1. Introduction
Over 1 million people were diagnosed with Gastric Cancer (GC) in 2020, making it the fifth most commonly diagnosed type of cancer [
1]. GC ranks fourth in mortality rates due to diagnosis at a late disease stage, where the median overall survival (OS) ranges from 4.6–13.1 months [
1,
2]. GC diagnosis is usually invasive, resorting to endoscopy with biopsy, and staging with endoscopic ultrasound and CT scans [
3]. Therefore, there is an increasing interest in less invasive molecular biology-based approaches both for GC diagnosis and prognosis [
4].
Detection of GC biomarkers in body fluids, such as blood or urine is already in test, with some studies showing promising results [
5]. For instance, detection of miR-21 in serum or in human peripheral blood mononuclear cells (PBMCs) is claimed to predict GC in 90% of the studied cases [
6]. In another study, a panel of six upregulated miRNAs was found both in serum and GC Extracellular Vesicles (GC-EVs), showing that GC-EV cargo can be a useful diagnosis marker [
7]. EVs are essential intercellular communication mediators, carrying mostly non-coding RNAs, mRNAs, DNAs, lipids, and proteins [
8]. This cargo is highly affected by cellular architecture and culture conditions, with EVs deriving from 3D cultured cancer cells recapitulating better EVs recovered from real cancer patients [
9,
10]. Studies using EVs are usually focused on miRNAs, however some studies report that up to 50% of their cargo is actually transfer RNAs (tRNAs) and tRNA-derived fragments (tRFs) [
9,
11,
12,
13,
14].
The interest in the role of tRNAs in cancer has risen in the past few years and, since then, tRNAs deregulation has been associated with tumor initiation, tumor growth, metastasis and bad prognosis [
15,
16,
17,
18] (reviewed in [
19]). There are two main species of non-coding RNAs derived from tRNAs: tRFs and tRNA halves (tiRNAs) [
20]. While tiRNAs result from cleavage at the anticodon loop under stress [
21,
22], tRFs are 14–30 nucleotides fragments that derive from mature or pre-tRNAs and can control gene expression similarly to miRNAs [
23]. tRFs can be further subdived in 3’-tRFs or 5’-tRFs if they include the terminal 5’ or 3’ portion of the original tRNA or i-tRFs if they originate from an internal tRNA sequence. Several tRFs have been implicated in many diseases, from cancer to neurodegenerative and metabolic disorders [
24,
25,
26]. Moreover, tRFs sorted into EVs are being evaluated for their value as non-invasive cancer diagnostic biomarkers [
27,
28]. A recent study has identified eight differentially expressed tRFs between GC tissues and adjacent tissues [
29], with tRF-24-V29K9UV3IU regulating the Wnt pathway to inhibit cell proliferation, migration and invasion, while promoting cell apoptosis. Additionally, it was observed a downregulation of tRF-Glu-TTC-027 in GC patients, which act as a tumor suppressor, inhibiting the MAPK pathway and negatively influencing tumor progression [
30]. On the other hand, tRF-3017A was shown to silence tumor-suppressor NELL2, promoting cell migration and invasion
in vitro, and it is associated with higher lymph node metastasis in GC patients [
31]. Moreover, expression levels of hsa_tsr016141 in serum could distinguish GC patients from healthy donors and gastritis patients with good sensitivity and specificity and holding potential to be used for dynamic monitoring of GC patients [
32].
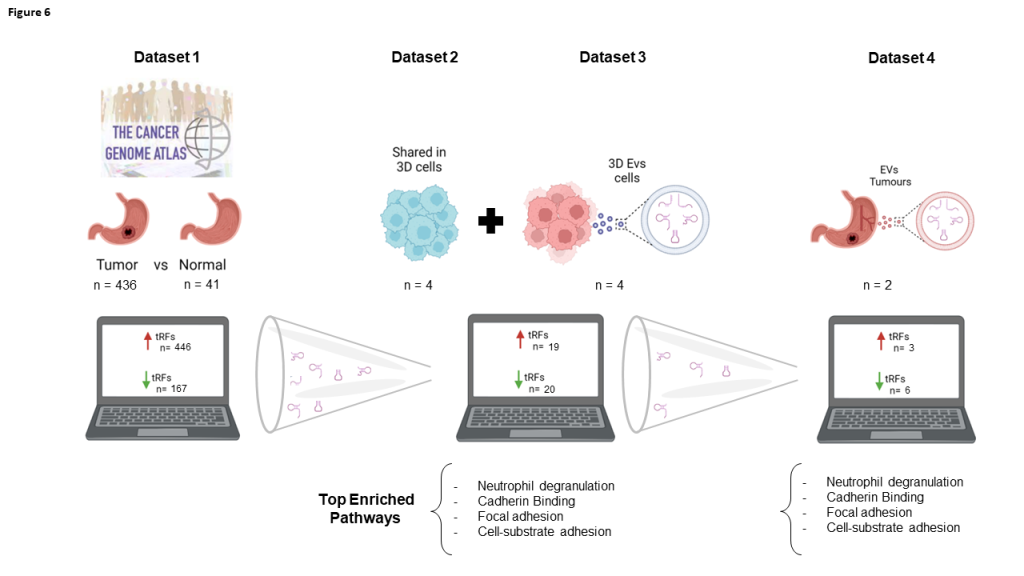
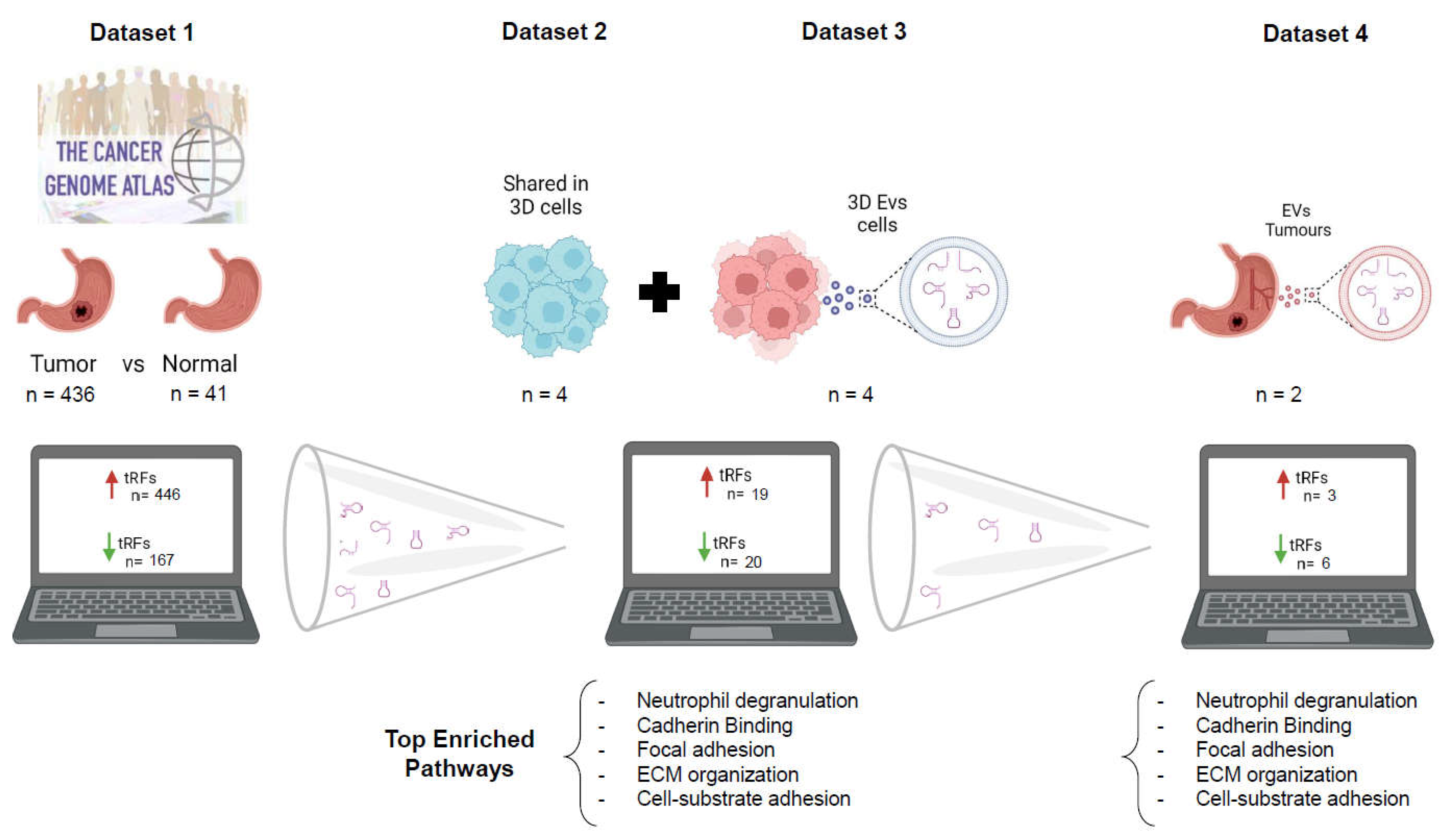
Due to the mandatory deposition of sequencing data in online repositories, there are numerous accessible datasets nowadays. These datasets can be explored to retrieve different information from what is depicted in corresponding initial publications. This is an interesting savings strategy, avoiding unnecessary experiments and costs. An example might be the use of small-RNAseq datasets used to evaluate miRNA deregulation, but containing information about other small RNAs such as tRFs, due to their similar RNA length. In this manuscript, we used miRNA datasets from the TCGA database to evaluate tRF modulation in GC patients. The deregulated tRFs were validated with another small-RNAseq dataset from our own laboratory, previously used to assess miRNA deregulation. We found tRFs with positive and negative correlations in TCGA tumors and normal counterparts, as well as in EVs produced by 3D-cultured GC-cell lines. Nine tRFs recurrently identified in the previous analysis were also detected in GC patient plasma-derived EVs. Besides their biomarker potential, these differentially expressed (DE)-tRFs were predicted to modulate tumor microenvironment and affect cell-cell adhesion.
2. Results
2.1. tRFs are highly expressed in GC tumors
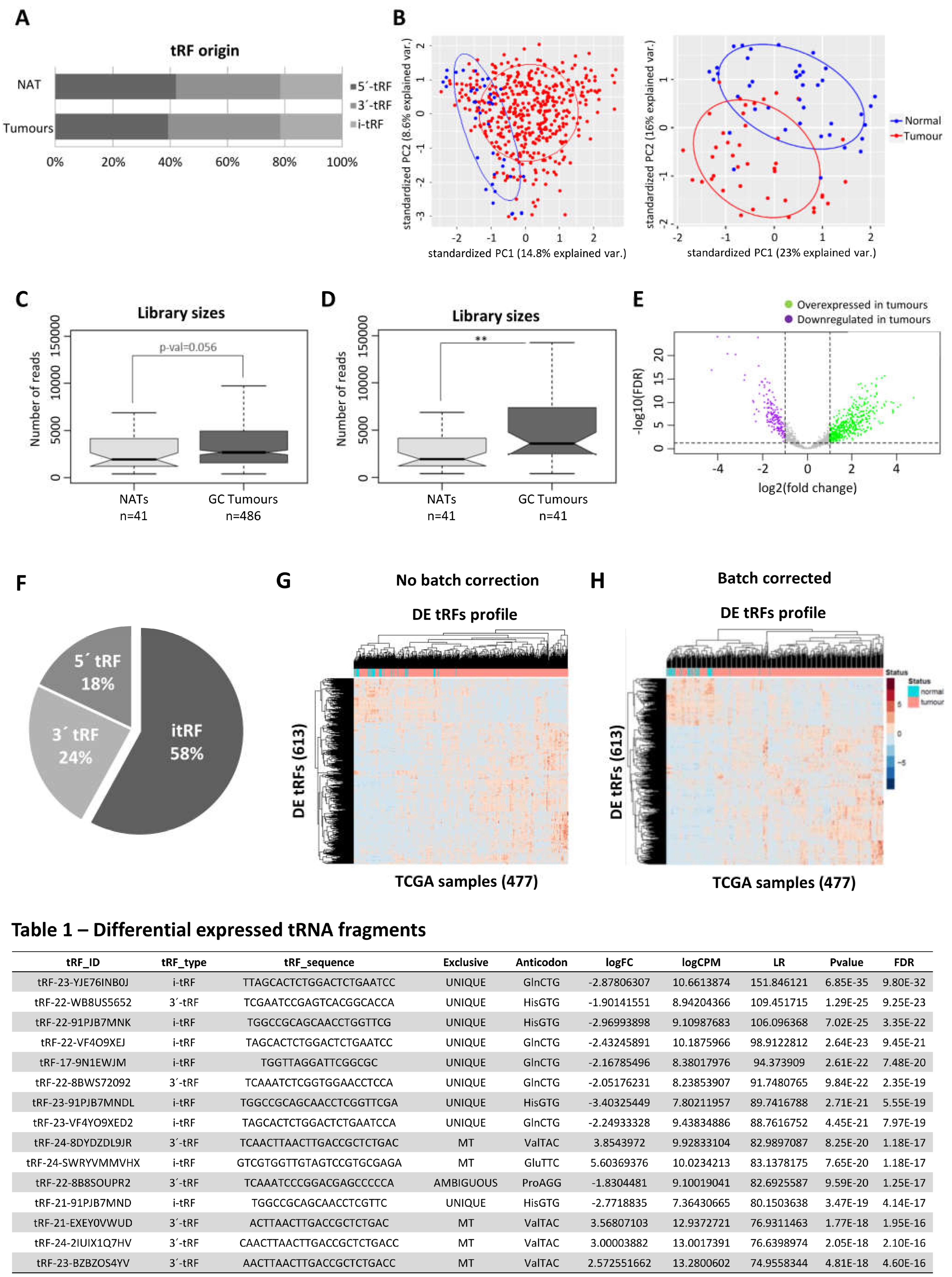
To have a broad perception of the expression of tRFs in GC, we analyzed the miRNAseq data of Stomach Adenocarcinomas (n = 436) and normal adjacent tissues (n = 41) deposited in the TCGA database. Our first finding was that the TCGA miRNAseq dataset is devoid of tRNA halves and enriched in 5’ and 3’ tRFs. The biotype distribution of tRFs is very similar between tumors and normal adjacent tissues (NATs) (
Figure 1A). PCA analysis, comparing tRF expression in all tumors against all NATs, hinted to a separation of tumors from NATs (
Figure 1B, left panel). This separation became more evident when tumors and matching NAT samples were analyzed (
Figure 1B, right panel). To increase confidence in resulting data, a tRF expression threshold of 5 Read per Million (RPMs) in at least 20% of samples from a given condition (tumor or NAT) was used. After application of this filter, we analyzed the total tRF reads and observed that tumors have an overall increase in tRF expression, especially if paired samples are considered (
p < 0.01) (
Figure 1C,D).
The differential expression (DE) analysis of tRFs considering |log2FC|>1 and an adjusted p-value < 0.05 unveiled 613 DE-tRFs, from which 446 were upregulated and 167 were downregulated in tumors
vs NATs (
Figure 1E). While 5-’ and 3’-tRFs were the most abundant species among tRFs from tumours and NATs (
Figure 1A), the most abundant biotype among DE-tRFs was i-tRFs (58%), followed by 3’-tRFs (24%) and 5’-tRFs (18%) (
Figure 1F). These 613 DE-tRFs when plotted into a heatmap unveiled a batch effect that has been previously described by other TCGA users [
33] (
Figure 1G). After batch effect correction, gastric tumors and NAT samples clustered quite distinctly based on the 613 tRFs expression (
Figure 1H). The top 15 DE-tRFs encompassed 10 downregulated tRFs (seven i-tRF and three 3’-tRFs) and five upregulated tRFs (one i-tRF and four 3’-tRFs) (Table 1).
2.2. tRF expression in GC cell lines and derived EVs
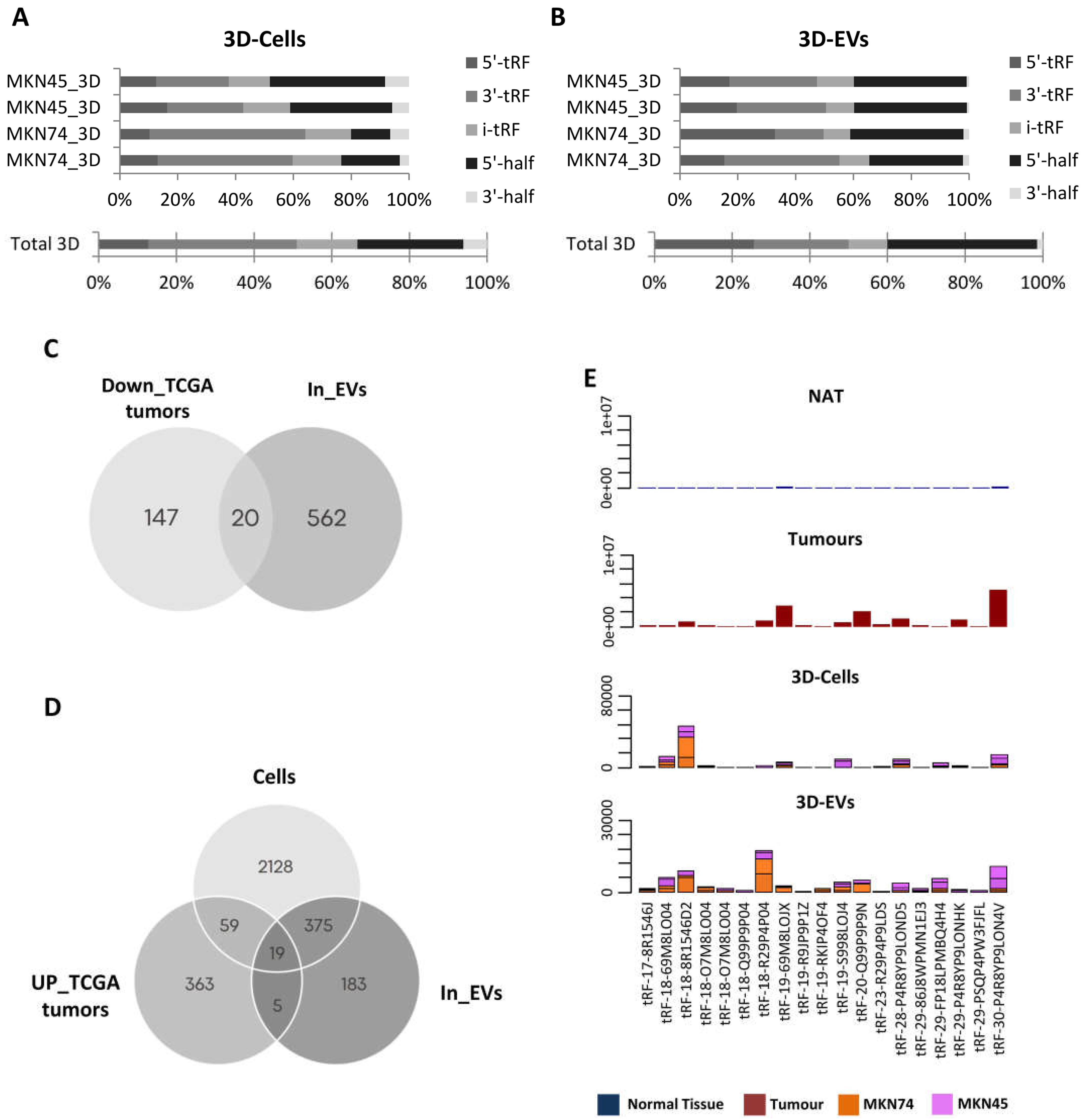
To assess a potential role for tRFs as biomarkers, we next assessed their expression in a small-RNAseq dataset from EVs produced by two different 3D-cultured GC cell lines. We chose to focus our analysis on GC cell lines and EVs grown in 3D cultures, as they resemble more the context of tumors growing in patients and often their expression patterns are closer to the
in vivo context. We detected a distinct landscape of tRF species from what we observed in TCGA, likely due to the use of a different type of sequencing. In 3D-cells, the most abundant tRF species was 3’-tRFs followed by 5-’halves, i-tRFs, 5’-tRFs and 3’-halves (
Figure 2A). In cell line-derived EVs, 5’-halves were the most abundant species, followed by 5’-tRFs, 3’-tRFs, itRFs and 3’-halves (
Figure 2B). These differences may reflect an asymmetric and orchestrated tRF packaging into EVs, in order to modulate the surrounding microenvironment, as it has been reported in T-cells [
34].
As RNA packaging into EVs does not seem to be random, we aimed at identifying tRFs that can be clinically useful to diagnose or stratify GC patients. For that, we used two strategies to find potentially relevant tRFs. First, we searched EVs from 3D-GC cells for the 167 tRFs found downregulated in TCGA tumors compared to NATs, assuming the hypothesis of tRFs active exclusion by tumor cells. In a second approach, we searched, in 3D-GC cells and their respective EVs, for the 446 tRFs found upregulated in TCGA tumors compared to NATs, assuming the hypothesis of active load of tRFs in cancer-derived EVs.
We found 20 tRFs which expression has a negative correlation between GC-EVs and TCGA tumors, and hypothetically packed into EVs to avoid a potential negative impact on the tumor cell fitness (
Figure 2C). Additionally, we found 19 tRFs that were concomitantly upregulated in TCGA tumors and present in 3D-cells and respective EVs (
Figure 2D). These tRFs were barely expressed in normal adjacent tissue from TCGA (NATs) and for most cases their expression levels have similar levels in 3D-GC cells and their derived EVs (
Figure 2E).
2.3. DE-tRFs are predicted to modulate immune response and cell adhesion
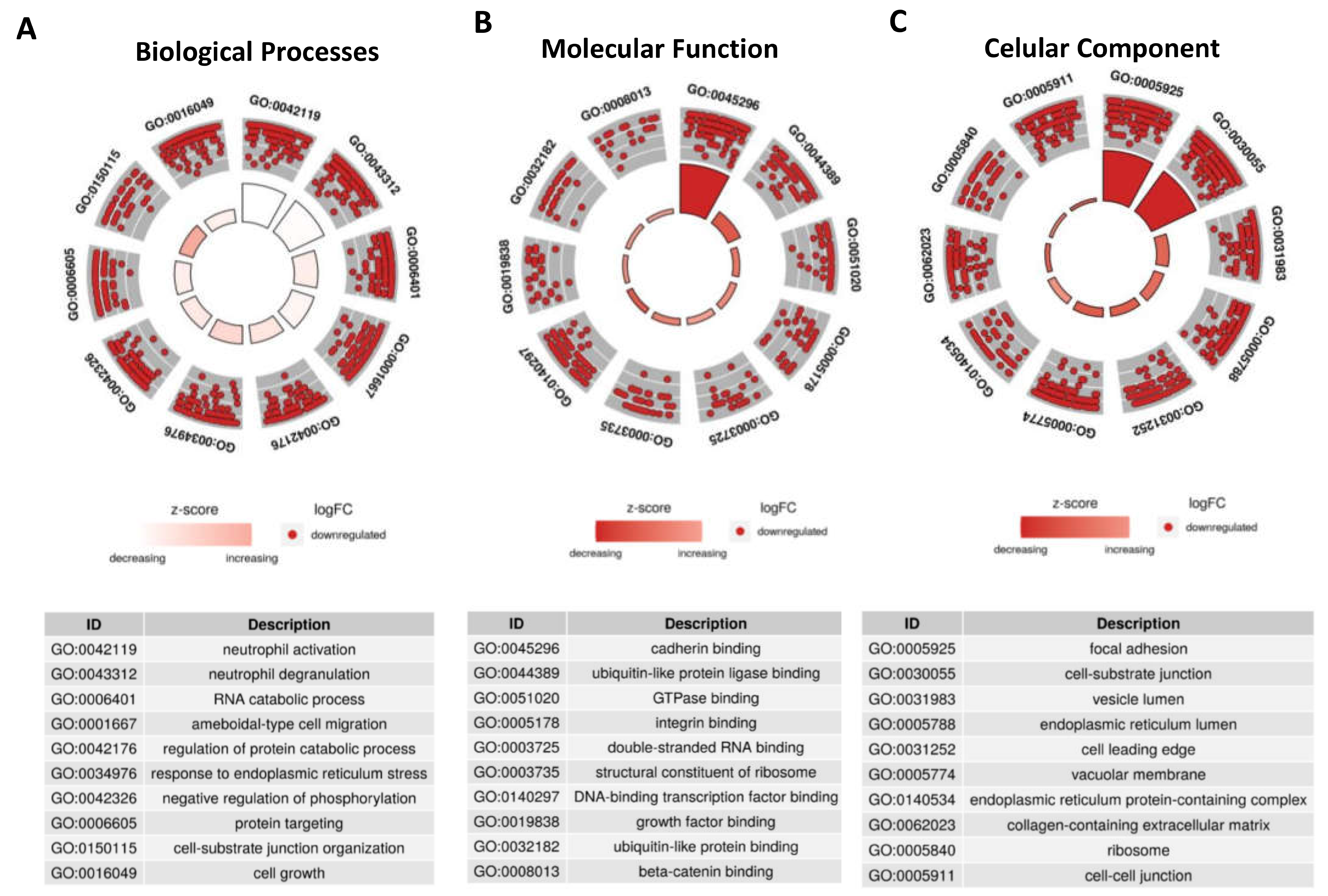
Since these 39 DE-tRFs have all been found within EVs from 3D-GC cells, we proceeded to predict their targets using the tRFTar tool. The predicted targets were then used to perform a GO-term analysis using
ClusterProfiler R package to assess their impact in cellular biology (
Figure 3).
Regarding biological processes, DE-tRFs were found to affect neutrophils and their degranulation (
Figure 3A). Interestingly, these tRFs also seem to affect cell adhesion properties, as they target genes that are involved in ameboidal-type cell migration (
Figure 3A), cadherin, β-catenin and integrin binding (
Figure 3B) among other adhesion related categories, such as focal adhesion and cell-substrate junction (
Figure 3C).
Besides their potential role as biomarkers for GC diagnosis, these DE-tRFs may also affect central pathways involved in gastric carcinogenesis and progression.
2.4. Nine DE-tRFs are also present in patients derived EVs
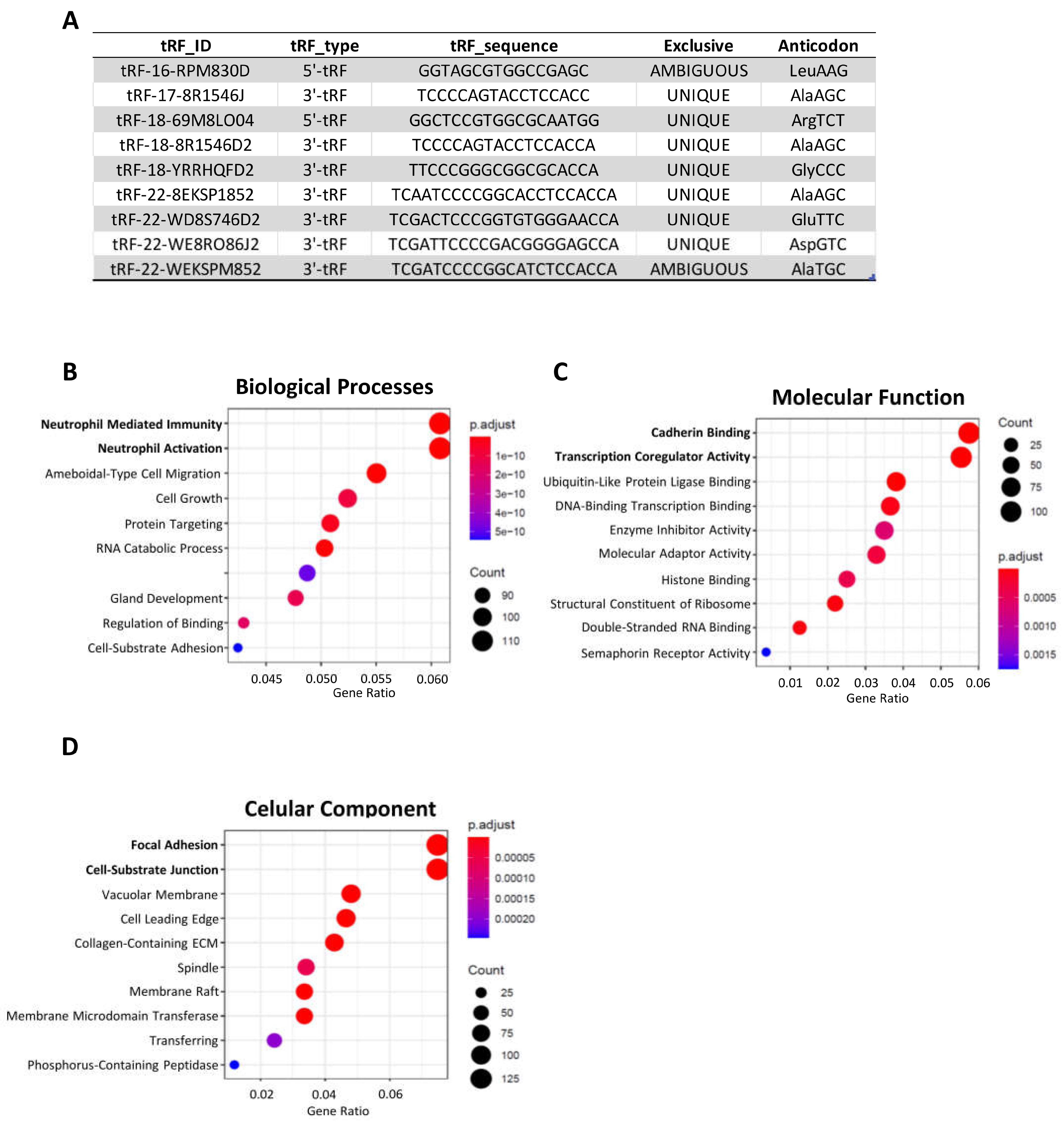
We explored whether any of the 39 tRFs, which were found to be differentially expressed in TCGA tumors vs NAT and present in GC-EVs, could be found in a small-RNAseq dataset from plasma EVs obtained from four GC-patients, prior to their GC surgery. Despite the low number of patients and low input material, we could validate the presence of 9 DE-tRFs in patient’s EVs. Six of these DE-tRFs were downregulated in TCGA tumors vs NAT, but present in EVs from 3D-GC cells; and three DE-tRFs were upregulated in TCGA tumors vs NAT and present both in 3D-GC cells and respective 3D-EVs. The tRF ID, sequence and type are described in
Figure 4A. This validates our strategy to find relevant biomarkers from previously available data. Interestingly, the targets of these 9 tRFs also affect neutrophil activation and degranulation, cadherin binding, focal adhesion and cell-substrate junction, highlighting these pathways as probable targets of EV-mediated crosstalk with the tumor microenvironment (TME) (
Figure 4B-D).
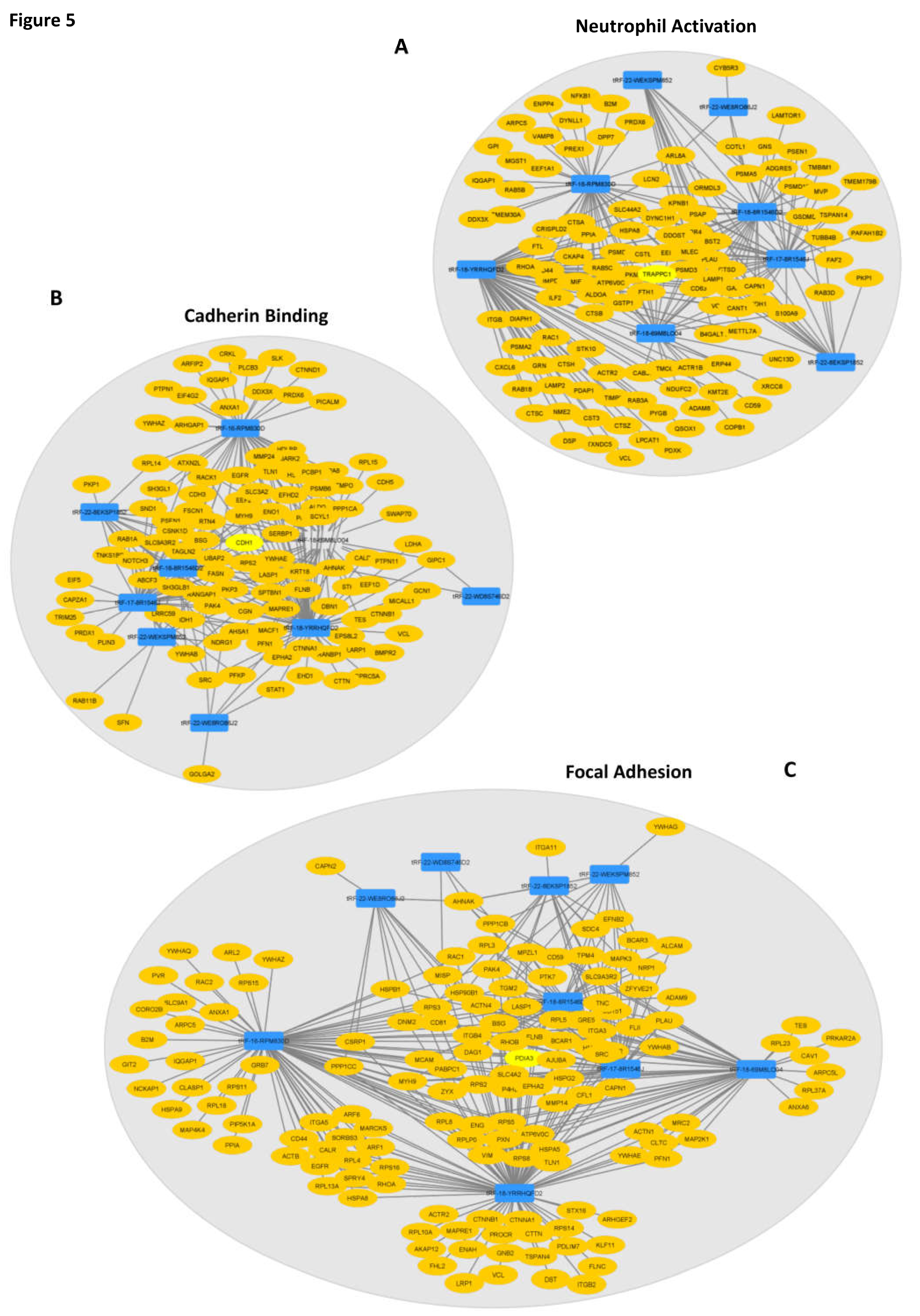
To understand how DE-tRFs and their targets are interacting to affect these biological processes, we generated interaction networks in Cytoscape and observed that almost all 9 DE-tRFs can target genes that affect the top GO-terms in each group (
Figure 5). These interaction networks are very complex, with each DE-tRF being able to target more than one gene, and several genes having seed sequences where distinct DE-tRFs can bind (
Figure 5). The top Biological Process affected is Neutrophil activation (GO:0042119) with 8 DE-tRFs being predicted to affect 110 genes in this process (
Figure 5A). Cadherin Binding (GO:0045296) is the most affected Molecular Function, with all the DE-tRFs affecting 100 genes. Interestingly,
CDH1, the gene coding for E-cadherin is the central node of this network, being predicted to be targeted by 5 out of 9 DE-tRFs (
Figure 5B). It is also remarkable that one of the central node of the “Focal Adhesion” network (GO:0005925) is PDIA3, a molecular chaperone reported to influence antigen presentation in gastric cancer, and thus likely to influence therapeutic response (
Figure 5C). In fact, low levels of this protein often correlate with immune evasion and worst overall survival [
35].
Overall, we identified 9 DE-tRFs across 5 different GC-related datasets, including patient-derived EVs, and with neglectable expression in normal gastric tissue, that may work both as diagnosis biomarkers and as modulators of GC behavior and related microenvironment.
3. Discussion
Our work demonstrated that it is possible to draw new and relevant hypothesis with the sole use of
in silico de novo data analysis. Using data available in public repositories and previously sequenced data in our lab, we could confirm that 9 DE-tRFs with a negative or positive correlation between tumors and GC cell line-derived EVs are also present in patient-derived EVs (
Figure 4A).
We started by using TCGA data to assess DE-tRFs between tumors and normal adjacent tissues. This analysis unveiled that GC tumors express more tRFs than NAT (
Figure 1C,D), in agreement with previous reports [
36]. Interestingly, a report showed that angiogenin, one of the enzymes responsible for cleaving tRNAs into tRFs, is more active in GC tumors than in NAT [
37]. In fact, we observed most DE-tRFs were overexpressed in tumors (
Figure 1E) likely reflecting a greater angiogenin activity. When plotting the DE-tRFs into a heatmap, we noticed a batch effect in our samples related to the center where the samples were sequenced (
Figure 1G), as previously reported [
33]. Therefore, we included cancer status and batch in our model for the differential expression analysis to exclude bias. After bias exclusion, we observed that the 613 DE-tRF set could discriminate GC samples from normal tissue fairly well (
Figure 1H).
As we were set to find tRFs with biomarker value, we validated the 613 DE-tRFs from TCGA with small RNA seq data from two different GC cell lines cultured in 3D and their respective EVs. Patient EVs can be obtained from liquid biopsies, an increasingly popular non-invasive method to monitor patients and people with increased risk to develop disease [
38]. We decided to use 3D-GC cell lines as a proxy, since EVs derived from 3D cell cultures resemble better the content of patient-derived EVs [
9,
39,
40]. We found 20 tRFs that were downregulated in TCGA tumors but present in EVs from 3D-GC cells, and 19 tRFs that were upregulated in TCGA tumors and present both in 3C-GC cells and in corresponding EVs (
Figure 2C,D). RNA packaging into EVs is not completely random, depending on cell type and physiological conditions [
41]. Although the mechanisms for active RNA loading into EVs still needs clarification, EXOmotifs were described in miRNAs that are enriched in neuron-derived EVs [
42]. Nevertheless, it is still controversial which proportion of EV content is selected for and which portion of their content is sampled by the nature of EV biogenesis process [
43]. Interestingly, the tRFs that we found to have a positive or negative expression correlation between GC-EVs and TCGA tumors, seem to have the potential to modulate the tumor microenvironment (
Figure 3). The crosstalk between tumor cells and the stromal cells in the TME mediated by EVs produces important functional changes that favor tumor development and progression [
44,
45,
46]. Indeed, we observed that the targets affected by these tRFs interfere with neutrophil activation, cell-substrate adhesion, focal adhesion, extracellular matrix organization and cadherin binding (
Figure 3), which are relevant in gastric cancer biology [
47,
48,
49,
50,
51]. Even if we consider only the 9 tRFs, which are also present in patient EVs, their targets modulate the exact same pathways (
Figure 4B–D), suggesting a highly selected biological process.
This work repurposing public and previously obtained data in our lab allowed the identification of a set of tRFs conserved across GC datasets that may work as GC diagnosis biomarkers (Fig.6). Remarkably, 9 tRFs alone were representative of the largest tRF set correlating tumors, 3D-GC cells and patient-derived EV data, and shown to target the same gene set responsible for TME modulation. Although this work needs experimental data validation regarding the effect of these vesicles in TME, the fact that this is predicted in four independent datasets, strengthens the validity of this analysis. Future studies following similar workflows and validating results obtained with repurposed data will represent a cost-effective approach to novel research questions.
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
4. Materials and Methods
tRF sequencing data collection and pre-processing
miRNA aligned reads (bam files) released by TCGA were obtained from the GDC data portal (
https://portal.gdc.cancer.gov/), filtering by TCGA-STAD project name. 491 bam files were downloaded and converted to sequencing reads (fastq files) using biobambam2 [
52]. Illumina Small RNA v1.5 adapter sequences were checked and clipped from all reads using cutadapt (version 2.8, with settings of “-a ATCTCGTATGCCGTCTTCTGCTTGT -q 20 -m 16”) [
53]. Low quality bases were trimmed from 3’ end of the reads. After this quality control, all sequence reads shorter than 16 bases were discarded.
On the other hand, 16 fastq files were generated corresponding to the small RNA sequencing reads from gastric cancer cell lines and EVs derived from them (Methods reported in [
9]). Ion Torrent adapter sequences were checked from all reads using cutadapt (version 2.8, with settings of “-b ATCACCGACTGCCCATAGAGAGG -q 20 -m 16 [
53]. Low quality bases were trimmed from 3’ end of the reads. After this quality control, all reads shorter than 16 bases were discarded.
In addition, we used 2 plasma samples of EVs from gastric cancer patients before undergoing surgery.
Blood sample collection from gastric cancer (GC) patients
The 4 patients with Gastric Cancer (GC) enrolled in this study were admitted at Instituto Português de Oncologia Francisco Gentil (Coimbra, Portugal). The study was approved by the hospital’s ethics committee, and written informed consent was obtained from all patients before sample collection.
Peripheral blood samples (5ml) were collected between April 2015 and May 2017 in K2 EDTA plasma preparation tubes (BD Vacutainer® PPT) and processed into plasma within 30 minutes following manufacturer’s instructions. Plasma samples were stored at -80°C until further processing.
EV isolation and characterization from plasma of GC patients
Thawed plasma samples (volume 1.5 – 3ml) were diluted with 0.9% NaCl (pH 7.4) to a final volume of 15ml and filtered through a 0.22µm filter. Filtered supernatants were centrifuged in a SW32 rotor (Beckman Coulter, Fullerton, CA, USA) at 100,000 g, for 14 hours at 4°C to pellet EVs. EV pellets were washed in 0.9% NaCl (pH 7.4), centrifuged at 100,000 g for 2 hours at 4°C and resuspended in a volume of 0.9% NaCl. EVs were stored at 4°C. EV size and concentration were further determined by Nanoparticle Tracking Analysis (NTA) using the NanoSight NS300 instrument (Malvern, Worcestershire, UK) with scientific CMOS sensor. Briefly, EVs were diluted (1:500) in 0.9% NaCl. Three technical measurements were recorded under a controlled fluid flow with a pump speed set to 40 and a camera focus level adjusted between 10 and 16. The three videos were further analyzed using the NTA 3.1 Build 3.1.54 software to calculate the concentration, mode, and mean size of EVs.
Small RNA library preparation and sequencing from human plasma EV-sRNA (GC)
Isolated sRNA was used for small RNA library preparation using the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. Briefly, 3′ and 5′ adapters were attached directionally, and simultaneously, to 3μl of sRNA input (0.94 - 5.23ng). Hybridized and ligated RNA was reversed transcribed using Ion RT primer v2 and SuperScript III Enzyme mix. Each cDNA sample was amplified and barcoded using Platinum PCR SuperMix High Fidelity, Ion Xpress RNA 3′ Barcode Primer, and a unique Ion Xpress RNA-Seq Barcode BC Primer, which allows sample identification and tracking. Size distribution of amplified cDNA was measured using the Agilent 2100 Bioanalyzer with Agilent DNA 1000 Kit (Agilent, Santa Clara, CA, USA). Due to the high amount of adapter dimers, an adapted protocol was implemented with the aim of reducing the quantity of adapter dimers and enriching the library of interest. Library selection and sequencing test run were performed. The first approach consisted of size selection with E-Gel® SizeSelect™ 2% Agarose Gel (Thermo Fisher Scientific), which decreased the proportion of adapter dimers. For less concentrated libraries size selection of the library was performed by using 4% Agarose Gel cut and purification of the band corresponding to ≈110bp sequences was performed with illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, New York, USA). Agilent 2200 TapeStation (S/N 3-PM-1173NA)—HS D1000 Screen Tape (P/N 5067-5584) was used for size distribution control. An equal volume (3µL) of each library was used to prepare the final pool. Pooled libraries were processed on Ion Chef System (S/N CHEF00657) using the Ion 540 Kit-Chef (P/N A27759) and the resulting 540 chip (P/N A27766) was sequenced on Ion S5 XL System (S/N 245717100156). Fastq files were generated using the Torrent Suit plugin FileExporter v5.0.
Pre-processing of human plasma EV-sRNA sequencing data (GC)
For the aim of this paper, we obtained 12 fastq files. FastQC (version 0.11.5) [
54] was used to perform quality control checks on sequencing data. Remaining Ion Torrent sRNA adapter sequences were checked and clipped from all reads using cutadapt (version 2.8, with settings of “-b ATCACCGACTGCCCATAGAGAGGAAAGCGG --error-rate 0.2 --times 1 -m 15 -q 20”)[
53]. Low quality bases (q
< 20) were trimmed from 3’ end of the reads. Finally, all sequence reads shorter than 15 bases were removed.
tRF expression estimation
The processed reads were aligned to human reference genome by MINTmap, specifically designed for tRF analysis [
55]. MINTmap considers in the alignment the complex representation of the tRNA-derived sequences in the genome (i.e. regions shared between tRNA isodecoders, sequences that resemble mitochondrially-encoded tRNAs in the nuclear genome, etc.) and, consequently, maps reads more exactly (no mismatches, insertions or deletions allowed), deterministically (reads matches are not based on probabilistic approaches), exhaustively (enumerates all possible alignments in genome) and specifically (labels a tRF sequence in
exclusive or
ambiguous depending, respectively, on whether the tRF only belongs to a tRNA gene/s or it is also present in other positions in the genome (i.e., partial tRNA sequences or non-tRNAs), thus informing about the possibility that the tRF may be a false positive [
56]. Only
exclusive tRFs where considered in this study. Alignment of tRFs with MINTmap included a classification of tRFs according to five structural types (5′-tRFs, i-tRFs, 3′-tRFs, 5′-tRNA halves (5′-tRHs) and 3′-tRNA halves (3′-tRHs))[
55].
tRF expression levels of 446 gastric primary tumour samples and 41 NAT were calculated by MINTmap [
55]. 10 replicates from 446 primary tumours were removed, leaving those ones with higher coverage, reaching a total of 436 tumours and 41 normal samples. Expression levels were tested for significant differential expression between the libraries of tumour
vs. normal samples using the edgeR package (version 3.32.0)[
57] in R (version 4.0.3)[
58]. Data were normalized using the TMM method (weighted trimmed mean of M-values)[
59]. Expression data from selected genes was used for heatmap construction depicting log
2(cpm) values scaled to z-score with supervised hierarchical clustering for dendrogram construction applying calculated Euclidean distances by R package “gplots”[
60]. Venn diagrams were performed by jvenn tool [
61].
A generalized linear model was fit after estimating the common, trended and tagwise dispersion. We noticed 2 batches in our samples related to the center where the samples were sequenced, as reported previously by [
62]; therefore, we included cancer status and batch as fixed effects in the model. The likelihood ratio test was used to evaluate differential expression between allergic and non-allergic donors. Differences in expression between conditions with |log
2(fold-change)| > 1 and adjusted
p-value
< 0.05 were considered to be significant.
P-values were adjusted for false discovery rate by the Benjamini–Hochberg method [
63].
tRF expression levels of 4 gastric cancer cell and 4 EVs samples were calculated by MINTmap [
55]. For cells, we considered those tRFs with >10 reads globally, and for EVs, those with >1 read were considered.
tRF expression levels of 12 gastric cancer EVs samples from blood were calculated by MINTmap [
55]. We considered those tRFs with > 1 read in samples collected before surgery from 2 patients, which had an acceptable sequencing quality.
Target prediction
We used tRFTar [
64] database to predict the targets of the tRF of interest.
ClusterProfiler R package was used for assessment of significantly-enriched GO terms and pathways (
padj < 5.00E-02). The hierarchical clustering was performed using the
heatmap.2 function from the
gplots R package, with the complete method as argument for the
hclust function that compute the hierarchical clustering. We used the euclidean method to compute the distance and the complete method to perform the hierarchical clustering. tRF-target gene interaction (TGI) networks were created by R package igraph (version 1.2.6) [
65]. Cytoscape (version 3.8.0) [
66] was used to depict the networks.
Author Contributions
“Conceptualization, M.S. and C.O..; methodology, J.J.M, M.F., S.M., S.R, M.R., J.C., N.S. and N.B; software, J.J.M, M.F.; validation, J.J.M., M.S., M.F. and C.O.; formal analysis, J.J.M., M.S., M.F. S.M.; investigation, J.J.M, M.S., M.F., S.M., S.R, M.R., J.C., N.S. and N.B ; resources, N.B. and C.O.; data curation, J.J.M., M.S., M.F. and C.O ; writing—J.J.M., M.S. and C.O.; writing—review and editing, J.J.M., M.S. and C.O; visualization, J.J.M., M.S., M.F. S.M. ; supervision, C.O.; project administration, C.O.; funding acquisition, M.S. and C.O. All authors have read and agreed to the published version of the manuscript.”
Funding
Research and salary to J.J.M. and M.S. was supported by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No. 722148 and the Fundação para a Ciência e a Tecnologia (FCT) through the project “MIStranslation: a novel trigger of genome instability In Cancer”- PTDC/MED-ONC/28834/2017, respectively. The Bioinformatics work has been developed in the frame of the project GenomePT (National Laboratory for Genome Sequencing and Analysis - POCI-01-0145-FEDER-022184), supported by COMPETE 2020 - Operational Programme for Competitiveness and Internationalisation (POCI2020), Lisboa Portugal Regional Operational Programme (Lisboa2020), Algarve Portugal Regional Operational Programme (CRESC Algarve2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF), and by Fundação para a Ciência e a Tecnologia (FCT). The human tumour work has been developed in the frame of the project “Tumour secreted factors in EMT/non-EMT cells for invasion and metastization” - NORTE-01-0145-FEDER-000029, supported by North Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF); and through projects references PTDC/BTMTEC/30164/2017 [3DChrome] and PTDC/BTM-TEC/6706/2020 [LEGOH], funded by Fundação para a Ciência e Tecnologia, Ministério da Ciência, Tecnologia e Inovação.
Institutional Review Board Statement
“The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Instituto Português de Oncologia Francisco Gentil (Coimbra, Portugal).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data will be made available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H. et al. (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 71, 209–249. [CrossRef]
- Li, S. and Zang, L. The Effectiveness of Gastrectomy With Chemoradiotherapy Among Stage IV Gastric Adenocarcinoma: A Population-Based Analysis. , Frontiers in Oncology, 10. (2020) , 630. [CrossRef]
- Van Cutsem, E. et al. (2016) Gastric cancer. Lancet 388, 2654–2664. [CrossRef]
- Pasechnikov, V. et al. (2014) Gastric cancer: prevention, screening and early diagnosis. World J. Gastroenterol. 20, 13842–13862. [CrossRef]
- Necula, L. et al. (2019) Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 25, 2029–2044. [CrossRef]
- Wu, J. et al. (2015) Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis. Markers 2015, 435656. [CrossRef]
- Huang, Z. et al. (2017) Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol. Biomarkers & Prev. 26, 188 LP – 196. [CrossRef]
- Maas, S.L.N. et al. (2017) Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 27, 172–188. [CrossRef]
- Rocha, S. et al. (2019) 3D Cellular Architecture Affects MicroRNA and Protein Cargo of Extracellular Vesicles. Adv. Sci. 6, 1800948. [CrossRef]
- Millan, C. et al. (2021) Extracellular Vesicles from 3D Engineered Microtissues Harbor Disease-Related Cargo Absent in EVs from 2D Cultures. Adv. Healthc. Mater. n/a, 2002067. [CrossRef]
- Chiou, N.-T. et al. (2018) Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Rep. 25, 3356-3370.e4. [CrossRef]
- Shurtleff, M.J. et al. (2017) Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. 114, E8987 LP-E8995. [CrossRef]
- Baglio, S.R. et al. (2015) Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 6, 127. [CrossRef]
- Li, M. et al. (2014) Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130502.
- Santos, M. et al. (2018) Codon misreading tRNAs promote tumor growth in mice. , RNA Biology, 1–14. [CrossRef]
- Goodarzi, H. et al. (2016) Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell. [CrossRef]
- Krishnan, P. et al. (2016) Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci. Rep. 6,. [CrossRef]
- Santos, M. et al. Upregulation of tRNA-Ser-AGA-2-1 Promotes Malignant Behavior in Normal Bronchial Cells . , Frontiers in Molecular Biosciences , 9. (2022). [CrossRef]
- Santos, M. et al. (2019) tRNA Deregulation and Its Consequences in Cancer. Trends Mol. Med. 25, 853–865. [CrossRef]
- Zhang, Y. et al. (2020) Mechanisms of tRNA-derived fragments and tRNA halves in cancer treatment resistance. Biomark. Res. 8, 52. [CrossRef]
- Thompson, D.M. et al. (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA DOI: 10.1261/rna.1232808. [CrossRef]
- Yamasaki, S. et al. (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42. [CrossRef]
- Kumar, P. et al. (2016) Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 41, 679–689. [CrossRef]
- Hogg, M.C. et al. (2019) Elevation of plasma tRNA fragments precedes seizures in human epilepsy. J. Clin. Invest. 129, 2946–2951. [CrossRef]
- Jacovetti, C. et al. (2021) Emerging Classes of Small Non-Coding RNAs With Potential Implications in Diabetes and Associated Metabolic Disorders. Front. Endocrinol. (Lausanne). 12, 503. [CrossRef]
- Yu, M. et al. (2020) tRNA-derived RNA fragments in cancer: current status and future perspectives. J. Hematol. Oncol. 13, 121. [CrossRef]
- Zhu, L. et al. (2019) Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18, 74. [CrossRef]
- Koi, Y. et al. (2020) Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 111, 2104–2115. [CrossRef]
- Dong, X. et al. (2020) Comprehensively Identifying the Key tRNA-Derived Fragments and Investigating Their Function in Gastric Cancer Processes. Onco. Targets. Ther. 13, 10931–10943.
- Xu, W. et al. tRNA-Derived Fragment tRF-Glu-TTC-027 Regulates the Progression of Gastric Carcinoma via MAPK Signaling Pathway. , Frontiers in Oncology, 11. (2021) , 3310. [CrossRef]
- Tong, L. et al. The tRNA-Derived Fragment-3017A Promotes Metastasis by Inhibiting NELL2 in Human Gastric Cancer. , Frontiers in Oncology, 10. (2021) , 3468. [CrossRef]
- Gu, X. et al. Serum hsa_tsr016141 as a Kind of tRNA-Derived Fragments Is a Novel Biomarker in Gastric Cancer. , Frontiers in Oncology, 11. (2021) , 1639. [CrossRef]
- Rasnic, R. et al. (2019) Substantial batch effects in TCGA exome sequences undermine pan-cancer analysis of germline variants. BMC Cancer 19, 783. [CrossRef]
- Torres, A.G. and Martí, E. Toward an Understanding of Extracellular tRNA Biology. , Frontiers in Molecular Biosciences, 8. (2021). [CrossRef]
- Shimoda, T. et al. (2019) Expression of protein disulfide isomerase A3 and its clinicopathological association in gastric cancer. Oncol Rep 41, 2265–2272. [CrossRef]
- Sun, X. et al. (2020) Global identification and characterization of tRNA-derived RNA fragment landscapes across human cancers. NAR Cancer 2, zcaa031. [CrossRef]
- Chen Y, Zhang S, Chen YP, L.J. (2006) Increased expression of angiogenin in gastric carcinoma in correlation with tumor angiogenesis and proliferation. World J Gastroenterol 12, 5135–5139. [CrossRef]
- Lengyel, C.G. et al. (2021) The Emerging Role of Liquid Biopsy in Gastric Cancer. J. Clin. Med. 10,. [CrossRef]
- Kusuma, G.D. et al. Effect of 2D and 3D Culture Microenvironments on Mesenchymal Stem Cell-Derived Extracellular Vesicles Potencies . , Frontiers in Cell and Developmental Biology , 10. (2022). [CrossRef]
- Thippabhotla, S. et al. (2019) 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 9, 13012. [CrossRef]
- Veziroglu, E.M. and Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents . , Frontiers in Genetics , 11. (2020). [CrossRef]
- Luo, X. et al. (2021) Differential RNA packaging into small extracellular vesicles by neurons and astrocytes. Cell Commun. Signal. 19, 75. [CrossRef]
- Pegtel, D.M. and Gould, S.J. (2019) Exosomes. Annu. Rev. Biochem. 88, 487–514. [CrossRef]
- Haga, H. et al. (2015) Tumour cell–derived extracellular vesicles interact with mesenchymal stem cells to modulate the microenvironment and enhance cholangiocarcinoma growth. J. Extracell. Vesicles 4, 24900. [CrossRef]
- Zhang, D.X. et al. (2021) Landscape of extracellular vesicles in the tumour microenvironment: Interactions with stromal cells and with non-cell components, and impacts on metabolic reprogramming, horizontal transfer of neoplastic traits, and the emergence of therapeutic resistance. Semin. Cancer Biol. 74, 24–44. [CrossRef]
- Yamada, N. et al. (2016) Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-β1-mediated suppression. Oncotarget 7, 27033–27043. [CrossRef]
- Fu, H. et al. (2016) Persisting and Increasing Neutrophil Infiltration Associates with Gastric Carcinogenesis and E-cadherin Downregulation. Sci. Rep. 6, 29762. [CrossRef]
- Mao, D. et al. Cross-Talk of Focal Adhesion-Related Gene Defines Prognosis and the Immune Microenvironment in Gastric Cancer . , Frontiers in Cell and Developmental Biology , 9. (2021). [CrossRef]
- Lee, Y. et al. (2020) Common and Unique Transcription Signatures of YAP and TAZ in Gastric Cancer Cells. Cancers (Basel). 12,. [CrossRef]
- Andreuzzi, E. et al. (2020) Role of Extracellular Matrix in Gastrointestinal Cancer-Associated Angiogenesis. Int. J. Mol. Sci. 21, 3686. [CrossRef]
- Oliveira, C. et al. (2015) Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 16, e60–e70. [CrossRef]
- Tischler, G. and Leonard, S. (2014) biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol. Med. 9, 13. [CrossRef]
- Martin, M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal; Vol 17, No 1 Next Gener. Seq. Data Anal. - 10.14806/ej.17.1.200 at < https://journal.embnet.org/index.php/embnetjournal/article/view/200>. [CrossRef]
- Andrews S, Krueger F, Seconds-Pichon A, Biggins F, Wingett S. FastQC. Babraham Bioinformatics [Internet]. Vol. 1, B.I. 2015. p. 1. A. from: https://www. bioinformatics. babraham. ac. uk/projects/fastqc/%0Ahttp://www. bioinformatics. bbsrc. ac. uk/projects/fastqc. A quality control tool for high throughput sequence data. Babraham Bioinforma. [Internet]. Vol. 1, Babraham Institute. 2015. p. 1. Available from https//www.bioinformatics.babraham.ac.uk/projects/fastqc/%0Ahttp//www.bioinformatics.bbsrc.ac.uk/projects/fastqc/.
- Loher, P. et al. (2017) MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 7, 41184. [CrossRef]
- Meseguer, S. et al. (2019) The MELAS mutation m.3243A>G alters the expression of mitochondrial tRNA fragments. Biochim. Biophys. acta. Mol. cell Res. 1866, 1433–1449. [CrossRef]
- Robinson, M.D. et al. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [CrossRef]
- R Core Team (2020) R: A language and environment for statistical computing. , R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2020).
- Robinson, M.D. and Oshlack, A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [CrossRef]
- Warnes, G.R. et al. (2019) gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1.1. http://CRAN.R-project.org/package = gplots.
- Bardou, P. et al. (2014) Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics 15,. [CrossRef]
- Rasnic, R. et al. (2019) Substantial batch effects in TCGA exome sequences undermine pan-cancer analysis of germline variants. BMC Cancer 19,. [CrossRef]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 57, 289–300. [CrossRef]
- Zhou, Y. et al. (2021) tRFTar: Prediction of tRF-target gene interactions via systemic re-analysis of Argonaute CLIP-seq datasets. Methods 187, 57–67. [CrossRef]
- Csardi, G. and Nepusz, T. (2006) The igraph software package for complex network research. InterJournal Complex Syst. [CrossRef]
- Shannon, P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).