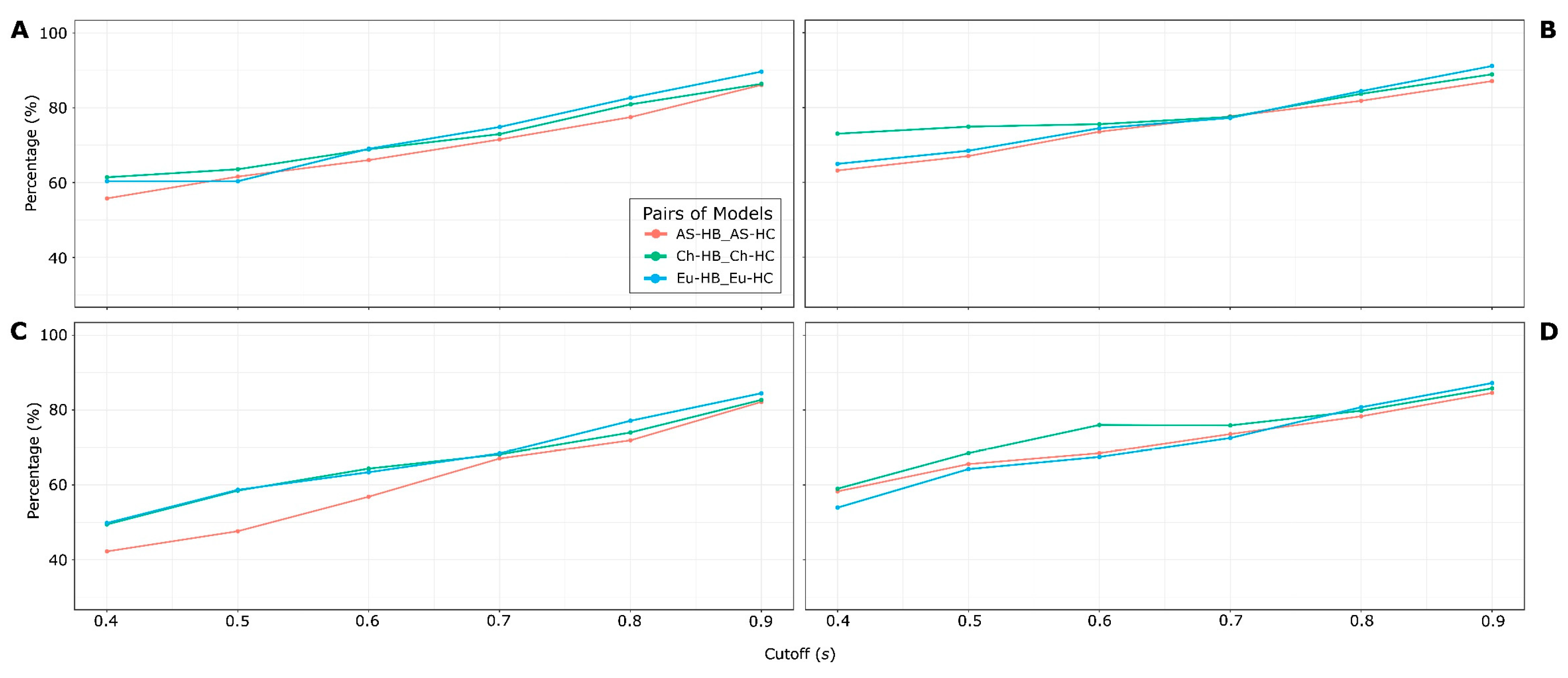
2.2.4. Selection of the best HSPNs
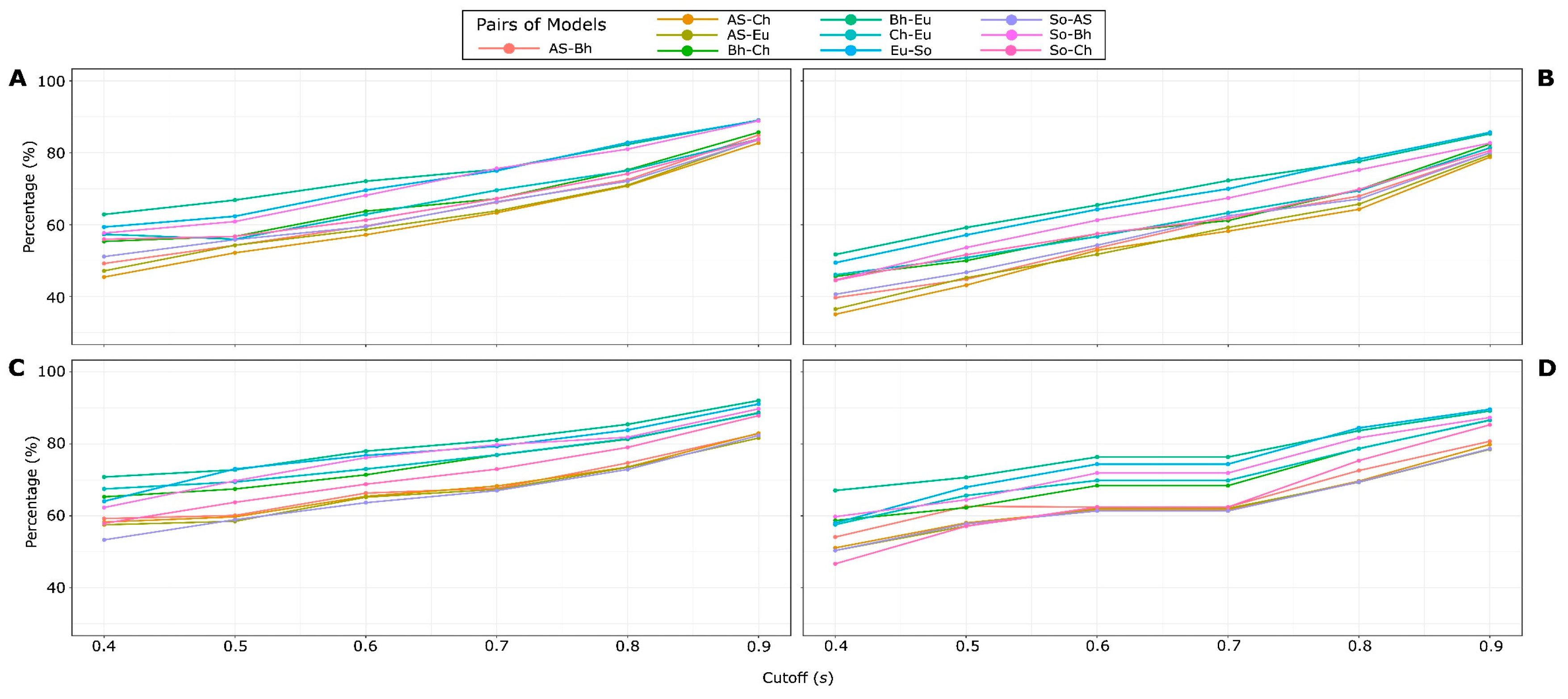
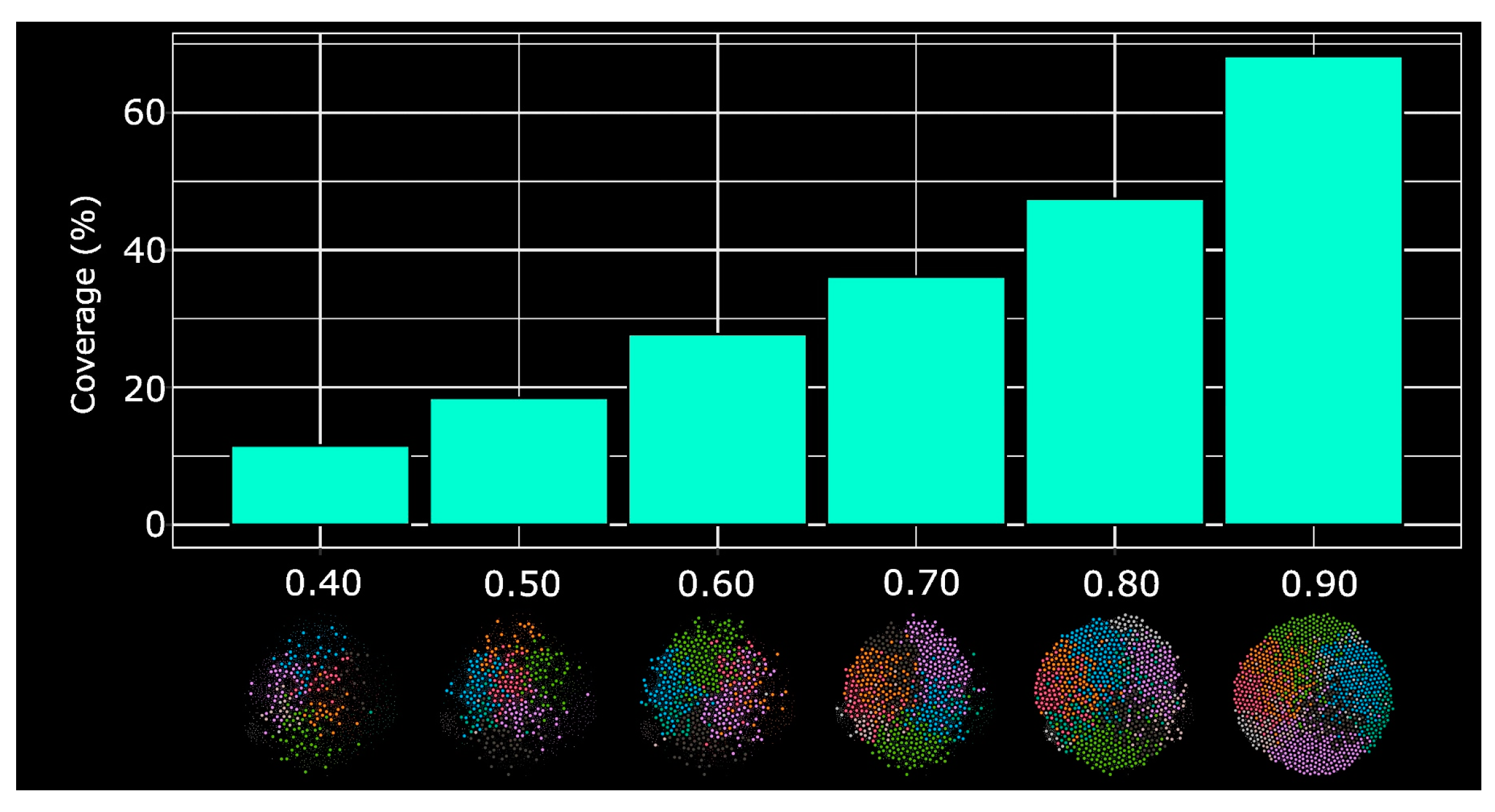
2.3 HSPNs Scaffold Extraction and Analysis
2.5. Motif Discovery and Enrichment
2.5.1. Motif Discovery
- Using the StarPep toolbox we extracted the sequences of peptides belonging to each cluster (community) and saved them as fasta files. Then these files were used as input sequences for motif discovery. For control sequences, we let the method use shuffled input sequences. Since our peptides contain non-standard AAs, we provided a customized alphabet (SM5.1.1). Motifs ranging from 3 to 6 letters, at least 20% present in the input sequences and with a p-value lower than 0.05 were retrieved.
2.5.2. Motif Enrichment
3. Results and Discussion
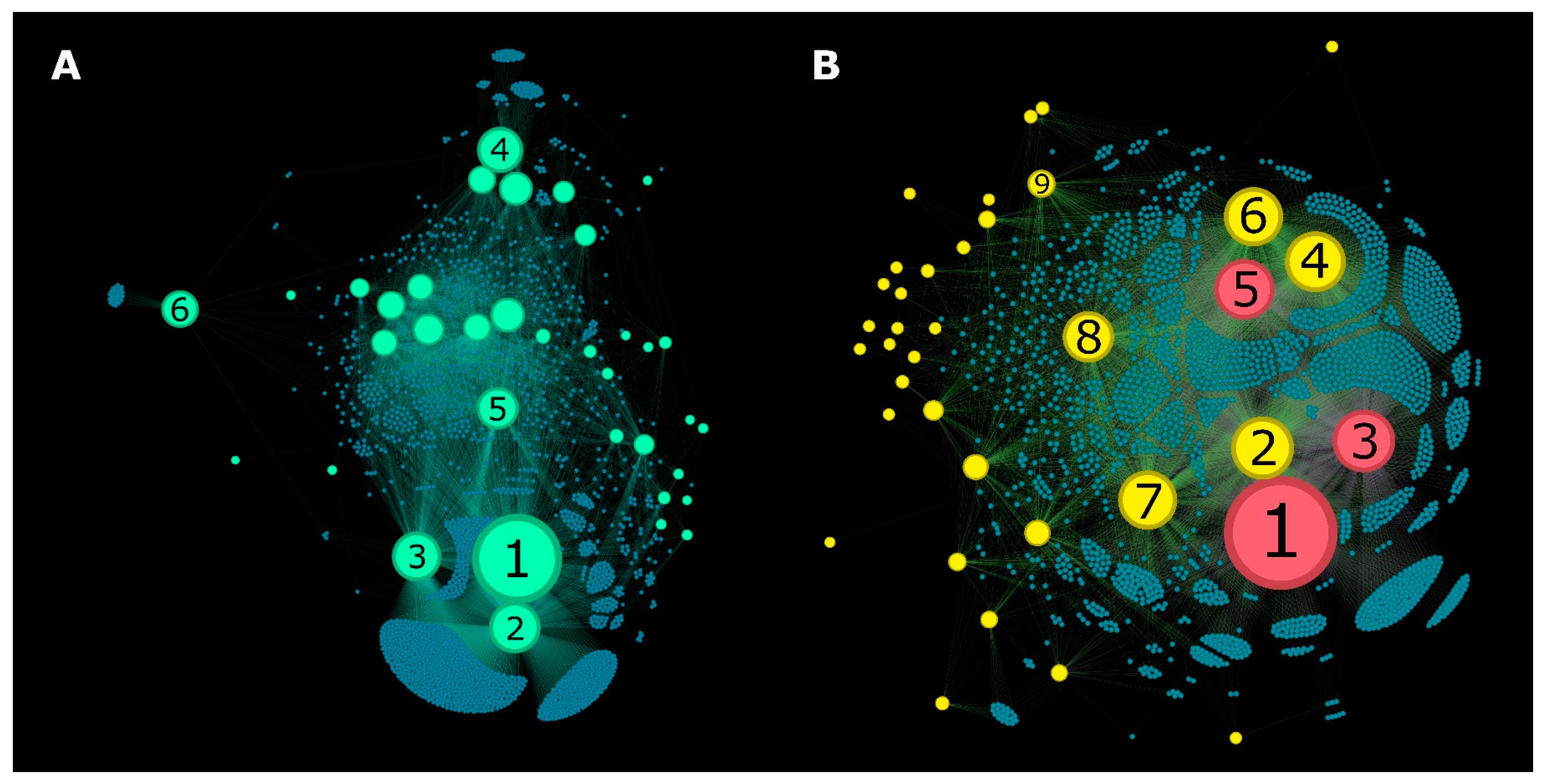
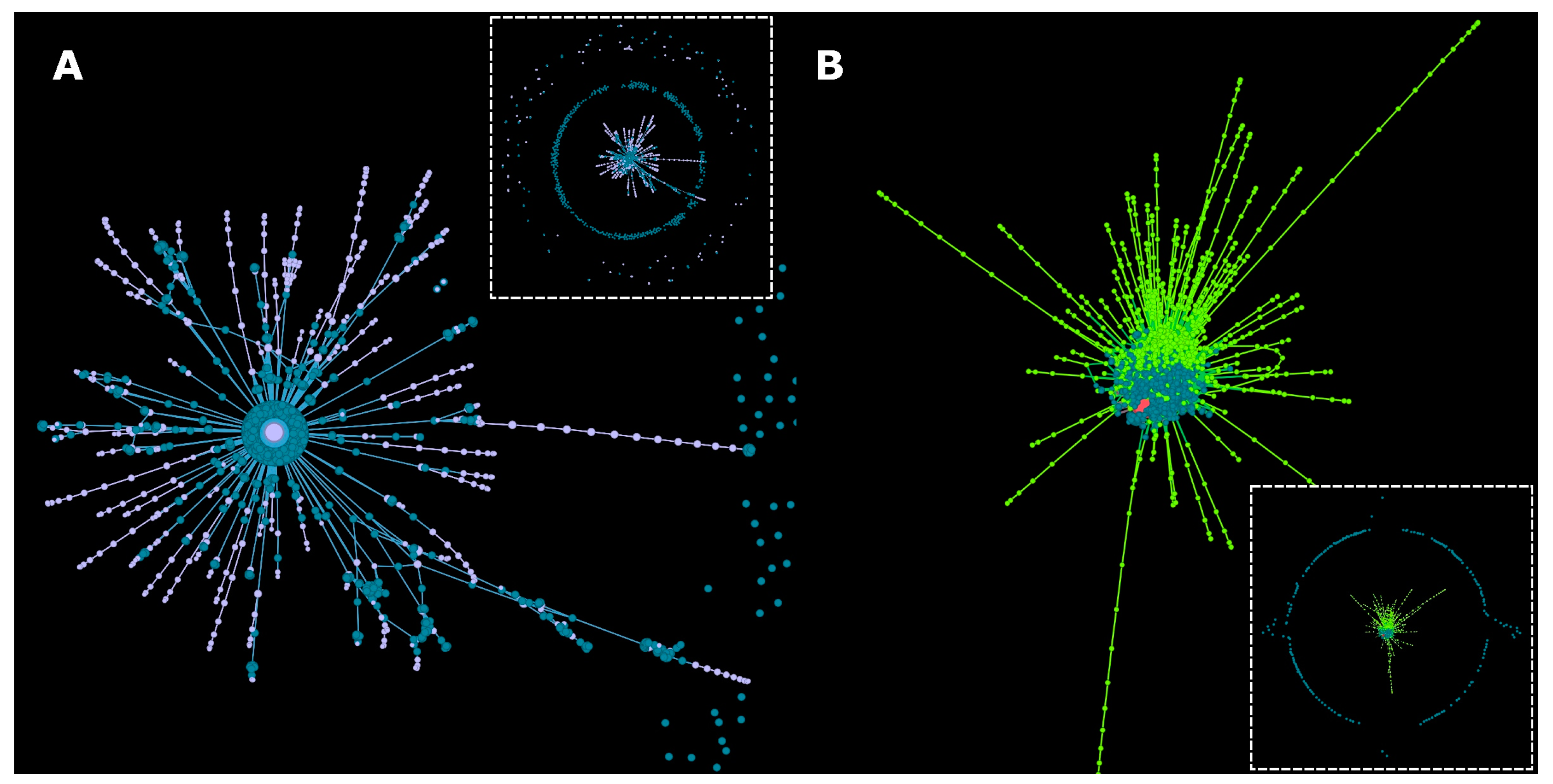
3.1. Metadata Networks (METNs)
3.2. Half-Space Proximal Networks (HSPNs)
3.3. HSPNs Scaffolds
3.4. Hemolytic Motif Discovery and Enrichment
- MFTLK, ALKAIS, GTCN, WKSFJK, VCGETC, WKK, AKKAL, GETCV, CYCR, LKKL, CVCV, ISWIK, RFC, LHTA[KL], FLHSAK, CSW, LWKT, FLGTI, GAVLKV,PGC, KKILG, KITK, KHI, LGKL, KWK, VNWK, K[GT]AGK, VCT, ALW, SWP, HIF,LLKK, [VI]LDTJ, CRR, KLL, JGKL, FKK, GAIA, VLK, GLP, PKIF, GKEV, GTIS, AAAK, GCS, IAS, MAL (Table 4).
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Timmons, P.B.; Hewage, C.M. HAPPENN Is a Novel Tool for Hemolytic Activity Prediction for Therapeutic Peptides Which Employs Neural Networks. Sci Rep 2020, 10, 10869. [Google Scholar] [CrossRef] [PubMed]
- Win, T.S.; Malik, A.A.; Prachayasittikul, V.; S Wikberg, J.E.; Nantasenamat, C.; Shoombuatong, W. HemoPred: A Web Server for Predicting the Hemolytic Activity of Peptides. Future Medicinal Chemistry 2017, 9, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.-F.; Jie, M.-M.; Li, B.-S.; Hu, C.-J.; Xie, R.; Tang, B.; Yang, S.-M. Peptide-Based Treatment: A Promising Cancer Therapy. Journal of Immunology Research 2015, 2015, e761820. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Schaduangrat, N.; Basith, S.; Lee, G.; Shoombuatong, W.; Manavalan, B. HLPpred-Fuse: Improved and Robust Prediction of Hemolytic Peptide and Its Activity by Fusing Multiple Feature Representation. Bioinformatics 2020, 36, 3350–3356. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, R.; Agrawal, P.; Patiyal, S.; Raghava, G.P.S. A Method for Predicting Hemolytic Potency of Chemically Modified Peptides From Its Structure. Front Pharmacol 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Plisson, F.; Ramírez-Sánchez, O.; Martínez-Hernández, C. Machine Learning-Guided Discovery and Design of Non-Hemolytic Peptides. Sci Rep 2020, 10, 16581. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic Peptides: Current Applications and Future Directions. Sig Transduct Target Ther 2022, 7, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci Rep 2016, 6, 22843. [Google Scholar] [CrossRef]
- Yaseen, A.; Gull, S.; Akhtar, N.; Amin, I.; Minhas, F. HemoNet: Predicting Hemolytic Activity of Peptides with Integrated Feature Learning. J Bioinform Comput Biol 2021, 19, 2150021. [Google Scholar] [CrossRef]
- Wilson, A.C.; Vadakkadath Meethal, S.; Bowen, R.L.; Atwood, C.S. Leuprolide Acetate: A Drug of Diverse Clinical Applications. Expert Opinion on Investigational Drugs 2007, 16, 1851–1863. [Google Scholar] [CrossRef]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. International Journal of Molecular Sciences 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P.S. SATPdb: A Database of Structurally Annotated Therapeutic Peptides. Nucleic Acids Research 2016, 44, D1119–D1126. [Google Scholar] [CrossRef] [PubMed]
- Van Avondt, K.; Nur, E.; Zeerleder, S. Mechanisms of Haemolysis-Induced Kidney Injury. Nat Rev Nephrol 2019, 15, 671–692. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Singh, S.; Joshi, A.; Anand, P.; Tuknait, A.; Mathur, D.; Varshney, G.C.; Raghava, G.P.S. Hemolytik: A Database of Experimentally Determined Hemolytic and Non-Hemolytic Peptides. Nucleic Acids Research 2014, 42, D444–D449. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Research 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Beltran, J.A.; Tellez Ibarra, R.; Guillen-Ramirez, H.A.; Brizuela, C.A. Graph-Based Data Integration from Bioactive Peptide Databases of Pharmaceutical Interest: Toward an Organized Collection Enabling Visual Network Analysis. Bioinformatics 2019, 35, 4739–4747. [Google Scholar] [CrossRef] [PubMed]
- Vespignani, A. Twenty Years of Network Science. Nature 2018, 558, 528–529. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective Dynamics of ‘Small-World’ Networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O.; Chialvo, D.R.; Kaiser, M.; Hilgetag, C.C. Organization, Development and Function of Complex Brain Networks. Trends in Cognitive Sciences 2004, 8, 418–425. [Google Scholar] [CrossRef]
- Roy, S.; Cherevko, A.; Chakraborty, S.; Ghosh, N.; Ghosh, P. Leveraging Network Science for Social Distancing to Curb Pandemic Spread. IEEE Access 2021, 9, 26196–26207. [Google Scholar] [CrossRef]
- Roy, S.; Biswas, P.; Ghosh, P. Effectiveness of Network Interdiction Strategies to Limit Contagion During a Pandemic. IEEE Access 2021, 9, 95862–95871. [Google Scholar] [CrossRef]
- Romero, M.; Marrero-Ponce, Y.; Rodríguez, H.; Agüero-Chapin, G.; Antunes, A.; Aguilera-Mendoza, L.; Martinez-Rios, F. A Novel Network Science and Similarity-Searching-Based Approach for Discovering Potential Tumor-Homing Peptides from Antimicrobials. Antibiotics 2022, 11, 401. [Google Scholar] [CrossRef]
- Ayala-Ruano, S.; Marrero-Ponce, Y.; Aguilera-Mendoza, L.; Pérez, N.; Agüero-Chapin, G.; Antunes, A.; Aguilar, A.C. Network Science and Group Fusion Similarity-Based Searching to Explore the Chemical Space of Antiparasitic Peptides. ACS Omega 2022, 7, 46012–46036. [Google Scholar] [CrossRef] [PubMed]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging Computational Approaches for Antimicrobial Peptide Discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; García-Jacas, C.R.; Chavez, E.; Beltran, J.A.; Guillen-Ramirez, H.A.; Brizuela, C.A. Automatic Construction of Molecular Similarity Networks for Visual Graph Mining in Chemical Space of Bioactive Peptides: An Unsupervised Learning Approach. Sci Rep 2020, 10, 18074. [Google Scholar] [CrossRef]
- Chavez, E.; Dobrev, S.; Kranakis, E.; Opatrny, J.; Stacho, L.; Tejeda, H.; Urrutia, J. Half-Space Proximal: A New Local Test for Extracting a Bounded Dilation Spanner of a Unit Disk Graph. In Proceedings of the Principles of Distributed Systems; Anderson, J.H., Prencipe, G., Wattenhofer, R., Eds.; Springer: Berlin, Heidelberg, 2006; pp. 235–245. [Google Scholar]
- Aggarwal, C.C.; Hinneburg, A.; Keim, D.A. On the Surprising Behavior of Distance Metrics in High Dimensional Space. In Proceedings of the Database Theory — ICDT 2001; Van den Bussche, J., Vianu, V., Eds.; Springer: Berlin, Heidelberg, 2001; pp. 420–434. [Google Scholar]
- Marrero-Ponce, Y.; García-Jacas, C.R.; Barigye, S.J.; Valdés-Martiní, J.R.; Rivera-Borroto, O.M.; Pino-Urias, R.W.; Cubillán, N.; Alvarado, Y.J.; Le-Thi-Thu, H. Optimum Search Strategies or Novel 3D Molecular Descriptors: Is There a Stalemate? Current Bioinformatics 2015, 10, 533–564. [Google Scholar] [CrossRef]
- Miranda-Quintana, R.A.; Bajusz, D.; Rácz, A.; Héberger, K. Differential Consistency Analysis: Which Similarity Measures Can Be Applied in Drug Discovery? Molecular Informatics 2021, 40, 2060017. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLOS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between Hemolytic Activity, Cytotoxicity and Systemic in Vivo Toxicity of Synthetic Antimicrobial Peptides. Sci Rep 2020, 10, 13206. [Google Scholar] [CrossRef]
- Inkscape. Inkscape Project 2023.
- Diestel, R. Graph Theory; Graduate Texts in Mathematics; Fifth Edition.; Springer: Berlin, Heidelberg, 2017; Volume 173, ISBN 978-3-662-53621-6. [Google Scholar]
- Smith, T.F.; Waterman, M.S. Identification of Common Molecular Subsequences. Journal of Molecular Biology 1981, 147, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Henikoff, J.G. Amino Acid Substitution Matrices from Protein Blocks. PNAS 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Brandes, U. A Faster Algorithm for Betweenness Centrality. The Journal of Mathematical Sociology 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. PLOS ONE 2014, 9, e98679. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast Unfolding of Communities in Large Networks. J. Stat. Mech. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Graph Drawing by Force-directed Placement - Fruchterman - 1991 - Software: Practice and Experience - Wiley Online Library Available online:. Available online: https://onlinelibrary.wiley.com/doi/10.1002/spe.4380211102 (accessed on 13 February 2023).
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proceedings of the International AAAI Conference on Web and Social Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J Mol Biol 1970, 48, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Reina, D.G.; Toral, S.L.; Johnson, P.; Barrero, F. Improving Discovery Phase of Reactive Ad Hoc Routing Protocols Using Jaccard Distance. J Supercomput 2014, 67, 131–152. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and Versatile Sequence Motif Discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523–D531. [CrossRef]
- Fan, L.; Sun, J.; Zhou, M.; Zhou, J.; Lao, X.; Zheng, H.; Xu, H. DRAMP: A Comprehensive Data Repository of Antimicrobial Peptides. Sci Rep 2016, 6, 24482. [Google Scholar] [CrossRef]
- Wang, C.K.L.; Kaas, Q.; Chiche, L.; Craik, D.J. CyBase: A Database of Cyclic Protein Sequences and Structures, with Applications in Protein Discovery and Engineering. Nucleic Acids Research 2008, 36, D206–D210. [Google Scholar] [CrossRef]
- Katsara, M.; Tselios, T.; Deraos, S.; Deraos, G.; Matsoukas, M.-T.; Lazoura, E.; Matsoukas, J.; Apostolopoulos, V. Round and Round We Go: Cyclic Peptides in Disease. Current Medicinal Chemistry 2006, 13, 2221–2232. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Yin, S.; Jang, R.; Wang, J.; Xue, Z.; Xu, T. NeuroPep: A Comprehensive Resource of Neuropeptides. Database 2015, 2015, bav038. [Google Scholar] [CrossRef]
- Seebah, S.; Suresh, A.; Zhuo, S.; Choong, Y.H.; Chua, H.; Chuon, D.; Beuerman, R.; Verma, C. Defensins Knowledgebase: A Manually Curated Database and Information Source Focused on the Defensins Family of Antimicrobial Peptides. Nucleic Acids Research 2007, 35, D265–D268. [Google Scholar] [CrossRef]
- de Jong, A.; van Heel, A.J.; Kok, J.; Kuipers, O.P. BAGEL2: Mining for Bacteriocins in Genomic Data. Nucleic Acids Research 2010, 38, W647–W651. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the Binding, Insertion and Destabilization of Phospholipid Bilayer Membranes by Alpha-Helical Antimicrobial and Cell Non-Selective Membrane-Lytic Peptides. Biochim Biophys Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Why and How Are Peptide-Lipid Interactions Utilized for Self Defence? Biochem Soc Trans 2001, 29, 598–601. [Google Scholar] [CrossRef]
- Saviello, M.R.; Malfi, S.; Campiglia, P.; Cavalli, A.; Grieco, P.; Novellino, E.; Carotenuto, A. New Insight into the Mechanism of Action of the Temporin Antimicrobial Peptides. Biochemistry 2010, 49, 1477–1485. [Google Scholar] [CrossRef]
- Kato, Y.; Aizawa, T.; Hoshino, H.; Kawano, K.; Nitta, K.; Zhang, H. Abf-1 and Abf-2, ASABF-Type Antimicrobial Peptide Genes in Caenorhabditis Elegans. Biochem J 2002, 361, 221–230. [Google Scholar] [CrossRef]
- Conlon, J.M. The Therapeutic Potential of Antimicrobial Peptides from Frog Skin. Reviews and Research in Medical Microbiology 2004, 15, 17. [Google Scholar] [CrossRef]
- Conlon, J.M.; Sonnevend, A.; Patel, M.; Al-Dhaheri, K.; Nielsen, P.F.; Kolodziejek, J.; Nowotny, N.; Iwamuro, S.; Pál, T. A Family of Brevinin-2 Peptides with Potent Activity against Pseudomonas Aeruginosa from the Skin of the Hokkaido Frog, Rana Pirica. Regul Pept 2004, 118, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, Z.; Hu, Y.; Yu, H.; Ran, R.; Xia, J.; Wang, D.; Yang, S.; Yang, X.; Liu, J. Molecular Cloning and Characterization of Antimicrobial Peptides from Skin of the Broad-Folded Frog, Hylarana Latouchii. Biochimie 2012, 94, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to Manage Pseudomonas Aeruginosa Infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Use R!; 1st ed.; Springer New York, NY, 2009; ISBN 978-0-387-98141-3.
- Zahoránszky-Kőhalmi, G.; Bologa, C.G.; Oprea, T.I. Impact of Similarity Threshold on the Topology of Molecular Similarity Networks and Clustering Outcomes. Journal of Cheminformatics 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Coscia, M. The Atlas for the Aspiring Network Scientist 2021.
- Hulsen, T. DeepVenn -- a Web Application for the Creation of Area-Proportional Venn Diagrams Using the Deep Learning Framework Tensorflow. Js 2022.
- Shin, S.Y.; Park, E.J.; Yang, S.T.; Jung, H.J.; Eom, S.H.; Song, W.K.; Kim, Y.; Hahm, K.S.; Kim, J.I. Structure-Activity Analysis of SMAP-29, a Sheep Leukocytes-Derived Antimicrobial Peptide. Biochem Biophys Res Commun 2001, 285, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhao, G.; Huang, Y.; Cai, M.; Shan, Y.; Wang, H.; Chen, Y. Specificity and Mechanism of Action of Alpha-Helical Membrane-Active Peptides Interacting with Model and Biological Membranes by Single-Molecule Force Spectroscopy. Sci Rep 2016, 6, 29145. [Google Scholar] [CrossRef] [PubMed]
- Dykes, G.A.; Aimoto, S.; Hastings, J.W. Modification of a Synthetic Antimicrobial Peptide (ESF1) for Improved Inhibitory Activity. Biochem Biophys Res Commun 1998, 248, 268–272. [Google Scholar] [CrossRef]
- Feder, R.; Dagan, A.; Mor, A. Structure-Activity Relationship Study of Antimicrobial Dermaseptin S4 Showing the Consequences of Peptide Oligomerization on Selective Cytotoxicity. J Biol Chem 2000, 275, 4230–4238. [Google Scholar] [CrossRef]
- Nikawa, H.; Fukushima, H.; Makihira, S.; Hamada, T.; Samaranayake, L.P. Fungicidal Effect of Three New Synthetic Cationic Peptides against Candida Albicans. Oral Dis 2004, 10, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Langham, A.A.; Khandelia, H.; Schuster, B.; Waring, A.J.; Lehrer, R.I.; Kaznessis, Y.N. Correlation between Simulated Physicochemical Properties and Hemolycity of Protegrin-like Antimicrobial Peptides: Predicting Experimental Toxicity. Peptides 2008, 29, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
| 1 | Do not confuse cuttof value t with cutoff value s. The former was used to construct networks whereas the latter was used for scaffold extraction. |

| Measure | Formulaa | Rangeb | Average | Range |
|---|---|---|---|---|
| Angular Separation/[1-Cosine (Ochiai)] (AS) | where, | |||
| Bhattacharyya (Bh) | ||||
| Chebyshev/Lagrange (Ch) | ||||
| Euclidean (Eu) | ||||
| Soergel (So) |
| No | Metrics | Cutoff (t) | Edges | Modularity | Density | ACC | Clusters (no D0) | Singletons (D0) | Diameter |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Angular Separation | 0.00 | 26471 | 0.490 | 0.020 | 0.183 | 8 | 0 | 5 |
| 2 | 0.90 | 25065 | 0.499 | 0.018 | 0.205 | 15 | 17 | 7 | |
| 3 | Bhattacharyya | 0.00 | 10555 | 0.456 | 0.008 | 0.025 | 8 | 0 | 6 |
| 4 | 0.75 | 9364 | 0.472 | 0.007 | 0.028 | 17 | 23 | 8 | |
| 5 | Chebyshev | 0.00 | 22431 | 0.313 | 0.017 | 0.021 | 7 | 0 | 3 |
| 6 | 0.65 | 16809 | 0.376 | 0.012 | 0.032 | 12 | 29 | 7 | |
| 7 | Euclidean | 0.00 | 10498 | 0.466 | 0.008 | 0.026 | 9 | 0 | 7 |
| 8 | 0.70 | 8482 | 0.494 | 0.006 | 0.030 | 20 | 21 | 10 | |
| 9 | Soergel | 0.00 | 12077 | 0.441 | 0.009 | 0.024 | 8 | 0 | 6 |
| 10 | 0.70 | 9521 | 0.496 | 0.007 | 0.028 | 17 | 27 | 11 |
| No | Metric | Motif | Cluster | Cluster size | Matches in positive seqs. | Matches in control seqs. | Sitesa (%) | p-value | E-value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Angular Separation (AS) | WKSFLK | 0 | 223 | 81 | 3 | 36.3 | 0.023 | 0.120 |
| 2 | SLCEZ | 1 | 140 | 61 | 0 | 43.6 | 0.005 | 0.048 | |
| 3 | GLPV | 3 | 61 | 45 | 0 | 73.8 | 0.017 | 0.140 | |
| 4 | CGETCV | 3 | 61 | 56 | 0 | 91.8 | 0.017 | 0.140 | |
| 5 | WKKI | 5 | 255 | 88 | 10 | 34.5 | 0.025 | 0.120 | |
| 6 | Chebyshev (Ch) | GILDTJ | 1 | 304 | 72 | 0 | 23.7 | 0.010 | 0.073 |
| 7 | MFTLK | 2 | 246 | 57 | 0 | 23.2 | 0.034 | 0.310 | |
| 8 | CSW | 4 | 59 | 44 | 0 | 74.6 | 0.024 | 0.190 | |
| 9 | VCGETC | 4 | 59 | 49 | 0 | 83.1 | 0.004 | 0.032 | |
| 10 | LCYCRR | 6 | 150 | 41 | 0 | 27.3 | 0.031 | 0.150 | |
| 11 | Euclidean (Eu) | LKGAGK | 0 | 339 | 74 | 0 | 21.8 | 0.004 | 0.047 |
| 12 | VCTRN | 1 | 101 | 76 | 0 | 75.2 | 0.004 | 0.038 | |
| 13 | WKSFJK | 5 | 220 | 45 | 0 | 20.5 | 0.015 | 0.092 | |
| 14 | LHTAKK | 5 | 220 | 54 | 0 | 24.5 | 0.002 | 0.011 | |
| 15 | CYCRR | 7 | 189 | 43 | 0 | 22.8 | 0.032 | 0.160 |
| HemoPI-1 | StarPepDB | Big-Hemo | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Motif | ERa | E-value | Rankb | ERa | E-value | Rankb | ERa | E-value | Rankb |
| 1 | ALKAIS | 3.66 | 1.92E-09 | 36 | 40.10 | 3.53E-21 | 35 | 3.32 | 8.48E-12 | 9 |
| 2 | WKSFJK | 19.20 | 2.80E-40 | 2 | 5.06 | 3.55E-158 | 1 | 4.94 | 6.01E-22 | 3 |
| 3 | AKKAL | 16.10 | 7.19E-29 | 11 | 3.33 | 6.66E-101 | 4 | 1.55 | 6.30E-04 | 24 |
| 4 | LKKL | 12.60 | 1.44E-31 | 4 | 3.62 | 2.65E-130 | 2 | 1.68 | 1.37E-08 | 12 |
| 5 | ISWIK | 7.86 | 5.69E-19 | 19 | 6.19 | 3.45E-59 | 15 | 2.51 | 4.45E-05 | 22 |
| 6 | LHTA[KL] | 3.94 | 1.90E-13 | 25 | 8.25 | 4.73E-27 | 29 | 3.74 | 1.76E-11 | 10 |
| 7 | FLHSAK | 7.04 | 1.82E-11 | 29 | 5.69 | 2.10E-45 | 21 | 1.95 | 1.14E-03 | 26 |
| 8 | LWKT | 7.25 | 4.60E-31 | 6 | 2.35 | 5.59E-55 | 18 | 3.50 | 2.59E-10 | 11 |
| 9 | FLGTI | 6.94 | 1.41E-14 | 22 | 2.15 | 1.18E-21 | 33 | 3.88 | 5.70E-24 | 2 |
| 10 | KKILG | 6.71 | 1.61E-26 | 13 | 3.29 | 3.56E-77 | 11 | 1.85 | 1.36E-07 | 13 |
| 11 | KITK | 6.99 | 5.48E-26 | 15 | 2.48 | 1.22E-57 | 16 | 2.05 | 1.68E-01 | 36 |
| 12 | LGKL | 5.47 | 1.14E-29 | 7 | 2.17 | 5.13E-87 | 8 | 3.34 | 5.48E-12 | 8 |
| 13 | KWK | 4.84 | 2.02E-31 | 5 | 3.97 | 1.22E-55 | 17 | 1.98 | 1.79E-07 | 15 |
| 14 | KGAGK | 5.13 | 2.35E-27 | 12 | 2.66 | 2.25E-43 | 22 | 2.81 | 2.43E-14 | 4 |
| 15 | SWP | 4.56 | 5.42E-26 | 14 | 3.76 | 7.67E-35 | 26 | 1.98 | 5.44E-03 | 28 |
| 16 | LLKK | 4.31 | 1.88E-34 | 3 | 3.82 | 1.35E-126 | 3 | 1.60 | 1.18E-01 | 35 |
| 17 | [VI]LDTJ | 3.02 | 4.39E-10 | 33 | 2.15 | 1.05E-40 | 23 | 4.27 | 1.58E-24 | 1 |
| 18 | JGKL | 4.07 | 1.38E-29 | 8 | 2.32 | 8.01E-90 | 7 | 1.71 | 2.12E-07 | 17 |
| 19 | VLK | 3.00 | 8.34E-17 | 20 | 2.06 | 9.64E-64 | 14 | 1.88 | 1.52E-07 | 14 |
| 20 | PKIF | 2.89 | 1.05E-14 | 21 | 2.19 | 3.22E-46 | 20 | 1.47 | 4.79E-03 | 27 |
| No. | Sequence | Length | No. Motifs | Consensus Motifs | Hemolytic Activity | Ref |
|---|---|---|---|---|---|---|
| 1 | RGLRRLGRKIAHGVKKYGPTVKRIKRKA | 28 | 0 | Not active at 100 µM | [63] | |
| 2 | KWKSFLKTFKSAAKTVLHTALKAISS | 28 | 4 | WKSFJK, LHTA[KL], KWK, ALKAIS | 50% hemolysis at 16 µM | [64] |
| 3 | MASRAARLAARLARLALRAL | 20 | 0 | 1% hemolysis at 92.95 µM | [65] | |
| 4 | ALWMTLLKKVLKAAAKAALN | 20 | 4 | LLKK, VLK, AAAK, ALW | 50% hemolysis at 5 ± 1 µM | [66] |
| 5 | KRLFRRWQWRMKKY | 14 | 0 | Not active up to 100 µM | [67] | |
| 6 | WCYCRRRFCVCVGR | 14 | 3 | RFC, CYCR, CRR | > 50% hemolytic at 44.3 µM | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





