Submitted:
18 September 2024
Posted:
20 September 2024
You are already at the latest version
Abstract
Keywords:
- Wnt related synergies in https://www.preprints.org/manuscript/202409.0453/v1
- NFκB related synergies in https://www.preprints.org/manuscript/202409.0696/v1
- TNF related synergies in https://www.preprints.org/manuscript/202409.0471/v1
- DNA repair related synergies in https://www.preprints.org/manuscript/202409.0885/v1
- ABC transporter related synergies in https://www.preprints.org/manuscript/202409.0908/v1
- Interleukin related synergies in https://www.preprints.org/manuscript/202409.1353/v1
- BCL related synergies in https://www.preprints.org/manuscript/202409.0855/v1
- ANTXR2 related synergies in https://www.preprints.org/manuscript/202409.0817/v1
1. Introduction
2. Materials and Methods
2.1. Combinatorial Search Problem and a Possible Solution
2.2. Wnt Signaling and Secretion
2.3. PORCN-WNT Inhibitors
3. Results & Discussion
3.1. WNT Related Synergies
3.1.1. WNT10B-ASCL2
3.1.2. ABC Transporters - WNT Cross Family Analysis
3.1.3. IL - WNT Cross Family Analysis
3.1.4. UBE2 - WNT Cross Family Analysis
| Unexplored combinatorial hypotheses | |
| UBE2 w.r.t WNT | |
| WNT-4 | UBE2-A |
| WNT-4/7 | UBE2-B |
| WNT-4/7B | UBE2-F |
| WNT-2B | UBE2-H |
| WNT-7B/9B | UBE2-J1 |
| WNT-2B/4/7B | UBE2-Z |
| WNT w.r.t UBE2 | |
| WNT-7B | UBE2-A |
| WNT-7B/9A | UBE2-B |
| WNT-7B/9A | UBE2-F |
| WNT-4 | UBE2-H |
| WNT-7B/9A | UBE2-J1 |
| WNT-7B | UBE2-Z |
3.1.5. EXOSC - WNT10B Cross Family Analysis
3.1.6. CASP - WNT Cross Family Analysis
| Ranking CASP family VS WNT family | |||||||
| Ranking of CASP4 w.r.t WNTs family | Ranking of CASP5 w.r.t WNTs family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - WNT2B | 2265 | 320 | 1517 | CASP5 - WNT2B | 975 | 2171 | 2366 |
| CASP4 - WNT4 | 1050 | 1081 | 558 | CASP5 - WNT4 | 1788 | 1356 | 569 |
| CASP4 - WNT7B | 622 | 9 | 632 | CASP5 - WNT7B | 716 | 978 | 606 |
| CASP4 - WNT9A | 446 | 1413 | 583 | CASP5 - WNT9A | 383 | 808 | 147 |
| Ranking of CASP7 w.r.t WNTs family | Ranking of CASP9 w.r.t WNTs family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - WNT2B | 1152 | 305 | 248 | CASP9 - WNT2B | 1345 | 1501 | 1328 |
| CASP7 - WNT4 | 936 | 1260 | 1787 | CASP9 - WNT4 | 1344 | 2472 | 2200 |
| CASP7 - WNT7B | 901 | 1403 | 1303 | CASP9 - WNT7B | 2196 | 1935 | 1713 |
| CASP7 - WNT9A | 1330 | 1527 | 2436 | CASP9 - WNT9A | 1863 | 428 | 2002 |
| Ranking of CASP10 w.r.t WNTs family | Ranking of CASP16 w.r.t WNTs family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - WNT2B | 1607 | 1108 | 739 | CASP16 - WNT2B | 240 | 621 | 193 |
| CASP10 - WNT4 | 432 | 689 | 132 | CASP16 - WNT4 | 2070 | 1783 | 711 |
| CASP10 - WNT7B | 1906 | 1171 | 1165 | CASP16 - WNT7B | 411 | 713 | 103 |
| CASP10 - WNT9A | 1611 | 2152 | 1451 | CASP16 - WNT9A | 14 | 2512 | 181 |
| Ranking of WNTs family w.r.t CASP4 | Ranking of WNTs family w.r.t CASP5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - WNT2B | 609 | 1317 | 2372 | CASP5 - WNT2B | 1849 | 1192 | 1590 |
| CASP4 - WNT4 | 105 | 711 | 1062 | CASP5 - WNT4 | 890 | 682 | 714 |
| CASP4 - WNT7B | 1093 | 2479 | 1739 | CASP5 - WNT7B | 2112 | 1919 | 2440 |
| CASP4 - WNT9A | 456 | 2278 | 1939 | CASP5 - WNT9A | 315 | 1880 | 1437 |
| Ranking of WNTs family w.r.t CASP7 | Ranking of WNTs family w.r.t CASP9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - WNT2B | 2505 | 1891 | 1120 | CASP9 - WNT2B | 282 | 639 | 1414 |
| CASP7 - WNT4 | 108 | 2456 | 2455 | CASP9 - WNT4 | 572 | 1788 | 378 |
| CASP7 - WNT7B | 1380 | 1559 | 1681 | CASP9 - WNT7B | 979 | 901 | 676 |
| CASP7 - WNT9A | 2183 | 1941 | 1632 | CASP9 - WNT9A | 2378 | 2396 | 2058 |
| Ranking of WNTs family w.r.t CASP10 | Ranking of WNTs family w.r.t CASP16 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - WNT2B | 625 | 1471 | 81 | CASP16 - WNT2B | 2197 | 2489 | 1775 |
| CASP10 - WNT4 | 1830 | 2229 | 1847 | CASP16 - WNT4 | 1382 | 954 | 1017 |
| CASP10 - WNT7B | 1965 | 937 | 147 | CASP16 - WNT7B | 2508 | 1820 | 1867 |
| CASP10 - WNT9A | 2185 | 1977 | 1350 | CASP16 - WNT9A | 1943 | 1154 | 1839 |
| Unexplored combinatorial hypotheses | |
| CASP w.r.t WNT | |
| CASP5 | WNT2B |
| CASP9 | WNT4/WNT7B/WNT9A |
| CASP16 | WNT4 |
| WNT w.r.t CASP | |
| WNT7B/WNT9A | CASP4 |
| WNT7B | CASP5 |
| WNT2B/WNT4/WNT9A | CASP7 |
| WNT9A | CASP9 |
| WNT4/WNT9A | CASP10 |
| WNT2B/WNT7B/WNT9A | CASP16 |
3.1.7. TP53 - WNT Cross Family Analysis
3.1.8. BCL - WNT Cross Family Analysis
| Ranking BCL family VS WNT | |||||||
| Ranking of BCL2L1 w.r.t WNT family | Ranking of WNT family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL2L1 | 1884 | 101 | 966 | WNT2B - BCL2L1 | 1854 | 1666 | 1699 |
| WNT4 - BCL2L1 | 98 | 1162 | 719 | WNT4 - BCL2L1 | 21 | 107 | 16 |
| WNT7B - BCL2L1 | 1434 | 1891 | 620 | WNT7B - BCL2L1 | 2213 | 2266 | 1511 |
| WNT9A - BCL2L1 | 1088 | 1020 | 1318 | WNT9A - BCL2L1 | 1019 | 1462 | 1345 |
| Ranking of BCL2L2 w.r.t WNT family | Ranking of WNT family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL2L2 | 625 | 2204 | 1677 | WNT2B - BCL2L2 | 1574 | 2206 | 955 |
| WNT4 - BCL2L2 | 2364 | 2042 | 1610 | WNT4 - BCL2L2 | 160 | 590 | 316 |
| WNT7B - BCL2L2 | 843 | 1877 | 2456 | WNT7B - BCL2L2 | 2456 | 2512 | 2286 |
| WNT9A - BCL2L2 | 1877 | 538 | 2447 | WNT9A - BCL2L2 | 1868 | 2333 | 990 |
| Ranking of BCL2L13 w.r.t WNT family | Ranking of WNT family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL2L13 | 201 | 1862 | 1353 | WNT2B - BCL2L13 | 1256 | 1254 | 1490 |
| WNT4 - BCL2L13 | 1938 | 2425 | 1900 | WNT4 - BCL2L13 | 922 | 270 | 187 |
| WNT7B - BCL2L13 | 1105 | 1993 | 2284 | WNT7B - BCL2L13 | 1610 | 1319 | 954 |
| WNT9A - BCL2L13 | 1855 | 268 | 2387 | WNT9A - BCL2L13 | 1858 | 2422 | 1934 |
| Ranking of BCL3 w.r.t WNT family | Ranking of WNT family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL3 | 950 | 1328 | 2482 | WNT2B - BCL3 | 1846 | 2056 | 1896 |
| WNT4 - BCL3 | 1228 | 1562 | 1353 | WNT4 - BCL3 | 591 | 359 | 1932 |
| WNT7B - BCL3 | 591 | 615 | 553 | WNT7B - BCL3 | 1687 | 2160 | 1428 |
| WNT9A - BCL3 | 1037 | 1410 | 1102 | WNT9A - BCL3 | 1539 | 1424 | 398 |
| Ranking of BCL6 w.r.t WNT family | Ranking of WNT family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL6 | 455 | 2426 | 1529 | WNT2B - BCL6 | 52 | 107 | 170 |
| WNT4 - BCL6 | 256 | 486 | 787 | WNT4 - BCL6 | 2483 | 2488 | 1273 |
| WNT7B - BCL6 | 2147 | 1466 | 1105 | WNT7B - BCL6 | 975 | 1893 | 2284 |
| WNT9A - BCL6 | 1547 | 734 | 2012 | WNT9A - BCL6 | 1558 | 2098 | 1905 |
| Ranking of BCL9L w.r.t WNT family | Ranking of WNT family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL9L | 2348 | 804 | 1558 | WNT2B - BCL9L | 1918 | 700 | 1882 |
| WNT4 - BCL9L | 1446 | 657 | 309 | WNT4 - BCL9L | 303 | 2498 | 2509 |
| WNT7B - BCL9L | 1539 | 253 | 1279 | WNT7B - BCL9L | 1608 | 811 | 2168 |
| WNT9A - BCL9L | 1923 | 677 | 688 | WNT9A - BCL9L | 941 | 1843 | 1238 |
| Ranking of BCL10 w.r.t WNT family | Ranking of WNT family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - BCL10 | 2321 | 69 | 2023 | WNT2B - BCL10 | 1951 | 1101 | 1599 |
| WNT4 - BCL10 | 285 | 1170 | 465 | WNT4 - BCL10 | 2032 | 34 | 406 |
| WNT7B - BCL10 | 1847 | 606 | 1252 | WNT7B - BCL10 | 1297 | 74 | 2009 |
| WNT9A - BCL10 | 217 | 798 | 1649 | WNT9A - BCL10 | 1771 | 335 | 861 |
| Unexplored combinatorial hypotheses | |
| BCL w.r.t WNT family | |
| WNT-4/7B/9A | BCL2L2 |
| WNT-4/7B | BCL2L13 |
| WNT-2B | BCL10 |
| WNT family w.r.t BCL | |
| WNT-7B | BCL2L1 |
| WNT-7B/9A | BCL2L2 |
| WNT-9A | BCL2L13 |
| WNT-2B | BCL3 |
| WNT-4/7B/9A | BCL6 |
| WNT-2B/4 | BCL9L |
3.2. NF-κB Related Synergies
3.2.1. CASP - RIPK Cross Family Analysis
| Ranking CASP family w.r.t RIPK family | |||||||
| Ranking of CASP4 w.r.t RIPK family | Ranking of CASP5 family w.r.t RIPK | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - RIPK1 | 1154 | 1259 | 147 | CASP5 - RIPK1 | 490 | 152 | 1818 |
| CASP4 - RIPK2 | 559 | 2147 | 434 | CASP5 - RIPK2 | 1274 | 2485 | 608 |
| CASP4 - RIPK3 | 111 | 131 | 41 | CASP5 - RIPK3 | 523 | 1047 | 317 |
| CASP4 - RIPK4 | 187 | 1048 | 1039 | CASP5 - RIPK4 | 1176 | 2361 | 1292 |
| Ranking of CASP7 w.r.t RIPK family | Ranking of CASP9 family w.r.t RIPK | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - RIPK1 | 2445 | 1289 | 1253 | CASP9 - RIPK1 | 1726 | 1304 | 1480 |
| CASP7 - RIPK2 | 1584 | 406 | 155 | CASP9 - RIPK2 | 2079 | 291 | 1647 |
| CASP7 - RIPK3 | 1406 | 1057 | 2091 | CASP9 - RIPK3 | 2133 | 2030 | 2295 |
| CASP7 - RIPK4 | 1739 | 231 | 2332 | CASP9 - RIPK4 | 2037 | 1627 | 363 |
| Ranking of CASP10 w.r.t RIPK family | Ranking of CASP16 family w.r.t RIPK | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - RIPK1 | 758 | 846 | 1405 | CASP16 - RIPK1 | 73 | 1046 | 1887 |
| CASP10 - RIPK2 | 1535 | 2312 | 884 | CASP16 - RIPK2 | 20 | 932 | 1189 |
| CASP10 - RIPK3 | 1530 | 250 | 2181 | CASP16 - RIPK3 | 30 | 359 | 717 |
| CASP10 - RIPK4 | 954 | 415 | 1547 | CASP16 - RIPK4 | 493 | 2507 | 519 |
| Ranking RIPK family w.r.t CASP family | |||||||
| Ranking of RIPK family w.r.t CASP4 | Ranking of RIPK family w.r.t CASP5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - RIPK1 | 2363 | 1374 | 1805 | CASP5 - RIPK1 | 7 | 82 | 131 |
| CASP4 - RIPK2 | 1713 | 2349 | 1261 | CASP5 - RIPK2 | 1577 | 1776 | 2247 |
| CASP4 - RIPK3 | 1397 | 768 | 1008 | CASP5 - RIPK3 | 574 | 14 | 30 |
| CASP4 - RIPK4 | 2215 | 1334 | 1425 | CASP5 - RIPK4 | 2448 | 1178 | 810 |
| Ranking of RIPK family w.r.t CASP7 | Ranking of RIPK family w.r.t CASP9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - RIPK1 | 1341 | 2005 | 1131 | CASP9 - RIPK1 | 820 | 140 | 611 |
| CASP7 - RIPK2 | 1287 | 727 | 1143 | CASP9 - RIPK2 | 2000 | 2476 | 2138 |
| CASP7 - RIPK3 | 579 | 595 | 775 | CASP9 - RIPK3 | 1550 | 430 | 97 |
| CASP7 - RIPK4 | 852 | 1586 | 595 | CASP9 - RIPK4 | 1565 | 862 | 209 |
| Ranking of RIPK family w.r.t CASP10 | Ranking of RIPK family w.r.t CASP16 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - RIPK1 | 2438 | 1915 | 1039 | CASP16 - RIPK1 | 924 | 686 | 587 |
| CASP10 - RIPK2 | 1526 | 1800 | 1228 | CASP16 - RIPK2 | 1613 | 2006 | 2046 |
| CASP10 - RIPK3 | 419 | 1481 | 2001 | CASP16 - RIPK3 | 827 | 494 | 328 |
| CASP10 - RIPK4 | 1303 | 947 | 785 | CASP16 - RIPK4 | 2273 | 2023 | 1698 |
| Unexplored combinatorial hypotheses | |
| CASP w.r.t RIKP family | |
| CASP9 | RIPK3 |
| RIPK w.r.t CASP family | |
| RIPK1 | CASP4/CASP10 |
| RIPK2 | CASP5/CASP9/CASP16 |
| RIPK4 | CASP16 |
3.2.2. MUC - RIPK Cross Family Analysis
| Ranking MUC family w.r.t RIPK family | |||||||
| Ranking of MUC1 w.r.t RIPK family | Ranking of MUCA3 w.r.t MUC3A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - RIPK1 | 2027 | 2249 | 218 | MUC3A - RIPK1 | 945 | 186 | 1508 |
| MUC1 - RIPK2 | 248 | 1802 | 389 | MUC3A - RIPK2 | 840 | 2390 | 1653 |
| MUC1 - RIPK3 | 342 | 410 | 342 | MUC3A - RIPK3 | 2208 | 2017 | 689 |
| MUC1 - RIPK4 | 176 | 162 | 853 | MUC3A - RIPK4 | 714 | 1494 | 797 |
| Ranking of MUC4 w.r.t RIPK family | Ranking of MUC12 w.r.t RIPK family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC4 - RIPK1 | 358 | 2384 | 690 | MUC12 - RIPK1 | 317 | 2437 | 167 |
| MUC4 - RIPK2 | 371 | 500 | 408 | MUC12 - RIPK2 | 286 | 2178 | 76 |
| MUC4 - RIPK3 | 809 | 371 | 1096 | MUC12 - RIPK3 | 747 | 366 | 136 |
| MUC4 - RIPK4 | 652 | 1863 | 1248 | MUC12 - RIPK4 | 176 | 2249 | 2130 |
| Ranking of MUC13 w.r.t RIPK family | Ranking of MUC17 w.r.t RIPK family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC13 - RIPK1 | 379 | 2241 | 227 | MUC17 - RIPK1 | 858 | 932 | 1503 |
| MUC13 - RIPK2 | 824 | 2483 | 227 | MUC17 - RIPK2 | 248 | 934 | 37 |
| MUC13 - RIPK3 | 1687 | 19 | 24 | MUC17 - RIPK3 | 342 | 64 | 329 |
| MUC13 - RIPK4 | 562 | 532 | 184 | MUC17 - RIPK4 | 209 | 2335 | 1080 |
| Ranking of MUC20 w.r.t RIPK family | |||||||
| laplace | linear | rbf | |||||
| MUC20 - RIPK1 | 1419 | 760 | 1794 | ||||
| MUC20 - RIPK2 | 948 | 2482 | 137 | ||||
| MUC20 - RIPK3 | 2192 | 2288 | 1796 | ||||
| MUC20 - RIPK4 | 1564 | 1619 | 2179 | ||||
| Ranking RIPK family w.r.t MUC family | |||||||
| Ranking of RIPK family w.r.t MUC1 | Ranking of RIPK family w.r.t MUC3A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - RIPK1 | 1839 | 58 | 2421 | MUC3A - RIPK1 | 783 | 1668 | 1842 |
| MUC1 - RIPK2 | 1913 | 2091 | 954 | MUC3A - RIPK2 | 758 | 2301 | 459 |
| MUC1 - RIPK3 | 1038 | 268 | 295 | MUC3A - RIPK3 | 268 | 1595 | 1893 |
| MUC1 - RIPK4 | 1385 | 2246 | 1298 | MUC3A - RIPK4 | 1770 | 1109 | 1461 |
| Ranking of RIPK family w.r.t MUC4 | Ranking of RIPK family w.r.t MUC12 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC4 - RIPK1 | 562 | 1621 | 2216 | MUC12 - RIPK1 | 1462 | 682 | 2351 |
| MUC4 - RIPK2 | 383 | 924 | 494 | MUC12 - RIPK2 | 989 | 597 | 1798 |
| MUC4 - RIPK3 | 541 | 43 | 129 | MUC12 - RIPK3 | 2158 | 1286 | 1636 |
| MUC4 - RIPK4 | 1981 | 1949 | 2028 | MUC12 - RIPK4 | 1577 | 975 | 976 |
| Ranking of RIPK family w.r.t MUC13 | Ranking of RIPK family w.r.t MUC17 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC13 - RIPK1 | 1961 | 1535 | 32 | MUC17 - RIPK1 | 260 | 446 | 260 |
| MUC13 - RIPK2 | 784 | 494 | 1467 | MUC17 - RIPK2 | 1021 | 1114 | 2355 |
| MUC13 - RIPK3 | 860 | 1514 | 1425 | MUC17 - RIPK3 | 427 | 223 | 128 |
| MUC13 - RIPK4 | 107 | 1387 | 1972 | MUC17 - RIPK4 | 1567 | 2225 | 2048 |
| Ranking of RIPK family w.r.t MUC20 | |||||||
| laplace | linear | rbf | |||||
| MUC20 - RIPK1 | 514 | 2042 | 420 | ||||
| MUC20 - RIPK2 | 1039 | 1751 | 1950 | ||||
| MUC20 - RIPK3 | 303 | 2504 | 280 | ||||
| MUC20 - RIPK4 | 794 | 1193 | 989 | ||||
3.2.3. TNF - NF-κB-2/I Cross Family Analysis
| Unexplored combinatorial hypotheses | |
| TNF w.r.t NFkB-2/I | |
| NFkB2 | TNF-RSF10A/RSF12A |
| NFkBI-A | TNF-AIP1/RSF10A/RSF10D/RSF14/SF10 |
| NFkBI-E | TNF-AIP2/RSF14 |
| NFkBI-Z | TNF-RSF10B/RSF10D/RSF12A |
| NFkB-2/I w.r.t TNF | |
| NFkB-2 | TNF-AIP1/AIP2/AIP3 |
| NFkBI-E | TNF-RSF10D |
3.2.4. NFkB-2/I - STAT Cross Family Analysis
| Ranking STAT family w.r.t NFkB-2/I family | |||||||
| Ranking of STAT2 w.r.t NFkB-2/I family | Ranking of STAT3 w.r.t NFkB-2/I family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - STAT2 | 2220 | 1068 | 1207 | NFkB2 - STAT3 | 2125 | 252 | 1453 |
| NFkBIA - STAT2 | 2211 | 1253 | 2402 | NFkBIA - STAT3 | 1614 | 702 | 1333 |
| NFkBIE - STAT2 | 1809 | 512 | 1207 | NFkBIE - STAT3 | 1493 | 211 | 1850 |
| NFkBIZ - STAT2 | 802 | 2121 | 1862 | NFkBIZ - STAT3 | 1633 | 1679 | 2122 |
| Ranking of NFkB-2/I family w.r.t STAT5A | |||||||
| laplace | linear | rbf | |||||
| NFkB2 - STAT5A | 2034 | 1321 | 1502 | ||||
| NFkBIA - STAT5A | 490 | 2215 | 283 | ||||
| NFkBIE - STAT5A | 578 | 1969 | 2485 | ||||
| NFkBIZ - STAT5A | 2286 | 473 | 1409 | ||||
| Ranking NFkB-2/I family w.r.t STAT family | |||||||
| Ranking of NFkB-2/I family w.r.t STAT2 | Ranking of NFkB-2/I family w.r.t STAT3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - STAT2 | 935 | 952 | 86 | NFkB2 - STAT3 | 858 | 606 | 162 |
| NFkBIA - STAT2 | 543 | 36 | 1180 | NFkBIA - STAT3 | 1547 | 88 | 476 |
| NFkBIE - STAT2 | 1449 | 1861 | 1262 | NFkBIE - STAT3 | 1731 | 1063 | 509 |
| NFkBIZ - STAT2 | 483 | 1150 | 262 | NFkBIZ - STAT3 | 1262 | 489 | 1145 |
| Ranking of NFkB-2/I family w.r.t STAT3 | |||||||
| laplace | linear | rbf | |||||
| NFkB2 - STAT5A | 558 | 1070 | 670 | ||||
| NFkBIA - STAT5A | 1509 | 1020 | 81 | ||||
| NFkBIE - STAT5A | 18 | 854 | 1052 | ||||
| NFkBIZ - STAT5A | 83 | 1208 | 240 | ||||
| Unexplored combinatorial hypotheses | |
| STAT w.r.t NFkB-2/I | |
| NFkBIA | STAT2 |
| NFkBIZ | STAT2 |
| NFkBIE | STAT5A |
3.2.5. IKBKE and STAT Cross Family Analysis
3.2.6. IKBKE - TRAF Cross Family Analysis
3.2.7. ABC Transporters - NFkB Cross Family Analysis
| Ranking ABC family w.r.t NFkB-2/I family | |||||||
| Ranking of ABC family w.r.t NFkB2 | Ranking of ABC family w.r.t NFkBI-A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - ABC-A5 | 851 | 1517 | 350 | ABC-A5 - NFkBIA | 398 | 365 | 1660 |
| ABC-B11 - NFkB2 | 1684 | 400 | 412 | ABC-B11 - NFkBIA | 1079 | 566 | 104 |
| NFkB2 - ABC-C3 | 127 | 2031 | 6 | ABC-C3 - NFkBIA | 601 | 1048 | 1760 |
| NFkB2 - ABC-C5 | 1035 | 1431 | 889 | NFkBIA - ABC-C5 | 1683 | 2404 | 1341 |
| NFkB2 - ABC-C13 | 1399 | 1951 | 747 | NFkBIA - ABC-C13 | 200 | 886 | 1275 |
| NFkB2 - ABC-D1 | 1317 | 1133 | 1773 | ABC-D1 - NFkBIA | 1361 | 1361 | 1432 |
| NFkB2 - ABC-G1 | 1983 | 1343 | 1140 | ABC-G1 - NFkBIA | 21 | 313 | 461 |
| NFkB2 - ABC-G2 | 1322 | 955 | 1292 | ABC-G2 - NFkBIA | 809 | 613 | 48 |
| Ranking of ABC family w.r.t NFkBI-E | Ranking of ABC family w.r.t NFkBI-Z | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - NFkBIE | 1445 | 1662 | 679 | ABC-A5 - NFkBIZ | 699 | 1806 | 1290 |
| ABC-B11 - NFkBIE | 2285 | 1154 | 54 | ABC-B11 - NFkBIZ | 1240 | 37 | 803 |
| ABC-C3 - NFkBIE | 1547 | 2168 | 355 | ABC-C3 - NFkBIZ | 468 | 1366 | 1571 |
| NFkBIE - ABC-C5 | 876 | 2048 | 1735 | ABC-C5 - NFkBIZ | 1278 | 1714 | 1065 |
| NFkBIE - ABC-C13 | 623 | 1992 | 2351 | ABC-C13 - NFkBIZ | 1083 | 1063 | 1386 |
| ABC-D1 - NFkBIE | 2380 | 1795 | 861 | ABC-D1 - NFkBIZ | 1677 | 1688 | 794 |
| ABC-G1 - NFkBIE | 2193 | 251 | 208 | ABC-G1 - NFkBIZ | 979 | 2373 | 590 |
| ABC-G2 - NFkBIE | 2124 | 383 | 766 | ABC-G2 - NFkBIZ | 86 | 77 | 845 |
| Ranking NFkB-2/I family w.r.t ABC family | |||||||
| Ranking of NFkB-2/I family w.r.t ABC-A5 | Ranking of NFkB-2/I family w.r.t ABC-B11 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - NFkB2 | 2097 | 1772 | 2086 | NFkB2 - ABC-B11 | 1916 | 1955 | 1020 |
| ABC-A5 - NFkBIA | 827 | 1142 | 379 | NFkBIA - ABC-B11 | 365 | 1702 | 602 |
| ABC-A5 - NFkBIE | 1276 | 1749 | 1795 | NFkBIE - ABC-B11 | 893 | 1285 | 1173 |
| ABC-A5 - NFkBIZ | 778 | 272 | 930 | NFkBIZ - ABC-B11 | 683 | 254 | 421 |
| Ranking of NFkB-2/I family w.r.t ABC-C3 | Ranking of NFkB-2/I family w.r.t ABC-C5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-C3 - NFkB2 | 1225 | 936 | 281 | NFkB2 - ABC-C5 | 1510 | 1712 | 939 |
| ABC-C3 - NFkBIA | 782 | 271 | 1996 | NFkBIA - ABC-C5 | 2017 | 953 | 1649 |
| ABC-C3 - NFkBIE | 1071 | 1094 | 308 | NFkBIE - ABC-C5 | 567 | 615 | 1600 |
| ABC-C3 - NFkBIZ | 546 | 653 | 841 | ABC-C5 - NFkBIZ | 1978 | 943 | 160 |
| Ranking of NFkB-2/I family w.r.t ABC-C13 | Ranking of NFkB-2/I family w.r.t TNF-ABC-D1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - ABC-C13 | 618 | 1423 | 1550 | NFkB2 - ABC-D1 | 2094 | 1655 | 318 |
| NFkBIA - ABC-C13 | 1499 | 1092 | 456 | NFkBIA - ABC-D1 | 613 | 1812 | 1581 |
| NFkBIE - ABC-C13 | 2318 | 586 | 2513 | NFkBIE - ABC-D1 | 806 | 2204 | 410 |
| NFkBIZ - ABC-C13 | 1799 | 2175 | 1068 | NFkBIZ - ABC-D1 | 16 | 1723 | 955 |
| Ranking of NFkB-2/I family w.r.t ABC-G1 | Ranking of NFkB-2/I family w.r.t ABC-G2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - ABC-G1 | 1951 | 2240 | 2215 | NFkB2 - ABC-G2 | 957 | 1427 | 788 |
| NFkBIA - ABC-G1 | 1155 | 258 | 238 | NFKBIA - ABC-G2 | 508 | 417 | 686 |
| NFkBIE - ABC-G1 | 2034 | 612 | 490 | NFKBIE - ABC-G2 | 2223 | 806 | 685 |
| NFkBIZ - ABC-G1 | 1146 | 324 | 900 | NFkBIZ - ABC-G2 | 229 | 221 | 1196 |
| Unexplored combinatorial hypotheses | |
| ABC w.r.t NFkB-2/I family | |
| NFkIBE | ABC-C13/ABC-D1 |
| NFkB-2/I w.r.t ABC family | |
| NFkB2 | ABC-A5/ABC-B11 |
| NFkBIE | ABC-C13 |
| NFkBIZ | ABC-C13 |
| NFkB2 | ABC-G1 |
3.2.8. IKBKE - UBA/UBE Cross Family Analysis
| Ranking UBA/E2 family vs IKBKE | |||||||
| Ranking of UBA/E2 family w.r.t IKBKE | Ranking of IKBKE w.r.t UBA/E2 family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IKBKE - UBA-1 | 1752 | 785 | 966 | UBA-1 - IKBKE | 2327 | 1807 | 2066 |
| IKBKE - UBA-7 | 2189 | 2271 | 1335 | IKBKE - UBA-7 | 1134 | 2326 | 2456 |
| IKBKE - UBA-P1 | 2262 | 1901 | 2341 | IKBKE - UBA-P1 | 2162 | 1817 | 1407 |
| IKBKE - UBA-LD2 | 2034 | 1773 | 1409 | IKBKE - UBA-LD2 | 1381 | 1647 | 556 |
| IKBKE - UBE2-A | 2293 | 2319 | 2396 | IKBKE - UBE2-A | 2422 | 536 | 2328 |
| IKBKE - UBE2-B | 2129 | 1516 | 1795 | IKBKE - UBE2-B | 680 | 2367 | 2427 |
| IKBKE - UBE2-F | 2494 | 2233 | 1896 | IKBKE - UBE2-F | 2309 | 181 | 24 |
| IKBKE - UBE2-H | 1265 | 1666 | 1257 | IKBKE - UBE2-H | 385 | 710 | 746 |
| IKBKE - UBE2-J1 | 905 | 1936 | 1046 | IKBKE - UBE2-J1 | 903 | 1729 | 2215 |
| IKBKE - UBE2-Z | 2016 | 2103 | 481 | IKBKE - UBE2-Z | 783 | 2366 | 1909 |
| Unexplored combinatorial hypotheses | |
| UBA/E2 w.r.t IKBKE | |
| IKBKE | UBA-1/7/P1 |
| IKBKE | UBE2-A/B/Z |
| IKBKE w.r.t UBE/A2 | |
| IKBKE | UBA-7/P1/LD2 |
| IKBKE | UBE2-A/B/F/Z |
3.2.9. REL-A/B - NF-kB Cross Family Analysis
| Ranking NFkB-2/I VS REL-A | |||||||
| Ranking of NFkB-2/I family w.r.t REL-A | Ranking of REL-A w.r.t NFkB-2/I family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - RELA | 664 | 420 | 271 | NFkB2 - RELA | 2454 | 794 | 2307 |
| NFKBIA - RELA | 198 | 205 | 190 | NFKBIA - RELA | 2106 | 2305 | 1153 |
| NFKBIE - RELA | 1503 | 2321 | 331 | NFKBIE - RELA | 1664 | 456 | 1926 |
| NFKBIZ - RELA | 323 | 1714 | 619 | NFKBIZ - RELA | 1924 | 1687 | 1584 |
| Ranking REL-B VS NFkB-2/I family | |||||||
| Ranking of NFKB-2/I w.r.t REL-B | Ranking of REL-B w.r.t NFKB-2/I | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - RELB | 503 | 2146 | 1788 | NFkB2 - RELB | 1156 | 1346 | 2184 |
| NFKBIA - RELB | 239 | 1576 | 924 | NFKBIA - RELB | 968 | 424 | 1725 |
| NFKBIE - RELB | 1203 | 714 | 2200 | NFKBIE - RELB | 1414 | 2228 | 800 |
| NFKBIZ - RELB | 1776 | 2244 | 1869 | NFKBIZ - RELB | 746 | 1281 | 1055 |
3.3. Tumor Necrosis Factor Related Synergies
3.3.1. TNF - WNT Cross Family Analysis
| Ranking TNF family vs WNT family | |||||||
| Ranking of TNF family w.r.t WNT2B | Ranking of WNT2B w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - WNT2B | 503 | 893 | 1656 | TNF - WNT2B | 1341 | 808 | 1366 |
| TNFAIP1 - WNT2B | 235 | 156 | 1811 | TNFAIP1 - WNT2B | 1671 | 1434 | 1404 |
| TNFAIP2 - WNT2B | 868 | 527 | 2439 | TNFAIP2 - WNT2B | 1130 | 218 | 1105 |
| TNFAIP3 - WNT2B | 1135 | 2381 | 1688 | TNFAIP3 - WNT2B | 997 | 1280 | 1902 |
| TNFRSF1A - WNT2B | 2170 | 2127 | 1628 | TNFRSF1A - WNT2B | 1747 | 1857 | 1550 |
| TNFRSF10A - WNT2B | 1861 | 2367 | 1800 | TNFRSF10A - WNT2B | 100 | 464 | 1162 |
| TNFRSF10B - WNT2B | 2020 | 615 | 1881 | TNFRSF10B - WNT2B | 1797 | 120 | 2056 |
| TNFRSF10D - WNT2B | 29 | 2515 | 1174 | TNFRSF10D - WNT2B | 1348 | 1989 | 2130 |
| TNFRSF12A - WNT2B | 1072 | 2061 | 1109 | TNFRSF12A - WNT2B | 1595 | 298 | 1432 |
| TNFRSF14 - WNT2B | 333 | 1585 | 1247 | TNFRSF14 - WNT2B | 1932 | 277 | 2399 |
| TNFRSF21 - WNT2B | 1275 | 648 | 1114 | TNFRSF21 - WNT2B | 1396 | 620 | 2136 |
| TNFSF10 - WNT2B | 1204 | 2287 | 1396 | TNFSF10 - WNT2B | 1732 | 738 | 1751 |
| TNFSF15 - WNT2B | 2476 | 359 | 2073 | TNFSF15 - WNT2B | 402 | 128 | 1875 |
| Ranking of TNF family w.r.t WNT4 | Ranking of WNT4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - WNT4 | 1982 | 1301 | 928 | TNF - WNT4 | 1021 | 420 | 864 |
| TNFAIP1 - WNT4 | 1434 | 1078 | 804 | TNFAIP1 - WNT4 | 1114 | 337 | 1015 |
| TNFAIP2 - WNT4 | 1810 | 1047 | 330 | TNFAIP2 - WNT4 | 1611 | 1341 | 423 |
| TNFAIP3 - WNT4 | 646 | 1955 | 1534 | TNFAIP3 - WNT4 | 2336 | 2511 | 2342 |
| TNFRSF1A - WNT4 | 915 | 545 | 829 | TNFRSF1A - WNT4 | 132 | 333 | 1321 |
| TNFRSF10A - WNT4 | 2509 | 2460 | 897 | TNFRSF10A - WNT4 | 535 | 202 | 582 |
| TNFRSF10B - WNT4 | 517 | 875 | 1365 | TNFRSF10B - WNT4 | 320 | 2105 | 2264 |
| TNFRSF10D - WNT4 | 1719 | 2233 | 2126 | TNFRSF10D - WNT4 | 660 | 49 | 341 |
| TNFRSF12A - WNT4 | 2294 | 1775 | 2384 | TNFRSF12A - WNT4 | 649 | 1756 | 780 |
| TNFRSF14 - WNT4 | 1608 | 2284 | 1436 | TNFRSF14 - WNT4 | 61 | 519 | 1542 |
| TNFRSF21 - WNT4 | 1915 | 1596 | 93 | TNFRSF21 - WNT4 | 201 | 533 | 657 |
| TNFSF10 - WNT4 | 1747 | 2451 | 1782 | TNFSF10 - WNT4 | 904 | 1511 | 2280 |
| TNFSF15 - WNT4 | 1542 | 806 | 2439 | TNFSF15 - WNT4 | 64 | 709 | 793 |
| Ranking of TNF family w.r.t WNT7 | Ranking of WNT7 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - WNT7B | 815 | 381 | 47 | TNF - WNT7B | 1530 | 2511 | 2210 |
| TNFAIP1 - WNT7B | 313 | 1438 | 992 | TNFAIP1 - WNT7B | 2196 | 519 | 1058 |
| TNFAIP2 - WNT7B | 1897 | 85 | 631 | TNFAIP2 - WNT7B | 2121 | 599 | 1313 |
| TNFAIP3 - WNT7B | 577 | 1807 | 1251 | TNFAIP3 - WNT7B | 1901 | 1357 | 830 |
| TNFRSF1A - WNT7B | 165 | 844 | 353 | TNFRSF1A - WNT7B | 2084 | 1975 | 2154 |
| TNFRSF10A - WNT7B | 1084 | 1341 | 2119 | TNFRSF10A - WNT7B | 1301 | 1120 | 1663 |
| TNFRSF10B - WNT7B | 1274 | 1980 | 744 | TNFRSF10B - WNT7B | 1209 | 908 | 1075 |
| TNFRSF10D - WNT7B | 1314 | 774 | 1928 | TNFRSF10D - WNT7B | 1252 | 2301 | 1250 |
| TNFRSF12A - WNT7B | 2100 | 1332 | 1983 | TNFRSF12A - WNT7B | 1104 | 22 | 1879 |
| TNFRSF14 - WNT7B | 1576 | 981 | 1811 | TNFRSF14 - WNT7B | 2079 | 1028 | 1928 |
| TNFRSF21 - WNT7B | 1565 | 798 | 720 | TNFRSF21 - WNT7B | 2114 | 1219 | 737 |
| TNFSF10 - WNT7B | 1598 | 2462 | 2179 | TNFSF10 - WNT7B | 2129 | 763 | 204 |
| TNFSF15 - WNT7B | 1026 | 756 | 621 | TNFSF15 - WNT7B | 130 | 1599 | 2504 |
| Ranking of TNF family w.r.t WNT9A | Ranking of WNT9A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - WNT9A | 1624 | 2025 | 799 | TNF - WNT9A | 1121 | 1930 | 1400 |
| TNFAIP1 - WNT9A | 1433 | 839 | 465 | TNFAIP1 - WNT9A | 1254 | 569 | 394 |
| TNFAIP2 - WNT9A | 40 | 2167 | 397 | TNFAIP2 - WNT9A | 2125 | 2437 | 1605 |
| TNFAIP3 - WNT9A | 1427 | 1109 | 2040 | TNFAIP3 - WNT9A | 1764 | 2460 | 1032 |
| TNFRSF1A - WNT9A | 1470 | 719 | 1933 | TNFRSF1A - WNT9A | 1645 | 58 | 1419 |
| TNFRSF10A - WNT9A | 2272 | 1234 | 918 | TNFRSF10A - WNT9A | 2259 | 2413 | 1204 |
| TNFRSF10B - WNT9A | 2249 | 1222 | 1071 | TNFRSF10B - WNT9A | 882 | 566 | 813 |
| TNFRSF10D - WNT9A | 410 | 2132 | 968 | TNFRSF10D - WNT9A | 1808 | 1055 | 568 |
| TNFRSF12A - WNT9A | 1080 | 373 | 1120 | TNFRSF12A - WNT9A | 2345 | 1211 | 2466 |
| TNFRSF14 - WNT9A | 1106 | 2166 | 198 | TNFRSF14 - WNT9A | 1127 | 1147 | 1191 |
| TNFRSF21 - WNT9A | 1805 | 1999 | 986 | TNFRSF21 - WNT9A | 1265 | 832 | 1098 |
| TNFSF10 - WNT9A | 1258 | 864 | 1839 | TNFSF10 - WNT9A | 2054 | 2338 | 1523 |
| TNFSF15 - WNT9A | 1621 | 1129 | 1139 | TNFSF15 - WNT9A | 37 | 1105 | 1076 |
3.3.2. MUC - TNF Cross Family Analysis
3.3.3. STEAP4 - TNF Cross Family Analysis
| Unexplored combinatorial hypotheses | |
| TNF w.r.t STEAP4 | |
| TNF, TNF-AIP1/AIP2/AIP3/RSF10B/RSF10D/SF10 | STEAP4 |
| STEAP4 w.r.t TNF | |
| TNF-RSF10A/RSF10D/RSF14 | STEAP4 |
3.3.4. TNF - UBE2 Cross Family Analysis
| Ranking TNF family vs UBE2 family | |||||||
| Ranking of UBE2A w.r.t TNF family | Ranking of TNF family w.r.t UBE2A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2A | 1360 | 2307 | 1720 | TNF - UBE2A | 499 | 1379 | 750 |
| TNFAIP1 - UBE2A | 498 | 2357 | 2455 | TNFAIP1 - UBE2A | 1340 | 2494 | 578 |
| TNFAIP2 - UBE2A | 524 | 1161 | 2385 | TNFAIP2 - UBE2A | 441 | 1852 | 691 |
| TNFAIP3 - UBE2A | 855 | 1642 | 812 | TNFAIP3 - UBE2A | 1157 | 1048 | 207 |
| TNFRSF1A - UBE2A | 2457 | 1087 | 2020 | TNFRSF1A - UBE2A | 1066 | 655 | 1701 |
| TNFRSF10A - UBE2A | 2164 | 2126 | 621 | TNFRSF10A - UBE2A | 2116 | 858 | 2376 |
| TNFRSF10B - UBE2A | 2284 | 1901 | 1203 | TNFRSF10B - UBE2A | 362 | 1083 | 756 |
| TNFRSF10D - UBE2A | 1989 | 2291 | 677 | TNFRSF10D - UBE2A | 1848 | 1336 | 903 |
| TNFRSF12A - UBE2A | 2484 | 2427 | 339 | TNFRSF12A - UBE2A | 1537 | 1304 | 629 |
| TNFRSF14 - UBE2A | 2301 | 2180 | 2323 | TNFRSF14 - UBE2A | 908 | 1519 | 1945 |
| TNFRSF21 - UBE2A | 2419 | 2035 | 1169 | TNFRSF21 - UBE2A | 605 | 2245 | 60 |
| TNFSF10 - UBE2A | 832 | 2202 | 1036 | TNFSF10 - UBE2A | 1520 | 44 | 2125 |
| TNFSF15 - UBE2A | 1768 | 1184 | 1942 | TNFSF15 - UBE2A | 545 | 580 | 1448 |
| Ranking of UBE2B w.r.t TNF family | Ranking of TNF family w.r.t UBE2B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2B | 1072 | 2046 | 1316 | TNF - UBE2B | 1719 | 218 | 346 |
| TNFAIP1 - UBE2B | 1097 | 744 | 1295 | TNFAIP1 - UBE2B | 920 | 90 | 1028 |
| TNFAIP2 - UBE2B | 669 | 1158 | 2407 | TNFAIP2 - UBE2B | 1680 | 147 | 45 |
| TNFAIP3 - UBE2B | 470 | 1528 | 1388 | TNFAIP3 - UBE2B | 2259 | 742 | 1610 |
| TNFRSF1A - UBE2B | 937 | 1473 | 2390 | TNFRSF1A - UBE2B | 1277 | 1454 | 1258 |
| TNFRSF10A - UBE2B | 2132 | 1128 | 2184 | TNFRSF10A - UBE2B | 551 | 2318 | 2265 |
| TNFRSF10B - UBE2B | 2399 | 2000 | 402 | TNFRSF10B - UBE2B | 2272 | 1268 | 1080 |
| TNFRSF10D - UBE2B | 1959 | 1562 | 2232 | TNFRSF10D - UBE2B | 1157 | 207 | 1729 |
| TNFRSF12A - UBE2B | 1632 | 12 | 2259 | TNFRSF12A - UBE2B | 1940 | 1868 | 1758 |
| TNFRSF14 - UBE2B | 1137 | 2297 | 2373 | TNFRSF14 - UBE2B | 1143 | 1657 | 1507 |
| TNFRSF21 - UBE2B | 1986 | 1439 | 1754 | TNFRSF21 - UBE2B | 1291 | 569 | 17 |
| TNFSF10 - UBE2B | 2265 | 1488 | 769 | TNFSF10 - UBE2B | 2208 | 2326 | 2470 |
| TNFSF15 - UBE2B | 1432 | 2460 | 1655 | TNFSF15 - UBE2B | 2055 | 1964 | 183 |
| Ranking of UBE2F w.r.t TNF family | Ranking of TNF family w.r.t UBE2F | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2F | 2162 | 2484 | 2500 | TNF - UBE2F | 638 | 435 | 1471 |
| TNFAIP1 - UBE2F | 1732 | 2239 | 2003 | TNFAIP1 - UBE2F | 447 | 1376 | 1357 |
| TNFAIP2 - UBE2F | 693 | 1446 | 1706 | TNFAIP2 - UBE2F | 900 | 208 | 883 |
| TNFAIP3 - UBE2F | 498 | 2265 | 1264 | TNFAIP3 - UBE2F | 1881 | 113 | 1185 |
| TNFRSF1A - UBE2F | 1980 | 2255 | 1872 | TNFRSF1A - UBE2F | 368 | 1756 | 266 |
| TNFRSF10A - UBE2F | 2085 | 2218 | 179 | TNFRSF10A - UBE2F | 1767 | 1599 | 781 |
| TNFRSF10B - UBE2F | 2432 | 2011 | 2144 | TNFRSF10B - UBE2F | 1413 | 1157 | 1510 |
| TNFRSF10D - UBE2F | 1164 | 1400 | 2150 | TNFRSF10D - UBE2F | 389 | 206 | 2481 |
| TNFRSF12A - UBE2F | 2458 | 2336 | 531 | TNFRSF12A - UBE2F | 581 | 2022 | 630 |
| TNFRSF14 - UBE2F | 1757 | 574 | 1070 | TNFRSF14 - UBE2F | 2324 | 1924 | 1954 |
| TNFRSF21 - UBE2F | 1056 | 2498 | 1418 | TNFRSF21 - UBE2F | 718 | 2123 | 1022 |
| TNFSF10 - UBE2F | 1710 | 2365 | 1691 | TNFSF10 - UBE2F | 1656 | 1584 | 810 |
| TNFSF15 - UBE2F | 1910 | 1171 | 2353 | TNFSF15 - UBE2F | 1224 | 1637 | 394 |
| Ranking TNF family vs UBE2 family | |||||||
| Ranking of UBE2H w.r.t TNF family | Ranking of TNF family w.r.t UBE2H | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2H | 967 | 1966 | 1018 | TNF - UBE2H | 2277 | 770 | 640 |
| TNFAIP1 - UBE2H | 1235 | 1484 | 817 | TNFAIP1 - UBE2H | 883 | 2396 | 608 |
| TNFAIP2 - UBE2H | 1251 | 978 | 2517 | TNFAIP2 - UBE2H | 762 | 1362 | 593 |
| TNFAIP3 - UBE2H | 889 | 1055 | 1837 | TNFAIP3 - UBE2H | 1942 | 1421 | 1467 |
| TNFRSF1A - UBE2H | 589 | 1498 | 1428 | TNFRSF1A - UBE2H | 1134 | 2154 | 182 |
| TNFRSF10A - UBE2H | 1317 | 905 | 2229 | TNFRSF10A - UBE2H | 1139 | 202 | 1061 |
| TNFRSF10B - UBE2H | 33 | 2128 | 1725 | TNFRSF10B - UBE2H | 1053 | 539 | 1207 |
| TNFRSF10D - UBE2H | 1326 | 1814 | 1657 | TNFRSF10D - UBE2H | 2227 | 926 | 791 |
| TNFRSF12A - UBE2H | 1950 | 1793 | 1851 | TNFRSF12A - UBE2H | 1347 | 776 | 1899 |
| TNFRSF14 - UBE2H | 2297 | 1601 | 2385 | TNFRSF14 - UBE2H | 2244 | 703 | 1208 |
| TNFRSF21 - UBE2H | 2022 | 1131 | 2231 | TNFRSF21 - UBE2H | 827 | 880 | 672 |
| TNFSF10 - UBE2H | 2387 | 7 | 760 | TNFSF10 - UBE2H | 1313 | 1169 | 2002 |
| TNFSF15 - UBE2H | 125 | 58 | 96 | TNFSF15 - UBE2H | 350 | 2416 | 1960 |
| Ranking of UBE2J1 w.r.t TNF family | Ranking of TNF family w.r.t UBE2J1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2J1 | 1289 | 2308 | 2336 | TNF - UBE2J1 | 1101 | 1549 | 105 |
| TNFAIP1 - UBE2J1 | 1109 | 2292 | 1756 | TNFAIP1 - UBE2J1 | 329 | 1971 | 252 |
| TNFAIP2 - UBE2J1 | 1379 | 1516 | 1696 | TNFAIP2 - UBE2J1 | 112 | 22 | 969 |
| TNFAIP3 - UBE2J1 | 187 | 1261 | 1065 | TNFAIP3 - UBE2J1 | 289 | 891 | 1202 |
| TNFRSF1A - UBE2J1 | 1992 | 326 | 2268 | TNFRSF1A - UBE2J1 | 1422 | 624 | 73 |
| TNFRSF10A - UBE2J1 | 1893 | 2090 | 2363 | TNFRSF10A - UBE2J1 | 2379 | 2213 | 2135 |
| TNFRSF10B - UBE2J1 | 1913 | 1299 | 1838 | TNFRSF10B - UBE2J1 | 807 | 1793 | 1231 |
| TNFRSF10D - UBE2J1 | 325 | 1500 | 588 | TNFRSF10D - UBE2J1 | 2393 | 1360 | 2102 |
| TNFRSF12A - UBE2J1 | 2401 | 1901 | 437 | TNFRSF12A - UBE2J1 | 380 | 1284 | 650 |
| TNFRSF14 - UBE2J1 | 2277 | 2347 | 1943 | TNFRSF14 - UBE2J1 | 1614 | 2133 | 2313 |
| TNFRSF21 - UBE2J1 | 1976 | 2333 | 1681 | TNFRSF21 - UBE2J1 | 1315 | 1266 | 1070 |
| TNFSF10 - UBE2J1 | 511 | 2508 | 506 | TNFSF10 - UBE2J1 | 1322 | 203 | 1148 |
| TNFSF15 - UBE2J1 | 2021 | 2013 | 2515 | TNFSF15 - UBE2J1 | 678 | 886 | 1128 |
| Ranking of UBE2Z w.r.t TNF family | Ranking of TNF family w.r.t UBE2Z | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - UBE2Z | 2264 | 2505 | 2479 | TNF - UBE2Z | 739 | 701 | 1241 |
| TNFAIP1 - UBE2Z | 1283 | 2055 | 2332 | TNFAIP1 - UBE2Z | 1198 | 1213 | 226 |
| TNFAIP2 - UBE2Z | 2404 | 1625 | 2139 | TNFAIP2 - UBE2Z | 1281 | 1431 | 492 |
| TNFAIP3 - UBE2Z | 1066 | 1152 | 1627 | TNFAIP3 - UBE2Z | 530 | 51 | 972 |
| TNFRSF1A - UBE2Z | 2473 | 2194 | 2405 | TNFRSF1A - UBE2Z | 692 | 43 | 1382 |
| TNFRSF10A - UBE2Z | 2234 | 2152 | 713 | TNFRSF10A - UBE2Z | 2410 | 2103 | 1513 |
| TNFRSF10B - UBE2Z | 1501 | 451 | 2081 | TNFRSF10B - UBE2Z | 948 | 1369 | 403 |
| TNFRSF10D - UBE2Z | 2264 | 2360 | 2278 | TNFRSF10D - UBE2Z | 1786 | 661 | 1746 |
| TNFRSF12A - UBE2Z | 2207 | 2149 | 353 | TNFRSF12A - UBE2Z | 1621 | 2010 | 1448 |
| TNFRSF14 - UBE2Z | 1683 | 1983 | 705 | TNFRSF14 - UBE2Z | 1779 | 1360 | 2100 |
| TNFRSF21 - UBE2Z | 994 | 604 | 219 | TNFRSF21 - UBE2Z | 459 | 1030 | 584 |
| TNFSF10 - UBE2Z | 516 | 2374 | 2235 | TNFSF10 - UBE2Z | 1100 | 2047 | 168 |
| TNFSF15 - UBE2Z | 2081 | 1037 | 2102 | TNFSF15 - UBE2Z | 1342 | 1180 | 536 |
| Unexplored combinatorial hypotheses | |
| UBE2 w.r.t TNF | |
| TNF-AIP1/RSF1A/RSF10A/RSF10B/RSF10D/RSF12A/RSF14/RSF21/SF15 | UBE2A |
| TNF-RSF10A/RSF10B/RSF10D/RSF14/RSF21 | UBE2B |
| TNF, TNF-AIP1/RSF1A/RSF10A/RSF10B/RSF12A/SF15 | UBE2F |
| TNF-RSF12A/RSF14/RSF21 | UBE2H |
| TNF, TNF-AIP1/RSF1A/RSF10A/RSF10B/RSF12A/RSF14/RSF21/SF15 | UBE2J1 |
| TNF, TNF-AIP1/AIP2/RSF1A/RSF10A/RSF10D/RSF12A/SF10/SF15 | UBE2Z |
| TNF w.r.t UBE2 | |
| TNF-RSF10A | UBE2A |
| TNF-RSF10A/RSF12A/SF10/SF15 | UBE2B |
| TNF-RSF14 | UBE2F |
| TNF-SF15 | UBE2H |
| TNF-RSF10A/RSF10D/RSF14 | UBE2J1 |
| TNF-RSF10A/RSF14 | UBE2Z |
3.3.5. TNF - BCL Cross Family Analysis
| Unexplored combinatorial hypotheses | |
| BCL w.r.t TNF | |
| TNF, TNF-AIP1/RSF1A/RSF10B/RSF10D/RSF12A/RSF14/RSF21/SF10/SF15 | BCL2L2 |
| TNF, TNF-AIP1/RSF1A/RSF10A/RSF10D/RSF12A/RSF14/RSF21/SF10/SF15 | BCL2L13 |
| TNFRSF10B | BCL3 |
| TNF, TNF-AIP1/AIP2/RSF1A/RSF10A/RSF10D/RSF21 | BCL6 |
| TNF-RSF10D/RSF12A | BCL10 |
| TNF w.r.t BCL | |
| TNF-RSF10A/RSF10B/RSF10D/RSF12A/RSF14/SF10 | BCL2L1 |
| TNF-RSF10B | BCL2L2 |
| TNF-RSF14 | BCL2L13 |
| TNF-RSF10A/SF10D/SF10 | BCL3 |
| TNF-RSF12A | BCL6 |
| TNF-AIP1 | BCL9L |
| TNF-SF10/SF15 | BCL9L |
| TNF-RSF14 | BCL10 |
3.4. DNA Repair Related Synergies
3.4.1. XRCC - RAD Cross Family Analysis
| Ranking XRCC family w.r.t RAD family | |||||||
| Ranking of XRCC w.r.t RAD1 | Ranking of XRCC w.r.t RAD18 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - XRCC1 | 1751 | 1808 | 793 | RAD18 - XRCC1 | 927 | 2669 | 200 |
| XRCC2 - RAD1 | 62 | 498 | 1231 | XRCC2 - RAD18 | 506 | 1844 | 1517 |
| XRCC6 - RAD1 | 2736 | 2511 | 1284 | RAD18 - XRCC6 | 279 | 2193 | 804 |
| RAD1 - XRCC6BP1 | 764 | 2108 | 1325 | RAD18 - XRCC6BP1 | 819 | 1954 | 1976 |
| Ranking of XRCC w.r.t RAD50 | Ranking of XRCC w.r.t RAD51 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| XRCC1 - RAD50 | 2573 | 2374 | 2497 | RAD51 - XRCC1 | 1673 | 1818 | 2611 |
| XRCC2 - RAD50 | 53 | 244 | 147 | XRCC2 - RAD51 | 472 | 2348 | 1973 |
| XRCC6 - RAD50 | 2615 | 2568 | 2582 | RAD51 - XRCC6 | 80 | 1244 | 2595 |
| RAD50 - XRCC6BP1 | 1962 | 1375 | 1366 | RAD51 - XRCC6BP1 | 792 | 951 | 1595 |
| Ranking of XRCC w.r.t RAD51AP1 | Ranking of XRCC w.r.t RAD51C | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| XRCC1 - RAD51AP1 | 1802 | 2732 | 801 | RAD51C - XRCC1 | 2282 | 1846 | 2026 |
| XRCC2 - RAD51AP1 | 78 | 112 | 351 | XRCC2 - RAD51C | 1695 | 932 | 520 |
| XRCC6 - RAD51AP1 | 2653 | 2439 | 347 | RAD51C - XRCC6 | 2545 | 1848 | 1858 |
| RAD51AP1 - XRCC6BP1 | 1790 | 936 | 974 | RAD51C - XRCC6BP1 | 2325 | 1070 | 1844 |
| Ranking of XRCC w.r.t RAD54B | Ranking of XRCC w.r.t RAD54L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| XRCC1 - RAD54B | 2475 | 2670 | 1824 | RAD54L - XRCC1 | 1834 | 657 | 525 |
| XRCC2 - RAD54B | 1554 | 744 | 620 | XRCC2 - RAD54L | 2564 | 167 | 565 |
| XRCC6 - RAD54B | 2505 | 2709 | 2604 | RAD54L - XRCC6 | 2597 | 496 | 1247 |
| RAD54B - XRCC6BP1 | 1932 | 2504 | 2170 | RAD54L - XRCC6BP1 | 1389 | 1227 | 1454 |
| Unexplored combinatorial hypotheses | |
| RAD w.r.t XRCC family | |
| RAD-18/51/51AP1/51C/54B/54L | XRCC1 |
| RAD-18/51/51AP1/51C/54B/54L | XRCC2 |
| RAD-18/51/51AP1/51C/54B/54L | XRCC6 |
| RAD-1/18/50/51/51AP1/51C/54B/54L | XRCC6BP1 |
| XRCC w.r.t RAD family | |
| RAD1 | XRCC-2/6BP1 |
| RAD18 | XRCC-1/2/6 |
| RAD50 | XRCC-2/6BP1 |
| RAD51 | XRCC-6/6BP1 |
| RAD51AP1 | XRCC-2/6BP1 |
| RAD51C | XRCC-2 |
| RAD54B | XRCC-2 |
| RAD54L | XRCC-1/2/6/6BP1 |
3.4.2. XRN2 - RAD Cross Family Analysis
| Unexplored combinatorial hypotheses | |
| RAD w.r.t XRN2 | |
| XRN2 | RAD-51AP1/51/54L/51C/18/54B |
| XRN2 w.r.t RAD | |
| XRN2 | RAD1 |
| XRN2 | RAD51AP1 |
| XRN2 | RAD54L |
| XRN2 | RAD51C |
3.4.3. NKRF - RAD Cross Family Analysis
| Ranking NKRF w.r.t RAD family | |||||||
| Ranking of NFRK w.r.t RAD family | Ranking of RAD family w.r.t NKRF | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD51AP1 - NKRF | 1724 | 1642 | 649 | RAD51AP1 - NKRF | 157 | 553 | 2561 |
| RAD51 - NKRF | 982 | 1724 | 1352 | RAD51 - NKRF | 439 | 1441 | 1606 |
| RAD54L - NKRF | 1727 | 1387 | 1120 | RAD54L - NKRF | 117 | 1175 | 1415 |
| RAD51C - NKRF | 1568 | 472 | 1505 | RAD51C - NKRF | 418 | 2178 | 1653 |
| RAD18 - NKRF | 1508 | 615 | 405 | RAD18 - NKRF | 164 | 2306 | 1509 |
| RAD1 - NKRF | 2667 | 2222 | 1181 | NKRF - RAD1 | 1391 | 1115 | 735 |
| RAD54B - NKRF | 1476 | 1189 | 1534 | RAD54B - NKRF | 207 | 1869 | 2244 |
| RAD50 - NKRF | 2003 | 2343 | 2511 | NKRF - RAD50 | 1354 | 851 | 824 |
3.4.4. RAD - BCL Cross Family Analysis
| Ranking RAD family VS BCL family | |||||||
| Ranking of BCL2L12 w.r.t RAD family | Ranking of RAD family w.r.t BCL2L12 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL2L12 | 1797 | 1530 | 1401 | RAD1 - BCL2L12 | 1958 | 2120 | 1957 |
| RAD18 - BCL2L12 | 675 | 2437 | 1312 | RAD18 - BCL2L12 | 779 | 652 | 1388 |
| RAD50 - BCL2L12 | 2080 | 1151 | 929 | RAD50 - BCL2L12 | 1668 | 2566 | 1703 |
| RAD51 - BCL2L12 | 1234 | 1334 | 2350 | RAD51 - BCL2L12 | 1164 | 365 | 1213 |
| RAD51AP1 - BCL2L12 | 2267 | 2500 | 2265 | RAD51AP1 - BCL2L12 | 306 | 57 | 28 |
| RAD51C - BCL2L12 | 1561 | 2384 | 1647 | RAD51C - BCL2L12 | 495 | 1191 | 429 |
| RAD54B - BCL2L12 | 1979 | 1329 | 1625 | RAD54B - BCL2L12 | 678 | 432 | 787 |
| RAD54L - BCL2L12 | 2446 | 821 | 210 | RAD54L - BCL2L12 | 901 | 1128 | 263 |
| Ranking of BCL6B w.r.t RAD family | Ranking of RAD family w.r.t BCL6B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL6B | 194 | 481 | 102 | RAD1 - BCL6B | 2110 | 2151 | 2059 |
| RAD18 - BCL6B | 1790 | 176 | 929 | RAD18 - BCL6B | 1113 | 640 | 482 |
| RAD50 - BCL6B | 860 | 87 | 74 | RAD50 - BCL6B | 2164 | 2412 | 2581 |
| RAD51 - BCL6B | 2324 | 263 | 58 | RAD51 - BCL6B | 287 | 681 | 497 |
| RAD51AP1 - BCL6B | 723 | 428 | 579 | RAD51AP1 - BCL6B | 1607 | 1638 | 916 |
| RAD51C - BCL6B | 660 | 521 | 1609 | RAD51C - BCL6B | 43 | 871 | 999 |
| RAD54B - BCL6B | 708 | 596 | 647 | RAD54B - BCL6B | 1212 | 1392 | 1170 |
| RAD54L - BCL6B | 108 | 2684 | 1326 | RAD54L - BCL6B | 1867 | 1009 | 785 |
| Ranking of BCL7A w.r.t RAD family | Ranking of RAD family w.r.t BCL7A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL7A | 690 | 1791 | 1202 | RAD1 - BCL7A | 1989 | 2101 | 1804 |
| RAD18 - BCL7A | 385 | 2366 | 185 | RAD18 - BCL7A | 1514 | 1515 | 783 |
| RAD50 - BCL7A | 137 | 601 | 417 | RAD50 - BCL7A | 2123 | 1771 | 2085 |
| RAD51 - BCL7A | 514 | 1694 | 2361 | RAD51 - BCL7A | 879 | 274 | 639 |
| RAD51AP1 - BCL7A | 2440 | 2609 | 774 | RAD51AP1 - BCL7A | 412 | 416 | 4 |
| RAD51C - BCL7A | 2726 | 2448 | 983 | RAD51C - BCL7A | 215 | 394 | 461 |
| RAD54B - BCL7A | 2729 | 1830 | 2743 | RAD54B - BCL7A | 809 | 1407 | 213 |
| RAD54L - BCL7A | 1519 | 418 | 842 | RAD54L - BCL7A | 435 | 783 | 1499 |
| Ranking of BCL9 w.r.t RAD family | Ranking of RAD family w.r.t BCL9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL9 | 1296 | 2418 | 1775 | RAD1 - BCL9 | 1749 | 2528 | 1391 |
| RAD18 - BCL9 | 461 | 1952 | 1453 | RAD18 - BCL9 | 656 | 1194 | 482 |
| RAD50 - BCL9 | 2338 | 2653 | 2559 | RAD50 - BCL9 | 2220 | 1441 | 1098 |
| RAD51 - BCL9 | 1748 | 1143 | 952 | RAD51 - BCL9 | 622 | 929 | 860 |
| RAD51AP1 - BCL9 | 1861 | 2280 | 786 | RAD51AP1 - BCL9 | 331 | 61 | 102 |
| RAD51C - BCL9 | 956 | 2741 | 376 | RAD51C - BCL9 | 1113 | 417 | 1154 |
| RAD54B - BCL9 | 2063 | 2375 | 1050 | RAD54B - BCL9 | 1045 | 53 | 650 |
| RAD54L - BCL9 | 1450 | 1096 | 400 | RAD54L - BCL9 | 636 | 602 | 934 |
| Ranking of BCL11A w.r.t RAD family | Ranking of RAD family w.r.t BCL11A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL11A | 1069 | 507 | 1267 | RAD1 - BCL11A | 1430 | 1475 | 1584 |
| RAD18 - BCL11A | 1561 | 169 | 692 | RAD18 - BCL11A | 465 | 164 | 1952 |
| RAD50 - BCL11A | 582 | 1144 | 1047 | RAD50 - BCL11A | 2649 | 875 | 1226 |
| RAD51 - BCL11A | 1722 | 2073 | 339 | RAD51 - BCL11A | 255 | 2064 | 2461 |
| RAD51AP1 - BCL11A | 1120 | 752 | 645 | RAD51AP1 - BCL11A | 659 | 388 | 496 |
| RAD51C - BCL11A | 1024 | 199 | 899 | RAD51C - BCL11A | 363 | 1673 | 97 |
| RAD54B - BCL11A | 1037 | 917 | 867 | RAD54B - BCL11A | 581 | 2743 | 799 |
| RAD54L - BCL11A | 172 | 2193 | 2318 | RAD54L - BCL11A | 846 | 2733 | 209 |
| Ranking of BCL11B w.r.t RAD family | Ranking of RAD family w.r.t BCL11B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - BCL11B | 2371 | 2360 | 43 | RAD1 - BCL11B | 2571 | 230 | 1373 |
| RAD18 - BCL11B | 1741 | 993 | 2677 | RAD18 - BCL11B | 1747 | 2028 | 14 |
| RAD50 - BCL11B | 2010 | 1198 | 903 | RAD50 - BCL11B | 919 | 860 | 2263 |
| RAD51 - BCL11B | 2067 | 449 | 971 | RAD51 - BCL11B | 1095 | 1238 | 2373 |
| RAD51AP1 - BCL11B | 1247 | 908 | 1671 | RAD51AP1 - BCL11B | 196 | 2646 | 987 |
| RAD51C - BCL11B | 1736 | 1234 | 2282 | RAD51C - BCL11B | 1122 | 1844 | 1161 |
| RAD54B - BCL11B | 1193 | 1192 | 832 | RAD54B - BCL11B | 363 | 2150 | 1561 |
| RAD54L - BCL11B | 1421 | 1385 | 1854 | RAD54L - BCL11B | 579 | 2543 | 159 |
3.4.5. RAD - EXOSC Cross Family Analysis
3.4.6. XRCC - EXOSC Cross Family Analysis
| Ranking XRCC family VS EXOSC family | |||||||
| Ranking of EXOSC2 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC2 - XRCC1 | 277 | 176 | 423 | EXOSC2 - XRCC1 | 2708 | 2386 | 2634 |
| EXOSC2 - XRCC2 | 8 | 38 | 100 | EXOSC2 - XRCC2 | 166 | 417 | 56 |
| EXOSC2 - XRCC6 | 1252 | 398 | 623 | EXOSC2 - XRCC6 | 2678 | 2504 | 2576 |
| EXOSC2 - XRCC6BP1 | 935 | 905 | 1755 | EXOSC2 - XRCC6BP1 | 1740 | 1842 | 2177 |
| Ranking of EXOSC3 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC3 - XRCC1 | 1551 | 2256 | 1974 | EXOSC3 - XRCC1 | 2217 | 1418 | 2041 |
| EXOSC3 - XRCC2 | 2462 | 2553 | 2329 | EXOSC3 - XRCC2 | 125 | 15 | 194 |
| EXOSC3 - XRCC6 | 1720 | 1716 | 2398 | EXOSC3 - XRCC6 | 2742 | 2608 | 2193 |
| EXOSC3 - XRCC6BP1 | 2506 | 1523 | 1356 | EXOSC3 - XRCC6BP1 | 2561 | 2154 | 2406 |
| Ranking of EXOSC5 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC5 - XRCC1 | 741 | 291 | 8 | EXOSC5 - XRCC1 | 2578 | 2568 | 1910 |
| EXOSC5 - XRCC2 | 1244 | 791 | 702 | EXOSC5 - XRCC2 | 1559 | 1857 | 866 |
| EXOSC5 - XRCC6 | 65 | 1064 | 322 | EXOSC5 - XRCC6 | 2410 | 2465 | 2190 |
| EXOSC5 - XRCC6BP1 | 416 | 880 | 1434 | EXOSC5 - XRCC6BP1 | 1907 | 2029 | 1394 |
| Ranking of EXOSC6 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC6 - XRCC1 | 1890 | 985 | 1163 | EXOSC6 - XRCC1 | 509 | 2373 | 1046 |
| EXOSC6 - XRCC2 | 1512 | 648 | 1458 | EXOSC6 - XRCC2 | 486 | 2564 | 1901 |
| EXOSC6 - XRCC6 | 2304 | 1719 | 2690 | EXOSC6 - XRCC6 | 2576 | 35 | 188 |
| EXOSC6 - XRCC6BP1 | 2428 | 492 | 2112 | EXOSC6 - XRCC6BP1 | 1753 | 1295 | 366 |
| Ranking of EXOSC7 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC7 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC7 - XRCC1 | 1907 | 1510 | 1603 | EXOSC7 - XRCC1 | 1844 | 1229 | 987 |
| EXOSC7 - XRCC2 | 1369 | 2555 | 2124 | EXOSC7 - XRCC2 | 176 | 436 | 788 |
| EXOSC7 - XRCC6 | 584 | 1523 | 1018 | EXOSC7 - XRCC6 | 1074 | 242 | 288 |
| EXOSC7 - XRCC6BP1 | 1419 | 1944 | 876 | EXOSC7 - XRCC6BP1 | 2144 | 1577 | 2038 |
| Ranking of EXOSC8 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC8 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC8 - XRCC1 | 1373 | 1515 | 2103 | EXOSC8 - XRCC1 | 1769 | 2151 | 1435 |
| EXOSC8 - XRCC2 | 1086 | 2309 | 2435 | EXOSC8 - XRCC2 | 13 | 1932 | 6 |
| EXOSC8 - XRCC6 | 1820 | 2542 | 2693 | EXOSC8 - XRCC6 | 1869 | 1233 | 2625 |
| EXOSC8 - XRCC6BP1 | 2112 | 1994 | 2699 | EXOSC8 - XRCC6BP1 | 2305 | 2461 | 2319 |
| Ranking of EXOSC9 w.r.t XRCC family | Ranking of XRCC family w.r.t EXOSC9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC9 - XRCC1 | 44 | 1214 | 1410 | EXOSC9 - XRCC1 | 1804 | 2696 | 1629 |
| EXOSC9 - XRCC2 | 496 | 672 | 840 | EXOSC9 - XRCC2 | 1793 | 655 | 1526 |
| EXOSC9 - XRCC6 | 1121 | 151 | 689 | EXOSC9 - XRCC6 | 1882 | 2188 | 2404 |
| EXOSC9 - XRCC6BP1 | 362 | 463 | 1741 | EXOSC9 - XRCC6BP1 | 1206 | 1776 | 1626 |
| Unexplored combinatorial hypotheses | |
| XRCC w.r.t EXOSC | |
| EXOSC-2 | XRCC-2 |
| EXOSC-3 | XRCC-2 |
| EXOSC-5 | XRCC-2 |
| EXOSC-6 | XRCC-6 |
| EXOSC-7 | XRCC-1/2/6 |
| EXOSC-8 | XRCC-2 |
| EXOSC-9 | XRCC-2/6BP1 |
| EXOSC w.r.t XRCC | |
| EXOSC-2 | XRCC-1/2/6/6BP1 |
| EXOSC-3 | XRCC-6/6BP1 |
| EXOSC-5 | XRCC-1/2/6/6BP1 |
| EXOSC-6 | XRCC-1/2 |
| EXOSC-7 | XRCC-1/6/6BP1 |
| EXOSC-8 | XRCC-1 |
| EXOSC-9 | XRCC-1/2/6/6BP1 |
3.4.7. RAD - FANC Cross Family Analysis
| Ranking RAD family VS FANC family | |||||||
| Ranking of RAD family w.r.t FANCB | Ranking of FANCB w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCB | 2431 | 400 | 2553 | RAD1 - FANCB | 1499 | 656 | 340 |
| RAD18 - FANCB | 10 | 2219 | 625 | RAD18 - FANCB | 2708 | 383 | 2298 |
| RAD50 - FANCB | 2419 | 915 | 2556 | RAD50 - FANCB | 133 | 234 | 73 |
| RAD51 - FANCB | 247 | 73 | 610 | RAD51 - FANCB | 2444 | 378 | 8 |
| RAD51AP1 - FANCB | 479 | 1667 | 663 | RAD51AP1 - FANCB | 89 | 562 | 2 |
| RAD51C - FANCB | 769 | 536 | 887 | RAD51C - FANCB | 460 | 187 | 86 |
| RAD54B - FANCB | 468 | 133 | 438 | RAD54B - FANCB | 486 | 891 | 568 |
| RAD54L - FANCB | 583 | 2131 | 160 | RAD54L - FANCB | 41 | 2675 | 692 |
| Ranking of RAD family w.r.t FANCD2 | Ranking of FANCD2 w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCD2 | 1935 | 332 | 2102 | RAD1 - FANCD2 | 1451 | 1605 | 796 |
| RAD18 - FANCD2 | 1035 | 1271 | 405 | RAD18 - FANCD2 | 2356 | 403 | 1299 |
| RAD50 - FANCD2 | 2109 | 436 | 2038 | RAD50 - FANCD2 | 646 | 357 | 769 |
| RAD51 - FANCD2 | 885 | 1995 | 1383 | RAD51 - FANCD2 | 591 | 1938 | 85 |
| RAD51AP1 - FANCD2 | 1734 | 644 | 1291 | RAD51AP1 - FANCD2 | 993 | 603 | 2684 |
| RAD51C - FANCD2 | 54 | 2399 | 2566 | RAD51C - FANCD2 | 629 | 656 | 620 |
| RAD54B - FANCD2 | 275 | 2460 | 478 | RAD54B - FANCD2 | 227 | 230 | 131 |
| RAD54L - FANCD2 | 493 | 2530 | 203 | RAD54L - FANCD2 | 2457 | 1369 | 1816 |
| Ranking of RAD family w.r.t FANCD2OS | Ranking of FANCD2OS w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCD2OS | 693 | 1926 | 1146 | RAD1 - FANCD2OS | 1455 | 2445 | 1624 |
| RAD18 - FANCD2OS | 1472 | 526 | 239 | RAD18 - FANCD2OS | 851 | 1457 | 653 |
| RAD50 - FANCD2OS | 178 | 1534 | 2141 | RAD50 - FANCD2OS | 1763 | 1477 | 1372 |
| RAD51 - FANCD2OS | 2061 | 1080 | 1226 | RAD51 - FANCD2OS | 2007 | 2336 | 1739 |
| RAD51AP1 - FANCD2OS | 637 | 2050 | 2660 | RAD51AP1 - FANCD2OS | 2209 | 2376 | 1722 |
| RAD51C - FANCD2OS | 1297 | 977 | 1237 | RAD51C - FANCD2OS | 1729 | 779 | 2596 |
| RAD54B - FANCD2OS | 475 | 1367 | 2571 | RAD54B - FANCD2OS | 2032 | 1241 | 1637 |
| RAD54L - FANCD2OS | 2557 | 1227 | 252 | RAD54L - FANCD2OS | 1671 | 1830 | 1839 |
| Ranking of RAD family w.r.t FANCF | Ranking of FANCF w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCF | 1817 | 1582 | 285 | RAD1 - FANCF | 529 | 2198 | 1997 |
| RAD18 - FANCF | 770 | 1329 | 1445 | RAD18 - FANCF | 1063 | 2186 | 196 |
| RAD50 - FANCF | 1403 | 1684 | 803 | RAD50 - FANCF | 2205 | 1419 | 1676 |
| RAD51 - FANCF | 209 | 1247 | 2221 | RAD51 - FANCF | 1222 | 1060 | 2251 |
| RAD51AP1 - FANCF | 1681 | 13 | 2619 | RAD51AP1 - FANCF | 1963 | 2372 | 107 |
| RAD51C - FANCF | 1493 | 224 | 2051 | RAD51C - FANCF | 2062 | 1904 | 2386 |
| RAD54B - FANCF | 401 | 143 | 2359 | RAD54B - FANCF | 1903 | 1936 | 2026 |
| RAD54L - FANCF | 690 | 829 | 2120 | RAD54L - FANCF | 2529 | 716 | 1262 |
| Ranking of RAD family w.r.t FANCG | Ranking of FANCG w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCG | 2013 | 2215 | 2328 | RAD1 - FANCG | 1938 | 825 | 843 |
| RAD18 - FANCG | 755 | 393 | 82 | RAD18 - FANCG | 2352 | 878 | 2574 |
| RAD50 - FANCG | 2652 | 2408 | 2663 | RAD50 - FANCG | 695 | 511 | 933 |
| RAD51 - FANCG | 345 | 114 | 295 | RAD51 - FANCG | 2163 | 1 | 397 |
| RAD51AP1 - FANCG | 1743 | 749 | 1984 | RAD51AP1 - FANCG | 661 | 400 | 23 |
| RAD51C - FANCG | 957 | 218 | 1360 | RAD51C - FANCG | 450 | 2319 | 1122 |
| RAD54B - FANCG | 17 | 182 | 423 | RAD54B - FANCG | 140 | 194 | 64 |
| RAD54L - FANCG | 1058 | 701 | 581 | RAD54L - FANCG | 2167 | 1968 | 2344 |
| Ranking of RAD family w.r.t FANCI | Ranking of FANCI w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - FANCI | 1919 | 2263 | 2286 | RAD1 - FANCI | 2496 | 897 | 664 |
| RAD18 - FANCI | 1693 | 2466 | 436 | RAD18 - FANCI | 1601 | 1161 | 1668 |
| RAD50 - FANCI | 1703 | 2074 | 1458 | RAD50 - FANCI | 1133 | 1211 | 1238 |
| RAD51 - FANCI | 1038 | 1668 | 310 | RAD51 - FANCI | 1612 | 2724 | 1187 |
| RAD51AP1 - FANCI | 2496 | 2517 | 383 | RAD51AP1 - FANCI | 1513 | 1211 | 65 |
| RAD51C - FANCI | 597 | 165 | 2447 | RAD51C - FANCI | 143 | 137 | 87 |
| RAD54B - FANCI | 557 | 84 | 2055 | RAD54B - FANCI | 178 | 350 | 76 |
| RAD54L - FANCI | 468 | 606 | 2461 | RAD54L - FANCI | 211 | 2304 | 1128 |
3.5. Telomerase Related Synergies
3.5.1. TERT - ABC Transporters Cross Family Analysis
3.6. ABC Transporter Related Synergies
3.6.1. ABC Transporters - UBE2 Cross Family Analysis
3.6.2. ABC Transporters Intra Cross Family Analysis
3.6.3. Interleukin - ABC Transporters Cross Family Analysis
| Ranking ABC family VS IL family | |||||||
| Ranking of IL family w.r.t ABCA5 | Ranking of ABCA5 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCA5 | 705 | 95 | 11 | IL1A - ABCA5 | 677 | 2069 | 871 |
| IL1B - ABCA5 | 240 | 35 | 353 | IL1B - ABCA5 | 2069 | 790 | 2301 |
| IL1RAP - ABCA5 | 1515 | 2354 | 514 | IL1RAP - ABCA5 | 1763 | 335 | 2345 |
| IL1RN - ABCA5 | 771 | 1093 | 1417 | IL1RN - ABCA5 | 892 | 2252 | 1482 |
| IL2RG - ABCA5 | 500 | 246 | 173 | IL2RG - ABCA5 | 993 | 750 | 1745 |
| IL6ST - ABCA5 | 2464 | 1564 | 1365 | IL6ST - ABCA5 | 155 | 266 | 1386 |
| IL8 - ABCA5 | 1676 | 1568 | 1111 | IL8 - ABCA5 | 104 | 1261 | 946 |
| IL10RB - ABCA5 | 492 | 146 | 643 | IL10RB - ABCA5 | 2230 | 2184 | 2240 |
| IL15 - ABCA5 | 638 | 1169 | 65 | IL15 - ABCA5 | 661 | 169 | 711 |
| IL15RA - ABCA5 | 2151 | 1672 | 740 | IL15RA - ABCA5 | 706 | 1300 | 2031 |
| IL17C - ABCA5 | 680 | 197 | 164 | IL17C - ABCA5 | 615 | 575 | 1518 |
| IL17REL - ABCA5 | 1014 | 2405 | 2202 | IL17REL - ABCA5 | 212 | 1024 | 146 |
| Ranking of IL family w.r.t ABCB11 | Ranking of ABCB11 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCB11 | 1962 | 465 | 648 | IL1A - ABCB11 | 551 | 140 | 385 |
| IL1B - ABCB11 | 1778 | 851 | 438 | IL1B - ABCB11 | 255 | 428 | 208 |
| IL1RAP - ABCB11 | 1427 | 1704 | 1318 | IL1RAP - ABCB11 | 1681 | 878 | 1709 |
| IL1RN - ABCB11 | 1832 | 539 | 297 | IL1RN - ABCB11 | 342 | 1912 | 779 |
| IL2RG - ABCB11 | 2182 | 2102 | 550 | IL2RG - ABCB11 | 814 | 67 | 584 |
| IL6ST - ABCB11 | 1793 | 2140 | 1938 | IL6ST - ABCB11 | 1347 | 1504 | 385 |
| IL8 - ABCB11 | 1607 | 2441 | 1028 | IL8 - ABCB11 | 349 | 846 | 1786 |
| IL10RB - ABCB11 | 341 | 1119 | 449 | IL10RB - ABCB11 | 2101 | 2419 | 1352 |
| IL15 - ABCB11 | 2438 | 2512 | 576 | IL15 - ABCB11 | 344 | 224 | 256 |
| IL15RA - ABCB11 | 2271 | 1288 | 1784 | IL15RA - ABCB11 | 1052 | 48 | 719 |
| IL17C - ABCB11 | 1262 | 69 | 706 | IL17C - ABCB11 | 653 | 316 | 437 |
| IL17REL - ABCB11 | 50 | 305 | 783 | IL17REL - ABCB11 | 1004 | 736 | 896 |
| Ranking of IL family w.r.t ABCC3 | Ranking of ABCC3 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCC3 | 1860 | 758 | 1538 | IL1A - ABCC3 | 1343 | 1798 | 2459 |
| IL1B - ABCC3 | 1764 | 749 | 896 | IL1B - ABCC3 | 1647 | 1369 | 569 |
| IL1RAP - ABCC3 | 1514 | 2294 | 1989 | IL1RAP - ABCC3 | 2074 | 1377 | 303 |
| IL1RN - ABCC3 | 647 | 607 | 1252 | IL1RN - ABCC3 | 1366 | 975 | 1354 |
| IL2RG - ABCC3 | 990 | 444 | 40 | IL2RG - ABCC3 | 1229 | 379 | 844 |
| IL6ST - ABCC3 | 98 | 1589 | 339 | IL6ST - ABCC3 | 970 | 712 | 1342 |
| IL8 - ABCC3 | 1767 | 1046 | 2419 | IL8 - ABCC3 | 937 | 1033 | 430 |
| IL10RB - ABCC3 | 1354 | 78 | 359 | IL10RB - ABCC3 | 1609 | 29 | 1830 |
| IL15 - ABCC3 | 1580 | 602 | 1560 | IL15 - ABCC3 | 1087 | 1191 | 1084 |
| IL15RA - ABCC3 | 189 | 2403 | 1795 | IL15RA - ABCC3 | 2153 | 163 | 1324 |
| IL17C - ABCC3 | 1587 | 778 | 2425 | IL17C - ABCC3 | 466 | 631 | 2237 |
| IL17REL - ABCC3 | 1135 | 403 | 54 | IL17REL - ABCC3 | 2089 | 2388 | 1618 |
| Ranking of IL family w.r.t ABCC5 | Ranking of ABCC5 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCC5 | 2004 | 681 | 60 | IL1A - ABCC5 | 2217 | 2022 | 1512 |
| IL1B - ABCC5 | 1948 | 112 | 251 | IL1B - ABCC5 | 1223 | 2137 | 942 |
| IL1RAP - ABCC5 | 1038 | 355 | 2023 | IL1RAP - ABCC5 | 1982 | 1892 | 2296 |
| IL1RN - ABCC5 | 709 | 430 | 1087 | IL1RN - ABCC5 | 1668 | 816 | 2142 |
| IL2RG - ABCC5 | 1421 | 264 | 601 | IL2RG - ABCC5 | 500 | 2018 | 1691 |
| IL6ST - ABCC5 | 1569 | 2010 | 845 | IL6ST - ABCC5 | 754 | 2326 | 874 |
| IL8 - ABCC5 | 1869 | 143 | 1589 | IL8 - ABCC5 | 855 | 1211 | 2434 |
| IL10RB - ABCC5 | 1162 | 70 | 434 | IL10RB - ABCC5 | 1337 | 736 | 958 |
| IL15 - ABCC5 | 1262 | 147 | 389 | IL15 - ABCC5 | 1947 | 1991 | 1584 |
| IL15RA - ABCC5 | 1083 | 2255 | 1861 | IL15RA - ABCC5 | 2457 | 1444 | 534 |
| IL17C - ABCC5 | 2447 | 96 | 116 | IL17C - ABCC5 | 1836 | 845 | 1802 |
| IL17REL - ABCC5 | 54 | 2462 | 2509 | IL17REL - ABCC5 | 1247 | 2149 | 1031 |
| Ranking ABC family VS IL family | |||||||
| Ranking of IL family w.r.t ABCC13 | Ranking of ABCC13 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCC13 | 512 | 135 | 1207 | IL1A - ABCC13 | 464 | 352 | 201 |
| IL1B - ABCC13 | 152 | 103 | 1553 | IL1B - ABCC13 | 635 | 974 | 60 |
| IL1RAP - ABCC13 | 1092 | 502 | 2442 | IL1RAP - ABCC13 | 2136 | 2392 | 33 |
| IL1RN - ABCC13 | 1753 | 559 | 323 | IL1RN - ABCC13 | 114 | 1016 | 1839 |
| IL2RG - ABCC13 | 2064 | 674 | 1076 | IL2RG - ABCC13 | 807 | 1079 | 938 |
| IL6ST - ABCC13 | 332 | 1416 | 2112 | IL6ST - ABCC13 | 119 | 1098 | 2323 |
| IL8 - ABCC13 | 551 | 1200 | 1680 | IL8 - ABCC13 | 592 | 984 | 907 |
| IL10RB - ABCC13 | 631 | 621 | 561 | IL10RB - ABCC13 | 2011 | 1272 | 1297 |
| IL15 - ABCC13 | 502 | 296 | 373 | IL15 - ABCC13 | 612 | 968 | 170 |
| IL15RA - ABCC13 | 2248 | 1955 | 2456 | IL15RA - ABCC13 | 2397 | 2485 | 790 |
| IL17C - ABCC13 | 25 | 140 | 123 | IL17C - ABCC13 | 924 | 308 | 711 |
| IL17REL - ABCC13 | 2339 | 2137 | 1497 | IL17REL - ABCC13 | 462 | 376 | 461 |
| Ranking of IL family w.r.t ABCC5 | Ranking of ABCC5 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCD1 | 1932 | 30 | 2203 | IL1A - ABCD1 | 530 | 2046 | 1196 |
| IL1B - ABCD1 | 569 | 109 | 1778 | IL1B - ABCD1 | 1400 | 605 | 453 |
| IL1RAP - ABCD1 | 2508 | 2006 | 1907 | IL1RAP - ABCD1 | 399 | 840 | 1548 |
| IL1RN - ABCD1 | 606 | 2003 | 789 | IL1RN - ABCD1 | 551 | 2025 | 60 |
| IL2RG - ABCD1 | 1064 | 284 | 2374 | IL2RG - ABCD1 | 311 | 1233 | 1322 |
| IL6ST - ABCD1 | 1347 | 1237 | 1220 | IL6ST - ABCD1 | 1581 | 507 | 612 |
| IL8 - ABCD1 | 2010 | 2315 | 1814 | IL8 - ABCD1 | 2501 | 2154 | 539 |
| IL10RB - ABCD1 | 631 | 825 | 85 | IL10RB - ABCD1 | 1795 | 1028 | 2325 |
| IL15 - ABCD1 | 890 | 325 | 1578 | IL15 - ABCD1 | 1795 | 302 | 1258 |
| IL15RA - ABCD1 | 2097 | 1765 | 1629 | IL15RA - ABCD1 | 580 | 1240 | 2342 |
| IL17C - ABCD1 | 1372 | 56 | 2509 | IL17C - ABCD1 | 687 | 1753 | 851 |
| IL17REL - ABCD1 | 5 | 2388 | 237 | IL17REL - ABCD1 | 1423 | 642 | 2164 |
| Ranking of IL family w.r.t ABCG1 | Ranking of ABCG1 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCG1 | 724 | 67 | 80 | IL1A - ABCG1 | 699 | 824 | 600 |
| IL1B - ABCG1 | 938 | 178 | 533 | IL1B - ABCG1 | 70 | 783 | 81 |
| IL1RAP - ABCG1 | 1263 | 2205 | 2339 | IL1RAP - ABCG1 | 2298 | 394 | 612 |
| IL1RN - ABCG1 | 1240 | 688 | 1396 | IL1RN - ABCG1 | 2465 | 834 | 1051 |
| IL2RG - ABCG1 | 1396 | 7 | 112 | IL2RG - ABCG1 | 587 | 24 | 21 |
| IL6ST - ABCG1 | 357 | 845 | 520 | IL6ST - ABCG1 | 1723 | 1345 | 177 |
| IL8 - ABCG1 | 977 | 1835 | 1099 | IL8 - ABCG1 | 1730 | 1748 | 382 |
| IL10RB - ABCG1 | 2244 | 349 | 840 | IL10RB - ABCG1 | 167 | 1315 | 61 |
| IL15 - ABCG1 | 1960 | 613 | 1279 | IL15 - ABCG1 | 2212 | 734 | 326 |
| IL15RA - ABCG1 | 785 | 651 | 2191 | IL15RA - ABCG1 | 1195 | 862 | 1876 |
| IL17C - ABCG1 | 2516 | 486 | 51 | IL17C - ABCG1 | 80 | 95 | 177 |
| IL17REL - ABCG1 | 2229 | 732 | 150 | IL17REL - ABCG1 | 1579 | 1025 | 452 |
| Ranking of IL family w.r.t ABCG2 | Ranking of ABCG2 family w.r.t IL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - ABCG2 | 745 | 716 | 1299 | IL1A - ABCG2 | 238 | 89 | 659 |
| IL1B - ABCG2 | 354 | 232 | 668 | IL1B - ABCG2 | 31 | 197 | 439 |
| IL1RAP - ABCG2 | 2184 | 2167 | 1384 | IL1RAP - ABCG2 | 1314 | 253 | 2434 |
| IL1RN - ABCG2 | 783 | 228 | 11 | IL1RN - ABCG2 | 552 | 1692 | 827 |
| IL2RG - ABCG2 | 444 | 463 | 1024 | IL2RG - ABCG2 | 261 | 87 | 1275 |
| IL6ST - ABCG2 | 1647 | 1827 | 55 | IL6ST - ABCG2 | 1792 | 1477 | 1222 |
| IL8 - ABCG2 | 2212 | 1362 | 563 | IL8 - ABCG2 | 448 | 441 | 1423 |
| IL10RB - ABCG2 | 31 | 80 | 667 | IL10RB - ABCG2 | 2144 | 2335 | 2434 |
| IL15 - ABCG2 | 76 | 312 | 187 | IL15 - ABCG2 | 247 | 590 | 832 |
| IL15RA - ABCG2 | 1910 | 2428 | 1921 | IL15RA - ABCG2 | 1116 | 1005 | 1059 |
| IL17C - ABCG2 | 649 | 692 | 61 | IL17C - ABCG2 | 784 | 462 | 775 |
| IL17REL - ABCG2 | 883 | 1435 | 35 | IL17REL - ABCG2 | 852 | 1606 | 1597 |
3.6.4. BCL - ABC Transporters Cross Family Analysis
| Ranking BCL family VS ABC family | |||||||
| Ranking of BCL6 w.r.t ABC family | Ranking of ABC family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL6 | 2045 | 557 | 1384 | ABCA5 - BCL6 | 211 | 283 | 1615 |
| ABCB11 - BCL6 | 1611 | 2010 | 2350 | ABCB11 - BCL6 | 841 | 427 | 2320 |
| ABCC3 - BCL6 | 1895 | 983 | 958 | ABCC3 - BCL6 | 1084 | 570 | 594 |
| ABCC5 - BCL6 | 615 | 597 | 567 | ABCC5 - BCL6 | 1370 | 1841 | 2389 |
| ABCC13 - BCL6 | 1097 | 2431 | 1731 | ABCC13 - BCL6 | 2172 | 2456 | 1063 |
| ABCD1 - BCL6 | 1446 | 1139 | 1953 | ABCD1 - BCL6 | 1097 | 1297 | 827 |
| ABCG1 - BCL6 | 1462 | 1688 | 1918 | ABCG1 - BCL6 | 192 | 27 | 1111 |
| ABCG2 - BCL6 | 947 | 1503 | 978 | ABCG2 - BCL6 | 129 | 745 | 719 |
| Ranking of BCL9L w.r.t ABC family | Ranking of ABC family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL9L | 67 | 1008 | 94 | ABCA5 - BCL9L | 1753 | 1167 | 2312 |
| ABCB11 - BCL9L | 1989 | 158 | 1705 | ABCB11 - BCL9L | 1033 | 494 | 48 |
| ABCC3 - BCL9L | 1307 | 2249 | 1357 | ABCC3 - BCL9L | 457 | 2296 | 971 |
| ABCC5 - BCL9L | 1694 | 432 | 477 | ABCC5 - BCL9L | 1775 | 1551 | 2073 |
| ABCC13 - BCL9L | 1724 | 1410 | 862 | ABCC13 - BCL9L | 110 | 2475 | 2325 |
| ABCD1 - BCL9L | 1366 | 2344 | 1666 | ABCD1 - BCL9L | 1016 | 2440 | 2411 |
| ABCG1 - BCL9L | 1248 | 1680 | 536 | ABCG1 - BCL9L | 1146 | 676 | 16 |
| ABCG2 - BCL9L | 2451 | 1119 | 224 | ABCG2 - BCL9L | 1263 | 1421 | 218 |
| Ranking of BCL10 w.r.t ABC family | Ranking of ABC family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL10 | 687 | 176 | 808 | ABCA5 - BCL10 | 1753 | 1167 | 2312 |
| ABCB11 - BCL10 | 2234 | 2382 | 322 | ABCB11 - BCL10 | 1033 | 494 | 48 |
| ABCC3 - BCL10 | 589 | 379 | 492 | ABCC3 - BCL10 | 457 | 2296 | 971 |
| ABCC5 - BCL10 | 1489 | 397 | 1643 | ABCC5 - BCL10 | 1775 | 1551 | 2073 |
| ABCC13 - BCL10 | 956 | 538 | 1491 | ABCC13 - BCL10 | 110 | 2475 | 2325 |
| ABCD1 - BCL10 | 1009 | 470 | 1597 | ABCD1 - BCL10 | 1016 | 2440 | 2411 |
| ABCG1 - BCL10 | 1613 | 310 | 1115 | ABCG1 - BCL10 | 1146 | 676 | 16 |
| ABCG2 - BCL10 | 361 | 676 | 2020 | ABCG2 - BCL10 | 1263 | 1421 | 218 |
3.6.5. CASPASE - ABC Transporters Cross Family Analysis
| Ranking ABC family w.r.t CASP family | |||||||
| Ranking of ABC family w.r.t CASP4 | Ranking of ABC family w.r.t CASP5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - ABC-A5 | 957 | 682 | 991 | CASP5 - ABC-A5 | 733 | 1986 | 421 |
| CASP4 - ABC-B11 | 19 | 727 | 158 | CASP5 - ABC-B11 | 513 | 406 | 355 |
| CASP4 - ABC-C3 | 1242 | 857 | 1848 | CASP5 - ABC-C3 | 685 | 1694 | 1558 |
| CASP4 - ABC-C5 | 2495 | 1316 | 2257 | CASP5 - ABC-C5 | 2475 | 1038 | 2234 |
| CASP4 - ABC-C13 | 154 | 1537 | 1206 | CASP5 - ABC-C13 | 1660 | 1581 | 853 |
| CASP4 - ABC-D1 | 1494 | 964 | 999 | CASP5 - ABC-D1 | 354 | 725 | 1304 |
| CASP4 - ABC-G1 | 1405 | 70 | 326 | CASP5 - ABC-G1 | 298 | 485 | 382 |
| CASP4 - ABC-G2 | 157 | 176 | 523 | CASP5 - ABC-G2 | 706 | 846 | 1598 |
| Ranking of ABC family w.r.t CASP7 | Ranking of ABC family w.r.t CASP9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - ABC-A5 | 2515 | 1742 | 25 | CASP9 - ABC-A5 | 1125 | 1863 | 694 |
| CASP7 - ABC-B11 | 1299 | 207 | 348 | CASP9 - ABC-B11 | 729 | 2001 | 2051 |
| CASP7 - ABC-C3 | 992 | 511 | 2222 | CASP9 - ABC-C3 | 1108 | 1470 | 1465 |
| CASP7 - ABC-C5 | 1232 | 1449 | 2154 | CASP9 - ABC-C5 | 2180 | 2343 | 1732 |
| CASP7 - ABC-C13 | 2489 | 2418 | 1623 | CASP9 - ABC-C13 | 2267 | 1472 | 2382 |
| CASP7 - ABC-D1 | 1544 | 2323 | 2004 | CASP9 - ABC-D1 | 1011 | 1086 | 174 |
| CASP7 - ABC-G1 | 665 | 382 | 670 | CASP9 - ABC-G1 | 580 | 1890 | 2286 |
| CASP7 - ABC-G2 | 1930 | 23 | 963 | CASP9 - ABC-G2 | 647 | 2374 | 310 |
| Ranking of ABC family w.r.t CASP10 | Ranking of ABC family w.r.t CASP16 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - ABC-A5 | 2292 | 2311 | 1108 | CASP16 - ABC-A5 | 165 | 408 | 113 |
| CASP10 - ABC-B11 | 2245 | 1467 | 1182 | CASP16 - ABC-B11 | 495 | 949 | 1417 |
| CASP10 - ABC-C3 | 760 | 2479 | 923 | CASP16 - ABC-C3 | 50 | 4 | 556 |
| CASP10 - ABC-C5 | 326 | 485 | 1429 | CASP16 - ABC-C5 | 1635 | 2487 | 1309 |
| CASP10 - ABC-C13 | 2139 | 2203 | 1524 | CASP16 - ABC-C13 | 1517 | 936 | 1236 |
| CASP10 - ABC-D1 | 2210 | 475 | 1655 | CASP16 - ABC-D1 | 1029 | 1210 | 1285 |
| CASP10 - ABC-G1 | 2337 | 128 | 71 | CASP16 - ABC-G1 | 350 | 756 | 109 |
| CASP10 - ABC-G2 | 2075 | 1693 | 1306 | CASP16 - ABC-G2 | 318 | 476 | 515 |
| Ranking CASP family w.r.t ABC family | |||||||
| Ranking of CASP4 w.r.t ABC family | Ranking of CASP5 w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - ABC-A5 | 791 | 586 | 753 | CASP5 - ABC-A5 | 696 | 427 | 48 |
| CASP4 - ABC-B11 | 462 | 263 | 427 | CASP5 - ABC-B11 | 1470 | 1300 | 242 |
| CASP4 - ABC-C3 | 1013 | 54 | 1140 | CASP5 - ABC-C3 | 821 | 286 | 459 |
| CASP4 - ABC-C5 | 2396 | 26 | 209 | CASP5 - ABC-C5 | 2368 | 665 | 171 |
| CASP4 - ABC-C13 | 1305 | 775 | 2193 | CASP5 - ABC-C13 | 2286 | 739 | 1905 |
| CASP4 - ABC-D1 | 1791 | 591 | 1954 | CASP5 - ABC-D1 | 653 | 440 | 972 |
| CASP4 - ABC-G1 | 593 | 99 | 173 | CASP5 - ABC-G1 | 2176 | 446 | 317 |
| CASP4 - ABC-G2 | 423 | 109 | 1364 | CASP5 - ABC-G2 | 332 | 122 | 533 |
| Ranking of CASP7 w.r.t ABC family | Ranking of CASP9 w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP7 - ABC-A5 | 1726 | 697 | 1874 | CASP9 - ABC-A5 | 2404 | 2374 | 1265 |
| CASP7 - ABC-B11 | 1549 | 189 | 1692 | CASP9 - ABC-B11 | 998 | 1258 | 2046 |
| CASP7 - ABC-C3 | 2331 | 1572 | 69 | CASP9 - ABC-C3 | 1398 | 2358 | 1445 |
| CASP7 - ABC-C5 | 2168 | 1881 | 2016 | CASP9 - ABC-C5 | 1023 | 965 | 1080 |
| CASP7 - ABC-C13 | 1822 | 865 | 1239 | CASP9 - ABC-C13 | 2449 | 1545 | 2506 |
| CASP7 - ABC-D1 | 111 | 813 | 2230 | CASP9 - ABC-D1 | 1858 | 2430 | 412 |
| CASP7 - ABC-G1 | 1609 | 983 | 1994 | CASP9 - ABC-G1 | 305 | 2342 | 2468 |
| CASP7 - ABC-G2 | 1094 | 952 | 102 | CASP9 - ABC-G2 | 1868 | 1621 | 1154 |
| Ranking of CASP10 w.r.t ABC family | Ranking of CASP16 w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP10 - ABC-A5 | 683 | 1453 | 1437 | CASP16 - ABC-A5 | 960 | 2477 | 2315 |
| CASP10 - ABC-B11 | 1301 | 774 | 558 | CASP16 - ABC-B11 | 402 | 1860 | 794 |
| CASP10 - ABC-C3 | 369 | 683 | 1453 | CASP16 - ABC-C3 | 171 | 825 | 23 |
| CASP10 - ABC-C5 | 1823 | 346 | 761 | CASP16 - ABC-C5 | 2467 | 585 | 258 |
| CASP10 - ABC-C13 | 1320 | 832 | 868 | CASP16 - ABC-C13 | 428 | 177 | 64 |
| CASP10 - ABC-D1 | 249 | 1440 | 387 | CASP16 - ABC-D1 | 651 | 153 | 2010 |
| CASP10 - ABC-G1 | 1687 | 1232 | 156 | CASP16 - ABC-G1 | 2398 | 421 | 1120 |
| CASP10 - ABC-G2 | 1151 | 651 | 464 | CASP16 - ABC-G2 | 1193 | 734 | 479 |
| Unexplored combinatorial hypotheses | |
| ABC w.r.t CASP | |
| CASP-4 | ABC-C5 |
| CASP-5 | ABC-C5 |
| CASP-7 | ABC-A5/C13/D1 |
| CASP-9 | ABC-B11/C5/D1/G1 |
| CASP-10 | ABC-A5/C13 |
| CASP w.r.t ABC | |
| CASP-4 | ABC-D1 |
| CASP-5 | ABC-C13 |
| CASP-7 | ABC-C5 |
| CASP-9 | ABC-C5/C13/D1/G1 |
| CASP-16 | ABC-A5 |
3.7. Interleukin Related Synergies
3.7.1. NFkB-2/I - Interleukin Cross Family Analysis
| Ranking Interleukin family vs NFkB-2 family | |||||||
| Ranking of IL family w.r.t NFkB-2 | Ranking of NFkB-2/I w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - NFkB2 | 1485 | 6 | 2494 | IL1A - NFkB2 | 616 | 276 | 1358 |
| IL1B - NFkB2 | 1852 | 638 | 1587 | IL1B - NFkB2 | 283 | 284 | 1088 |
| IL1RAP - NFkB2 | 1369 | 1849 | 1463 | IL1RAP - NFkB2 | 967 | 377 | 161 |
| IL1RN - NFkB2 | 1285 | 1963 | 1604 | IL1RN - NFkB2 | 1386 | 2086 | 52 |
| IL2RG - NFkB2 | 486 | 1077 | 1300 | IL2RG - NFkB2 | 1436 | 1123 | 2163 |
| IL6ST - NFkB2 | 493 | 814 | 283 | IL6ST - NFkB2 | 2177 | 343 | 2255 |
| IL8 - NFkB2 | 1907 | 865 | 335 | IL8 - NFkB2 | 303 | 2355 | 1152 |
| IL10RB - NFkB2 | 707 | 1607 | 595 | IL10RB - NFkB2 | 2282 | 2381 | 1897 |
| IL15 - NFkB2 | 792 | 1113 | 1434 | IL15 - NFkB2 | 2112 | 1214 | 1217 |
| IL15RA - NFkB2 | 1787 | 233 | 1957 | IL15RA - NFkB2 | 1289 | 1235 | 1913 |
| IL17C - NFkB2 | 2288 | 305 | 2018 | IL17C - NFkB2 | 380 | 529 | 1492 |
| IL17REL - NFkB2 | 9 | 2464 | 167 | IL17REL - NFkB2 | 115 | 1540 | 308 |
| Ranking of IL family w.r.t NFkBI-A | Ranking of NFkB-2/I w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - NFkBIA | 116 | 46 | 1885 | IL1A - NFkBIA | 989 | 1179 | 705 |
| IL1B - NFkBIA | 328 | 56 | 1228 | IL1B - NFkBIA | 611 | 397 | 1378 |
| IL1RAP - NFkBIA | 1376 | 778 | 359 | IL1RAP - NFkBIA | 1131 | 515 | 1887 |
| IL1RN - NFkBIA | 1753 | 1906 | 267 | IL1RN - NFkBIA | 2357 | 578 | 382 |
| IL2RG - NFkBIA | 32 | 6 | 898 | IL2RG - NFkBIA | 132 | 684 | 784 |
| IL6ST - NFkBIA | 1011 | 2400 | 2094 | IL6ST - NFkBIA | 2008 | 533 | 90 |
| IL8 - NFkBIA | 1988 | 1234 | 1232 | IL8 - NFkBIA | 183 | 993 | 1109 |
| IL10RB - NFkBIA | 864 | 2239 | 8 | IL10RB - NFkBIA | 616 | 1251 | 107 |
| IL15 - NFkBIA | 1181 | 453 | 462 | IL15 - NFkBIA | 2227 | 958 | 165 |
| IL15RA - NFkBIA | 2251 | 2390 | 1652 | IL15RA - NFkBIA | 765 | 291 | 2301 |
| IL17C - NFkBIA | 538 | 229 | 330 | IL17C - NFkBIA | 450 | 178 | 19 |
| IL17REL - NFkBIA | 643 | 16 | 4 | IL17REL - NFkBIA | 1275 | 403 | 2190 |
| Ranking of IL family w.r.t NFkBI-E | Ranking of NFkBI-E w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - NFkBIE | 2486 | 27 | 76 | IL1A - NFkBIE | 433 | 1574 | 953 |
| IL1B - NFkBIE | 2089 | 39 | 311 | IL1B - NFkBIE | 1103 | 507 | 1931 |
| IL1RAP - NFkBIE | 201 | 2221 | 1807 | IL1RAP - NFkBIE | 474 | 1404 | 875 |
| IL1RN - NFkBIE | 2025 | 610 | 1153 | IL1RN - NFkBIE | 2051 | 381 | 468 |
| IL2RG - NFkBIE | 1141 | 986 | 654 | IL2RG - NFkBIE | 1327 | 1464 | 983 |
| IL6ST - NFkBIE | 1155 | 2381 | 2277 | IL6ST - NFkBIE | 309 | 143 | 939 |
| IL8 - NFkBIE | 259 | 2198 | 2133 | IL8 - NFkBIE | 1507 | 911 | 67 |
| IL10RB - NFkBIE | 1730 | 191 | 310 | IL10RB - NFkBIE | 305 | 478 | 1960 |
| IL15 - NFkBIE | 1922 | 365 | 117 | IL15 - NFkBIE | 2476 | 783 | 1302 |
| IL15RA - NFkBIE | 1912 | 839 | 1385 | IL15RA - NFkBIE | 424 | 526 | 1423 |
| IL17C - NFkBIE | 2179 | 105 | 404 | IL17C - NFkBIE | 2231 | 1205 | 321 |
| IL17REL - NFkBIE | 13 | 2216 | 2168 | IL17REL - NFkBIE | 333 | 831 | 949 |
| Ranking of IL family w.r.t NFkBI-Z | Ranking of NFkBI-Z w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - NFkBIZ | 2381 | 2049 | 1578 | IL1A - NFkBIZ | 157 | 792 | 1460 |
| IL1B - NFkBIZ | 1241 | 2210 | 463 | IL1B - NFkBIZ | 586 | 217 | 1617 |
| IL1RAP - NFkBIZ | 694 | 1077 | 936 | IL1RAP - NFkBIZ | 1326 | 240 | 1080 |
| IL1RN - NFkBIZ | 860 | 2151 | 231 | IL1RN - NFkBIZ | 2463 | 739 | 579 |
| IL2RG - NFkBIZ | 1362 | 2054 | 68 | IL2RG - NFkBIZ | 68 | 829 | 1212 |
| IL6ST - NFkBIZ | 2279 | 980 | 2431 | IL6ST - NFkBIZ | 996 | 1223 | 140 |
| IL8 - NFkBIZ | 992 | 1732 | 966 | IL8 - NFkBIZ | 816 | 1510 | 119 |
| IL10RB - NFkBIZ | 717 | 2275 | 571 | IL10RB - NFkBIZ | 2271 | 42 | 2082 |
| IL15 - NFkBIZ | 1780 | 2098 | 626 | IL15 - NFkBIZ | 2155 | 200 | 245 |
| IL15RA - NFkBIZ | 633 | 1726 | 2422 | IL15RA - NFkBIZ | 834 | 1284 | 1785 |
| IL17C - NFkBIZ | 1716 | 2430 | 1098 | IL17C - NFkBIZ | 848 | 1282 | 1391 |
| IL17REL - NFkBIZ | 14 | 75 | 314 | IL17REL - NFkBIZ | 289 | 1883 | 1830 |
3.7.2. Potassium Channel - Interleukin Cross Family Analysis
3.7.3. Mucin - Interleukin Cross Family Analysis
| Ranking Interleukin family vs MUC family | |||||||
| Ranking of IL family w.r.t MUC1 | Ranking of MUC1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC1 | 1961 | 1711 | 107 | IL1A - MUC1 | 111 | 879 | 535 |
| IL1B - MUC1 | 2218 | 1757 | 228 | IL1B - MUC1 | 1847 | 520 | 2049 |
| IL1RAP - MUC1 | 837 | 604 | 146 | IL1RAP - MUC1 | 1968 | 589 | 439 |
| IL1RN - MUC1 | 1084 | 918 | 1859 | IL1RN - MUC1 | 1752 | 353 | 507 |
| IL2RG - MUC1 | 1872 | 272 | 1281 | IL2RG - MUC1 | 1769 | 1009 | 285 |
| IL6ST - MUC1 | 2415 | 1115 | 1633 | IL6ST - MUC1 | 296 | 801 | 245 |
| IL8 - MUC1 | 1276 | 544 | 1055 | IL8 - MUC1 | 2079 | 1320 | 82 |
| IL10RB - MUC1 | 291 | 1638 | 1710 | IL10RB - MUC1 | 973 | 1691 | 924 |
| IL15 - MUC1 | 212 | 1003 | 1060 | IL15 - MUC1 | 160 | 205 | 942 |
| IL15RA - MUC1 | 213 | 1346 | 1067 | IL15RA - MUC1 | 1127 | 1057 | 1521 |
| IL17C - MUC1 | 1215 | 1841 | 2003 | IL17C - MUC1 | 3 | 236 | 7 |
| IL17REL - MUC1 | 19 | 44 | 2069 | IL17REL - MUC1 | 1142 | 541 | 1464 |
| Ranking of IL family w.r.t MUC3A | Ranking of MUC3A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC3A | 2513 | 2480 | 194 | IL1A - MUC3A | 1426 | 1017 | 1484 |
| IL1B - MUC3A | 1820 | 2308 | 1086 | IL1B - MUC3A | 816 | 1157 | 908 |
| IL1RAP - MUC3A | 753 | 1270 | 526 | IL1RAP - MUC3A | 1403 | 1402 | 102 |
| IL1RN - MUC3A | 2138 | 2270 | 313 | IL1RN - MUC3A | 1123 | 360 | 1333 |
| IL2RG - MUC3A | 1816 | 2115 | 1900 | IL2RG - MUC3A | 480 | 1560 | 514 |
| IL6ST - MUC3A | 283 | 1126 | 1229 | IL6ST - MUC3A | 1601 | 908 | 889 |
| IL8 - MUC3A | 356 | 760 | 1517 | IL8 - MUC3A | 2350 | 587 | 80 |
| IL10RB - MUC3A | 1401 | 729 | 157 | IL10RB - MUC3A | 520 | 458 | 2324 |
| IL15 - MUC3A | 850 | 2391 | 2288 | IL15 - MUC3A | 1385 | 1351 | 959 |
| IL15RA - MUC3A | 1304 | 1949 | 959 | IL15RA - MUC3A | 1538 | 1685 | 584 |
| IL17C - MUC3A | 2443 | 2512 | 647 | IL17C - MUC3A | 2153 | 623 | 1349 |
| IL17REL - MUC3A | 200 | 243 | 2048 | IL17REL - MUC3A | 1274 | 1250 | 1387 |
| Ranking of IL family w.r.t MUC4 | Ranking of MUC4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC4 | 1268 | 489 | 112 | IL1A - MUC4 | 42 | 1449 | 331 |
| IL1B - MUC4 | 779 | 1142 | 393 | IL1B - MUC4 | 780 | 301 | 393 |
| IL1RAP - MUC4 | 1672 | 1203 | 926 | IL1RAP - MUC4 | 460 | 358 | 883 |
| IL1RN - MUC4 | 2010 | 438 | 1960 | IL1RN - MUC4 | 1681 | 1164 | 51 |
| IL2RG - MUC4 | 161 | 292 | 36 | IL2RG - MUC4 | 581 | 659 | 1056 |
| IL6ST - MUC4 | 2204 | 1116 | 1765 | IL6ST - MUC4 | 977 | 1555 | 873 |
| IL8 - MUC4 | 619 | 741 | 1030 | IL8 - MUC4 | 222 | 1341 | 1552 |
| IL10RB - MUC4 | 1818 | 1343 | 599 | IL10RB - MUC4 | 87 | 1511 | 95 |
| IL15 - MUC4 | 434 | 1268 | 602 | IL15 - MUC4 | 440 | 806 | 276 |
| IL15RA - MUC4 | 2190 | 1814 | 2061 | IL15RA - MUC4 | 427 | 1145 | 305 |
| IL17C - MUC4 | 255 | 60 | 558 | IL17C - MUC4 | 167 | 152 | 159 |
| IL17REL - MUC4 | 222 | 482 | 52 | IL17REL - MUC4 | 2266 | 419 | 160 |
| Ranking of IL family w.r.t MUC12 | Ranking of MUC12 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC12 | 1806 | 166 | 2396 | IL1A - MUC12 | 706 | 84 | 570 |
| IL1B - MUC12 | 1004 | 113 | 2086 | IL1B - MUC12 | 1352 | 167 | 445 |
| IL1RAP - MUC12 | 1906 | 1588 | 517 | IL1RAP - MUC12 | 52 | 272 | 1955 |
| IL1RN - MUC12 | 2209 | 669 | 235 | IL1RN - MUC12 | 2505 | 1891 | 567 |
| IL2RG - MUC12 | 2195 | 751 | 2089 | IL2RG - MUC12 | 1913 | 1833 | 939 |
| IL6ST - MUC12 | 1115 | 1522 | 1031 | IL6ST - MUC12 | 2100 | 1759 | 1508 |
| IL8 - MUC12 | 1814 | 1554 | 2497 | IL8 - MUC12 | 439 | 121 | 1635 |
| IL10RB - MUC12 | 2467 | 1114 | 1044 | IL10RB - MUC12 | 381 | 1863 | 12 |
| IL15 - MUC12 | 2408 | 192 | 2340 | IL15 - MUC12 | 2400 | 1307 | 1408 |
| IL15RA - MUC12 | 612 | 1636 | 203 | IL15RA - MUC12 | 137 | 127 | 468 |
| IL17C - MUC12 | 2436 | 484 | 2416 | IL17C - MUC12 | 411 | 182 | 283 |
| IL17REL - MUC12 | 2421 | 331 | 611 | IL17REL - MUC12 | 1452 | 678 | 651 |
| Ranking Interleukin family vs MUC family contd. | |||||||
| Ranking of IL family w.r.t MUC13 | Ranking of MUC13 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC13 | 655 | 2323 | 826 | IL1A - MUC13 | 1176 | 148 | 803 |
| IL1B - MUC13 | 2250 | 298 | 185 | IL1B - MUC13 | 833 | 30 | 8 |
| IL1RAP - MUC13 | 386 | 490 | 360 | IL1RAP - MUC13 | 1887 | 1142 | 2263 |
| IL1RN - MUC13 | 904 | 1614 | 698 | IL1RN - MUC13 | 1749 | 1607 | 313 |
| IL2RG - MUC13 | 1043 | 59 | 27 | IL2RG - MUC13 | 434 | 852 | 1140 |
| IL6ST - MUC13 | 635 | 1774 | 730 | IL6ST - MUC13 | 1901 | 535 | 163 |
| IL8 - MUC13 | 225 | 510 | 1130 | IL8 - MUC13 | 2328 | 722 | 555 |
| IL10RB - MUC13 | 944 | 491 | 1631 | IL10RB - MUC13 | 1459 | 1841 | 342 |
| IL15 - MUC13 | 1773 | 609 | 1047 | IL15 - MUC13 | 315 | 465 | 302 |
| IL15RA - MUC13 | 1884 | 1360 | 1067 | IL15RA - MUC13 | 2109 | 158 | 2402 |
| IL17C - MUC13 | 562 | 106 | 149 | IL17C - MUC13 | 73 | 4 | 84 |
| IL17REL - MUC13 | 1808 | 83 | 59 | IL17REL - MUC13 | 694 | 676 | 586 |
| Ranking of IL family w.r.t MUC17 | Ranking of MUC17 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC17 | 1573 | 2431 | 1622 | IL1A - MUC17 | 881 | 311 | 254 |
| IL1B - MUC17 | 1122 | 514 | 1035 | IL1B - MUC17 | 676 | 1243 | 174 |
| IL1RAP - MUC17 | 1634 | 1148 | 1469 | IL1RAP - MUC17 | 136 | 369 | 2512 |
| IL1RN - MUC17 | 38 | 260 | 911 | IL1RN - MUC17 | 361 | 22 | 690 |
| IL2RG - MUC17 | 754 | 218 | 403 | IL2RG - MUC17 | 1379 | 530 | 177 |
| IL6ST - MUC17 | 1616 | 554 | 1381 | IL6ST - MUC17 | 1782 | 668 | 270 |
| IL8 - MUC17 | 241 | 583 | 402 | IL8 - MUC17 | 1612 | 436 | 1984 |
| IL10RB - MUC17 | 401 | 464 | 51 | IL10RB - MUC17 | 1707 | 1305 | 1857 |
| IL15 - MUC17 | 307 | 438 | 878 | IL15 - MUC17 | 466 | 366 | 596 |
| IL15RA - MUC17 | 2265 | 2064 | 1458 | IL15RA - MUC17 | 63 | 376 | 849 |
| IL17C - MUC17 | 1045 | 581 | 2291 | IL17C - MUC17 | 1530 | 285 | 1449 |
| IL17REL - MUC17 | 656 | 657 | 456 | IL17REL - MUC17 | 380 | 580 | 1306 |
| Ranking of IL family w.r.t MUC20 | Ranking of MUC20 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - MUC20 | 103 | 1729 | 18 | IL1A - MUC20 | 2218 | 1499 | 2260 |
| IL1B - MUC20 | 85 | 1810 | 30 | IL1B - MUC20 | 1313 | 1719 | 735 |
| IL1RAP - MUC20 | 974 | 2025 | 2251 | IL1RAP - MUC20 | 1784 | 859 | 1705 |
| IL1RN - MUC20 | 1176 | 2264 | 246 | IL1RN - MUC20 | 1265 | 726 | 823 |
| IL2RG - MUC20 | 405 | 2168 | 335 | IL2RG - MUC20 | 2152 | 165 | 1400 |
| IL6ST - MUC20 | 1475 | 1093 | 2233 | IL6ST - MUC20 | 1743 | 203 | 1643 |
| IL8 - MUC20 | 1820 | 538 | 2303 | IL8 - MUC20 | 1875 | 883 | 488 |
| IL10RB - MUC20 | 394 | 1884 | 312 | IL10RB - MUC20 | 889 | 1883 | 1947 |
| IL15 - MUC20 | 244 | 2241 | 166 | IL15 - MUC20 | 1412 | 2057 | 1669 |
| IL15RA - MUC20 | 589 | 1406 | 1406 | IL15RA - MUC20 | 1450 | 1902 | 1570 |
| IL17C - MUC20 | 228 | 2278 | 46 | IL17C - MUC20 | 2212 | 1843 | 255 |
| IL17REL - MUC20 | 2121 | 962 | 2267 | IL17REL - MUC20 | 1130 | 1000 | 1868 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t MUC | |
| IL-1B/17C | MUC1 |
| IL-1A/1B/1RN/2RG/15/17C | MUC3A |
| IL-1RN/6ST/15RA | MUC4 |
| IL-1A/2RG/8/15/17C | MUC12 |
| IL-15RA | MUC17 |
| IL-1RAP/8/17REL | MUC20 |
| MUC w.r.t IL | |
| IL-1B | MUC1 |
| IL-1RN/2RG/6ST | MUC12 |
| IL-1RAP/15RA | MUC13 |
| IL-1A/10RB/17C | MUC20 |
3.7.4. Interleukin - TP53 Cross Family Analysis
| Ranking Interleukin family vs TP53 family | |||||||
| Ranking of IL family w.r.t TP53BP2 | Ranking of TP53BP2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TP53BP2 | 2396 | 1377 | 302 | IL1A - TP53BP2 | 390 | 2306 | 2483 |
| IL1B - TP53BP2 | 1868 | 1606 | 16 | IL1B - TP53BP2 | 2003 | 1319 | 2317 |
| IL1RAP - TP53BP2 | 154 | 1863 | 1166 | IL1RAP - TP53BP2 | 1565 | 1196 | 133 |
| IL1RN - TP53BP2 | 320 | 1676 | 1920 | IL1RN - TP53BP2 | 1559 | 1149 | 2489 |
| IL2RG - TP53BP2 | 755 | 377 | 644 | IL2RG - TP53BP2 | 1842 | 1888 | 1791 |
| IL6ST - TP53BP2 | 2237 | 581 | 1526 | IL6ST - TP53BP2 | 1862 | 1530 | 2234 |
| IL8 - TP53BP2 | 1135 | 1279 | 2250 | IL8 - TP53BP2 | 2356 | 2336 | 325 |
| IL10RB - TP53BP2 | 645 | 977 | 289 | IL10RB - TP53BP2 | 420 | 705 | 2040 |
| IL15 - TP53BP2 | 1715 | 281 | 973 | IL15 - TP53BP2 | 879 | 2029 | 1896 |
| IL15RA - TP53BP2 | 1225 | 727 | 567 | IL15RA - TP53BP2 | 2086 | 2287 | 2198 |
| IL17C - TP53BP2 | 2286 | 1214 | 617 | IL17C - TP53BP2 | 1158 | 1243 | 2313 |
| IL17REL - TP53BP2 | 76 | 1873 | 2403 | IL17REL - TP53BP2 | 1526 | 1463 | 1600 |
| Ranking of IL family w.r.t TP53I3 | Ranking of TP53I3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TP53I3 | 1140 | 1547 | 1558 | IL1A - TP53I3 | 283 | 157 | 341 |
| IL1B - TP53I3 | 759 | 333 | 1392 | IL1B - TP53I3 | 156 | 164 | 463 |
| IL1RAP - TP53I3 | 1521 | 885 | 1978 | IL1RAP - TP53I3 | 432 | 605 | 818 |
| IL1RN - TP53I3 | 737 | 340 | 1797 | IL1RN - TP53I3 | 1504 | 1674 | 16 |
| IL2RG - TP53I3 | 7 | 3 | 328 | IL2RG - TP53I3 | 836 | 637 | 134 |
| IL6ST - TP53I3 | 524 | 363 | 981 | IL6ST - TP53I3 | 2157 | 897 | 778 |
| IL8 - TP53I3 | 579 | 485 | 697 | IL8 - TP53I3 | 1921 | 290 | 1265 |
| IL10RB - TP53I3 | 185 | 137 | 758 | IL10RB - TP53I3 | 345 | 1080 | 326 |
| IL15 - TP53I3 | 240 | 244 | 428 | IL15 - TP53I3 | 353 | 1153 | 456 |
| IL15RA - TP53I3 | 2069 | 2079 | 2228 | IL15RA - TP53I3 | 106 | 644 | 1794 |
| IL17C - TP53I3 | 74 | 114 | 647 | IL17C - TP53I3 | 49 | 75 | 37 |
| IL17REL - TP53I3 | 597 | 326 | 1290 | IL17REL - TP53I3 | 2268 | 429 | 2220 |
| Ranking of IL family w.r.t TP53INP1 | Ranking of TP53INP1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TP53INP1 | 2309 | 746 | 7 | IL1A - TP53INP1 | 1049 | 1135 | 1138 |
| IL1B - TP53INP1 | 2281 | 21 | 461 | IL1B - TP53INP1 | 1395 | 1370 | 1684 |
| IL1RAP - TP53INP1 | 531 | 1274 | 2407 | IL1RAP - TP53INP1 | 2223 | 1460 | 680 |
| IL1RN - TP53INP1 | 2482 | 1911 | 891 | IL1RN - TP53INP1 | 1473 | 1252 | 2399 |
| IL2RG - TP53INP1 | 2152 | 1798 | 932 | IL2RG - TP53INP1 | 2063 | 1864 | 1956 |
| IL6ST - TP53INP1 | 591 | 790 | 1740 | IL6ST - TP53INP1 | 537 | 404 | 2042 |
| IL8 - TP53INP1 | 573 | 2388 | 2343 | IL8 - TP53INP1 | 1671 | 1787 | 1014 |
| IL10RB - TP53INP1 | 2510 | 2293 | 1664 | IL10RB - TP53INP1 | 1000 | 2339 | 218 |
| IL15 - TP53INP1 | 663 | 878 | 1116 | IL15 - TP53INP1 | 2147 | 588 | 429 |
| IL15RA - TP53INP1 | 663 | 149 | 169 | IL15RA - TP53INP1 | 1266 | 2264 | 1636 |
| IL17C - TP53INP1 | 2455 | 220 | 435 | IL17C - TP53INP1 | 823 | 523 | 438 |
| IL17REL - TP53INP1 | 83 | 2505 | 2509 | IL17REL - TP53INP1 | 1085 | 1476 | 1393 |
| Ranking of IL family w.r.t TP53INP2 | Ranking of TP53INP2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TP53INP2 | 1481 | 41 | 2490 | IL1A - TP53INP2 | 952 | 505 | 487 |
| IL1B - TP53INP2 | 489 | 310 | 267 | IL1B - TP53INP2 | 200 | 26 | 146 |
| IL1RAP - TP53INP2 | 1159 | 684 | 1263 | IL1RAP - TP53INP2 | 1168 | 757 | 1827 |
| IL1RN - TP53INP2 | 2374 | 779 | 110 | IL1RN - TP53INP2 | 1735 | 1927 | 264 |
| IL2RG - TP53INP2 | 2118 | 103 | 995 | IL2RG - TP53INP2 | 1151 | 539 | 380 |
| IL6ST - TP53INP2 | 261 | 1459 | 333 | IL6ST - TP53INP2 | 2512 | 1952 | 113 |
| IL8 - TP53INP2 | 82 | 679 | 779 | IL8 - TP53INP2 | 2349 | 85 | 1561 |
| IL10RB - TP53INP2 | 865 | 1991 | 67 | IL10RB - TP53INP2 | 653 | 2479 | 236 |
| IL15 - TP53INP2 | 1354 | 989 | 161 | IL15 - TP53INP2 | 1105 | 449 | 1506 |
| IL15RA - TP53INP2 | 1574 | 1545 | 2295 | IL15RA - TP53INP2 | 345 | 488 | 825 |
| IL17C - TP53INP2 | 449 | 56 | 221 | IL17C - TP53INP2 | 1065 | 260 | 116 |
| IL17REL - TP53INP2 | 1325 | 93 | 593 | IL17REL - TP53INP2 | 1251 | 643 | 1832 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t TP53 | |
| IL17REL | TP53BP2 |
| IL15RA | TP53I3 |
| IL-1RN/2RG/8/10RB/17REL | TP53INP1 |
| TP53 w.r.t IL | |
| IL-1A/1B/2RG/6ST/8/15/15RA | TP53BP2 |
| IL17REL | TP53I3 |
| IL2RG | TP53INP1 |
| IL6ST | TP53INP2 |
3.7.5. Interleukin - STAT Cross Family Analysis
| Ranking Interleukin family vs STAT family | |||||||
| Ranking of IL family w.r.t STAT2 | Ranking of STAT2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - STAT2 | 171 | 207 | 709 | IL1A - STAT2 | 1000 | 687 | 1941 |
| IL1B - STAT2 | 347 | 559 | 188 | IL1B - STAT2 | 1629 | 1019 | 2351 |
| IL1RAP - STAT2 | 2111 | 2258 | 2012 | IL1RAP - STAT2 | 1826 | 2005 | 70 |
| IL1RN - STAT2 | 828 | 1942 | 1226 | IL1RN - STAT2 | 2050 | 2082 | 1030 |
| IL2RG - STAT2 | 939 | 1424 | 272 | IL2RG - STAT2 | 1986 | 2021 | 2031 |
| IL6ST - STAT2 | 2167 | 2313 | 1042 | IL6ST - STAT2 | 1532 | 1766 | 696 |
| IL8 - STAT2 | 806 | 1012 | 69 | IL8 - STAT2 | 397 | 1015 | 2349 |
| IL10RB - STAT2 | 1093 | 2401 | 1260 | IL10RB - STAT2 | 1566 | 1241 | 467 |
| IL15 - STAT2 | 929 | 197 | 446 | IL15 - STAT2 | 1875 | 1724 | 940 |
| IL15RA - STAT2 | 537 | 415 | 1916 | IL15RA - STAT2 | 1406 | 1988 | 1863 |
| IL17C - STAT2 | 175 | 78 | 514 | IL17C - STAT2 | 1199 | 2473 | 1883 |
| IL17REL - STAT2 | 2508 | 2488 | 2172 | IL17REL - STAT2 | 244 | 1890 | 1885 |
| Ranking of IL family w.r.t STAT3 | Ranking of STAT3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - STAT3 | 2516 | 173 | 7 | IL1A - STAT3 | 1872 | 1289 | 2350 |
| IL1B - STAT3 | 1628 | 127 | 613 | IL1B - STAT3 | 1367 | 2391 | 901 |
| IL1RAP - STAT3 | 23 | 2252 | 2211 | IL1RAP - STAT3 | 2169 | 1483 | 179 |
| IL1RN - STAT3 | 2309 | 300 | 488 | IL1RN - STAT3 | 2090 | 2312 | 1440 |
| IL2RG - STAT3 | 1168 | 397 | 611 | IL2RG - STAT3 | 2233 | 2146 | 1387 |
| IL6ST - STAT3 | 1355 | 1217 | 381 | IL6ST - STAT3 | 2400 | 2491 | 1953 |
| IL8 - STAT3 | 2353 | 740 | 1176 | IL8 - STAT3 | 1371 | 942 | 2018 |
| IL10RB - STAT3 | 2494 | 1257 | 1320 | IL10RB - STAT3 | 1118 | 406 | 1299 |
| IL15 - STAT3 | 2164 | 903 | 62 | IL15 - STAT3 | 2015 | 2412 | 1356 |
| IL15RA - STAT3 | 1140 | 1572 | 1618 | IL15RA - STAT3 | 1724 | 1638 | 1963 |
| IL17C - STAT3 | 2437 | 30 | 20 | IL17C - STAT3 | 554 | 1446 | 1428 |
| IL17REL - STAT3 | 339 | 2282 | 2517 | IL17REL - STAT3 | 573 | 2181 | 521 |
| Ranking of IL family w.r.t STAT5A | Ranking of STAT5A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - STAT5A | 1631 | 1110 | 2210 | IL1A - STAT5A | 275 | 464 | 1645 |
| IL1B - STAT5A | 1194 | 1561 | 2223 | IL1B - STAT5A | 416 | 240 | 1659 |
| IL1RAP - STAT5A | 1768 | 1680 | 2149 | IL1RAP - STAT5A | 1852 | 391 | 432 |
| IL1RN - STAT5A | 119 | 285 | 908 | IL1RN - STAT5A | 86 | 2026 | 960 |
| IL2RG - STAT5A | 1136 | 1088 | 1435 | IL2RG - STAT5A | 1367 | 1832 | 2149 |
| IL6ST - STAT5A | 1441 | 2022 | 1697 | IL6ST - STAT5A | 1903 | 436 | 317 |
| IL8 - STAT5A | 1932 | 1543 | 1069 | IL8 - STAT5A | 2000 | 2386 | 4 |
| IL10RB - STAT5A | 897 | 87 | 2033 | IL10RB - STAT5A | 2103 | 1292 | 1326 |
| IL15 - STAT5A | 1116 | 801 | 1653 | IL15 - STAT5A | 436 | 2139 | 1041 |
| IL15RA - STAT5A | 2342 | 2350 | 788 | IL15RA - STAT5A | 621 | 1185 | 1537 |
| IL17C - STAT5A | 984 | 1386 | 2045 | IL17C - STAT5A | 1760 | 2060 | 2201 |
| IL17REL - STAT5A | 1308 | 755 | 3 | IL17REL - STAT5A | 477 | 369 | 992 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t STAT | |
| IL-1RAP/6ST/17REL | STAT2 |
| IL-1RAP/17REL | STAT3 |
| IL-1RAP/15RA | STAT5A |
| STAT w.r.t IL | |
| IL-1RN/2RG | STAT2 |
| IL-1A/1RN/2RG/6ST/15 | STAT3 |
| IL-2RG/8/17C | STAT5A |
3.7.6. Interleukin - TRAF Cross Family Analysis
3.7.7. Interleukin - Metalloreductase STEAP4 Cross Family Analysis
| Ranking Interleukin family vs STEAP4 family | |||||||
| Ranking of IL family w.r.t STEAP4 | Ranking of STEAP4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - STEAP4 | 422 | 482 | 992 | IL1A - STEAP4 | 71 | 2358 | 2223 |
| IL1B - STEAP4 | 423 | 814 | 982 | IL1B - STEAP4 | 240 | 1570 | 1863 |
| IL1RAP - STEAP4 | 2092 | 262 | 661 | IL1RAP - STEAP4 | 1871 | 1898 | 2077 |
| IL1RN - STEAP4 | 404 | 1602 | 370 | IL1RN - STEAP4 | 195 | 2043 | 1763 |
| IL2RG - STEAP4 | 1293 | 1458 | 1323 | IL2RG - STEAP4 | 299 | 1562 | 1284 |
| IL6ST - STEAP4 | 920 | 1641 | 2424 | IL6ST - STEAP4 | 1374 | 504 | 1628 |
| IL8 - STEAP4 | 2204 | 1987 | 1558 | IL8 - STEAP4 | 794 | 1049 | 1615 |
| IL10RB - STEAP4 | 2422 | 2310 | 1179 | IL10RB - STEAP4 | 476 | 254 | 906 |
| IL15 - STEAP4 | 700 | 1154 | 2320 | IL15 - STEAP4 | 288 | 1965 | 2283 |
| IL15RA - STEAP4 | 2277 | 1114 | 1528 | IL15RA - STEAP4 | 1170 | 1334 | 1347 |
| IL17C - STEAP4 | 433 | 2103 | 1889 | IL17C - STEAP4 | 17 | 2426 | 1108 |
| IL17REL - STEAP4 | 33 | 1965 | 2297 | IL17REL - STEAP4 | 2439 | 715 | 100 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t STEAP4 | |
| IL-8/10RB/17C/17REL | STEAP4 |
| STEAP4 w.r.t IL | |
| IL-1A/1RAP/1RN/15 | STEAP4 |
3.7.8. Interleukin - Metalloreductase STEAP3 Cross Family Analysis
| Ranking Interleukin family vs STEAP3 family | |||||||
| Ranking of IL family w.r.t STEAP3 | Ranking of STEAP3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1RL2 - STEAP3 | 619 | 1471 | 2246 | IL1RL2 - STEAP3 | 835 | 2234 | 1733 |
| IL17D - STEAP3 | 1338 | 1275 | 458 | IL17D - STEAP3 | 596 | 705 | 2273 |
| IL17RB - STEAP3 | 1101 | 2302 | 239 | IL17RB - STEAP3 | 208 | 2462 | 404 |
| IL17RD - STEAP3 | 1323 | 810 | 1834 | IL17RD - STEAP3 | 2352 | 589 | 2233 |
| IL33 - STEAP3 | 1589 | 781 | 1210 | IL33 - STEAP3 | 1070 | 57 | 2098 |
| ILF2 - STEAP3 | 1571 | 811 | 579 | ILF2 - STEAP3 | 1986 | 1029 | 2474 |
| ILF3 - STEAP3 | 261 | 1866 | 1953 | ILF3 - STEAP3 | 121 | 2314 | 926 |
| ILF3.AS1 - STEAP3 | 947 | 2255 | 926 | ILF3.AS1 - STEAP3 | 1592 | 678 | 1094 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t STEAP3 | |
| IL-1RL2/17D/17RB/17RD/33/F2/F3.AS1 - STEAP3 | |
| STEAP3 w.r.t IL | |
| IL-1RL2/17D/17RB/33/F3/F3.AS1 - STEAP3 | |
3.7.9. Interleukin - ATP-Binding Cassette Transporters
| Ranking Interleukin family vs ABC family | |||||||
| Ranking of IL family w.r.t ABCA2 | Ranking of ABCA2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1RL2 - ABCA2 | 2055 | 2097 | 405 | IL1RL2 - ABCA2 | 2022 | 2490 | 1234 |
| IL17D - ABCA2 | 1778 | 2160 | 1120 | IL17D - ABCA2 | 540 | 227 | 1006 |
| IL17RB - ABCA2 | 2419 | 1404 | 1727 | IL17RB - ABCA2 | 2146 | 1543 | 1991 |
| IL17RD - ABCA2 | 2202 | 1799 | 358 | IL17RD - ABCA2 | 1717 | 1671 | 517 |
| IL33 - ABCA2 | 1076 | 1707 | 1854 | IL33 - ABCA2 | 1507 | 497 | 743 |
| ILF2 - ABCA2 | 944 | 1054 | 2607 | ILF2 - ABCA2 | 831 | 822 | 752 |
| ILF3 - ABCA2 | 1380 | 1369 | 1702 | ILF3 - ABCA2 | 1691 | 2094 | 2275 |
| ILF3.AS1 - ABCA2 | 2243 | 1006 | 1924 | ILF3.AS1 - ABCA2 | 2058 | 1664 | 2165 |
| Ranking of IL family w.r.t ABCE1 | Ranking of ABCE1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1RL2 - ABCE1 | 906 | 1403 | 2365 | IL1RL2 - ABCE1 | 525 | 2034 | 723 |
| IL17D - ABCE1 | 1531 | 636 | 753 | IL17D - ABCE1 | 1432 | 2146 | 1401 |
| IL17RB - ABCE1 | 459 | 2056 | 1993 | IL17RB - ABCE1 | 1090 | 2618 | 263 |
| IL17RD - ABCE1 | 1030 | 1332 | 1565 | IL17RD - ABCE1 | 1523 | 727 | 2185 |
| IL33 - ABCE1 | 1649 | 719 | 937 | IL33 - ABCE1 | 2619 | 808 | 2025 |
| ILF2 - ABCE1 | 20 | 310 | 560 | ILF2 - ABCE1 | 2650 | 331 | 2103 |
| ILF3 - ABCE1 | 2410 | 2409 | 1826 | ILF3 - ABCE1 | 1767 | 2674 | 19 |
| ILF3.AS1 - ABCE1 | 1154 | 2222 | 786 | ILF3.AS1 - ABCE1 | 1788 | 1948 | 820 |
| Ranking of IL family w.r.t ABCF2 | Ranking of ABCF2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1RL2 - ABCF2 | 1031 | 1806 | 2002 | IL1RL2 - ABCF2 | 2257 | 818 | 1274 |
| IL17D - ABCF2 | 2481 | 2016 | 1006 | IL17D - ABCF2 | 796 | 2104 | 568 |
| IL17RB - ABCF2 | 509 | 1294 | 2302 | IL17RB - ABCF2 | 1271 | 621 | 1631 |
| IL17RD - ABCF2 | 610 | 1935 | 1084 | IL17RD - ABCF2 | 957 | 2276 | 1431 |
| IL33 - ABCF2 | 735 | 2050 | 1855 | IL33 - ABCF2 | 421 | 1781 | 252 |
| ILF2 - ABCF2 | 2093 | 1104 | 2073 | ILF2 - ABCF2 | 683 | 2304 | 529 |
| ILF3 - ABCF2 | 812 | 1686 | 1080 | ILF3 - ABCF2 | 1243 | 585 | 1452 |
| ILF3.AS1 - ABCF2 | 430 | 2416 | 1983 | ILF3.AS1 - ABCF2 | 2272 | 1169 | 862 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t ABC | |
| IL-1RB/33/F2/F3 | ABCA2 |
| IL-1RL2/17D/17RD/33/F2/F3.AS1 | ABCE1 |
| IL-17RB/17RD/F3 | ABCF2 |
| ABC w.r.t IL | |
| IL-17D/17RD/33/F2 | ABCA2 |
| IL-1RL2/17D/17RB/17RD | ABCE1 |
| IL-1RL2/17D/17RB/17RD/33/F2/F3/F3.AS1 | ABCF2 |
3.7.10. Interleukin - TNF Cross Family Analysis
| Ranking Interleukin family vs TNF family | |||||||
| Ranking of IL family w.r.t TNF | Ranking of TNF w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNF | 1382 | 727 | 725 | IL1A - TNF | 172 | 660 | 230 |
| IL1B - TNF | 519 | 539 | 187 | IL1B - TNF | 443 | 458 | 244 |
| IL1RAP - TNF | 1475 | 1995 | 2255 | IL1RAP - TNF | 564 | 550 | 1500 |
| IL1RN - TNF | 163 | 106 | 609 | IL1RN - TNF | 292 | 462 | 276 |
| IL2RG - TNF | 276 | 820 | 340 | IL2RG - TNF | 419 | 708 | 1035 |
| IL6ST - TNF | 2374 | 2037 | 2003 | IL6ST - TNF | 2410 | 1901 | 666 |
| IL8 - TNF | 921 | 1325 | 1148 | IL8 - TNF | 1072 | 206 | 118 |
| IL10RB - TNF | 346 | 595 | 339 | IL10RB - TNF | 2065 | 2120 | 2296 |
| IL15 - TNF | 242 | 944 | 616 | IL15 - TNF | 265 | 828 | 279 |
| IL15RA - TNF | 2341 | 1843 | 2195 | IL15RA - TNF | 131 | 914 | 1488 |
| IL17C - TNF | 906 | 1573 | 776 | IL17C - TNF | 2148 | 568 | 280 |
| IL17REL - TNF | 296 | 804 | 677 | IL17REL - TNF | 1223 | 1901 | 11 |
| Ranking of IL family w.r.t TNFAIP1 | Ranking of TNFAIP1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFAIP1 | 2515 | 549 | 1534 | IL1A - TNFAIP1 | 533 | 1901 | 1548 |
| IL1B - TNFAIP1 | 2398 | 440 | 2449 | IL1B - TNFAIP1 | 1324 | 756 | 1062 |
| IL1RAP - TNFAIP1 | 326 | 866 | 2226 | IL1RAP - TNFAIP1 | 1555 | 1284 | 1291 |
| IL1RN - TNFAIP1 | 1952 | 649 | 1453 | IL1RN - TNFAIP1 | 1567 | 307 | 979 |
| IL2RG - TNFAIP1 | 1791 | 104 | 2482 | IL2RG - TNFAIP1 | 421 | 973 | 1169 |
| IL6ST - TNFAIP1 | 156 | 1415 | 1062 | IL6ST - TNFAIP1 | 1281 | 104 | 2086 |
| IL8 - TNFAIP1 | 456 | 682 | 1389 | IL8 - TNFAIP1 | 2293 | 2126 | 752 |
| IL10RB - TNFAIP1 | 97 | 425 | 2020 | IL10RB - TNFAIP1 | 716 | 2092 | 569 |
| IL15 - TNFAIP1 | 367 | 1392 | 159 | IL15 - TNFAIP1 | 24 | 436 | 324 |
| IL15RA - TNFAIP1 | 1860 | 1979 | 611 | IL15RA - TNFAIP1 | 873 | 2141 | 1853 |
| IL17C - TNFAIP1 | 2382 | 1072 | 2446 | IL17C - TNFAIP1 | 961 | 2143 | 791 |
| IL17REL - TNFAIP1 | 307 | 79 | 161 | IL17REL - TNFAIP1 | 1603 | 1462 | 1764 |
| Ranking of IL family w.r.t TNFAIP2 | Ranking of TNFAIP2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFAIP2 | 219 | 1815 | 790 | IL1A - TNFAIP2 | 450 | 1041 | 465 |
| IL1B - TNFAIP2 | 210 | 1123 | 538 | IL1B - TNFAIP2 | 1923 | 557 | 944 |
| IL1RAP - TNFAIP2 | 1535 | 660 | 1525 | IL1RAP - TNFAIP2 | 105 | 229 | 845 |
| IL1RN - TNFAIP2 | 1769 | 2475 | 683 | IL1RN - TNFAIP2 | 957 | 868 | 839 |
| IL2RG - TNFAIP2 | 1358 | 576 | 188 | IL2RG - TNFAIP2 | 415 | 1132 | 613 |
| IL6ST - TNFAIP2 | 2007 | 633 | 1704 | IL6ST - TNFAIP2 | 1649 | 929 | 1558 |
| IL8 - TNFAIP2 | 769 | 331 | 368 | IL8 - TNFAIP2 | 1262 | 1412 | 1595 |
| IL10RB - TNFAIP2 | 2319 | 2497 | 719 | IL10RB - TNFAIP2 | 93 | 1583 | 204 |
| IL15 - TNFAIP2 | 1362 | 2383 | 795 | IL15 - TNFAIP2 | 537 | 749 | 120 |
| IL15RA - TNFAIP2 | 2032 | 821 | 1502 | IL15RA - TNFAIP2 | 519 | 737 | 1146 |
| IL17C - TNFAIP2 | 868 | 1684 | 1770 | IL17C - TNFAIP2 | 199 | 424 | 687 |
| IL17REL - TNFAIP2 | 279 | 563 | 299 | IL17REL - TNFAIP2 | 2057 | 437 | 2008 |
| Ranking Interleukin family vs TNF family | |||||||
| Ranking of IL family w.r.t TNFAIP3 | Ranking of TNFAIP3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFAIP3 | 2307 | 319 | 108 | IL1A - TNFAIP3 | 78 | 51 | 2058 |
| IL1B - TNFAIP3 | 495 | 98 | 339 | IL1B - TNFAIP3 | 140 | 146 | 520 |
| IL1RAP - TNFAIP3 | 30 | 2428 | 1376 | IL1RAP - TNFAIP3 | 1802 | 1610 | 903 |
| IL1RN - TNFAIP3 | 579 | 277 | 299 | IL1RN - TNFAIP3 | 60 | 1610 | 1320 |
| IL2RG - TNFAIP3 | 1705 | 330 | 125 | IL2RG - TNFAIP3 | 1056 | 1608 | 2333 |
| IL6ST - TNFAIP3 | 2068 | 2432 | 2282 | IL6ST - TNFAIP3 | 1652 | 1470 | 1507 |
| IL8 - TNFAIP3 | 1918 | 2255 | 1587 | IL8 - TNFAIP3 | 2224 | 1717 | 118 |
| IL10RB - TNFAIP3 | 1576 | 666 | 1377 | IL10RB - TNFAIP3 | 1073 | 417 | 943 |
| IL15 - TNFAIP3 | 732 | 254 | 273 | IL15 - TNFAIP3 | 907 | 628 | 684 |
| IL15RA - TNFAIP3 | 727 | 1547 | 1476 | IL15RA - TNFAIP3 | 1340 | 445 | 1031 |
| IL17C - TNFAIP3 | 1675 | 222 | 138 | IL17C - TNFAIP3 | 1105 | 1887 | 866 |
| IL17REL - TNFAIP3 | 2364 | 2503 | 2283 | IL17REL - TNFAIP3 | 2040 | 1143 | 1486 |
| Ranking of IL family w.r.t TNFRSF1A | Ranking of TNFRSF1A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF1A | 1556 | 2184 | 1375 | IL1A - TNFRSF1A | 2028 | 113 | 226 |
| IL1B - TNFRSF1A | 1621 | 1917 | 446 | IL1B - TNFRSF1A | 147 | 2027 | 2247 |
| IL1RAP - TNFRSF1A | 1236 | 2500 | 2293 | IL1RAP - TNFRSF1A | 1339 | 1003 | 2062 |
| IL1RN - TNFRSF1A | 411 | 1571 | 755 | IL1RN - TNFRSF1A | 1713 | 387 | 102 |
| IL2RG - TNFRSF1A | 565 | 2350 | 574 | IL2RG - TNFRSF1A | 1191 | 597 | 1479 |
| IL6ST - TNFRSF1A | 2221 | 1465 | 561 | IL6ST - TNFRSF1A | 1143 | 291 | 225 |
| IL8 - TNFRSF1A | 1536 | 750 | 304 | IL8 - TNFRSF1A | 1483 | 669 | 673 |
| IL10RB - TNFRSF1A | 620 | 35 | 1791 | IL10RB - TNFRSF1A | 230 | 1510 | 385 |
| IL15 - TNFRSF1A | 345 | 489 | 384 | IL15 - TNFRSF1A | 157 | 838 | 425 |
| IL15RA - TNFRSF1A | 442 | 1155 | 697 | IL15RA - TNFRSF1A | 682 | 322 | 1575 |
| IL17C - TNFRSF1A | 1113 | 284 | 149 | IL17C - TNFRSF1A | 5 | 169 | 122 |
| IL17REL - TNFRSF1A | 766 | 336 | 249 | IL17REL - TNFRSF1A | 1547 | 452 | 22 |
| Ranking of IL family w.r.t TNFRSF10A | Ranking of TNFRSF10A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF10A | 366 | 73 | 48 | IL1A - TNFRSF10A | 1972 | 1805 | 2504 |
| IL1B - TNFRSF10A | 317 | 45 | 367 | IL1B - TNFRSF10A | 2375 | 2373 | 2320 |
| IL1RAP - TNFRSF10A | 2104 | 1342 | 2027 | IL1RAP - TNFRSF10A | 981 | 1665 | 2504 |
| IL1RN - TNFRSF10A | 1739 | 346 | 173 | IL1RN - TNFRSF10A | 1261 | 2287 | 2469 |
| IL2RG - TNFRSF10A | 645 | 1448 | 1009 | IL2RG - TNFRSF10A | 1244 | 2246 | 2467 |
| IL6ST - TNFRSF10A | 1307 | 823 | 1778 | IL6ST - TNFRSF10A | 2128 | 2320 | 1738 |
| IL8 - TNFRSF10A | 402 | 1615 | 1908 | IL8 - TNFRSF10A | 566 | 733 | 2117 |
| IL10RB - TNFRSF10A | 1243 | 689 | 2119 | IL10RB - TNFRSF10A | 389 | 532 | 723 |
| IL15 - TNFRSF10A | 321 | 1602 | 358 | IL15 - TNFRSF10A | 2414 | 2260 | 1705 |
| IL15RA - TNFRSF10A | 2126 | 2342 | 148 | IL15RA - TNFRSF10A | 2398 | 1970 | 2088 |
| IL17C - TNFRSF10A | 981 | 269 | 1027 | IL17C - TNFRSF10A | 1831 | 2025 | 1718 |
| IL17REL - TNFRSF10A | 2497 | 2470 | 2109 | IL17REL - TNFRSF10A | 1034 | 1482 | 2068 |
| Ranking Interleukin family vs TNF family | |||||||
| Ranking of IL family w.r.t TNFRSF10B | Ranking of TNFRSF10B w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF10B | 771 | 190 | 110 | IL1A - TNFRSF10B | 294 | 1870 | 1471 |
| IL1B - TNFRSF10B | 2301 | 109 | 19 | IL1B - TNFRSF10B | 829 | 626 | 1465 |
| IL1RAP - TNFRSF10B | 752 | 2148 | 1579 | IL1RAP - TNFRSF10B | 2102 | 1685 | 405 |
| IL1RN - TNFRSF10B | 840 | 2005 | 443 | IL1RN - TNFRSF10B | 2087 | 1403 | 1966 |
| IL2RG - TNFRSF10B | 1868 | 1485 | 57 | IL2RG - TNFRSF10B | 1616 | 2134 | 1376 |
| IL6ST - TNFRSF10B | 788 | 1851 | 1038 | IL6ST - TNFRSF10B | 1149 | 510 | 1603 |
| IL8 - TNFRSF10B | 1494 | 1467 | 2312 | IL8 - TNFRSF10B | 1769 | 1763 | 196 |
| IL10RB - TNFRSF10B | 461 | 1770 | 1497 | IL10RB - TNFRSF10B | 1212 | 994 | 1542 |
| IL15 - TNFRSF10B | 360 | 1028 | 620 | IL15 - TNFRSF10B | 1712 | 815 | 2039 |
| IL15RA - TNFRSF10B | 2330 | 932 | 1932 | IL15RA - TNFRSF10B | 1640 | 1375 | 2210 |
| IL17C - TNFRSF10B | 557 | 1911 | 91 | IL17C - TNFRSF10B | 1594 | 969 | 1624 |
| IL17REL - TNFRSF10B | 457 | 1701 | 2422 | IL17REL - TNFRSF10B | 1074 | 2117 | 347 |
| Ranking of IL family w.r.t TNFRSF10D | Ranking of TNFRSF10D w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF10D | 143 | 625 | 21 | IL1A - TNFRSF10D | 2415 | 2517 | 1894 |
| IL1B - TNFRSF10D | 185 | 142 | 191 | IL1B - TNFRSF10D | 2513 | 2300 | 2430 |
| IL1RAP - TNFRSF10D | 1106 | 1750 | 1376 | IL1RAP - TNFRSF10D | 811 | 1241 | 1946 |
| IL1RN - TNFRSF10D | 881 | 520 | 337 | IL1RN - TNFRSF10D | 2512 | 1658 | 857 |
| IL2RG - TNFRSF10D | 713 | 413 | 905 | IL2RG - TNFRSF10D | 2514 | 2419 | 2043 |
| IL6ST - TNFRSF10D | 752 | 2009 | 1617 | IL6ST - TNFRSF10D | 2324 | 2515 | 460 |
| IL8 - TNFRSF10D | 1267 | 903 | 629 | IL8 - TNFRSF10D | 463 | 446 | 2468 |
| IL10RB - TNFRSF10D | 1072 | 1050 | 1031 | IL10RB - TNFRSF10D | 1822 | 1959 | 982 |
| IL15 - TNFRSF10D | 108 | 842 | 333 | IL15 - TNFRSF10D | 2490 | 2234 | 2019 |
| IL15RA - TNFRSF10D | 2197 | 943 | 2126 | IL15RA - TNFRSF10D | 1895 | 1048 | 24 |
| IL17C - TNFRSF10D | 11 | 268 | 7 | IL17C - TNFRSF10D | 2493 | 2062 | 2488 |
| IL17REL - TNFRSF10D | 54 | 638 | 278 | IL17REL - TNFRSF10D | 2514 | 100 | 2452 |
| Ranking of IL family w.r.t TNFRSF12A | Ranking of TNFRSF12A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF12A | 52 | 2189 | 374 | IL1A - TNFRSF12A | 239 | 2080 | 1330 |
| IL1B - TNFRSF12A | 709 | 1592 | 1066 | IL1B - TNFRSF12A | 1422 | 516 | 1025 |
| IL1RAP - TNFRSF12A | 606 | 1030 | 1639 | IL1RAP - TNFRSF12A | 165 | 1595 | 1273 |
| IL1RN - TNFRSF12A | 122 | 1173 | 1182 | IL1RN - TNFRSF12A | 2176 | 529 | 1135 |
| IL2RG - TNFRSF12A | 206 | 1875 | 756 | IL2RG - TNFRSF12A | 1705 | 1060 | 2416 |
| IL6ST - TNFRSF12A | 2128 | 898 | 1092 | IL6ST - TNFRSF12A | 707 | 2213 | 2187 |
| IL8 - TNFRSF12A | 1132 | 1827 | 2355 | IL8 - TNFRSF12A | 461 | 1199 | 1587 |
| IL10RB - TNFRSF12A | 51 | 37 | 238 | IL10RB - TNFRSF12A | 852 | 781 | 910 |
| IL15 - TNFRSF12A | 281 | 1535 | 686 | IL15 - TNFRSF12A | 1984 | 1469 | 530 |
| IL15RA - TNFRSF12A | 2138 | 2090 | 1981 | IL15RA - TNFRSF12A | 1065 | 576 | 1568 |
| IL17C - TNFRSF12A | 326 | 2512 | 52 | IL17C - TNFRSF12A | 1497 | 1898 | 2209 |
| IL17REL - TNFRSF12A | 2475 | 587 | 2496 | IL17REL - TNFRSF12A | 148 | 1299 | 410 |
| Ranking of IL family w.r.t TNFRSF14 | Ranking of TNFRSF14 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF14 | 208 | 29 | 683 | IL1A - TNFRSF14 | 2061 | 1969 | 693 |
| IL1B - TNFRSF14 | 70 | 664 | 924 | IL1B - TNFRSF14 | 592 | 1647 | 1743 |
| IL1RAP - TNFRSF14 | 1356 | 2249 | 756 | IL1RAP - TNFRSF14 | 2103 | 1414 | 1691 |
| IL1RN - TNFRSF14 | 1001 | 794 | 745 | IL1RN - TNFRSF14 | 1898 | 2414 | 975 |
| IL2RG - TNFRSF14 | 1619 | 1780 | 1158 | IL2RG - TNFRSF14 | 2009 | 1949 | 1367 |
| IL6ST - TNFRSF14 | 2248 | 221 | 619 | IL6ST - TNFRSF14 | 1033 | 1923 | 2175 |
| IL8 - TNFRSF14 | 517 | 299 | 1301 | IL8 - TNFRSF14 | 1776 | 578 | 2205 |
| IL10RB - TNFRSF14 | 1595 | 156 | 943 | IL10RB - TNFRSF14 | 763 | 1457 | 834 |
| IL15 - TNFRSF14 | 1265 | 550 | 1692 | IL15 - TNFRSF14 | 2039 | 954 | 1230 |
| IL15RA - TNFRSF14 | 2378 | 1929 | 1577 | IL15RA - TNFRSF14 | 2440 | 2031 | 253 |
| IL17C - TNFRSF14 | 11 | 40 | 605 | IL17C - TNFRSF14 | 1856 | 1836 | 671 |
| IL17REL - TNFRSF14 | 46 | 306 | 293 | IL17REL - TNFRSF14 | 2312 | 72 | 1623 |
| Ranking Interleukin family vs TNF family | |||||||
| Ranking of IL family w.r.t TNFRSF21 | Ranking of TNFRSF21 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFRSF21 | 904 | 2313 | 1127 | IL1A - TNFRSF21 | 322 | 1745 | 688 |
| IL1B - TNFRSF21 | 1862 | 2164 | 2305 | IL1B - TNFRSF21 | 1336 | 157 | 829 |
| IL1RAP - TNFRSF21 | 1446 | 1762 | 2163 | IL1RAP - TNFRSF21 | 563 | 22 | 497 |
| IL1RN - TNFRSF21 | 1593 | 2373 | 627 | IL1RN - TNFRSF21 | 1626 | 1341 | 320 |
| IL2RG - TNFRSF21 | 403 | 2297 | 2351 | IL2RG - TNFRSF21 | 618 | 719 | 981 |
| IL6ST - TNFRSF21 | 1372 | 1894 | 753 | IL6ST - TNFRSF21 | 2019 | 1123 | 1143 |
| IL8 - TNFRSF21 | 1204 | 1944 | 1585 | IL8 - TNFRSF21 | 2493 | 999 | 1513 |
| IL10RB - TNFRSF21 | 238 | 845 | 1081 | IL10RB - TNFRSF21 | 2502 | 842 | 1641 |
| IL15 - TNFRSF21 | 1591 | 1905 | 1740 | IL15 - TNFRSF21 | 65 | 1459 | 96 |
| IL15RA - TNFRSF21 | 421 | 1934 | 1269 | IL15RA - TNFRSF21 | 98 | 1109 | 1259 |
| IL17C - TNFRSF21 | 2130 | 1039 | 1676 | IL17C - TNFRSF21 | 2272 | 1163 | 266 |
| IL17REL - TNFRSF21 | 557 | 765 | 61 | IL17REL - TNFRSF21 | 1846 | 704 | 2381 |
| Ranking of IL family w.r.t TNFRS10 | Ranking of TNFRS10 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFSF10 | 120 | 1575 | 2499 | IL1A - TNFSF10 | 2369 | 1086 | 1034 |
| IL1B - TNFSF10 | 972 | 2448 | 1993 | IL1B - TNFSF10 | 2348 | 1544 | 1076 |
| IL1RAP - TNFSF10 | 754 | 1045 | 2015 | IL1RAP - TNFSF10 | 1613 | 2470 | 966 |
| IL1RN - TNFSF10 | 740 | 1535 | 570 | IL1RN - TNFSF10 | 1035 | 75 | 1074 |
| IL2RG - TNFSF10 | 2272 | 1447 | 1285 | IL2RG - TNFSF10 | 1032 | 882 | 1271 |
| IL6ST - TNFSF10 | 1978 | 227 | 778 | IL6ST - TNFSF10 | 1647 | 1602 | 2369 |
| IL8 - TNFSF10 | 818 | 1702 | 791 | IL8 - TNFSF10 | 1161 | 790 | 2265 |
| IL10RB - TNFSF10 | 744 | 1146 | 2257 | IL10RB - TNFSF10 | 1496 | 2252 | 1864 |
| IL15 - TNFSF10 | 967 | 1382 | 1910 | IL15 - TNFSF10 | 1400 | 1383 | 486 |
| IL15RA - TNFSF10 | 346 | 2163 | 2059 | IL15RA - TNFSF10 | 1458 | 790 | 1428 |
| IL17C - TNFSF10 | 460 | 2337 | 2431 | IL17C - TNFSF10 | 558 | 1004 | 942 |
| IL17REL - TNFSF10 | 1728 | 145 | 989 | IL17REL - TNFSF10 | 1664 | 718 | 250 |
| Ranking of IL family w.r.t TNFRS15 | Ranking of TNFRS15 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TNFSF15 | 1177 | 2494 | 979 | IL1A - TNFSF15 | 1014 | 613 | 1449 |
| IL1B - TNFSF15 | 1435 | 1529 | 1571 | IL1B - TNFSF15 | 1898 | 1032 | 767 |
| IL1RAP - TNFSF15 | 271 | 1665 | 2368 | IL1RAP - TNFSF15 | 890 | 843 | 793 |
| IL1RN - TNFSF15 | 2319 | 377 | 566 | IL1RN - TNFSF15 | 414 | 1457 | 1704 |
| IL2RG - TNFSF15 | 316 | 874 | 487 | IL2RG - TNFSF15 | 2332 | 1362 | 1632 |
| IL6ST - TNFSF15 | 1834 | 1004 | 1471 | IL6ST - TNFSF15 | 771 | 1171 | 1445 |
| IL8 - TNFSF15 | 1266 | 1571 | 1141 | IL8 - TNFSF15 | 2422 | 515 | 966 |
| IL10RB - TNFSF15 | 1488 | 326 | 1367 | IL10RB - TNFSF15 | 1611 | 2041 | 1635 |
| IL15 - TNFSF15 | 1356 | 1508 | 737 | IL15 - TNFSF15 | 201 | 1922 | 1756 |
| IL15RA - TNFSF15 | 2124 | 956 | 2365 | IL15RA - TNFSF15 | 1551 | 668 | 864 |
| IL17C - TNFSF15 | 2222 | 2328 | 954 | IL17C - TNFSF15 | 2403 | 1049 | 1338 |
| IL17REL - TNFSF15 | 1214 | 177 | 208 | IL17REL - TNFSF15 | 513 | 1515 | 1943 |
3.8. BCL Related Synergies
3.8.1. Interleukin - BCL Cross Family Analysis
3.8.2. Selenbp1 - BCL Cross Family Analysis
| Ranking SELENBP1 vs BCL family | |||||||
| Ranking of BCL family w.r.t SELENBP1 | Ranking of SELENBP1 w.r.t BCL | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| SELENBP1 - BCL2L12 | 2426 | 2033 | 2629 | SELENBP1 - BCL2L12 | 2589 | 2195 | 2082 |
| SELENBP1 - BCL6B | 2446 | 2575 | 1956 | SELENBP1 - BCL6B | 182 | 110 | 494 |
| SELENBP1 - BCL7A | 2620 | 1326 | 2006 | SELENBP1 - BCL7A | 2015 | 1799 | 767 |
| SELENBP1 - BCL9 | 2453 | 1568 | 1738 | SELENBP1 - BCL9 | 2538 | 1916 | 1793 |
| SELENBP1 - BCL11A | 1921 | 2463 | 1566 | SELENBP1 - BCL11A | 905 | 931 | 401 |
| SELENBP1 - BCL11B | 1896 | 299 | 1385 | SELENBP1 - BCL11B | 2496 | 2636 | 2510 |
| Unexplored combinatorial hypotheses | |
| SELENBP1 w.r.t BCL | |
| SELENBP1 | BCL-9/11B |
| BCL w.r.t SELENBP1 | |
| SELENBP1 | BCL-6B/11A |
3.8.3. TP53 - BCL Cross Family Analysis
| Ranking TP53 family vs BCL family | |||||||
| Ranking of BCL2L1 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL2L1 | 2431 | 1529 | 1728 | TP53BP2 - BCL2L1 | 1786 | 1961 | 1225 |
| TP53I3 - BCL2L1 | 799 | 554 | 728 | TP53I3 - BCL2L1 | 1980 | 1752 | 756 |
| TP53INP1 - BCL2L1 | 1064 | 1154 | 1414 | TP53INP1 - BCL2L1 | 1193 | 258 | 1850 |
| TP53INP2 - BCL2L1 | 282 | 2371 | 851 | TP53INP2 - BCL2L1 | 830 | 1477 | 1512 |
| Ranking of BCL2L2 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL2L2 | 1471 | 34 | 1367 | TP53BP2 - BCL2L2 | 1076 | 2168 | 1658 |
| TP53I3 - BCL2L2 | 2423 | 2377 | 2452 | TP53I3 - BCL2L2 | 1911 | 245 | 378 |
| TP53INP1 - BCL2L2 | 1693 | 180 | 987 | TP53INP1 - BCL2L2 | 482 | 1653 | 1130 |
| TP53INP2 - BCL2L2 | 1688 | 1827 | 2035 | TP53INP2 - BCL2L2 | 85 | 376 | 1146 |
| Ranking of BCL2L13 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL2L13 | 1515 | 1261 | 1842 | TP53BP2 - BCL2L13 | 1128 | 1827 | 1613 |
| TP53I3 - BCL2L13 | 1264 | 1501 | 1963 | TP53I3 - BCL2L13 | 419 | 1088 | 959 |
| TP53INP1 - BCL2L13 | 759 | 387 | 205 | TP53INP1 - BCL2L13 | 1550 | 1616 | 1245 |
| TP53INP2 - BCL2L13 | 507 | 2427 | 2008 | TP53INP2 - BCL2L13 | 1190 | 573 | 513 |
| Ranking of BCL3 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL3 | 1754 | 335 | 226 | TP53BP2 - BCL3 | 1177 | 1625 | 423 |
| TP53I3 - BCL3 | 388 | 392 | 25 | TP53I3 - BCL3 | 921 | 1151 | 233 |
| TP53INP1 - BCL3 | 2350 | 766 | 472 | TP53INP1 - BCL3 | 1126 | 2259 | 2043 |
| TP53INP2 - BCL3 | 266 | 1184 | 379 | TP53INP2 - BCL3 | 325 | 609 | 726 |
| Ranking of BCL6 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL6 | 1172 | 1783 | 1120 | TP53BP2 - BCL6 | 1667 | 1140 | 185 |
| TP53I3 - BCL6 | 2275 | 2312 | 2146 | TP53I3 - BCL6 | 979 | 71 | 859 |
| TP53INP1 - BCL6 | 201 | 1818 | 1572 | TP53INP1 - BCL6 | 1458 | 1200 | 2503 |
| TP53INP2 - BCL6 | 1681 | 2329 | 2352 | TP53INP2 - BCL6 | 346 | 833 | 1557 |
| Ranking of BCL9L w.r.t TP53 family | Ranking of TP53 family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL9L | 263 | 2320 | 2197 | TP53BP2 - BCL9L | 2093 | 2217 | 1010 |
| TP53I3 - BCL9L | 819 | 635 | 789 | TP53I3 - BCL9L | 1249 | 927 | 107 |
| TP53INP1 - BCL9L | 2090 | 1740 | 1179 | TP53INP1 - BCL9L | 2113 | 854 | 1711 |
| TP53INP2 - BCL9L | 640 | 951 | 316 | TP53INP2 - BCL9L | 2222 | 1900 | 151 |
| Ranking of BCL10 w.r.t TP53 family | Ranking of TP53 family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - BCL10 | 2230 | 2418 | 73 | TP53BP2 - BCL10 | 493 | 1999 | 351 |
| TP53I3 - BCL10 | 727 | 1159 | 1301 | TP53I3 - BCL10 | 519 | 1572 | 446 |
| TP53INP1 - BCL10 | 543 | 1223 | 1275 | TP53INP1 - BCL10 | 1094 | 1120 | 1848 |
| TP53INP2 - BCL10 | 632 | 1910 | 2087 | TP53INP2 - BCL10 | 789 | 1566 | 848 |
| Unexplored combinatorial hypotheses | |
| BCL w.r.t TP53 | |
| TP53-I3/INP2 | BCL2L2 |
| TP53-INP2 | BCL2L13 |
| TP53-I3/INP2 | BCL6 |
| TP53-BP2 | BCL9L |
| TP53-BP2/INP2 | BCL10 |
| TP53 w.r.t BCL | |
| TP53-BP2/I3 | BCL2L1 |
| TP53-INP1 | BCL3 |
| TP53-BP2/INP2 | BCL9L |
3.8.4. CASP - BCL Cross Family Analysis
| Ranking CASP family vs BCL family | |||||||
| Ranking of BCL2L1 w.r.t CASP family | Ranking of CASP family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL2L1 | 170 | 1441 | 1555 | CASP4 - BCL2L1 | 355 | 1603 | 202 |
| CASP5 - BCL2L1 | 1236 | 766 | 1261 | CASP5 - BCL2L1 | 1992 | 2053 | 291 |
| CASP7 - BCL2L1 | 2235 | 1161 | 1252 | CASP7 - BCL2L1 | 657 | 2203 | 1750 |
| CASP9 - BCL2L1 | 291 | 984 | 692 | CASP9 - BCL2L1 | 833 | 1386 | 1855 |
| CASP10 - BCL2L1 | 1162 | 2043 | 218 | CASP10 - BCL2L1 | 721 | 2088 | 101 |
| CASP16 - BCL2L1 | 239 | 34 | 305 | CASP16 - BCL2L1 | 43 | 489 | 351 |
| Ranking of BCL2L2 w.r.t CASP family | Ranking of CASP family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL2L2 | 1144 | 1441 | 2348 | CASP4 - BCL2L2 | 988 | 966 | 1400 |
| CASP5 - BCL2L2 | 1896 | 766 | 914 | CASP5 - BCL2L2 | 401 | 174 | 1136 |
| CASP7 - BCL2L2 | 895 | 1161 | 1604 | CASP7 - BCL2L2 | 2371 | 1352 | 1312 |
| CASP9 - BCL2L2 | 1414 | 984 | 1933 | CASP9 - BCL2L2 | 863 | 720 | 102 |
| CASP10 - BCL2L2 | 1335 | 2043 | 1809 | CASP10 - BCL2L2 | 1630 | 1912 | 884 |
| CASP16 - BCL2L2 | 2263 | 34 | 1863 | CASP16 - BCL2L2 | 2 | 151 | 114 |
| Ranking of BCL2L13 w.r.t CASP family | Ranking of CASP family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL2L13 | 1873 | 1096 | 2415 | CASP4 - BCL2L13 | 1257 | 1902 | 1979 |
| CASP5 - BCL2L13 | 1962 | 2514 | 2493 | CASP5 - BCL2L13 | 1438 | 1376 | 664 |
| CASP7 - BCL2L13 | 601 | 1195 | 756 | CASP7 - BCL2L13 | 1877 | 1646 | 2216 |
| CASP9 - BCL2L13 | 1592 | 2371 | 1376 | CASP9 - BCL2L13 | 447 | 1618 | 844 |
| CASP10 - BCL2L13 | 489 | 384 | 987 | CASP10 - BCL2L13 | 1403 | 1048 | 354 |
| CASP16 - BCL2L13 | 1762 | 2492 | 2166 | CASP16 - BCL2L13 | 1927 | 376 | 510 |
| Ranking of BCL3 w.r.t CASP family | Ranking of CASP family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL3 | 18 | 844 | 1229 | CASP4 - BCL3 | 335 | 172 | 1629 |
| CASP5 - BCL3 | 728 | 953 | 1616 | CASP5 - BCL3 | 343 | 498 | 628 |
| CASP7 - BCL3 | 737 | 574 | 580 | CASP7 - BCL3 | 1313 | 1804 | 1556 |
| CASP9 - BCL3 | 1478 | 284 | 242 | CASP9 - BCL3 | 2392 | 1123 | 1394 |
| CASP10 - BCL3 | 2409 | 2011 | 1425 | CASP10 - BCL3 | 156 | 838 | 1678 |
| CASP16 - BCL3 | 868 | 103 | 715 | CASP16 - BCL3 | 361 | 162 | 2505 |
| Ranking of BCL6 w.r.t CASP family | Ranking of CASP family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL6 | 1311 | 2266 | 1297 | CASP4 - BCL6 | 27 | 507 | 944 |
| CASP5 - BCL6 | 1787 | 2124 | 2309 | CASP5 - BCL6 | 760 | 10 | 770 |
| CASP7 - BCL6 | 996 | 1314 | 2322 | CASP7 - BCL6 | 1478 | 1230 | 2366 |
| CASP9 - BCL6 | 1022 | 824 | 2021 | CASP9 - BCL6 | 1855 | 903 | 1296 |
| CASP10 - BCL6 | 469 | 1559 | 1085 | CASP10 - BCL6 | 591 | 787 | 1410 |
| CASP16 - BCL6 | 2397 | 2166 | 2387 | CASP16 - BCL6 | 1514 | 54 | 1881 |
| Ranking of BCL9L w.r.t CASP family | Ranking of CASP family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL9L | 578 | 897 | 325 | CASP4 - BCL9L | 1758 | 1346 | 1584 |
| CASP5 - BCL9L | 1075 | 791 | 1134 | CASP5 - BCL9L | 363 | 1731 | 632 |
| CASP7 - BCL9L | 2279 | 1347 | 632 | CASP7 - BCL9L | 1813 | 853 | 1980 |
| CASP9 - BCL9L | 98 | 1126 | 455 | CASP9 - BCL9L | 1472 | 717 | 940 |
| CASP10 - BCL9L | 24 | 841 | 2358 | CASP10 - BCL9L | 675 | 1449 | 699 |
| CASP16 - BCL9L | 591 | 666 | 233 | CASP16 - BCL9L | 12 | 2499 | 2027 |
| Ranking of BCL10 w.r.t CASP family | Ranking of CASP family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| CASP4 - BCL10 | 1272 | 1457 | 619 | CASP4 - BCL10 | 244 | 1637 | 426 |
| CASP5 - BCL10 | 1732 | 1092 | 1293 | CASP5 - BCL10 | 667 | 2488 | 522 |
| CASP7 - BCL10 | 1448 | 1028 | 681 | CASP7 - BCL10 | 2489 | 1516 | 1945 |
| CASP9 - BCL10 | 612 | 553 | 205 | CASP9 - BCL10 | 1644 | 1117 | 956 |
| CASP10 - BCL10 | 2289 | 1694 | 1401 | CASP10 - BCL10 | 664 | 917 | 84 |
| CASP16 - BCL10 | 27 | 102 | 301 | CASP16 - BCL10 | 2192 | 3 | 387 |
| Unexplored combinatorial hypotheses | |
| BCL w.r.t CASP | |
| CASP-10/16 | BCL2L2 |
| CASP-4/5/16 | BCL2L13 |
| CASP-10 | BCL3 |
| CASP-5/16 | BCL6 |
| CASP w.r.t BCL | |
| CASP-5/7 | BCL2L1 |
| CASP-4/7 | BCL2L13 |
| CASP-7/16 | BCL9L |
| CASP-7 | BCL10 |
3.8.5. MUC - BCL Cross Family Analysis
| Ranking MUC family vs BCL family | |||||||
| Ranking of BCL2L1 w.r.t MUC family | Ranking of MUC family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL2L1 | 2055 | 2297 | 1854 | MUC1 - BCL2L1 | 1226 | 1681 | 986 |
| MUC3A - BCL2L1 | 603 | 2089 | 1637 | MUC3A - BCL2L1 | 759 | 1107 | 678 |
| MUC4 - BCL2L1 | 531 | 1137 | 711 | MUC4 - BCL2L1 | 1758 | 999 | 487 |
| MUC12 - BCL2L1 | 882 | 810 | 1305 | MUC12 - BCL2L1 | 1591 | 900 | 272 |
| MUC13 - BCL2L1 | 1927 | 1201 | 2108 | MUC13 - BCL2L1 | 98 | 2160 | 1099 |
| MUC17 - BCL2L1 | 1170 | 917 | 743 | MUC17 - BCL2L1 | 2500 | 93 | 148 |
| MUC20 - BCL2L1 | 1810 | 700 | 1627 | MUC20 - BCL2L1 | 270 | 343 | 423 |
| Ranking of BCL2L2 w.r.t MUC family | Ranking of MUC family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL2L2 | 1578 | 1425 | 1826 | MUC1 - BCL2L2 | 2476 | 903 | 739 |
| MUC3A - BCL2L2 | 1542 | 370 | 159 | MUC3A - BCL2L2 | 2099 | 241 | 2397 |
| MUC4 - BCL2L2 | 1323 | 2506 | 1988 | MUC4 - BCL2L2 | 797 | 727 | 851 |
| MUC12 - BCL2L2 | 602 | 2504 | 815 | MUC12 - BCL2L2 | 516 | 38 | 1688 |
| MUC13 - BCL2L2 | 2084 | 2402 | 1200 | MUC13 - BCL2L2 | 2201 | 717 | 233 |
| MUC17 - BCL2L2 | 2283 | 2212 | 1279 | MUC17 - BCL2L2 | 903 | 295 | 913 |
| MUC20 - BCL2L2 | 890 | 1886 | 480 | MUC20 - BCL2L2 | 1892 | 569 | 1040 |
| Ranking of BCL2L13 w.r.t MUC family | Ranking of MUC family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL2L13 | 2029 | 2347 | 550 | MUC1 - BCL2L13 | 1838 | 903 | 739 |
| MUC3A - BCL2L13 | 2140 | 1123 | 1100 | MUC3A - BCL2L13 | 173 | 241 | 2397 |
| MUC4 - BCL2L13 | 1497 | 1918 | 1579 | MUC4 - BCL2L13 | 1906 | 727 | 851 |
| MUC12 - BCL2L13 | 581 | 2353 | 1997 | MUC12 - BCL2L13 | 2096 | 38 | 1688 |
| MUC13 - BCL2L13 | 1210 | 2185 | 1658 | MUC13 - BCL2L13 | 1688 | 717 | 233 |
| MUC17 - BCL2L13 | 1079 | 1270 | 1254 | MUC17 - BCL2L13 | 1167 | 295 | 913 |
| MUC20 - BCL2L13 | 187 | 2081 | 535 | MUC20 - BCL2L13 | 1653 | 569 | 1040 |
| Ranking of BCL3 w.r.t MUC family | Ranking of MUC family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL3 | 458 | 1016 | 1881 | MUC1 - BCL3 | 273 | 360 | 1683 |
| MUC3A - BCL3 | 1642 | 668 | 588 | MUC3A - BCL3 | 1044 | 860 | 1452 |
| MUC4 - BCL3 | 427 | 321 | 457 | MUC4 - BCL3 | 624 | 1360 | 585 |
| MUC12 - BCL3 | 1813 | 311 | 1623 | MUC12 - BCL3 | 1193 | 1092 | 132 |
| MUC13 - BCL3 | 2151 | 641 | 1407 | MUC13 - BCL3 | 279 | 65 | 603 |
| MUC17 - BCL3 | 1106 | 531 | 2310 | MUC17 - BCL3 | 305 | 1285 | 257 |
| MUC20 - BCL3 | 2512 | 63 | 2440 | MUC20 - BCL3 | 16 | 539 | 2198 |
| Ranking of BCL6 w.r.t MUC family | Ranking of MUC family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL6 | 1652 | 2294 | 173 | MUC1 - BCL6 | 1550 | 595 | 788 |
| MUC3A - BCL6 | 2323 | 1435 | 187 | MUC3A - BCL6 | 407 | 809 | 318 |
| MUC4 - BCL6 | 723 | 711 | 1403 | MUC4 - BCL6 | 176 | 203 | 1963 |
| MUC12 - BCL6 | 184 | 1024 | 1267 | MUC12 - BCL6 | 1126 | 26 | 229 |
| MUC13 - BCL6 | 158 | 1083 | 2198 | MUC13 - BCL6 | 1633 | 1052 | 603 |
| MUC17 - BCL6 | 2411 | 2153 | 1808 | MUC17 - BCL6 | 242 | 719 | 1026 |
| MUC20 - BCL6 | 925 | 840 | 2153 | MUC20 - BCL6 | 1132 | 1669 | 652 |
| Ranking of BCL9L w.r.t MUC family | Ranking of MUC family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL9L | 2194 | 744 | 1112 | MUC1 - BCL9L | 1144 | 1999 | 896 |
| MUC3A - BCL9L | 2114 | 1441 | 1359 | MUC3A - BCL9L | 901 | 2180 | 2106 |
| MUC4 - BCL9L | 882 | 466 | 1526 | MUC4 - BCL9L | 658 | 1152 | 781 |
| MUC12 - BCL9L | 1547 | 526 | 2391 | MUC12 - BCL9L | 1733 | 1510 | 366 |
| MUC13 - BCL9L | 1545 | 1891 | 796 | MUC13 - BCL9L | 1529 | 502 | 602 |
| MUC17 - BCL9L | 1282 | 1160 | 1362 | MUC17 - BCL9L | 955 | 1788 | 99 |
| MUC20 - BCL9L | 2101 | 116 | 2408 | MUC20 - BCL9L | 307 | 1516 | 1042 |
| Ranking of BCL10 w.r.t MUC family | Ranking of MUC family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - BCL10 | 1325 | 1524 | 1900 | MUC1 - BCL10 | 547 | 1319 | 284 |
| MUC3A - BCL10 | 1298 | 1004 | 1509 | MUC3A - BCL10 | 1681 | 751 | 2250 |
| MUC4 - BCL10 | 304 | 1632 | 1050 | MUC4 - BCL10 | 591 | 570 | 151 |
| MUC12 - BCL10 | 1019 | 1093 | 2239 | MUC12 - BCL10 | 38 | 1155 | 817 |
| MUC13 - BCL10 | 358 | 1687 | 2004 | MUC13 - BCL10 | 517 | 2229 | 455 |
| MUC17 - BCL10 | 524 | 2038 | 1579 | MUC17 - BCL10 | 216 | 803 | 132 |
| MUC20 - BCL10 | 1380 | 619 | 2081 | MUC20 - BCL10 | 97 | 465 | 239 |
| Unexplored combinatorial hypotheses | |
| MUC w.r.t BCL | |
| MUC-3A | BCL2L2 |
| MUC-3A | BCL9L |
| BCL w.r.t MUC | |
| MUC-1/13 | BCL2L1 |
| MUC-4/13/17 | BCL2L2 |
| MUC-1/12 | BCL2L13 |
| MUC-20 | BCL3 |
| MUC-17 | BCL6 |
| MUC-20 | BCL9L |
3.8.6. EXOSC - BCL Cross Family Analysis
3.9. Poliovirus-Receptor Related Synergies
3.9.1. PVR - Interferon Cross Family Analysis
| Ranking PVR family vs IFN family | |||||||
| Ranking of PVR w.r.t IFN family | Ranking of IFN family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVR - IFNAR2 | 1378 | 1651 | 1539 | PVR - IFNAR2 | 1630 | 161 | 930 |
| PVR - IFNE | 1305 | 2044 | 1975 | PVR - IFNE | 1071 | 1486 | 362 |
| PVR - IFNGR1 | 1331 | 268 | 1000 | PVR - IFNGR1 | 2268 | 2040 | 2235 |
| PVR - IFNGR2 | 1911 | 1871 | 1426 | PVR - IFNGR2 | 598 | 1059 | 832 |
| PVR - IFNLR1 | 717 | 2212 | 1884 | PVR - IFNLR1 | 2119 | 1918 | 1499 |
| PVR - IFNWP19 | 1648 | 1438 | 1547 | PVR - IFNWP19 | 1699 | 168 | 2218 |
| Ranking of PVRL2 w.r.t IFN family | Ranking of IFN family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVRL2 - IFNAR2 | 1877 | 1085 | 492 | PVRL2 - IFNAR2 | 623 | 1259 | 2073 |
| PVRL2 - IFNE | 1851 | 990 | 2120 | PVRL2 - IFNE | 683 | 328 | 1416 |
| PVRL2 - IFNGR1 | 1174 | 510 | 526 | PVRL2 - IFNGR1 | 1352 | 1885 | 1433 |
| PVRL2 - IFNGR2 | 1608 | 173 | 1036 | PVRL2 - IFNGR2 | 2049 | 490 | 2056 |
| PVRL2 - IFNLR1 | 1577 | 701 | 1333 | PVRL2 - IFNLR1 | 535 | 1258 | 295 |
| PVRL2 - IFNWP19 | 1455 | 954 | 182 | PVRL2 - IFNWP19 | 870 | 1803 | 1394 |
| Ranking of PVRL4 w.r.t IFN family | Ranking of IFN family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVRL4 - IFNAR2 | 1555 | 227 | 2433 | PVRL4 - IFNAR2 | 490 | 2303 | 1701 |
| PVRL4 - IFNE | 64 | 781 | 1466 | PVRL4 - IFNE | 2026 | 1908 | 465 |
| PVRL4 - IFNGR1 | 2218 | 651 | 188 | PVRL4 - IFNGR1 | 560 | 793 | 889 |
| PVRL4 - IFNGR2 | 220 | 31 | 873 | PVRL4 - IFNGR2 | 213 | 2079 | 31 |
| PVRL4 - IFNLR1 | 284 | 958 | 683 | PVRL4 - IFNLR1 | 766 | 1432 | 2153 |
| PVRL4 - IFNWP19 | 138 | 2271 | 384 | PVRL4 - IFNWP19 | 788 | 1046 | 921 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t IFN | |
| PVR | IFN-E/GR2/LR1 |
| PVRL2 | IFN-E |
| IFN w.r.t PVR | |
| IFN-GR1/LR1 | PVR |
| IFN-GR2 | PVRL2 |
| IFN-E | PVRL4 |
3.9.2. Interferon - Wnt Cross Family Analysis
| Ranking IFN family vs WNT family | |||||||
| Ranking of IFNAR2 w.r.t WNT family | Ranking of WNT family w.r.t IFNAR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNAR2 - WNT2B | 826 | 829 | 1463 | IFNAR2 - WNT2B | 785 | 1146 | 1642 |
| IFNAR2 - WNT4 | 680 | 658 | 802 | IFNAR2 - WNT4 | 1969 | 130 | 126 |
| IFNAR2 - WNT7B | 1009 | 1252 | 581 | IFNAR2 - WNT7B | 2208 | 1635 | 1647 |
| IFNAR2 - WNT9A | 532 | 180 | 1737 | IFNAR2 - WNT9A | 1223 | 1422 | 1632 |
| Ranking of IFNE w.r.t WNT family | Ranking of WNT family w.r.t IFNE | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNE - WNT2B | 1612 | 1973 | 519 | IFNE - WNT2B | 1057 | 1146 | 239 |
| IFNE - WNT4 | 1609 | 2262 | 1320 | IFNE - WNT4 | 585 | 440 | 6 |
| IFNE - WNT7B | 1872 | 1240 | 2341 | IFNE - WNT7B | 2055 | 941 | 936 |
| IFNE - WNT9A | 2114 | 1029 | 267 | IFNE - WNT9A | 124 | 458 | 708 |
| Ranking of IFNGR1 w.r.t WNT family | Ranking of WNT family w.r.t IFNGR1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNGR1 - WNT2B | 1623 | 144 | 361 | IFNGR1 - WNT2B | 1057 | 1849 | 2282 |
| IFNGR1 - WNT4 | 225 | 455 | 1773 | IFNGR1 - WNT4 | 2428 | 1226 | 2479 |
| IFNGR1 - WNT7B | 1004 | 1259 | 1135 | IFNGR1 - WNT7B | 710 | 2278 | 2164 |
| IFNGR1 - WNT9A | 601 | 958 | 1864 | IFNGR1 - WNT9A | 1668 | 1725 | 1462 |
| Ranking of IFNGR2 w.r.t WNT family | Ranking of WNT family w.r.t IFNGR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNGR2 - WNT2B | 1224 | 1322 | 2156 | IFNGR2 - WNT2B | 828 | 1599 | 400 |
| IFNGR2 - WNT4 | 584 | 1117 | 59 | IFNGR2 - WNT4 | 498 | 33 | 168 |
| IFNGR2 - WNT7B | 1185 | 745 | 242 | IFNGR2 - WNT7B | 1964 | 1020 | 638 |
| IFNGR2 - WNT9A | 754 | 501 | 676 | IFNGR2 - WNT9A | 261 | 1711 | 654 |
| Ranking of IFNLR1 w.r.t WNT family | Ranking of WNT family w.r.t IFNLR1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNLR1 - WNT2B | 1621 | 851 | 510 | IFNLR1 - WNT2B | 2263 | 1683 | 2216 |
| IFNLR1 - WNT4 | 1538 | 250 | 220 | IFNLR1 - WNT4 | 2364 | 231 | 503 |
| IFNLR1 - WNT7B | 1012 | 173 | 506 | IFNLR1 - WNT7B | 406 | 573 | 446 |
| IFNLR1 - WNT9A | 347 | 134 | 2160 | IFNLR1 - WNT9A | 1815 | 1709 | 106 |
| Ranking of IFNWP19 w.r.t WNT family | Ranking of WNT family w.r.t IFNWP19 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IFNWP19 - WNT2B | 826 | 176 | 787 | IFNWP19 - WNT2B | 1600 | 2096 | 1650 |
| IFNWP19 - WNT4 | 680 | 1507 | 391 | IFNWP19 - WNT4 | 1441 | 2423 | 2330 |
| IFNWP19 - WNT7B | 1009 | 1101 | 1327 | IFNWP19 - WNT7B | 1838 | 966 | 1478 |
| IFNWP19 - WNT9A | 532 | 1404 | 431 | IFNWP19 - WNT9A | 1354 | 1968 | 703 |
| Unexplored combinatorial hypotheses | |
| IFN w.r.t WNT | |
| IFNE | WNT7B |
| WNT w.r.t IFN | |
| IFNGR1 | WNT2B |
| IFNGR1 | WNT4/WNT7B |
| IFNLR1 | WNT2B |
| IFNWP19 | WNT4 |
3.9.3. PVR - WNT Cross Family Analysis
| Ranking PVR family vs WNT family | |||||||
| Ranking of PVR w.r.t WNT family | Ranking of WNT family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVR - WNT2B | 2204 | 733 | 1 | PVR - WNT2B | 1205 | 2257 | 607 |
| PVR - WNT4 | 1295 | 970 | 878 | PVR - WNT4 | 38 | 2470 | 1094 |
| PVR - WNT7B | 1237 | 770 | 1887 | PVR - WNT7B | 2216 | 1844 | 2096 |
| PVR - WNT9A | 2322 | 649 | 2202 | PVR - WNT9A | 2152 | 2120 | 1131 |
| Ranking of PVRL2 w.r.t WNT family | Ranking of WNT family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVRL2 - WNT2B | 616 | 375 | 2381 | PVRL2 - WNT2B | 1901 | 1044 | 621 |
| PVRL2 - WNT4 | 1110 | 1584 | 1391 | PVRL2 - WNT4 | 2324 | 216 | 2462 |
| PVRL2 - WNT7B | 2186 | 1122 | 349 | PVRL2 - WNT7B | 560 | 560 | 953 |
| PVRL2 - WNT9A | 110 | 1367 | 1858 | PVRL2 - WNT9A | 1044 | 1502 | 794 |
| Ranking of PVRL4 w.r.t WNT family | Ranking of WNT family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| PVRL4 - WNT2B | 949 | 565 | 95 | PVRL4 - WNT2B | 78 | 966 | 1938 |
| PVRL4 - WNT4 | 885 | 1672 | 2149 | PVRL4 - WNT4 | 611 | 1922 | 1488 |
| PVRL4 - WNT7B | 299 | 241 | 798 | PVRL4 - WNT7B | 1192 | 1159 | 2505 |
| PVRL4 - WNT9A | 1375 | 1306 | 492 | PVRL4 - WNT9A | 1383 | 634 | 224 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t WNT | |
| PVR | WNT9A |
| WNT w.r.t PVR | |
| WNT-7B/9A | PVR |
| WNT4 | PVRL2 |
3.9.4. PVR - Integrin Cross Family Analysis
| Ranking PVR family vs ITG family | |||||||
| Ranking of PVR w.r.t ITG family | Ranking of ITG family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ITGA2-PVR | 294 | 564 | 1996 | ITGA2-PVR | 627 | 2062 | 2106 |
| ITGA3-PVR | 1739 | 117 | 1420 | ITGA3-PVR | 2172 | 99 | 827 |
| ITGA6-PVR | 214 | 1328 | 435 | ITGA6-PVR | 576 | 2199 | 817 |
| ITGB1-PVR | 1896 | 136 | 1121 | ITGB1-PVR | 1506 | 1093 | 2203 |
| ITGB1BP1-PVR | 1876 | 1724 | 1505 | ITGB1BP1-PVR | 1241 | 1108 | 535 |
| ITGB4-PVR | 783 | 1495 | 1044 | ITGB4-PVR | 1499 | 120 | 873 |
| ITGB5-PVR | 1719 | 981 | 490 | ITGB5-PVR | 1269 | 1433 | 914 |
| ITGB6-PVR | 1457 | 664 | 1744 | ITGB6-PVR | 1686 | 988 | 879 |
| ITGB8-PVR | 283 | 290 | 334 | ITGB8-PVR | 1407 | 2498 | 2366 |
| Ranking of PVRL2 w.r.t ITG family | Ranking of ITG family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ITGA2-PVRL2 | 1072 | 501 | 851 | ITGA2-PVRL2 | 1883 | 327 | 1141 |
| ITGA3-PVRL2 | 960 | 1905 | 1160 | ITGA3-PVRL2 | 1199 | 1937 | 39 |
| ITGA6-PVRL2 | 352 | 993 | 212 | ITGA6-PVRL2 | 2102 | 709 | 1337 |
| ITGB1-PVRL2 | 720 | 1751 | 836 | ITGB1-PVRL2 | 922 | 568 | 1546 |
| ITGB1BP1-PVRL2 | 1436 | 1313 | 88 | ITGB1BP1-PVRL2 | 168 | 2470 | 2408 |
| ITGB4-PVRL2 | 1857 | 1269 | 1750 | ITGB4-PVRL2 | 565 | 440 | 1197 |
| ITGB5-PVRL2 | 238 | 100 | 1314 | ITGB5-PVRL2 | 543 | 1738 | 1605 |
| ITGB6-PVRL2 | 1873 | 582 | 1492 | ITGB6-PVRL2 | 1052 | 2428 | 2364 |
| ITGB8-PVRL2 | 695 | 612 | 1500 | ITGB8-PVRL2 | 2046 | 2385 | 2110 |
| Ranking of PVRL4 w.r.t ITG family | Ranking of ITG family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ITGA2-PVRL4 | 69 | 951 | 917 | ITGA2-PVRL4 | 2154 | 666 | 1266 |
| ITGA3-PVRL4 | 66 | 1648 | 825 | ITGA3-PVRL4 | 2355 | 357 | 801 |
| ITGA6-PVRL4 | 994 | 528 | 109 | ITGA6-PVRL4 | 2359 | 299 | 157 |
| ITGB1-PVRL4 | 1631 | 1724 | 917 | ITGB1-PVRL4 | 2100 | 1282 | 526 |
| ITGB1BP1-PVRL4 | 1369 | 90 | 462 | ITGB1BP1-PVRL4 | 1815 | 1287 | 2362 |
| ITGB4-PVRL4 | 743 | 1602 | 2443 | ITGB4-PVRL4 | 126 | 1844 | 703 |
| ITGB5-PVRL4 | 2418 | 1802 | 119 | ITGB5-PVRL4 | 1818 | 834 | 2256 |
| ITGB6-PVRL4 | 500 | 1187 | 122 | ITGB6-PVRL4 | 1618 | 2425 | 402 |
| ITGB8-PVRL4 | 861 | 699 | 780 | ITGB8-PVRL4 | 1641 | 394 | 1282 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t ITG | |
| PVRL2 | ITGB4 |
| PVRL4 | ITGB5 |
| ITG w.r.t PVR | |
| ITG-A2/B8 | PVR |
| ITG-B1BP1/B6/B8 | PVRL2 |
| ITG-B1BP1/B5 | PVRL4 |
3.9.5. PVR - TNF Cross Family Analysis
| Ranking PVR vs TNF family | |||||||
| Ranking of PVR w.r.t TNF family | Ranking of TNF family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF-PVR | 1963 | 2422 | 1822 | TNF-PVR | 451 | 54 | 209 |
| TNFAIP1-PVR | 88 | 2210 | 2243 | TNFAIP1-PVR | 527 | 474 | 743 |
| TNFAIP2-PVR | 2028 | 300 | 2451 | TNFAIP2-PVR | 1422 | 632 | 1486 |
| TNFAIP3-PVR | 2454 | 1065 | 1293 | TNFAIP3-PVR | 517 | 1476 | 1611 |
| TNFRSF1A-PVR | 2029 | 500 | 2078 | TNFRSF1A-PVR | 1530 | 778 | 1865 |
| TNFRSF10A-PVR | 1140 | 1978 | 1942 | TNFRSF10A-PVR | 2124 | 1648 | 1420 |
| TNFRSF10B-PVR | 1529 | 1608 | 463 | TNFRSF10B-PVR | 151 | 1266 | 649 |
| TNFRSF10D-PVR | 1321 | 2136 | 1561 | TNFRSF10D-PVR | 1997 | 732 | 1614 |
| TNFRSF12A-PVR | 507 | 93 | 1816 | TNFRSF12A-PVR | 1149 | 1358 | 2417 |
| TNFRSF14-PVR | 983 | 1419 | 409 | TNFRSF14-PVR | 2351 | 2289 | 1577 |
| TNFRSF21-PVR | 485 | 541 | 1910 | TNFRSF21-PVR | 1414 | 969 | 1247 |
| TNFSF10-PVR | 1482 | 317 | 297 | TNFSF10-PVR | 681 | 1150 | 1983 |
| TNFSF15-PVR | 210 | 194 | 56 | TNFSF15-PVR | 635 | 2086 | 1054 |
| Ranking of PVRL2 w.r.t TNF family | Ranking of TNF family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF-PVRL2 | 831 | 367 | 526 | TNF-PVRL2 | 2494 | 652 | 966 |
| TNFAIP1-PVRL2 | 867 | 1568 | 1962 | TNFAIP1-PVRL2 | 2244 | 27 | 1932 |
| TNFAIP2-PVRL2 | 2515 | 2423 | 1062 | TNFAIP2-PVRL2 | 2337 | 2483 | 2401 |
| TNFAIP3-PVRL2 | 973 | 595 | 988 | TNFAIP3-PVRL2 | 1334 | 777 | 896 |
| TNFRSF1A-PVRL2 | 742 | 1326 | 1798 | TNFRSF1A-PVRL2 | 1741 | 2355 | 1810 |
| TNFRSF10A-PVRL2 | 830 | 1008 | 478 | TNFRSF10A-PVRL2 | 1027 | 1672 | 3 |
| TNFRSF10B-PVRL2 | 27 | 2160 | 1210 | TNFRSF10B-PVRL2 | 253 | 1794 | 168 |
| TNFRSF10D-PVRL2 | 312 | 1154 | 1229 | TNFRSF10D-PVRL2 | 564 | 1719 | 170 |
| TNFRSF12A-PVRL2 | 1282 | 382 | 1056 | TNFRSF12A-PVRL2 | 594 | 1870 | 1376 |
| TNFRSF14-PVRL2 | 288 | 922 | 264 | TNFRSF14-PVRL2 | 2148 | 1496 | 232 |
| TNFRSF21-PVRL2 | 1590 | 771 | 1034 | TNFRSF21-PVRL2 | 2120 | 734 | 1782 |
| TNFSF10-PVRL2 | 472 | 1160 | 1056 | TNFSF10-PVRL2 | 98 | 695 | 714 |
| TNFSF15-PVRL2 | 373 | 2154 | 420 | TNFSF15-PVRL2 | 768 | 296 | 2448 |
| Ranking of PVRL4 w.r.t TNF family | Ranking of TNF family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF-PVRL4 | 540 | 680 | 284 | TNF-PVRL4 | 983 | 2010 | 1345 |
| TNFAIP1-PVRL4 | 1885 | 5 | 131 | TNFAIP1-PVRL4 | 965 | 1896 | 1196 |
| TNFAIP2-PVRL4 | 72 | 2496 | 120 | TNFAIP2-PVRL4 | 2270 | 2429 | 239 |
| TNFAIP3-PVRL4 | 1926 | 599 | 794 | TNFAIP3-PVRL4 | 1442 | 1809 | 1269 |
| TNFRSF1A-PVRL4 | 615 | 1904 | 641 | TNFRSF1A-PVRL4 | 1328 | 2481 | 1316 |
| TNFRSF10A-PVRL4 | 337 | 974 | 1802 | TNFRSF10A-PVRL4 | 1375 | 324 | 1822 |
| TNFRSF10B-PVRL4 | 1762 | 1126 | 483 | TNFRSF10B-PVRL4 | 76 | 40 | 1225 |
| TNFRSF10D-PVRL4 | 2232 | 1056 | 92 | TNFRSF10D-PVRL4 | 1799 | 1327 | 2430 |
| TNFRSF12A-PVRL4 | 601 | 1722 | 1566 | TNFRSF12A-PVRL4 | 2386 | 1670 | 2064 |
| TNFRSF14-PVRL4 | 444 | 1007 | 193 | TNFRSF14-PVRL4 | 1514 | 402 | 447 |
| TNFRSF21-PVRL4 | 12 | 552 | 1875 | TNFRSF21-PVRL4 | 2441 | 1917 | 1689 |
| TNFSF10-PVRL4 | 1149 | 1554 | 341 | TNFSF10-PVRL4 | 150 | 1907 | 743 |
| TNFSF15-PVRL4 | 936 | 1057 | 1160 | TNFSF15-PVRL4 | 2019 | 1452 | 760 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t TNF | |
| PVR | TNF, TNF-AIP1/AIP2/RSF1A/RSF10A |
| PVRL2 | TNF-AIP2 |
| TNF w.r.t PVR | |
| TNFRSF14 | PVR |
| TNF-AIP1/AIP2/RSF1A/RSF10B/RSF21 | PVRL2 |
| TNF-AIP2/RSF10D/RSF12A/RSF21 | PVRL4 |
3.9.6. PVR - IL Cross Family Analysis
| Ranking PVR vs IL family | |||||||
| Ranking of PVR w.r.t IL family | Ranking of IL family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A-PVR | 295 | 854 | 2060 | IL1A-PVR | 2419 | 822 | 1099 |
| IL1B-PVR | 708 | 1282 | 1092 | IL1B-PVR | 1739 | 405 | 155 |
| IL1RAP-PVR | 980 | 714 | 197 | IL1RAP-PVR | 370 | 2194 | 2026 |
| IL1RN-PVR | 1273 | 25 | 2065 | IL1RN-PVR | 2025 | 266 | 796 |
| IL2RG-PVR | 2007 | 704 | 2476 | IL2RG-PVR | 250 | 740 | 988 |
| IL6ST-PVR | 1125 | 356 | 1368 | IL6ST-PVR | 706 | 2434 | 1767 |
| IL8-PVR | 2429 | 2507 | 257 | IL8-PVR | 92 | 1468 | 812 |
| IL10RB-PVR | 2310 | 2190 | 1140 | IL10RB-PVR | 1362 | 798 | 881 |
| IL15-PVR | 2065 | 2008 | 2385 | IL15-PVR | 1612 | 134 | 658 |
| IL15RA-PVR | 376 | 1621 | 1736 | IL15RA-PVR | 752 | 1865 | 2405 |
| IL17C-PVR | 2114 | 2301 | 1160 | IL17C-PVR | 894 | 26 | 350 |
| IL17REL-PVR | 482 | 2317 | 1971 | IL17REL-PVR | 1010 | 2408 | 2028 |
| Ranking of PVRL2 w.r.t IL family | Ranking of IL family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A-PVRL2 | 493 | 849 | 1517 | IL1A-PVRL2 | 2168 | 2431 | 774 |
| IL1B-PVRL2 | 1234 | 180 | 1177 | IL1B-PVRL2 | 1425 | 2390 | 2067 |
| IL1RAP-PVRL2 | 2029 | 1039 | 943 | IL1RAP-PVRL2 | 1393 | 1104 | 670 |
| IL1RN-PVRL2 | 1168 | 295 | 347 | IL1RN-PVRL2 | 959 | 2382 | 481 |
| IL2RG-PVRL2 | 356 | 1752 | 1964 | IL2RG-PVRL2 | 1908 | 990 | 1959 |
| IL6ST-PVRL2 | 243 | 770 | 513 | IL6ST-PVRL2 | 2349 | 384 | 2158 |
| IL8-PVRL2 | 1138 | 641 | 532 | IL8-PVRL2 | 612 | 489 | 1686 |
| IL10RB-PVRL2 | 1746 | 639 | 2502 | IL10RB-PVRL2 | 1968 | 839 | 2084 |
| IL15-PVRL2 | 1107 | 1263 | 759 | IL15-PVRL2 | 1841 | 1579 | 399 |
| IL15RA-PVRL2 | 1216 | 1194 | 330 | IL15RA-PVRL2 | 473 | 811 | 1472 |
| IL17C-PVRL2 | 507 | 920 | 402 | IL17C-PVRL2 | 1167 | 1469 | 43 |
| IL17REL-PVRL2 | 79 | 1306 | 2210 | IL17REL-PVRL2 | 2137 | 962 | 2004 |
| Ranking of PVRL4 w.r.t IL family | Ranking of IL family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A-PVRL4 | 338 | 1487 | 666 | IL1A-PVRL4 | 1409 | 1219 | 2410 |
| IL1B-PVRL4 | 1713 | 110 | 481 | IL1B-PVRL4 | 908 | 305 | 399 |
| IL1RAP-PVRL4 | 864 | 269 | 523 | IL1RAP-PVRL4 | 1704 | 2070 | 309 |
| IL1RN-PVRL4 | 2496 | 643 | 144 | IL1RN-PVRL4 | 964 | 1264 | 697 |
| IL2RG-PVRL4 | 858 | 1229 | 2101 | IL2RG-PVRL4 | 18 | 1504 | 727 |
| IL6ST-PVRL4 | 117 | 605 | 744 | IL6ST-PVRL4 | 1954 | 976 | 204 |
| IL8-PVRL4 | 1041 | 1291 | 975 | IL8-PVRL4 | 1441 | 234 | 1826 |
| IL10RB-PVRL4 | 73 | 523 | 278 | IL10RB-PVRL4 | 2137 | 1296 | 506 |
| IL15-PVRL4 | 2062 | 549 | 556 | IL15-PVRL4 | 1029 | 1377 | 281 |
| IL15RA-PVRL4 | 302 | 1519 | 2186 | IL15RA-PVRL4 | 2429 | 1246 | 1758 |
| IL17C-PVRL4 | 25 | 110 | 14 | IL17C-PVRL4 | 1782 | 2245 | 54 |
| IL17REL-PVRL4 | 1487 | 1107 | 1148 | IL17REL-PVRL4 | 1934 | 2140 | 107 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t IL | |
| PVR | IL-2RG/8/10RB/15/17C/17REL |
| IL w.r.t PVR | |
| IL-1RAP/6ST/15RA/17REL | PVR |
| IL-1A/1B/2RG/6ST | PVRL2 |
| IL-15RA/17C/17REL | PVRL4 |
3.9.7. PVR - Collagen Cross Family Analysis
| Ranking PVR vs COL family | |||||||
| Ranking of PVR w.r.t COL family | Ranking of COL family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| COL5A3-PVR | 193 | 790 | 616 | COL5A3-PVR | 2341 | 1172 | 2472 |
| COL6A1-PVR | 2259 | 1740 | 2385 | COL6A1-PVR | 2213 | 70 | 1280 |
| COL7A1-PVR | 1448 | 1166 | 424 | COL7A1-PVR | 144 | 1008 | 1701 |
| COL9A2-PVR | 218 | 166 | 1375 | COL9A2-PVR | 244 | 2501 | 351 |
| COL17A1-PVR | 1800 | 1167 | 1528 | COL17A1-PVR | 1145 | 559 | 685 |
| COL28A1-PVR | 263 | 1273 | 177 | COL28A1-PVR | 1255 | 2266 | 1034 |
| Ranking of PVRL2 w.r.t COL family | Ranking of COL family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| COL5A3-PVRL2 | 1275 | 1132 | 515 | COL5A3-PVRL2 | 962 | 1588 | 640 |
| COL6A1-PVRL2 | 533 | 1954 | 826 | COL6A1-PVRL2 | 2372 | 850 | 1193 |
| COL7A1-PVRL2 | 594 | 2111 | 1299 | COL7A1-PVRL2 | 22 | 662 | 2168 |
| COL9A2-PVRL2 | 1336 | 939 | 970 | COL9A2-PVRL2 | 2483 | 1548 | 2363 |
| COL17A1-PVRL2 | 1157 | 1080 | 1232 | COL17A1-PVRL2 | 173 | 1103 | 728 |
| COL28A1-PVRL2 | 991 | 348 | 1618 | COL28A1-PVRL2 | 955 | 864 | 1981 |
| Ranking of PVRL4 w.r.t COL family | Ranking of COL family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| COL5A3-PVRL4 | 692 | 1155 | 1446 | COL5A3-PVRL4 | 499 | 405 | 331 |
| COL6A1-PVRL4 | 221 | 1906 | 571 | COL6A1-PVRL4 | 1701 | 2059 | 1315 |
| COL7A1-PVRL4 | 859 | 1320 | 1088 | COL7A1-PVRL4 | 1364 | 2205 | 65 |
| COL9A2-PVRL4 | 1893 | 754 | 1155 | COL9A2-PVRL4 | 1397 | 1797 | 1053 |
| COL17A1-PVRL4 | 1124 | 1647 | 431 | COL17A1-PVRL4 | 1401 | 1174 | 596 |
| COL28A1-PVRL4 | 417 | 1536 | 433 | COL28A1-PVRL4 | 642 | 2446 | 1540 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t COL | |
| PVR | COL6A1 |
| COL w.r.t PVR | |
| COL5A3 | PVR |
| COL9A2 | PVRL2 |
3.9.8. PVR - MUCIN Cross Family Analysis
| Ranking PVR vs MUC family | |||||||
| Ranking of PVR w.r.t MUC family | Ranking of MUC family w.r.t PVR | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1-PVR | 2173 | 1184 | 1021 | MUC1-PVR | 2106 | 377 | 426 |
| MUC3A-PVR | 64 | 130 | 1828 | MUC3A-PVR | 136 | 1217 | 1004 |
| MUC4-PVR | 2000 | 804 | 1026 | MUC4-PVR | 2494 | 366 | 586 |
| MUC12-PVR | 1449 | 1589 | 1899 | MUC12-PVR | 2370 | 224 | 18 |
| MUC13-PVR | 1701 | 1292 | 1226 | MUC13-PVR | 1230 | 144 | 59 |
| MUC17-PVR | 209 | 684 | 881 | MUC17-PVR | 2388 | 320 | 1048 |
| MUC20-PVR | 1772 | 1242 | 2085 | MUC20-PVR | 1380 | 188 | 1039 |
| Ranking of PVRL2 w.r.t MUC family | Ranking of MUC family w.r.t PVRL2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1-PVRL2 | 828 | 454 | 2362 | MUC1-PVRL2 | 1690 | 923 | 1060 |
| MUC3A-PVRL2 | 584 | 708 | 2332 | MUC3A-PVRL2 | 2031 | 164 | 546 |
| MUC4-PVRL2 | 91 | 1214 | 478 | MUC4-PVRL2 | 1315 | 1150 | 230 |
| MUC12-PVRL2 | 1129 | 1655 | 1439 | MUC12-PVRL2 | 1820 | 1417 | 94 |
| MUC13-PVRL2 | 1052 | 179 | 329 | MUC13-PVRL2 | 471 | 1760 | 240 |
| MUC17-PVRL2 | 328 | 2098 | 1869 | MUC17-PVRL2 | 873 | 2111 | 1462 |
| MUC20-PVRL2 | 1407 | 1350 | 1612 | MUC20-PVRL2 | 1348 | 325 | 133 |
| Ranking of PVRL4 w.r.t MUC family | Ranking of MUC family w.r.t PVRL4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1-PVRL4 | 569 | 99 | 155 | MUC1-PVRL4 | 2272 | 1827 | 1457 |
| MUC3A-PVRL4 | 1088 | 1214 | 2498 | MUC3A-PVRL4 | 1698 | 2103 | 1835 |
| MUC4-PVRL4 | 18 | 13 | 714 | MUC4-PVRL4 | 1518 | 1251 | 789 |
| MUC12-PVRL4 | 526 | 449 | 1420 | MUC12-PVRL4 | 311 | 1059 | 2065 |
| MUC13-PVRL4 | 2160 | 1404 | 1937 | MUC13-PVRL4 | 1540 | 45 | 1570 |
| MUC17-PVRL4 | 731 | 221 | 234 | MUC17-PVRL4 | 1736 | 2206 | 219 |
| MUC20-PVRL4 | 22 | 1820 | 361 | MUC20-PVRL4 | 626 | 722 | 2232 |
| Unexplored combinatorial hypotheses | |
| PVR w.r.t MUC | |
| PVR | MUC20 |
| PVRL2 | MUC17 |
| PVRL4 | MUC13 |
| MUC w.r.t PVR | |
| MUC1 | PVRL4 |
| MUC3A | PVRL4 |
3.10. Anthrax Toxin Receptor Related Synergies
3.10.1. ANTXR2 - Collagen Cross Family Analysis
3.10.2. ANTXR2 - Integrin Cross Family Analysis
| Ranking ANTRX2 vs ITG family | |||||||
| Ranking of ANTRX2 w.r.t ITG family | Ranking of ITG family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ITGA2-ANTXR2 | 2261 | 1129 | 2444 | ITGA2-ANTXR2 | 657 | 1662 | 215 |
| ITGA3-ANTXR2 | 2027 | 2134 | 179 | ITGA3-ANTXR2 | 305 | 1402 | 278 |
| ITGA6-ANTXR2 | 1065 | 1850 | 1660 | ITGA6-ANTXR2 | 352 | 2029 | 583 |
| ITGB1-ANTXR2 | 2192 | 1273 | 1431 | ITGB1-ANTXR2 | 1538 | 987 | 1593 |
| ITGB1BP1-ANTXR2 | 2444 | 498 | 2128 | ITGB1BP1-ANTXR2 | 1743 | 152 | 534 |
| ITGB4-ANTXR2 | 1484 | 699 | 249 | ITGB4-ANTXR2 | 123 | 1420 | 2116 |
| ITGB5-ANTXR2 | 1318 | 1860 | 2315 | ITGB5-ANTXR2 | 2216 | 718 | 1182 |
| ITGB6-ANTXR2 | 1205 | 1262 | 1244 | ITGB6-ANTXR2 | 1200 | 2000 | 1896 |
| ITGB8-ANTXR2 | 1710 | 2354 | 2136 | ITGB8-ANTXR2 | 296 | 1724 | 1485 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t ITG | |
| ANTXR2 | ITG-A2/A3 |
| ANTXR2 | ITG-B1BP1/B5/B8 |
| ITG w.r.t ANTXR2 | |
| ITGB6 | ANTXR2 |
3.10.3. ANTXR2 - MMP Cross Family Analysis
| Ranking ANTRX2 vs MMP family | |||||||
| Ranking of ANTRX2 w.r.t MMP family | Ranking of MMP family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MMP1-ANTXR2 | 1428 | 1407 | 1620 | MMP1-ANTXR2 | 244 | 2009 | 2142 |
| MMP14-ANTXR2 | 1067 | 1141 | 900 | MMP14-ANTXR2 | 866 | 971 | 443 |
| MMP15-ANTXR2 | 1457 | 740 | 1881 | MMP15-ANTXR2 | 121 | 2219 | 1926 |
| MMP28-ANTXR2 | 2468 | 1765 | 1202 | MMP28-ANTXR2 | 11 | 1857 | 2092 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t MMP | |
| ANTXR2 | MMP28 |
| MMP w.r.t ANTXR2 | |
| MMP-1/15/28 | ANTXR2 |
3.10.4. ANTXR2 - WNT Cross Family Analysis
| Ranking ANTRX2 vs WNT family | |||||||
| Ranking of ANTRX2 w.r.t WNT family | Ranking of WNT family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B-ANTXR2 | 1160 | 1013 | 2286 | WNT2B-ANTXR2 | 1577 | 1367 | 944 |
| WNT4-ANTXR2 | 1735 | 1833 | 2341 | WNT4-ANTXR2 | 175 | 1643 | 97 |
| WNT7B-ANTXR2 | 2453 | 304 | 1196 | WNT7B-ANTXR2 | 2106 | 242 | 1144 |
| WNT9A-ANTXR2 | 1618 | 487 | 1766 | WNT9A-ANTXR2 | 2317 | 162 | 845 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t WNT | |
| ANTXR2 | WNT4 |
3.10.5. ANTXR2 - TNF Cross Family Analysis
| Ranking ANTRX2 vs TNF family | |||||||
| Ranking of ANTRX2 w.r.t TNF family | Ranking of TNF family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF-ANTXR2 | 1439 | 1568 | 1285 | TNF-ANTXR2 | 709 | 1758 | 1479 |
| TNFAIP1-ANTXR2 | 1552 | 1769 | 1946 | TNFAIP1-ANTXR2 | 1252 | 2177 | 218 |
| TNFAIP2-ANTXR2 | 125 | 962 | 2134 | TNFAIP2-ANTXR2 | 659 | 1156 | 2109 |
| TNFAIP3-ANTXR2 | 1184 | 1253 | 1558 | TNFAIP3-ANTXR2 | 1429 | 2485 | 1731 |
| TNFRSF1A-ANTXR2 | 1063 | 310 | 2145 | TNFRSF1A-ANTXR2 | 1557 | 2471 | 935 |
| TNFRSF10A-ANTXR2 | 351 | 1358 | 1280 | TNFRSF10A-ANTXR2 | 2260 | 32 | 2377 |
| TNFRSF10B-ANTXR2 | 2278 | 2218 | 982 | TNFRSF10B-ANTXR2 | 852 | 715 | 216 |
| TNFRSF10D-ANTXR2 | 1352 | 891 | 1685 | TNFRSF10D-ANTXR2 | 2258 | 454 | 2363 |
| TNFRSF12A-ANTXR2 | 551 | 1283 | 1794 | TNFRSF12A-ANTXR2 | 2190 | 1150 | 2061 |
| TNFRSF14-ANTXR2 | 999 | 442 | 498 | TNFRSF14-ANTXR2 | 2370 | 1777 | 1014 |
| TNFRSF21-ANTXR2 | 897 | 997 | 298 | TNFRSF21-ANTXR2 | 1474 | 343 | 510 |
| TNFRSF10-ANTXR2 | 2151 | 966 | 324 | TNFRSF10-ANTXR2 | 2065 | 112 | 339 |
| TNFRSF15-ANTXR2 | 868 | 967 | 1590 | TNFRSF15-ANTXR2 | 664 | 1211 | 1669 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t TNF | |
| ANTXR2 | TNFAIP1 |
| ANTXR2 | TNFRSF10B |
| TNF w.r.t ANTXR2 | |
| TNFRSF10A | ANTXR2 |
| TNFRSF10D | ANTXR2 |
| TNFRSF12A | ANTXR2 |
| TNFRSF14 | ANTXR2 |
3.10.6. ANTXR2 - IL Cross Family Analysis
| Ranking ANTRX2 vs IL family | |||||||
| Ranking of ANTRX2 w.r.t IL family | Ranking of IL family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A-ANTXR2 | 1733 | 454 | 2253 | IL1A-ANTXR2 | 275 | 2356 | 1859 |
| IL1B-ANTXR2 | 1222 | 1302 | 714 | IL1B-ANTXR2 | 330 | 2011 | 1762 |
| IL1RAP-ANTXR2 | 1288 | 367 | 80 | IL1RAP-ANTXR2 | 2339 | 442 | 747 |
| IL1RN-ANTXR2 | 1389 | 1914 | 1894 | IL1RN-ANTXR2 | 349 | 1031 | 1919 |
| IL2RG-ANTXR2 | 1897 | 25 | 432 | IL2RG-ANTXR2 | 368 | 1867 | 450 |
| IL6ST-ANTXR2 | 1944 | 2219 | 1914 | IL6ST-ANTXR2 | 46 | 1780 | 1865 |
| IL8-ANTXR2 | 1169 | 1281 | 1398 | IL8-ANTXR2 | 1343 | 2002 | 434 |
| IL10RB-ANTXR2 | 1737 | 496 | 1545 | IL10RB-ANTXR2 | 1403 | 800 | 754 |
| IL15-ANTXR2 | 787 | 1812 | 927 | IL15-ANTXR2 | 1002 | 1340 | 481 |
| IL15RA-ANTXR2 | 840 | 800 | 1695 | IL15RA-ANTXR2 | 1924 | 636 | 1901 |
| IL17C-ANTXR2 | 1832 | 2334 | 1191 | IL17C-ANTXR2 | 339 | 2121 | 2437 |
| IL17REL-ANTXR2 | 29 | 1889 | 2303 | IL17REL-ANTXR2 | 2406 | 111 | 960 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t IL | |
| ANTXR2 | IL1RN |
| ANTXR2 | IL6ST |
| ANTXR2 | IL17C |
| ANTXR2 | IL17REL |
| IL w.r.t ANTXR2 | |
| IL1A | ANTXR2 |
| IL1B | ANTXR2 |
| IL6ST | ANTXR2 |
| IL17C | ANTXR2 |
Conclusion
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Choice of Sensitivity Indices
- sensiFdiv - conducts a density-based sensitivity analysis where the impact of an input variable is defined in terms of dissimilarity between the original output density function and the output density function when the input variable is fixed. The dissimilarity between density functions is measured with Csiszar f-divergences. Estimation is performed through kernel density estimation and the function kde of the package ks. Borgonovo [271] and Da Veiga [272]
- sensiHSIC - conducts a sensitivity analysis where the impact of an input variable is defined in terms of the distance between the input/output joint probability distribution and the product of their marginals when they are embedded in a Reproducing Kernel Hilbert Space (RKHS). This distance corresponds to HSIC proposed by Gretton et al. [273] and serves as a dependence measure between random variables.
- sobol2002 - implements the Monte Carlo estimation of the Sobol indices for both first-order and total indices at the same time (all together 2p indices), at a total cost of (p+2) × n model evaluations. These are called the Saltelli estimators. This estimator suffers from a conditioning problem when estimating the variances behind the indices computations. This can seriously affect the Sobol indices estimates in case of largely non-centered output. To avoid this effect, you have to center the model output before applying "sobol2002". Functions ”soboljansen" and "sobolmartinez" do not suffer from this problem. Saltelli [276]
- sobol2007 - implements the Monte Carlo estimation of the Sobol indices for both first-order and total indices at the same time (all together 2p indices), at a total cost of (p+2) × n model evaluations. These are called the Mauntz estimators. Saltelli and Annoni [277]
- sobolmartinez - implements the Monte Carlo estimation of the Sobol indices for both first-order and total indices using correlation coefficients-based formulas, at a total cost of (p + 2) × n model evaluations. These are called the Martinez estimators.
- sobol - implements the Monte Carlo estimation of the Sobol sensitivity indices. Allows the estimation of the indices of the variance decomposition up to a given order, at a total cost of (N + 1) × n where N is the number of indices to estimate. Sobol’ [278]
References
- Brancati, F.; Fortugno, P.; Bottillo, I.; Lopez, M.; Josselin, E.; Boudghene-Stambouli, O.; Agolini, E.; Bernardini, L.; Bellacchio, E.; Iannicelli, M.; et al. Mutations in PVRL4, encoding cell adhesion molecule nectin-4, cause ectodermal dysplasia-syndactyly syndrome. The American Journal of Human Genetics 2010, 87, 265–273. [Google Scholar] [CrossRef]
- Sinha, S. Inchoative Discovery of Plausible (Un)explored Synergistic Combinatorial Biological Hypotheses for Static/Time Series Wnt Measurements via Ranking Search Engine: BioSearch Engine Design. Preprints 2018. [Google Scholar]
- Sinha, S. Sensitivity analysis based ranking reveals unknown biological hypotheses for down regulated genes in time buffer during administration of PORCN-WNT inhibitor ETC-1922159 in CRC. bioRxiv 2017, 180927. [Google Scholar]
- Sinha, S. Prioritizing 2nd order interactions via support vector ranking using sensitivity indices on time series Wnt measurements. bioRxiv 2017, 060228. [Google Scholar]
- Sharma, R. Wingless a new mutant in Drosophila melanogaster. Drosophila information service 1973, 50, 134–134. [Google Scholar]
- Thorstensen, L.; Lind, G.E.; Løvig, T.; Diep, C.B.; Meling, G.I.; Rognum, T.O.; Lothe, R.A. Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia 2005, 7, 99–108. [Google Scholar] [CrossRef]
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nature medicine 2013, 19, 179–192. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/[β]-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Sokol, S. Wnt Signaling in Embryonic Development; Elsevier, 2011; Volume 17. [Google Scholar]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes & development 2003, 17, 1709–1713. [Google Scholar]
- Zhong, Z.; Ethen, N.J.; Williams, B.O. WNT signaling in bone development and homeostasis. Wiley Interdisciplinary Reviews: Developmental Biology 2014, 3, 489–500. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell International 2010, 10, 1–5. [Google Scholar] [CrossRef]
- Kahn, M. Can we safely target the WNT pathway? Nature Reviews Drug Discovery 2014, 13, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. Drugging the Wnt pathway: problems and progress. Journal of the National Cancer Institute 2009, 101, 548–550. [Google Scholar] [CrossRef]
- Voronkov, A.; Krauss, S. Wnt/beta-catenin signaling and small molecule inhibitors. Current pharmaceutical design 2012, 19, 634. [Google Scholar] [CrossRef]
- Blagodatski, A.; Poteryaev, D.; Katanaev, V. Targeting the Wnt pathways for therapies. Mol Cell Ther 2014, 2, 28. [Google Scholar] [CrossRef]
- Curtin, J.C.; Lorenzi, M.V. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget 2010, 1, 552. [Google Scholar] [CrossRef]
- Tanaka, K.; Okabayashi, K.; Asashima, M.; Perrimon, N.; Kadowaki, T. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. The FEBS Journal 2000, 267, 4300–4311. [Google Scholar] [CrossRef]
- Bänziger, C.; Soldini, D.; Schütt, C.; Zipperlen, P.; Hausmann, G.; Basler, K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 2006, 125, 509–522. [Google Scholar] [CrossRef]
- Bartscherer, K.; Pelte, N.; Ingelfinger, D.; Boutros, M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 2006, 125, 523–533. [Google Scholar] [CrossRef]
- Kurayoshi, M.; Yamamoto, H.; Izumi, S.; Kikuchi, A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochemical Journal 2007, 402, 515–523. [Google Scholar] [CrossRef]
- Gao, X.; Hannoush, R.N. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nature chemical biology 2014, 10, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Dodge, M.E.; Tang, W.; Lu, J.; Ma, Z.; Fan, C.W.; Wei, S.; Hao, W.; Kilgore, J.; Williams, N.S.; et al. Small molecule–mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nature chemical biology 2009, 5, 100–107. [Google Scholar] [CrossRef]
- Wang, X.; Moon, J.; Dodge, M.E.; Pan, X.; Zhang, L.; Hanson, J.M.; Tuladhar, R.; Ma, Z.; Shi, H.; Williams, N.S.; et al. The development of highly potent inhibitors for porcupine. Journal of medicinal chemistry 2013, 56, 2700–2704. [Google Scholar] [CrossRef]
- Proffitt, K.D.; Madan, B.; Ke, Z.; Pendharkar, V.; Ding, L.; Lee, M.A.; Hannoush, R.N.; Virshup, D.M. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer research 2013, 73, 502–507. [Google Scholar] [CrossRef]
- Liu, J.; Pan, S.; Hsieh, M.H.; Ng, N.; Sun, F.; Wang, T.; Kasibhatla, S.; Schuller, A.G.; Li, A.G.; Cheng, D.; et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences 2013, 110, 20224–20229. [Google Scholar] [CrossRef] [PubMed]
- Duraiswamy, A.J.; Lee, M.A.; Madan, B.; Ang, S.H.; Tan, E.S.W.; Cheong, W.W.V.; Ke, Z.; Pendharkar, V.; Ding, L.J.; Chew, Y.S.; et al. Discovery and optimization of a porcupine inhibitor. Journal of medicinal chemistry 2015, 58, 5889–5899. [Google Scholar] [CrossRef]
- Wu, X.D.; Bie, Q.L.; Zhang, B.; Yan, Z.H.; Han, Z.J. Wnt10B is critical for the progression of gastric cancer. Oncology Letters 2017, 13, 4231–4237. [Google Scholar] [CrossRef]
- Stevens, J.R.; Miranda-Carboni, G.A.; Singer, M.A.; Brugger, S.M.; Lyons, K.M.; Lane, T.F. Wnt10b deficiency results in age-dependent loss of bone mass and progressive reduction of mesenchymal progenitor cells. Journal of Bone and Mineral Research 2010, 25, 2138–2147. [Google Scholar] [CrossRef]
- Tassew, N.G.; Charish, J.; Shabanzadeh, A.P.; Luga, V.; Harada, H.; Farhani, N.; D’Onofrio, P.; Choi, B.; Ellabban, A.; Nickerson, P.E.; et al. Exosomes Mediate Mobilization of Autocrine Wnt10b to Promote Axonal Regeneration in the Injured CNS. Cell reports 2017, 20, 99–111. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Bree, A.J.; Yao, Y.; Du, B.; Hemati, N.; Martinez-Santibañez, G.; MacDougald, O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 2012, 50, 477–489. [Google Scholar] [CrossRef]
- Collins, F.L.; Rios-Arce, N.D.; McCabe, L.R.; Parameswaran, N. Cytokine and hormonal regulation of bone marrow immune cell Wnt10b expression. PloS one 2017, 12, e0181979. [Google Scholar] [CrossRef]
- Wu, G.; Fan, X.; Sun, L. Silencing of Wnt10B reduces viability of heptocellular carcinoma HepG2 cells. American journal of cancer research 2015, 5, 1911. [Google Scholar]
- Wend, P.; Runke, S.; Wend, K.; Anchondo, B.; Yesayan, M.; Jardon, M.; Hardie, N.; Loddenkemper, C.; Ulasov, I.; Lesniak, M.S.; et al. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO molecular medicine 2013, 5, 264–279. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, C.; Zhan, Y.; Wang, H.; Jiang, X.; Li, W. Aberrant low expression of p85α in stromal fibroblasts promotes breast cancer cell metastasis through exosome-mediated paracrine Wnt10b. Oncogene 2017, 36, 4692. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Xue, F. Expression and the clinical significance of Wnt10a and Wnt10b in endometrial cancer are associated with the Wnt/β-catenin pathway. Oncology reports 2013, 29, 507–514. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Matsubara, K.; Zhou, X.; Okamura, S.; Kubo, T.; Murase, Y.; Shikauchi, Y.; Esteller, M.; Herman, J.G.; Wang, X.W.; et al. WNT10B functional dualism: β-catenin/Tcf-dependent growth promotion or independent suppression with deregulated expression in cancer. Molecular biology of the cell 2007, 18, 4292–4303. [Google Scholar] [CrossRef]
- Zhu, R.; Yang, Y.; Tian, Y.; Bai, J.; Zhang, X.; Li, X.; Peng, Z.; He, Y.; Chen, L.; Pan, Q.; et al. Ascl2 knockdown results in tumor growth arrest by miRNA-302b-related inhibition of colon cancer progenitor cells. PloS one 2012, 7, e32170. [Google Scholar] [CrossRef]
- Giakountis, A.; Moulos, P.; Zarkou, V.; Oikonomou, C.; Harokopos, V.; Hatzigeorgiou, A.G.; Reczko, M.; Hatzis, P. A Positive regulatory loop between a Wnt-Regulated non-coding RNA and ASCL2 controls intestinal stem cell fate. Cell reports 2016, 15, 2588–2596. [Google Scholar] [CrossRef]
- Schuijers, J.; Junker, J.P.; Mokry, M.; Hatzis, P.; Koo, B.K.; Sasselli, V.; Van Der Flier, L.G.; Cuppen, E.; van Oudenaarden, A.; Clevers, H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell stem cell 2015, 16, 158–170. [Google Scholar] [CrossRef]
- Reddy, V.K.; Short, S.P.; Barrett, C.W.; Mittal, M.K.; Keating, C.E.; Thompson, J.J.; Harris, E.I.; Revetta, F.; Bader, D.M.; Brand, T.; et al. BVES regulates intestinal stem cell programs and intestinal crypt viability after radiation. Stem Cells 2016, 34, 1626–1636. [Google Scholar] [CrossRef]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef]
- Kobayashi, M.; Funayama, R.; Ohnuma, S.; Unno, M.; Nakayama, K. Wnt-β-catenin signaling regulates ABCC3 (MRP3) transporter expression in colorectal cancer. Cancer science 2016, 107, 1776–1784. [Google Scholar] [CrossRef]
- Kaler, P.; Godasi, B.N.; Augenlicht, L.; Klampfer, L. The NF-κB/AKT-dependent induction of Wnt signaling in colon cancer cells by macrophages and IL-1β. Cancer Microenvironment 2009, 2, 69. [Google Scholar] [CrossRef]
- Zhong, L.; Schivo, S.; Huang, X.; Leijten, J.; Karperien, M.; Post, J.N. Nitric Oxide Mediates Crosstalk between Interleukin 1β and WNT Signaling in Primary Human Chondrocytes by Reducing DKK1 and FRZB Expression. International journal of molecular sciences 2017, 18, 2491. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Su, H.; Zhong, W.; Yuan, Y.; Yu, Z.; Fang, Y.; Zhou, H.; Li, C.; Huang, K. Interleukin-17 is a favorable prognostic marker for colorectal cancer. Clinical and Translational Oncology 2015, 17, 50–56. [Google Scholar] [CrossRef]
- Housseau, F.; Wu, S.; Wick, E.C.; Fan, H.; Wu, X.; Llosa, N.J.; Smith, K.N.; Tam, A.; Ganguly, S.; Wanyiri, J.W.; et al. Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer research 2016, 76, 2115–2124. [Google Scholar] [CrossRef]
- Starnes, T.; Broxmeyer, H.E.; Robertson, M.J.; Hromas, R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. The Journal of Immunology 2002, 169, 642–646. [Google Scholar] [CrossRef]
- Ma, B.; Hottiger, M.O. Crosstalk between Wnt/β-catenin and NF-κB signaling pathway during inflammation. Frontiers in immunology 2016, 7, 378. [Google Scholar] [CrossRef]
- Ma, B.; van Blitterswijk, C.A.; Karperien, M. A Wnt/β-catenin negative feedback loop inhibits interleukin-1–induced matrix metalloproteinase expression in human articular chondrocytes. Arthritis & Rheumatism 2012, 64, 2589–2600. [Google Scholar]
- Masckauchán, T.N.H.; Shawber, C.J.; Funahashi, Y.; Li, C.M.; Kitajewski, J. Wnt/β-catenin signaling induces proliferation, survival and interleukin-8 in human endothelial cells. Angiogenesis 2005, 8, 43–51. [Google Scholar] [CrossRef]
- Pfalzer, A.C.; Crott, J.W.; Koh, G.Y.; Smith, D.E.; Garcia, P.E.; Mason, J.B. Interleukin-1 Signaling Mediates Obesity-Promoted Elevations in Inflammatory Cytokines, Wnt Activation, and Epithelial Proliferation in the Mouse Colon. Journal of Interferon & Cytokine Research 2018, 38, 445–451. [Google Scholar]
- Aumiller, V.; Balsara, N.; Wilhelm, J.; Günther, A.; Königshoff, M. WNT/β-catenin signaling induces IL-1β expression by alveolar epithelial cells in pulmonary fibrosis. American journal of respiratory cell and molecular biology 2013, 49, 96–104. [Google Scholar] [CrossRef]
- Chen, D.; Li, W.; Liu, S.; Su, Y.; Han, G.; Xu, C.; Liu, H.; Zheng, T.; Zhou, Y.; Mao, C. Interleukin-23 promotes the epithelial-mesenchymal transition of oesophageal carcinoma cells via the Wnt/β-catenin pathway. Scientific reports 2015, 5, 8604. [Google Scholar] [CrossRef]
- Malysheva, K.; Rooij, K.d.; WGM Löwik, C.; L Baeten, D.; Rose-John, S. Interleukin 6/Wnt interactions in rheumatoid arthritis: interleukin 6 inhibits Wnt signaling in synovial fibroblasts and osteoblasts. Croatian medical journal 2016, 57, 89–98. [Google Scholar] [CrossRef]
- Mukai, A.; Yamamoto-Hino, M.; Awano, W.; Watanabe, W.; Komada, M.; Goto, S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. The EMBO journal 2010, 29, 2114–2125. [Google Scholar] [CrossRef]
- Zhang, L.; Wrana, J.L. The emerging role of exosomes in Wnt secretion and transport. Current opinion in genetics & development 2014, 27, 14–19. [Google Scholar]
- Tassew, N.G.; Charish, J.; Shabanzadeh, A.P.; Luga, V.; Harada, H.; Farhani, N.; D’Onofrio, P.; Choi, B.; Ellabban, A.; Nickerson, P.E.; et al. Exosomes mediate mobilization of autocrine Wnt10b to promote axonal regeneration in the injured CNS. Cell reports 2017, 20, 99–111. [Google Scholar] [CrossRef]
- Koles, K.; Budnik, V. Exosomes go with the Wnt. Cellular logistics 2012, 2, 169–173. [Google Scholar] [CrossRef]
- Wu, X.; Deng, G.; Hao, X.; Li, Y.; Zeng, J.; Ma, C.; He, Y.; Liu, X.; Wang, Y. A caspase-dependent pathway is involved in Wnt/β-catenin signaling promoted apoptosis in Bacillus Calmette-Guerin infected RAW264. 7 macrophages. International journal of molecular sciences 2014, 15, 5045–5062. [Google Scholar] [CrossRef]
- Abdul-Ghani, M.; Dufort, D.; Stiles, R.; De Repentigny, Y.; Kothary, R.; Megeney, L.A. Wnt11 promotes cardiomyocyte development by caspase-mediated suppression of canonical Wnt signals. Molecular and cellular biology 2011, 31, 163–178. [Google Scholar] [CrossRef]
- Bisson, J.A.; Mills, B.; Helt, J.C.P.; Zwaka, T.P.; Cohen, E.D. Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT. Developmental biology 2015, 398, 80–96. [Google Scholar] [CrossRef]
- Singh, V.; Holla, S.; Ramachandra, S.G.; Balaji, K.N. WNT-inflammasome signaling mediates NOD2-induced development of acute arthritis in mice. The Journal of Immunology 2015, 194, 3351–3360. [Google Scholar] [CrossRef]
- Flanagan, L.; Meyer, M.; Fay, J.; Curry, S.; Bacon, O.; Duessmann, H.; John, K.; Boland, K.C.; McNamara, D.A.; Kay, E.W.; et al. Low levels of Caspase-3 predict favourable response to 5FU-based chemotherapy in advanced colorectal cancer: Caspase-3 inhibition as a therapeutic approach. Cell death & disease 2016, 7, e2087. [Google Scholar]
- Yao, Q.; Wang, W.; Jin, J.; Min, K.; Yang, J.; Zhong, Y.; Xu, C.; Deng, J.; Zhou, Y. Synergistic role of Caspase-8 and Caspase-3 expressions: Prognostic and predictive biomarkers in colorectal cancer. Cancer Biomarkers 2018, 1–10. [Google Scholar] [CrossRef]
- Sadot, E.; Geiger, B.; Oren, M.; Ben-Ze’ev, A. Down-regulation of β-catenin by activated p53. Molecular and cellular biology 2001, 21, 6768–6781. [Google Scholar] [CrossRef]
- Peng, X.; Yang, L.; Chang, H.; Dai, G.; Wang, F.; Duan, X.; Guo, L.; Zhang, Y.; Chen, G. Wnt/β-catenin signaling regulates the proliferation and differentiation of mesenchymal progenitor cells through the p53 pathway. PloS one 2014, 9, e97283. [Google Scholar] [CrossRef]
- Zhukova, N.; Ramaswamy, V.; Remke, M.; Martin, D.C.; Castelo-Branco, P.; Zhang, C.H.; Fraser, M.; Tse, K.; Poon, R.; Shih, D.J.; et al. WNT activation by lithium abrogates TP53 mutation associated radiation resistance in medulloblastoma. Acta neuropathologica communications 2014, 2, 174. [Google Scholar] [CrossRef]
- Liu, W.; Xu, X.; Fan, Z.; Sun, G.; Han, Y.; Zhang, D.; Xu, L.; Wang, M.; Wang, X.; Zhang, S.; et al. Wnt Signaling Activates TP53-Induced Glycolysis and Apoptosis Regulator and Protects Against Cisplatin-Induced Spiral Ganglion Neuron Damage in the Mouse Cochlea. Antioxidants & redox signaling 2018. [Google Scholar]
- Okayama, S.; Kopelovich, L.; Balmus, G.; Weiss, R.S.; Herbert, B.S.; Dannenberg, A.J.; Subbaramaiah, K. p53 protein regulates Hsp90 ATPase activity and thereby Wnt signaling by modulating Aha1 expression. Journal of Biological Chemistry 2014, 289, 6513–6525. [Google Scholar] [CrossRef]
- Wang, H.; Fan, L.; Xia, X.; Rao, Y.; Ma, Q.; Yang, J.; Lu, Y.; Wang, C.; Ma, D.; Huang, X. Silencing Wnt2B by siRNA interference inhibits metastasis and enhances chemotherapy sensitivity in ovarian cancer. International Journal of Gynecological Cancer 2012, 22, 755–761. [Google Scholar] [CrossRef]
- Takada, K.; Zhu, D.; Bird, G.H.; Sukhdeo, K.; Zhao, J.J.; Mani, M.; Lemieux, M.; Carrasco, D.E.; Ryan, J.; Horst, D.; et al. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Science translational medicine 2012, 4, 148ra117. [Google Scholar] [CrossRef]
- Chen, J.; Rajasekaran, M.; Xia, H.; Kong, S.N.; Deivasigamani, A.; Sekar, K.; Gao, H.; Swa, H.L.; Gunaratne, J.; Ooi, L.L.; et al. CDK1-mediated BCL9 phosphorylation inhibits clathrin to promote mitotic Wnt signalling. The EMBO journal 2018, 37, e99395. [Google Scholar] [CrossRef]
- Green, D.R.; Oberst, A.; Dillon, C.P.; Weinlich, R.; Salvesen, G.S. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Molecular cell 2011, 44, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Devin, A.; Rodriguez, Y.; Liu, Z.g. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes & development 1999, 13, 2514–2526. [Google Scholar]
- Estornes, Y.; Aguileta, M.; Dubuisson, C.; De Keyser, J.; Goossens, V.; Kersse, K.; Samali, A.; Vandenabeele, P.; Bertrand, M. RIPK1 promotes death receptor-independent caspase-8-mediated apoptosis under unresolved ER stress conditions. Cell death & disease 2014, 5, e1555. [Google Scholar]
- Weng, D.; Marty-Roix, R.; Ganesan, S.; Proulx, M.K.; Vladimer, G.I.; Kaiser, W.J.; Mocarski, E.S.; Pouliot, K.; Chan, F.K.M.; Kelliher, M.A.; et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proceedings of the National Academy of Sciences 2014, 111, 7391–7396. [Google Scholar] [CrossRef]
- Moriwaki, K.; Bertin, J.; Gough, P.J.; Chan, F.K.M. A RIPK3–caspase 8 complex mediates atypical pro–IL-1β processing. The Journal of Immunology 2015, 194, 1938–1944. [Google Scholar] [CrossRef]
- Declercq, W.; Berghe, T.V.; Vandenabeele, P. RIP kinases at the crossroads of cell death and survival. Cell 2009, 138, 229–232. [Google Scholar] [CrossRef]
- Chaudhary, P.M.; Eby, M.T.; Jasmin, A.; Kumar, A.; Liu, L.; Hood, L. Activation of the NF-κB pathway by caspase 8 and its homologs. Oncogene 2000, 19, 4451. [Google Scholar] [CrossRef]
- Sheng, Y.H.; He, Y.; Hasnain, S.Z.; Wang, R.; Tong, H.; Clarke, D.T.; Lourie, R.; Oancea, I.; Wong, K.; Lumley, J.W.; et al. MUC13 protects colorectal cancer cells from death by activating the NF-κB pathway and is a potential therapeutic target. Oncogene 2017, 36, 700. [Google Scholar] [CrossRef]
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. cell 1986, 46, 705–716. [Google Scholar] [CrossRef]
- Tanaka, S.; Nakano, H. NF-κB2 (p100) limits TNF-α–induced osteoclastogenesis. The Journal of clinical investigation 2009, 119, 2879–2881. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Yoshina, S.; Ikari, K.; Miyazawa, K.; Momohara, S.; Mitani, S. Impaired NFKBIE gene function decreases cellular uptake of methotrexate by down-regulating SLC19A1 expression in a human rheumatoid arthritis cell line. Modern rheumatology 2016, 26, 507–516. [Google Scholar] [CrossRef]
- Lee, R.E.; Walker, S.R.; Savery, K.; Frank, D.A.; Gaudet, S. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Molecular cell 2014, 53, 867–879. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine & growth factor reviews 2010, 21, 11–19. [Google Scholar]
- Liu, Z.; Hazan-Halevy, I.; Harris, D.M.; Li, P.; Ferrajoli, A.; Faderl, S.; Keating, M.J.; Estrov, Z. STAT-3 activates NF-κB in chronic lymphocytic leukemia cells. Molecular Cancer Research 2011, 9, 507–515. [Google Scholar] [CrossRef]
- Lam, L.T.; Wright, G.; Davis, R.E.; Lenz, G.; Farinha, P.; Dang, L.; Chan, J.W.; Rosenwald, A.; Gascoyne, R.D.; Staudt, L.M. Cooperative signaling through the STAT3 and NF-κB pathways in subtypes of diffuse large B cell lymphoma. Blood 2007. [Google Scholar]
- Lee, T.L.; Yeh, J.; Friedman, J.; Yan, B.; Yang, X.; Yeh, N.T.; Van Waes, C.; Chen, Z. A signal network involving coactivated NF-κB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. International journal of cancer 2008, 122, 1987–1998. [Google Scholar] [CrossRef]
- Ng, S.L.; Friedman, B.A.; Schmid, S.; Gertz, J.; Myers, R.M.; Maniatis, T.; et al. IκB kinase ε (IKKε) regulates the balance between type I and type II interferon responses. Proceedings of the National Academy of Sciences 2011, 108, 21170–21175. [Google Scholar] [CrossRef]
- Guo, J.; Kim, D.; Gao, J.; Kurtyka, C.; Chen, H.; Yu, C.; Wu, D.; Mittal, A.; Beg, A.; Chellappan, S.; et al. IKBKE is induced by STAT3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene 2013, 32, 151. [Google Scholar] [CrossRef]
- Shen, R.R.; Zhou, A.Y.; Kim, E.; Lim, E.; Habelhah, H.; Hahn, W.C. IκB kinase ε phosphorylates TRAF2 to promote mammary epithelial cell transformation. Molecular and cellular biology 2012, 32, 4756–4768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.Y.; Shen, R.R.; Kim, E.; Lock, Y.J.; Xu, M.; Chen, Z.J.; Hahn, W.C. IKKε-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell reports 2013, 3, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Akira, S. NF-κB activation through IKK-i-dependent I-TRAF/TANK phosphorylation. Genes to Cells 2000, 5, 191–202. [Google Scholar]
- Gerbod-Giannone, M.C.; Li, Y.; Holleboom, A.; Han, S.; Hsu, L.C.; Tabas, I.; Tall, A.R. TNFα induces ABCA1 through NF-κB in macrophages and in phagocytes ingesting apoptotic cells. Proceedings of the National Academy of Sciences 2006, 103, 3112–3117. [Google Scholar] [CrossRef]
- Van Eck, M.; Bos, I.S.T.; Kaminski, W.E.; Orsó, E.; Rothe, G.; Twisk, J.; Böttcher, A.; Van Amersfoort, E.S.; Christiansen-Weber, T.A.; Fung-Leung, W.P.; et al. Leukocyte ABCA1 controls susceptibility to atherosclerosis and macrophage recruitment into tissues. Proceedings of the National Academy of Sciences 2002, 99, 6298–6303. [Google Scholar] [CrossRef]
- Tang, C.; Liu, Y.; Kessler, P.S.; Vaughan, A.M.; Oram, J.F. The macrophage cholesterol exporter ABCA1 functions as an anti-inflammatory receptor. Journal of Biological Chemistry 2009, 284, 32336–32343. [Google Scholar] [CrossRef]
- Zhu, X.; Owen, J.S.; Wilson, M.D.; Li, H.; Griffiths, G.L.; Thomas, M.J.; Hiltbold, E.M.; Fessler, M.B.; Parks, J.S. Macrophage ABCA1 reduces MyD88-dependent Toll-like receptor trafficking to lipid rafts by reduction of lipid raft cholesterol. Journal of lipid research 2010, 51, 3196–3206. [Google Scholar] [CrossRef]
- Tian, B.; Widen, S.G.; Yang, J.; Wood, T.G.; Kudlicki, A.; Zhao, Y.; Brasier, A.R. The NFκB subunit RELA is a master transcriptional regulator of the committed epithelial-mesenchymal transition in airway epithelial cells. Journal of Biological Chemistry 2018, 293, 16528–16545. [Google Scholar] [CrossRef]
- Ke, B.; Zhao, Z.; Ye, X.; Gao, Z.; Manganiello, V.; Wu, B.; Ye, J. Inactivation of NF-κB p65 (RelA) in liver improves insulin sensitivity and inhibits cAMP/PKA pathway. Diabetes 2015, 64, 3355–3362. [Google Scholar] [CrossRef]
- Weichert, W.; Boehm, M.; Gekeler, V.; Bahra, M.; Langrehr, J.; Neuhaus, P.; Denkert, C.; Imre, G.; Weller, C.; Hofmann, H.; et al. High expression of RelA/p65 is associated with activation of nuclear factor-κB-dependent signaling in pancreatic cancer and marks a patient population with poor prognosis. British journal of cancer 2007, 97, 523. [Google Scholar] [CrossRef]
- Brooks, R.S.; Ciappio, E.D.; Bennett, G.; Crott, J.W.; Mason, J.B.; Liu, Z. TNF-α induced alterations in the Wnt signaling cascade: a potential mechanism for obesity-associated colorectal tumorigenesis. The FASEB Journal 2010, 24, lb384. [Google Scholar] [CrossRef]
- Adami, G.; Orsolini, G.; Adami, S.; Viapiana, O.; Idolazzi, L.; Gatti, D.; Rossini, M. Effects of TNF inhibitors on parathyroid hormone and Wnt signaling antagonists in rheumatoid arthritis. Calcified tissue international 2016, 99, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, A.; Yokoyama, K.; Nukaga, T.; Sakai, D.; Mochida, J. A complex interaction between Wnt signaling and TNF-α in nucleus pulposus cells. Arthritis research & therapy 2013, 15, R189. [Google Scholar]
- Roubert, A.; Gregory, K.; Li, Y.; Pfalzer, A.C.; Li, J.; Schneider, S.S.; Wood, R.J.; Liu, Z. The influence of tumor necrosis factor-α on the tumorigenic Wnt-signaling pathway in human mammary tissue from obese women. Oncotarget 2017, 8, 36127. [Google Scholar] [CrossRef]
- Jang, J.; Jung, Y.; Chae, S.; Chung, S.I.; Kim, S.M.; Yoon, Y. WNT/β-catenin pathway modulates the TNF-α-induced inflammatory response in bronchial epithelial cells. Biochemical and biophysical research communications 2017, 484, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Dharmani, P.; Leung, P.; Chadee, K. Tumor necrosis factor-α and Muc2 mucin play major roles in disease onset and progression in dextran sodium sulphate-induced colitis. PloS one 2011, 6, e25058. [Google Scholar] [CrossRef]
- Levine, S.J.; Larivee, P.; Logun, C.; Angus, C.W.; Ognibene, F.P.; Shelhamer, J.H. Tumor necrosis factor-alpha induces mucin hypersecretion and MUC-2 gene expression by human airway epithelial cells. American Journal of Respiratory Cell and Molecular Biology 1995, 12, 196–204. [Google Scholar] [CrossRef]
- Sikder, M.A.; Lee, H.J.; Mia, M.Z.; Park, S.H.; Ryu, J.; Kim, J.H.; Min, S.Y.; Hong, J.H.; Seok, J.H.; Lee, C.J. Inhibition of TNF-α-Induced MUC5AC Mucin Gene Expression and Production by Wogonin Through the Inactivation of NF-κB Signaling in Airway Epithelial Cells. Phytotherapy Research 2014, 28, 62–68. [Google Scholar] [CrossRef]
- Shao, M.X.; Nadel, J.A. Neutrophil elastase induces MUC5AC mucin production in human airway epithelial cells via a cascade involving protein kinase C, reactive oxygen species, and TNF-α-converting enzyme. The Journal of Immunology 2005, 175, 4009–4016. [Google Scholar] [CrossRef]
- Lora, J.M.; Zhang, D.M.; Liao, S.M.; Burwell, T.; King, A.M.; Barker, P.A.; Singh, L.; Keaveney, M.; Morgenstern, J.; Gutiérrez-Ramos, J.C.; et al. Tumor necrosis factor-α triggers mucus production in airway epithelium through an IκB kinase β-dependent mechanism. Journal of Biological Chemistry 2005, 280, 36510–36517. [Google Scholar] [CrossRef]
- Fischer, B.M.; Rochelle, L.G.; Voynow, J.A.; Akley, N.J.; Adler, K.B. Tumor necrosis factor-α stimulates mucin secretion and cyclic GMP production by guinea pig tracheal epithelial cells in vitro. American Journal of Respiratory Cell and Molecular Biology 1999, 20, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Haruta, A.; Kawano, H.; Ho, S.B.; Adams, G.L.; Juhn, S.K.; Kim, Y. Induction of Mucin Gene Expression in Middle Ear of Rats by Tumor Necrosis Factor—α: Potential Cause for Mucoid Otitis Media. The Journal of infectious diseases 2000, 182, 882–887. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kim, K.S.; Kim, H.U.; Linton, J.A.; Lee, J.G. Effects of TNF-a and IL-1 ß on Mucin, Lysozyme, IL-6 and IL-8 in passage-2 Normal human nasal epithelial cells. Acta oto-laryngologica 1999, 119, 905–910. [Google Scholar] [PubMed]
- Mercogliano, M.F.; De Martino, M.; Venturutti, L.; Rivas, M.A.; Proietti, C.J.; Inurrigarro, G.; Frahm, I.; Allemand, D.H.; Deza, E.G.; Ares, S.; et al. TNFα-induced mucin 4 expression elicits trastuzumab resistance in HER2-positive breast cancer. Clinical Cancer Research 2017, 23, 636–648. [Google Scholar] [CrossRef]
- Scarl, R.T.; Lawrence, C.M.; Gordon, H.M.; Nunemaker, C.S. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. Journal of Endocrinology 2017, 234, R123–R134. [Google Scholar] [CrossRef]
- Moldes, M.; Lasnier, F.; Gauthereau, X.; Klein, C.; Pairault, J.; Fève, B.; Chambaut-Guérin, A.M. Tumor necrosis factor-α-induced adipose-related protein (TIARP), a cell-surface protein that is highly induced by tumor necrosis factor-α and adipose conversion. Journal of Biological Chemistry 2001, 276, 33938–33946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Tao, Y.; Zhang, Z.; Guo, X.; An, P.; Shen, Y.; Wu, Q.; Yu, Y.; Wang, F. Metalloreductase Steap3 coordinates the regulation of iron homeostasis and inflammatory responses. Haematologica 2012, 97, 1826–1835. [Google Scholar] [CrossRef]
- Tanaka, Y.; Matsumoto, I.; Iwanami, K.; Inoue, A.; Umeda, N.; Sugihara, M.; Hayashi, T.; Ito, S.; Sumida, T. Six-transmembrane epithelial antigen of prostate 4 (STEAP4) is expressed on monocytes/neutrophils, and is regulated by TNF antagonist in patients with rheumatoid arthritis. Clinical and experimental rheumatology 2012, 30, 99–102. [Google Scholar]
- Tanaka, Y.; Matsumoto, I.; Iwanami, K.; Inoue, A.; Minami, R.; Umeda, N.; Kanamori, A.; Ochiai, N.; Miyazawa, K.; Sugihara, M. Six-transmembrane epithelial antigen of prostate4 (STEAP4) is a tumor necrosis factor alpha-induced protein that regulates IL-6, IL-8, and cell proliferation in synovium from patients with rheumatoid arthritis. Modern rheumatology 2012, 22, 128–136. [Google Scholar] [CrossRef]
- Gauss, G.H.; Kleven, M.D.; Sendamarai, A.K.; Fleming, M.D.; Lawrence, C.M. The crystal structure of six-transmembrane epithelial antigen of the prostate 4 (Steap4), a ferri/cuprireductase, suggests a novel interdomain flavin-binding site. Journal of Biological Chemistry 2013, 288, 20668–20682. [Google Scholar] [CrossRef]
- ZHANG, C.m.; Chi, X.; Wang, B.; Zhang, M.; NI, Y.h.; CHEN, R.h.; LI, X.n.; GUO, X.r. Downregulation of STEAP4, a highly-expressed TNF-α-inducible gene in adipose tissue, is associated with obesity in humans 1. Acta Pharmacologica Sinica 2008, 29, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xing, X.; Beamer, M.A.; Swindell, W.R.; Sarkar, M.K.; Roberts, L.W.; Voorhees, J.J.; Kahlenberg, J.M.; Harms, P.W.; Johnston, A.; et al. Six-transmembrane epithelial antigens of the prostate comprise a novel inflammatory nexus in patients with pustular skin disorders. Journal of Allergy and Clinical Immunology 2017, 139, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP proteins: from structure to applications in cancer therapy. Molecular Cancer Research 2012, 10, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Li, S.; Wang, L.; Berman, M.A.; Dorf, M.E. The ubiquitin conjugating enzyme UBE2L3 regulates TNFα-induced linear ubiquitination. Cell research 2014, 24, 376. [Google Scholar] [CrossRef]
- Li, Y.P.; Lecker, S.H.; Chen, Y.; Waddell, I.D.; Goldberg, A.L.; Reid, M.B. TNF-α increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. The FASEB Journal 2003, 17, 1048–1057. [Google Scholar] [CrossRef]
- Shembade, N.; Ma, A.; Harhaj, E.W. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 2010, 327, 1135–1139. [Google Scholar] [CrossRef]
- Tamatani, M.; Che, Y.H.; Matsuzaki, H.; Ogawa, S.; Okado, H.; Miyake, S.i.; Mizuno, T.; Tohyama, M. Tumor necrosis factor induces Bcl-2 and Bcl-x expression through NFκB activation in primary hippocampal neurons. Journal of Biological Chemistry 1999, 274, 8531–8538. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.; Winkler-Lowen, B.; Morrish, D.; Dakour, J.; Li, H.; Guilbert, L. The role of Bcl-2 expression in EGF inhibition of TNF-α/IFN-γ-induced villous trophoblast apoptosis. Placenta 1999, 20, 423–430. [Google Scholar] [CrossRef]
- Genestier, L.; Bonnefoy-Berard, N.; Rouault, J.P.; Flacher, M.; Revillard, J.P. Tumor necrosis factor-α up-regulates Bcl-2 expression and decreases calcium-dependent apoptosis in human B cell lines. International immunology 1995, 7, 533–540. [Google Scholar] [CrossRef]
- Jäättelä, M.; Benedict, M.; Tewari, M.; Shayman, J.; Dixit, V. Bcl-x and Bcl-2 inhibit TNF and Fas-induced apoptosis and activation of phospholipase A2 in breast carcinoma cells. Oncogene 1995, 10, 2297–2305. [Google Scholar]
- Kim, J.; Lee, S.; Park, J.; Yoo, Y. TNF-α-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X L. Cell death and differentiation 2010, 17, 1420. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, H.; Watanabe, Y.; Miyoshi, H.; Yamamoto, M.; Kaisho, T.; Takeda, K.; Akira, S. IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-α production in macrophages. Blood 2003, 102, 4123–4129. [Google Scholar] [CrossRef] [PubMed]
- Esche, C.; Shurin, G.V.; Kirkwood, J.M.; Wang, G.Q.; Rabinowich, H.; Pirtskhalaishvili, G.; Shurin, M.R. Tumor necrosis factor-α-promoted expression of Bcl-2 and inhibition of mitochondrial cytochrome c release mediate resistance of mature dendritic cells to melanoma-induced apoptosis. Clinical cancer research 2001, 7, 974s–979s. [Google Scholar] [PubMed]
- Thacker, J.; Zdzienicka, M.Z. The mammalian XRCC genes: their roles in DNA repair and genetic stability. DNA repair 2003, 2, 655–672. [Google Scholar] [CrossRef]
- Thacker, J.; Zdzienicka, M.Z. The XRCC genes: expanding roles in DNA double-strand break repair. DNA repair 2004, 3, 1081–1090. [Google Scholar] [CrossRef]
- Sultana, R.; Abdel-Fatah, T.; Perry, C.; Moseley, P.; Albarakti, N.; Mohan, V.; Seedhouse, C.; Chan, S.; Madhusudan, S. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PloS one 2013, 8, e57098. [Google Scholar] [CrossRef]
- Della-Maria, J.; Zhou, Y.; Tsai, M.S.; Kuhnlein, J.; Carney, J.P.; Paull, T.T.; Tomkinson, A.E. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIα/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. Journal of Biological Chemistry 2011, 286, 33845–33853. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 links transcription termination to DNA damage and replication stress. PLoS genetics 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Saintigny, Y.; Dumay, A.; Lambert, S.; Lopez, B.S. A novel role for the Bcl-2 protein family: specific suppression of the RAD51 recombination pathway. The EMBO journal 2001, 20, 2596–2607. [Google Scholar] [CrossRef]
- Meng, J.; Liu, X.; Cao, X. A new cytosolic DNA-recognition pathway for DNA-induced inflammatory responses. Cellular & Molecular Immunology 2014, 11, 506. [Google Scholar]
- Marin-Vicente, C.; Domingo-Prim, J.; Eberle, A.B.; Visa, N. RRP6/EXOSC10 is required for the repair of DNA double-strand breaks by homologous recombination. J Cell Sci 2015, 128, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Communication and Signaling 2013, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Walden, H.; Deans, A.J. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annual review of biophysics 2014, 43, 257–278. [Google Scholar] [CrossRef]
- Cohn, M.A.; D’Andrea, A.D. Chromatin recruitment of DNA repair proteins: lessons from the fanconi anemia and double-strand break repair pathways. Molecular cell 2008, 32, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Romick-Rosendale, L.E.; Lui, V.W.; Grandis, J.R.; Wells, S.I. The Fanconi anemia pathway: repairing the link between DNA damage and squamous cell carcinoma. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2013, 743, 78–88. [Google Scholar] [CrossRef]
- Michl, J.; Zimmer, J.; Tarsounas, M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. The EMBO journal 2016, 35, 909–923. [Google Scholar] [CrossRef]
- Liang, F.; Longerich, S.; Miller, A.S.; Tang, C.; Buzovetsky, O.; Xiong, Y.; Maranon, D.G.; Wiese, C.; Kupfer, G.M.; Sung, P. Promotion of RAD51-mediated homologous DNA pairing by the RAD51AP1-UAF1 complex. Cell reports 2016, 15, 2118–2126. [Google Scholar] [CrossRef]
- Taniguchi, T.; Garcia-Higuera, I.; Andreassen, P.R.; Gregory, R.C.; Grompe, M.; D’Andrea, A.D. S-phase–specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 2002, 100, 2414–2420. [Google Scholar] [CrossRef]
- Zadorozhny, K.; Sannino, V.; Beláň, O.; Mlčoušková, J.; Špírek, M.; Costanzo, V.; Krejčí, L. Fanconi-anemia-associated mutations destabilize RAD51 filaments and impair replication fork protection. Cell reports 2017, 21, 333–340. [Google Scholar] [CrossRef]
- Geng, L.; Huntoon, C.J.; Karnitz, L.M. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. The Journal of cell biology 2010, 191, 249–257. [Google Scholar] [CrossRef]
- Palle, K.; Vaziri, C. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle 2011, 10, 1625–1638. [Google Scholar] [CrossRef]
- García-Luis, J.; Machín, F. Fanconi Anaemia-Like Mph1 Helicase Backs up Rad54 and Rad5 to Circumvent Replication Stress-Driven Chromosome Bridges. Genes 2018, 9, 558. [Google Scholar] [CrossRef]
- Lipinska, N.; Romaniuk, A.; Paszel-Jaworska, A.; Toton, E.; Kopczynski, P.; Rubis, B. Telomerase and drug resistance in cancer. Cellular and Molecular Life Sciences 2017, 74, 4121–4132. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Fang, J. Expression and clinical significance of telomerase catalytic subunit gene in lung cancer and its correlations with genes related to drug resistance and apoptosis. Zhonghua zhong liu za zhi [Chinese journal of oncology] 1999, 21, 350–353. [Google Scholar]
- Sakin, V.; Eskiocak, U.; Kars, M.; Iseri, Ö.; Gunduz, U. hTERT gene expression levels and telomerase activity in drug resistant MCF-7 cells. Experimental oncology 2008. [Google Scholar]
- Keshet, G.I.; Goldstein, I.; Itzhaki, O.; Cesarkas, K.; Shenhav, L.; Yakirevitch, A.; Treves, A.J.; Schachter, J.; Amariglio, N.; Rechavi, G. MDR1 expression identifies human melanoma stem cells. Biochemical and biophysical research communications 2008, 368, 930–936. [Google Scholar] [CrossRef]
- Zhou, X.; Engel, T.; Goepfert, C.; Erren, M.; Assmann, G.; von Eckardstein, A. The ATP binding cassette transporter A1 contributes to the secretion of interleukin 1β from macrophages but not from monocytes. Biochemical and biophysical research communications 2002, 291, 598–604. [Google Scholar] [CrossRef]
- Haskó, G.; Deitch, E.A.; Németh, Z.H.; Kuhel, D.G.; Szabó, C. Inhibitors of ATP-binding cassette transporters suppress interleukin-12 p40 production and major histocompatibility complex II up-regulation in macrophages. Journal of Pharmacology and Experimental Therapeutics 2002, 301, 103–110. [Google Scholar] [CrossRef]
- Park, S.Y.; Han, J.; Kim, J.B.; Yang, M.G.; Kim, Y.J.; Lim, H.J.; An, S.Y.; Kim, J.H. Interleukin-8 is related to poor chemotherapeutic response and tumourigenicity in hepatocellular carcinoma. European Journal of Cancer 2014, 50, 341–350. [Google Scholar] [CrossRef]
- Marty, V.; Médina, C.; Combe, C.; Parnet, P.; Amédée, T. ATP binding cassette transporter ABC1 is required for the release of interleukin-1β by P2X7-stimulated and lipopolysaccharide-primed mouse Schwann cells. Glia 2005, 49, 511–519. [Google Scholar] [CrossRef]
- Lottaz, D.; Beleznay, Z.; Bickel, M. Inhibition of ATP-binding cassette transporter downregulates interleukin-1β-mediated autocrine activation of human dermal fibroblasts. Journal of investigative dermatology 2001, 117, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Ruzickova, E.; Janska, R.; Dolezel, P.; Mlejnek, P. Clinically relevant interactions of anti-apoptotic Bcl-2 protein inhibitors with ABC transporters. Die Pharmazie-An International Journal of Pharmaceutical Sciences 2017, 72, 751–758. [Google Scholar]
- Alla, V.; Kowtharapu, B.S.; Engelmann, D.; Emmrich, S.; Schmitz, U.; Steder, M.; Pützer, B.M. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell cycle 2012, 11, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Yasui, K.; Mihara, S.; Zhao, C.; Okamoto, H.; Saito-Ohara, F.; Tomida, A.; Funato, T.; Yokomizo, A.; Naito, S.; Imoto, I.; et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer research 2004, 64, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, M.; Guan, X.; Yuan, Z.; Liu, Z.; Zou, C.; Wang, G.; Gao, X.; Wang, X. Loss of ABCB4 attenuates the caspase-dependent apoptosis regulating resistance to 5-Fu in colorectal cancer. Bioscience reports 2018, 38, BSR20171428. [Google Scholar] [CrossRef]
- Ihlefeld, K.; Vienken, H.; Claas, R.F.; Blankenbach, K.; Rudowski, A.; Ter Braak, M.; Koch, A.; Van Veldhoven, P.P.; Pfeilschifter, J.; zu Heringdorf, D.M. Upregulation of ABC transporters contributes to chemoresistance of sphingosine 1-phosphate lyase-deficient fibroblasts. Journal of lipid research 2015, 56, 60–69. [Google Scholar] [CrossRef]
- Hörber, S.; Hildebrand, D.G.; Lieb, W.S.; Lorscheid, S.; Hailfinger, S.; Schulze-Osthoff, K.; Essmann, F. The atypical inhibitor of NF-κB, IκBζ, controls macrophage interleukin-10 expression. Journal of Biological Chemistry 2016, 291, 12851–12861. [Google Scholar] [CrossRef]
- Yamazaki, S.; Muta, T.; Matsuo, S.; Takeshige, K. Stimulus-specific induction of a novel nuclear factor-κB regulator, IκB-ζ, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. Journal of Biological Chemistry 2005, 280, 1678–1687. [Google Scholar] [CrossRef]
- Kurzrock, R.; Estrov, Z.; Ku, S.; Leonard, M.; Talpaz, M. Interleukin-1 increases expression of the LYT-10 (NFκB2) proto-oncogene/transcription factor in renal cell carcinoma lines. Journal of Laboratory and Clinical Medicine 1998, 131, 261–268. [Google Scholar] [CrossRef]
- Lee, S.; Sabath, D.; Deutsch, C.; Prystowsky, M.B. Increased voltage-gated potassium conductance during interleukin 2-stimulated proliferation of a mouse helper T lymphocyte clone. The Journal of cell biology 1986, 102, 1200–1208. [Google Scholar] [CrossRef]
- Martin, G.; O’connell, R.J.; Pietrzykowski, A.Z.; Treistman, S.N.; Ethier, M.F.; Madison, J.M. Interleukin-4 activates large-conductance, calcium-activated potassium (BKCa) channels in human airway smooth muscle cells. Experimental physiology 2008, 93, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Kerschner, J.E.; Meyer, T.K.; Yang, C.; Burrows, A. Middle ear epithelial mucin production in response to interleukin-6 exposure in vitro. Cytokine 2004, 26, 30–36. [Google Scholar] [CrossRef]
- Chen, Y.; Thai, P.; Zhao, Y.H.; Ho, Y.S.; DeSouza, M.M.; Wu, R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. Journal of Biological Chemistry 2003, 278, 17036–17043. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.S.; Hsu, H.P.; Lai, M.D.; Yen, M.C.; FANg, J.H.; Weng, T.Y.; Chen, Y.L. Suppression of mucin 2 promotes interleukin-6 secretion and tumor growth in an orthotopic immune-competent colon cancer animal model. Oncology reports 2014, 32, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Yokoigawa, N.; Takeuchi, N.; Toda, M.; Inoue, M.; Kaibori, M.; Yanagida, H.; Tanaka, H.; Ogura, T.; Takada, H.; Okumura, T.; et al. Enhanced production of interleukin 6 in peripheral blood monocytes stimulated with mucins secreted into the bloodstream. Clinical cancer research 2005, 11, 6127–6132. [Google Scholar] [CrossRef]
- Gray, T.; Nettesheim, P.; Loftin, C.; Koo, J.S.; Bonner, J.; Peddada, S.; Langenbach, R. Interleukin-1β–Induced Mucin Production in Human Airway Epithelium Is Mediated by Cyclooxygenase-2, Prostaglandin E2 Receptors, and Cyclic AMP-Protein Kinase A Signaling. Molecular Pharmacology 2004, 66, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.P.; Lai, M.D.; Lee, J.C.; Yen, M.C.; Weng, T.Y.; Chen, W.C.; Fang, J.H.; Chen, Y.L. Mucin 2 silencing promotes colon cancer metastasis through interleukin-6 signaling. Scientific reports 2017, 7, 5823. [Google Scholar] [CrossRef]
- Brighenti, E.; Calabrese, C.; Liguori, G.; Giannone, F.; Trere, D.; Montanaro, L.; Derenzini, M. Interleukin 6 downregulates p53 expression and activity by stimulating ribosome biogenesis: a new pathway connecting inflammation to cancer. Oncogene 2014, 33, 4396. [Google Scholar] [CrossRef]
- Tan, X.; Carretero, J.; Chen, Z.; Zhang, J.; Wang, Y.; Chen, J.; Li, X.; Ye, H.; Tang, C.; Cheng, X.; et al. Loss of p53 attenuates the contribution of IL-6 deletion on suppressed tumor progression and extended survival in Kras-driven murine lung cancer. PloS one 2013, 8, e80885. [Google Scholar] [CrossRef]
- Pützer, B.; Bramson, J.; Addison, C.; Hitt, M.; Siegel, P.; Muller, W.; Graham, F. Combination therapy with interleukin-2 and wild-type p53 expressed by adenoviral vectors potentiates tumor regression in a murine model of breast cancer. Human gene therapy 1998, 9, 707–718. [Google Scholar] [CrossRef]
- Dijsselbloem, N.; Goriely, S.; Albarani, V.; Gerlo, S.; Francoz, S.; Marine, J.C.; Goldman, M.; Haegeman, G.; Berghe, W.V. A critical role for p53 in the control of NF-κB-dependent gene expression in TLR4-stimulated dendritic cells exposed to genistein. The Journal of immunology 2007, 178, 5048–5057. [Google Scholar] [CrossRef] [PubMed]
- Schauer, I.G.; Zhang, J.; Xing, Z.; Guo, X.; Mercado-Uribe, I.; Sood, A.K.; Huang, P.; Liu, J. Interleukin-1β promotes ovarian tumorigenesis through a p53/NF-κB-mediated inflammatory response in stromal fibroblasts. Neoplasia 2013, 15, 409–IN18. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Quinton, L.J.; Simms, B.T.; Lupa, M.M.; Kogan, M.S.; Mizgerd, J.P. Roles of interleukin-6 in activation of STAT proteins and recruitment of neutrophils during Escherichia coli pneumonia. The Journal of infectious diseases 2006, 193, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Kotanides, H.; Moczygemba, M.; White, M.F.; Reich, N.C. Characterization of the interleukin-4 nuclear activated factor/STAT and its activation independent of the insulin receptor substrate proteins. Journal of Biological Chemistry 1995, 270, 19481–19486. [Google Scholar] [CrossRef]
- Adam, N.; Rabe, B.; Suthaus, J.; Grötzinger, J.; Rose-John, S.; Scheller, J. Unraveling viral interleukin-6 binding to gp130 and activation of STAT-signaling pathways independently of the interleukin-6 receptor. Journal of virology 2009, 83, 5117–5126. [Google Scholar] [CrossRef]
- Frank, D.A.; Robertson, M.J.; Bonni, A.; Ritz, J.; Greenberg, M.E. Interleukin 2 signaling involves the phosphorylation of Stat proteins. Proceedings of the National Academy of Sciences 1995, 92, 7779–7783. [Google Scholar] [CrossRef]
- Boyd, Z.S.; Kriatchko, A.; Yang, J.; Agarwal, N.; Wax, M.B.; Patil, R.V. Interleukin-10 receptor signaling through STAT-3 regulates the apoptosis of retinal ganglion cells in response to stress. Investigative ophthalmology & visual science 2003, 44, 5206–5211. [Google Scholar]
- Kurgonaite, K.; Gandhi, H.; Kurth, T.; Pautot, S.; Schwille, P.; Weidemann, T.; Bökel, C. Essential role of endocytosis for interleukin-4-receptor-mediated JAK/STAT signalling. J Cell Sci 2015, 128, 3781–3795. [Google Scholar] [CrossRef]
- Kondo, M.; Yamaoka, K.; Sakata, K.; Sonomoto, K.; Lin, L.; Nakano, K.; Tanaka, Y. Contribution of the interleukin-6/STAT-3 signaling pathway to chondrogenic differentiation of human mesenchymal stem cells. Arthritis & rheumatology 2015, 67, 1250–1260. [Google Scholar]
- Tanaka, N.; Hoshino, Y.; Gold, J.; Hoshino, S.; Martiniuk, F.; Kurata, T.; Pine, R.; Levy, D.; Rom, W.N.; Weiden, M. Interleukin-10 induces inhibitory C/EBPβ through STAT-3 and represses HIV-1 transcription in macrophages. American journal of respiratory cell and molecular biology 2005, 33, 406–411. [Google Scholar] [CrossRef]
- Jobst, B.; Weigl, J.; Michl, C.; Vivarelli, F.; Pinz, S.; Amslinger, S.; Rascle, A. Inhibition of interleukin-3-and interferon-α-induced JAK/STAT signaling by the synthetic α-X-2’, 3, 4, 4’-tetramethoxychalcones α-Br-TMC and α-CF3-TMC. Biological chemistry 2016, 397, 1187–1204. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.; O’Neill, L. Interleukin-1 receptor-associated kinase and TRAF-6 mediate the transcriptional regulation of interleukin-2 by interleukin-1 via NFκB but unlike interleukin-1 are unable to stabilise interleukin-2 mRNA. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1999, 1451, 109–121. [Google Scholar] [CrossRef]
- Cao, Z.; Xiong, J.; Takeuchi, M.; Kurama, T.; Goeddel, D.V. TRAF6 is a signal transducer for interleukin-1. Nature 1996, 383, 443. [Google Scholar] [CrossRef] [PubMed]
- Schwandner, R.; Yamaguchi, K.; Cao, Z. Requirement of tumor necrosis factor receptor–associated factor (TRAF) 6 in interleukin 17 signal transduction. Journal of Experimental Medicine 2000, 191, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Lomaga, M.A.; Yeh, W.C.; Sarosi, I.; Duncan, G.S.; Furlonger, C.; Ho, A.; Morony, S.; Capparelli, C.; Van, G.; Kaufman, S.; et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes & development 1999, 13, 1015–1024. [Google Scholar]
- Jefferies, C.; Bowie, A.; Brady, G.; Cooke, E.L.; Li, X.; O’Neill, L.A. Transactivation by the p65 subunit of NF-κB in response to interleukin-1 (IL-1) involves MyD88, IL-1 receptor-associated kinase 1, TRAF-6, and Rac1. Molecular and cellular biology 2001, 21, 4544–4552. [Google Scholar] [CrossRef]
- Wu, H.; Arron, J.R. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays 2003, 25, 1096–1105. [Google Scholar] [CrossRef]
- Ramadoss, P.; Chiappini, F.; Bilban, M.; Hollenberg, A.N. Regulation of hepatic six transmembrane epithelial antigen of prostate 4 (STEAP4) expression by STAT3 and CCAAT/enhancer-binding protein α. Journal of Biological Chemistry 2010, 285, 16453–16466. [Google Scholar] [CrossRef]
- Hamon, Y.; Luciani, M.F.; Becq, F.; Verrier, B.; Rubartelli, A.; Chimini, G. Interleukin-1β secretion is impaired by inhibitors of the ATP binding cassette transporter, ABC1. Blood 1997, 90, 2911–2915. [Google Scholar] [CrossRef]
- Neta, R.; Sayers, T.; Oppenheim, J. Relationship of TNF to interleukins. Immunology series 1992, 56, 499–566. [Google Scholar]
- Rieckmann, P.; Tuscano, J.; Kehrl, J. Tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) in B-lymphocyte function. Methods 1997, 11, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Bethea, J.R.; Gillespie, G.Y.; Benveniste, E.N. Interleukin-1β induction of TNF-α gene expression: Involvement of protein kinase C. Journal of cellular physiology 1992, 152, 264–273. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, F.; Hayes, B.P.; Horgan, C.M.; Beesley, J.E.; Campbell, C.J.; Randi, A.M. Tumor necrosis factor (TNF)-α and interleukin (IL)-1β down-regulate intercellular adhesion molecule (ICAM)-2 expression on the endothelium. Cell adhesion and communication 1998, 6, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Liu, G.; Ge, X.; Bao, W.; Wu, C.; Yang, C.; Liang, D. Serum levels of tumor necrosis factor-α (TNF-α), interleukin 6 (IL-6), and their soluble receptors in coal workers’ pneumoconiosis. Respiratory medicine 2002, 96, 829–834. [Google Scholar] [CrossRef]
- Havlir, D.V.; Torriani, F.J.; Schrier, R.D.; Huang, J.Y.; Lederman, M.M.; Chervenak, K.A.; Boom, W.H. Serum interleukin-6 (IL-6), IL-10, tumor necrosis factor (TNF) alpha, soluble type II TNF receptor, and transforming growth factor beta levels in human immunodeficiency virus type 1-infected individuals with Mycobacterium avium complex disease. Journal of clinical microbiology 2001, 39, 298–303. [Google Scholar] [CrossRef]
- Tissi, L.; Puliti, M.; Barluzzi, R.; Orefici, G.; von Hunolstein, C.; Bistoni, F. Role of tumor necrosis factor alpha, interleukin-1β, and interleukin-6 in a mouse model of group B streptococcal arthritis. Infection and immunity 1999, 67, 4545–4550. [Google Scholar] [CrossRef]
- Ismail, N.; Stevenson, H.L.; Walker, D.H. Role of tumor necrosis factor alpha (TNF-α) and interleukin-10 in the pathogenesis of severe murine monocytotropic ehrlichiosis: increased resistance of TNF receptor p55-and p75-deficient mice to fatal ehrlichial infection. Infection and immunity 2006, 74, 1846–1856. [Google Scholar] [CrossRef] [PubMed]
- Yap, S.; Moshage, H.; Hazenberg, B.; Roelofs, H.; Bijzet, J.; Limburg, P.; Aarden, L.; Van Rijswijk, M. Tumor necrosis factor (TNF) inhibits interleukin (IL)-1 and/or IL-6 stimulated synthesis of C-reactive protein (CRP) and serum amyloid A (SAA) in primary cultures of human hepatocytes. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1991, 1091, 405–408. [Google Scholar] [CrossRef]
- Qin, B.; Zhou, Z.; He, J.; Yan, C.; Ding, S. IL-6 inhibits starvation-induced autophagy via the STAT3/Bcl-2 signaling pathway. Scientific reports 2015, 5, 15701. [Google Scholar] [CrossRef]
- Gabellini, C.; Gómez-Abenza, E.; Ibáñez-Molero, S.; Tupone, M.G.; Pérez-Oliva, A.B.; de Oliveira, S.; Del Bufalo, D.; Mulero, V. Interleukin 8 mediates bcl-xL-induced enhancement of human melanoma cell dissemination and angiogenesis in a zebrafish xenograft model. International journal of cancer 2018, 142, 584–596. [Google Scholar] [CrossRef]
- Guruprasath, P.; Kim, J.; Gunassekaran, G.R.; Chi, L.; Kim, S.; Park, R.W.; Kim, S.H.; Baek, M.C.; Bae, S.M.; Kim, S.Y.; et al. Interleukin-4 receptor-targeted delivery of Bcl-xL siRNA sensitizes tumors to chemotherapy and inhibits tumor growth. Biomaterials 2017, 142, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Maraskovsky, E.; O’Reilly, L.A.; Teepe, M.; Corcoran, L.M.; Peschon, J.J.; Strasser, A. Bcl-2 can rescue t lymphocyte development in interleukin-7 receptor–deficient mice but not in mutant rag-1-/- mice. Cell 1997, 89, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Kondo, M.; von Freeden-Jeffry, U.; Murray, R.; Weissman, I.L. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor–deficient mice. Cell 1997, 89, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Weber-Nordt, R.; Henschler, R.; Schott, E.; Wehinger, J.; Behringer, D.; Mertelsmann, R.; Finke, J. Interleukin-10 increases Bcl-2 expression and survival in primary human CD34+ hematopoietic progenitor cells. Blood 1996, 88, 2549–2558. [Google Scholar] [CrossRef]
- Qin, J.Z.; Zhang, C.L.; Kamarashev, J.; Dummer, R.; Burg, G.; Döbbeling, U. Interleukin-7 and interleukin-15 regulate the expression of thebcl-2 and c-myb genes in cutaneous T-cell lymphoma cells. Blood 2001, 98, 2778–2783. [Google Scholar] [CrossRef]
- Escandell, J.; Recio, M.; Giner, R.; Máñez, S.; Ríos, J. Bcl-2 is a negative regulator of interleukin-1β secretion in murine macrophages in pharmacological-induced apoptosis. British journal of pharmacology 2010, 160, 1844–1856. [Google Scholar] [CrossRef]
- Alas, S.; Emmanouilides, C.; Bonavida, B. Inhibition of interleukin 10 by rituximab results in down-regulation of bcl-2 and sensitization of B-cell non-Hodgkin’s lymphoma to apoptosis. Clinical Cancer Research 2001, 7, 709–723. [Google Scholar]
- Deng, Z.; Fu, H.; Xiao, Y.; Zhang, B.; Sun, G.; Wei, Q.; Ai, B.; Hu, Q. Effects of selenium on lead-induced alterations in Aβ production and Bcl-2 family proteins. Environmental toxicology and pharmacology 2015, 39, 221–228. [Google Scholar] [CrossRef]
- Yaming, W.; Hai, B.; Haijian, Y.; Xiyan, Z. Effects of selenium dioxide on apoptosis, Bcl-2 and p53 expression, intracellular reactive oxygen species and calcium level in three human lung cancer cell lines. The Chinese-German Journal of Clinical Oncology 2004, 3, 141–146. [Google Scholar]
- Mauro, M.; Sartori, D.; Oliveira, R.J.; Ishii, P.L.; Mantovani, M.S.; Ribeiro, L.R. Activity of selenium on cell proliferation, cytotoxicity, and apoptosis and on the expression of CASP9, BCL-XL and APC in intestinal adenocarcinoma cells. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2011, 715, 7–12. [Google Scholar] [CrossRef]
- Hemann, M.; Lowe, S. The p53–Bcl-2 connection. 2006. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Marchenko, N.; Erster, S.; Nemajerova, A.; Dehner, A.; Klein, C.; Pan, H.; Kessler, H.; Pancoska, P.; Moll, U.M. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. Journal of Biological Chemistry 2006, 281, 8600–8606. [Google Scholar] [CrossRef]
- Jiang, M.; Milner, J. Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes & development 2003, 17, 832–837. [Google Scholar]
- Li, X.; Miao, X.; Wang, H.; Xu, Z.; Li, B. The tissue dependent interactions between p53 and Bcl-2 in vivo. Oncotarget 2015, 6, 35699. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Ruvolo, V.; Mu, H.; Leverson, J.D.; Nichols, G.; Reed, J.C.; Konopleva, M.; Andreeff, M. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell 2017, 32, 748–760. [Google Scholar] [CrossRef]
- Zaidi, A.U.; McDonough, J.S.; Klocke, B.J.; Latham, C.B.; Korsmeyer, S.J.; Flavell, R.A.; Schmidt, R.E.; Roth, K.A. Chloroquine-induced neuronal cell death is p53 and Bcl-2 family-dependent but caspase-independent. Journal of Neuropathology & Experimental Neurology 2001, 60, 937–945. [Google Scholar]
- Wilson, W.H.; Teruya-Feldstein, J.; Fest, T.; Harris, C.; Steinberg, S.M.; Jaffe, E.S.; Raffeld, M. Relationship of p53, bcl-2, and tumor proliferation to clinical drug resistance in non-Hodgkin’s lymphomas. Blood 1997, 89, 601–609. [Google Scholar] [CrossRef]
- Exley, G.E.; Tang, C.; McElhinny, A.S.; Warner, C.M. Expression of caspase and BCL-2 apoptotic family members in mouse preimplantation embryos. Biology of reproduction 1999, 61, 231–239. [Google Scholar] [CrossRef]
- Swanton, E.; Savory, P.; Cosulich, S.; Clarke, P.; Woodman, P. Bcl-2 regulates a caspase-3/caspase-2 apoptotic cascade in cytosolic extracts. Oncogene 1999, 18, 1781. [Google Scholar] [CrossRef]
- Pellegrini, M.; Strasser, A. Caspases, Bcl-2 family proteins and other components of the death machinery: their role in the regulation of the immune response. In Madame Curie Bioscience Database [Internet]; Landes Bioscience, 2013. [Google Scholar]
- Moriishi, K.; Huang, D.C.; Cory, S.; Adams, J.M. Bcl-2 family members do not inhibit apoptosis by binding the caspase activator Apaf-1. Proceedings of the National Academy of Sciences 1999, 96, 9683–9688. [Google Scholar] [CrossRef]
- Demirag, F.; Cakir, E.; Bayiz, H.; Eren Yazici, U. MUC1 and bcl-2 expression in preinvasive lesions and adenosquamous carcinoma of the lung. Acta Chirurgica Belgica 2013, 113, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.H.; He, Y.; Hasnain, S.Z.; Wang, R.; Tong, H.; Clarke, D.T.; Lourie, R.; Oancea, I.; Wong, K.; Lumley, J.W.; et al. MUC13 protects colorectal cancer cells from death by activating the NF-κB pathway and is a potential therapeutic target. Oncogene 2017, 36, 700. [Google Scholar] [CrossRef]
- Deng, M.; Yuan, H.; Liu, S.; Hu, Z.; Xiao, H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy 2019, 21, 96–106. [Google Scholar] [CrossRef]
- Xu, X.D.; Wu, X.H.; Fan, Y.R.; Tan, B.; Quan, Z.; Luo, C.L. Exosome-derived microRNA-29c induces apoptosis of BIU-87 cells by down regulating BCL-2 and MCL-1. Asian Pacific Journal of Cancer Prevention 2014, 15, 3471–3476. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, X.H.; Wang, D.; Luo, C.L.; Chen, L.X. Bladder cancer cell-derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Molecular medicine reports 2013, 8, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Zhuang, Y.; Fu, J.; Li, X.; Shi, Q.; Ju, X. Exosomes derived from bone marrow stromal cells decrease the sensitivity of leukemic cells to etoposide. Oncology letters 2017, 14, 3082–3088. [Google Scholar] [CrossRef]
- Brown, M.C.; Holl, E.K.; Boczkowski, D.; Dobrikova, E.; Mosaheb, M.; Chandramohan, V.; Bigner, D.D.; Gromeier, M.; Nair, S.K. Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen–specific CTLs. Science translational medicine 2017, 9, eaan4220. [Google Scholar] [CrossRef]
- Stamm, H.; Klingler, F.; Grossjohann, E.M.; Muschhammer, J.; Vettorazzi, E.; Heuser, M.; Mock, U.; Thol, F.; Vohwinkel, G.; Latuske, E.; et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene 2018, 37, 5269. [Google Scholar] [CrossRef]
- Stamm, H.; Klingler, F.; Pende, D.; Vettorazzi, E.; Heuser, M.; Mock, U.; Bokemeyer, C.; Kischel, R.; Stienen, S.; Friedrich, M.; et al. Expression of Novel Immune Checkpoint Molecules PVR and PVRL2 Confers a Negative Prognosis to Patients with Acute Myeloid Leukemia and Their Blockade Augments T-Cell Mediated Lysis of AML Cells Alone or in Combination with the BiTE® Antibody Construct AMG 330. Blood 2015, 789. [Google Scholar]
- Whelan, S.; Ophir, E.; Kotturi, M.F.; Levy, O.; Ganguly, S.; Leung, L.; Vaknin, I.; Kumar, S.; Dassa, L.; Hansen, K.; et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8+ T-cell Function. Cancer immunology research 2019, 7, 257–268. [Google Scholar] [CrossRef]
- Bai, M.; Li, W.; Yu, N.; Zhang, H.; Long, F.; Zeng, A. The crosstalk between β-catenin signaling and type I, type II and type III interferons in lung cancer cells. American journal of translational research 2017, 9, 2788. [Google Scholar] [PubMed]
- Hillesheim, A.; Nordhoff, C.; Boergeling, Y.; Ludwig, S.; Wixler, V. β-catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-κB signaling. Cell communication and signaling 2014, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Ohsugi, T.; Yamaguchi, K.; Zhu, C.; Ikenoue, T.; Furukawa, Y. Decreased expression of interferon-induced protein 2 (IFIT2) by Wnt/β-catenin signaling confers anti-apoptotic properties to colorectal cancer cells. Oncotarget 2017, 8, 100176. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Pallasch, C.; Elia, A.E.; Braun, C.J.; Westbrook, T.F.; Hemann, M.; Elledge, S.J. A role for PVRL4-driven cell–cell interactions in tumorigenesis. Elife 2013, 2, e00358. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: bidirectional, allosteric signaling machines. cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Fuchs, A.; Cella, M.; Giurisato, E.; Shaw, A.S.; Colonna, M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). The Journal of Immunology 2004, 172, 3994–3998. [Google Scholar] [CrossRef]
- Wikipedia contributors. Ferroptosis — Wikipedia, The Free Encyclopedia. 2019. Available online: https://en.wikipedia.org/w/index.php?title=Ferroptosis&oldid=882553314 (accessed on 11 May 2019).
- Brown, C.W.; Amante, J.J.; Mercurio, A.M. Cell clustering mediated by the adhesion protein PVRL4 is necessary for α6β4 integrin–promoted ferroptosis resistance in matrix-detached cells. Journal of Biological Chemistry 2018, 293, 12741–12748. [Google Scholar] [CrossRef]
- Abdullah, H.; Brankin, B.; Brady, C.; Cosby, S.L. Wild-type Measles Virus Infection Upregulates Poliovirus Receptor-Related 4 and Causes Apoptosis in Brain Endothelial Cells by Induction of Tumor Necrosis Factor-Related Apoptosis-lnducing Ligand. Journal of Neuropathology & Experimental Neurology 2013, 72, 681–696. [Google Scholar]
- Fabre-Lafay, S.; Garrido-Urbani, S.; Reymond, N.; Gonçalves, A.; Dubreuil, P.; Lopez, M. Nectin-4, a new serological breast cancer marker, is a substrate for tumor necrosis factor-α-converting enzyme (TACE)/ADAM-17. Journal of Biological Chemistry 2005, 280, 19543–19550. [Google Scholar] [CrossRef]
- Besschetnova, T.Y.; Ichimura, T.; Katebi, N.; Croix, B.S.; Bonventre, J.V.; Olsen, B.R. Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology 2015, 42, 56–73. [Google Scholar] [CrossRef]
- Nanda, A.; Carson-Walter, E.B.; Seaman, S.; Barber, T.D.; Stampfl, J.; Singh, S.; Vogelstein, B.; Kinzler, K.W.; Croix, B.S. TEM8 interacts with the cleaved C5 domain of collagen α3 (VI). Cancer Research 2004, 64, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, K.A.; Basile, C.M.; Spring, S.C.; Bonuccelli, G.; Lisanti, M.P.; Terman, B.I. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell–matrix interactions on collagen. Experimental cell research 2005, 305, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.E.; Mavila, A.; Salazar, R.; Bayless, K.J.; Kanagala, S.; Maxwell, S.A.; Davis, G.E. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. Journal of cell science 2001, 114, 2755–2773. [Google Scholar] [CrossRef] [PubMed]
- Bürgi, J.; Kunz, B.; Abrami, L.; Deuquet, J.; Piersigilli, A.; Scholl-Bürgi, S.; Lausch, E.; Unger, S.; Superti-Furga, A.; Bonaldo, P.; et al. CMG2/ANTXR2 regulates extracellular collagen VI which accumulates in hyaline fibromatosis syndrome. Nature communications 2017, 8, 15861. [Google Scholar] [CrossRef]
- Shieh, J.T.; Swidler, P.; Martignetti, J.A.; Ramirez, M.C.M.; Balboni, I.; Kaplan, J.; Kennedy, J.; Abdul-Rahman, O.; Enns, G.M.; Sandborg, C.; et al. Systemic hyalinosis: a distinctive early childhood–onset disorder characterized by mutations in the anthrax toxin receptor 2 gene (ANTRX2). Pediatrics 2006, 118, e1485–e1492. [Google Scholar] [CrossRef]
- Werner, E.; Kowalczyk, A.P.; Faundez, V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. Journal of Biological Chemistry 2006, 281, 23227–23236. [Google Scholar] [CrossRef]
- Go, M.Y.; Chow, E.M.; Mogridge, J. The cytoplasmic domain of anthrax toxin receptor 1 affects binding of the protective antigen. Infection and immunity 2009, 77, 52–59. [Google Scholar] [CrossRef]
- Scobie, H.M.; Rainey, G.J.A.; Bradley, K.A.; Young, J.A. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proceedings of the National Academy of Sciences 2003, 100, 5170–5174. [Google Scholar] [CrossRef]
- Löffek, S.; Schilling, O.; Franzke, C.W. Biological role of matrix metalloproteinases: a critical balance, 2011. European Respiratory Journal 2011, 38, 191–208. [Google Scholar] [CrossRef]
- Wikipedia contributors. Matrix metallopeptidase — Wikipedia, The Free Encyclopedia. 2019. Available online: https://en.wikipedia.org/w/index.php?title=Matrix_metallopeptidase&oldid=893034699 (accessed on 11 May 2019).
- Abrami, L.; Kunz, B.; Deuquet, J.; Bafico, A.; Davidson, G.; Van Der Goot, F.G. Functional interactions between anthrax toxin receptors and the WNT signalling protein LRP6. Cellular microbiology 2008, 10, 2509–2519. [Google Scholar] [CrossRef]
- Wei, W.; Lu, Q.; Chaudry, G.J.; Leppla, S.H.; Cohen, S.N. The LDL receptor-related protein LRP6 mediates internalization and lethality of anthrax toxin. Cell 2006, 124, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Gu, J.; Werner, E. Tumor endothelial marker 8 amplifies canonical Wnt signaling in blood vessels. PloS one 2011, 6, e22334. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, S.Y.; Rajasekaran, N.; Joe, H.E.; Shin, Y.K.; Kim, S.M.; Park, S.G.; Kang, T.J.; Kim, J.C. Human aortic endothelial cells compare favourably with macrophages for the study of anthrax toxins. International Journal of Nanotechnology 2013, 10. [Google Scholar] [CrossRef]
- Madan, B.; Ke, Z.; Harmston, N.; Ho, S.Y.; Frois, A.; Alam, J.; Jeyaraj, D.A.; Pendharkar, V.; Ghosh, K.; Virshup, I.H.; et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016, 35, 2197. [Google Scholar] [CrossRef]
- Faivre, R.; Iooss, B.; Mahévas, S.; Makowski, D.; Monod, H. Analyse de sensibilité et exploration de modèles: application aux sciences de la nature et de l’environnement; Editions Quae; 2013. [Google Scholar]
- Iooss, B.; Lemaître, P. A review on global sensitivity analysis methods. arXiv 2014, arXiv:1404.2405. [Google Scholar]
- Borgonovo, E. A new uncertainty importance measure. Reliability Engineering & System Safety 2007, 92, 771–784. [Google Scholar]
- Da Veiga, S. Global sensitivity analysis with dependence measures. Journal of Statistical Computation and Simulation 2015, 85, 1283–1305. [Google Scholar] [CrossRef]
- Gretton, A.; Bousquet, O.; Smola, A.; Schölkopf, B. Measuring statistical dependence with Hilbert-Schmidt norms. Algorithmic learning theory. Springer, 2005; pp. 63–77. [Google Scholar]
- Jansen, M.J. Analysis of variance designs for model output. Computer Physics Communications 1999, 117, 35–43. [Google Scholar] [CrossRef]
- Saltelli, A.; Annoni, P.; Azzini, I.; Campolongo, F.; Ratto, M.; Tarantola, S. Variance based sensitivity analysis of model output. Design and estimator for the total sensitivity index. Computer Physics Communications 2010, 181, 259–270. [Google Scholar] [CrossRef]
- Saltelli, A. Making best use of model evaluations to compute sensitivity indices. Computer Physics Communications 2002, 145, 280–297. [Google Scholar] [CrossRef]
- Saltelli, A.; Annoni, P. How to avoid a perfunctory sensitivity analysis. Environmental Modelling & Software 2010, 25, 1508–1517. [Google Scholar]
- Sobol’, I.M. On sensitivity estimation for nonlinear mathematical models. Matematicheskoe Modelirovanie 1990, 2, 112–118. [Google Scholar]
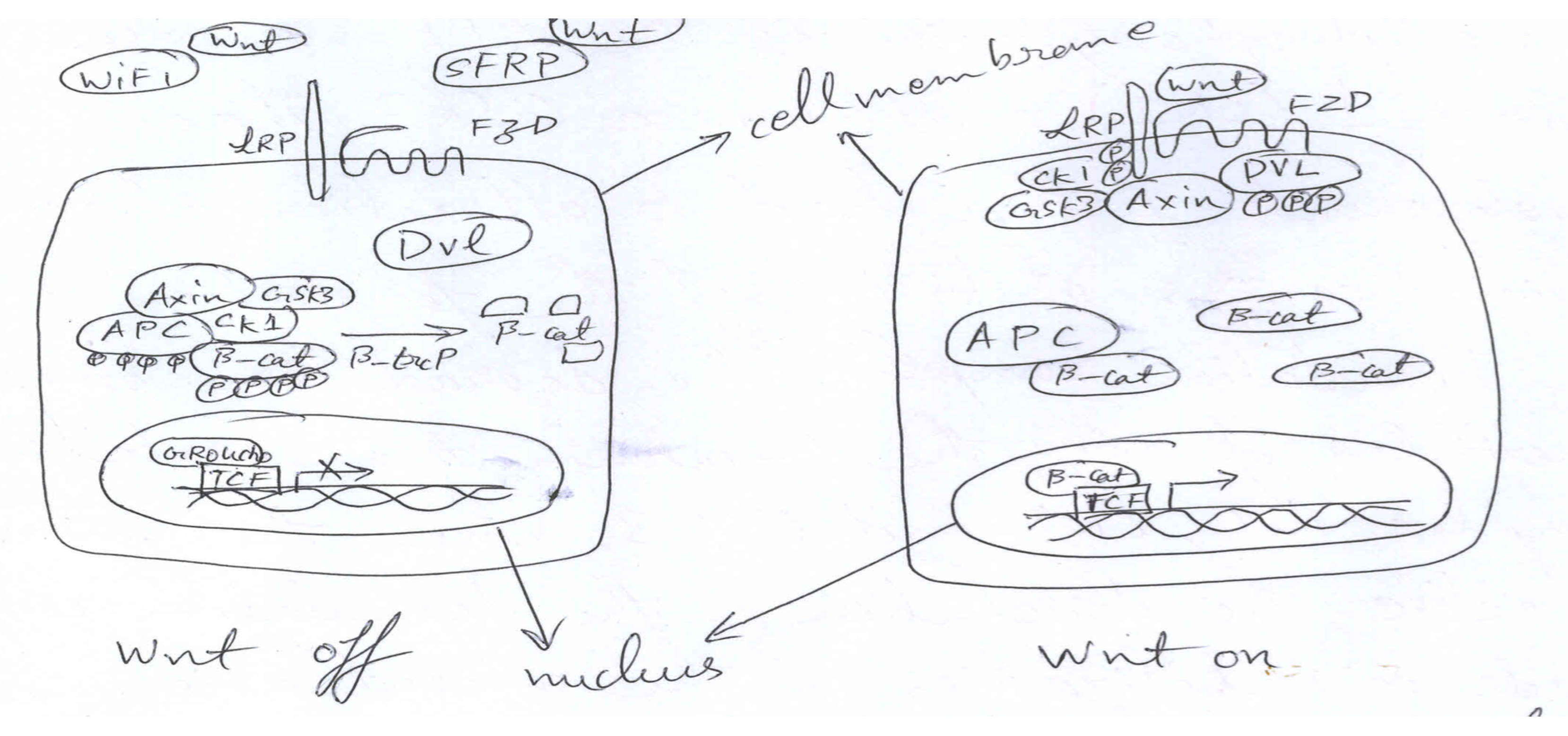
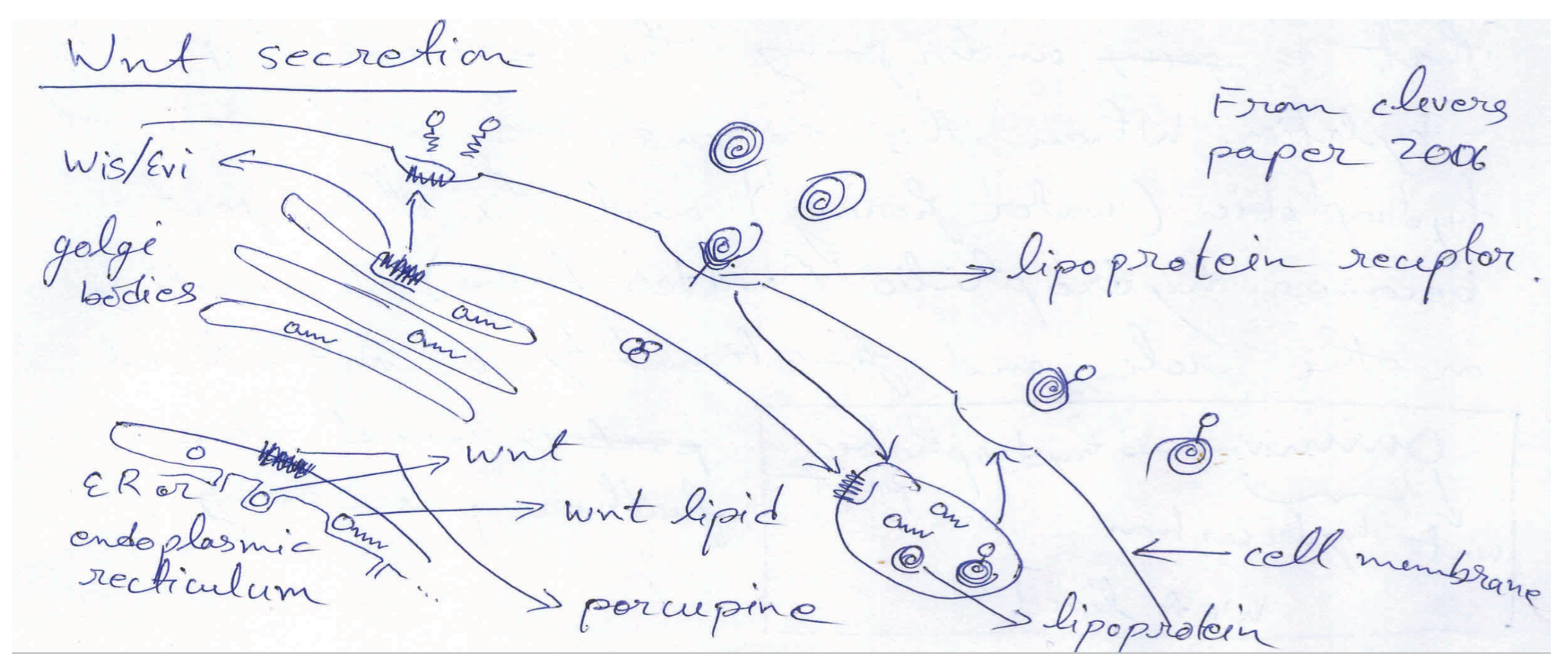
| Ranking ABC family w.r.t WNT family | |||||||
| Ranking of ABC family w.r.t WNT-2B | Ranking of ABC family w.r.t WNT4 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - ABC-A5 | 2108 | 310 | 72 | ABC-A5 - WNT4 | 359 | 1285 | 433 |
| ABC-B11 - WNT2B | 319 | 2132 | 18 | ABC-B11 - WNT4 | 872 | 1284 | 867 |
| WNT2B - ABC-C3 | 1853 | 262 | 2498 | ABC-C3 - WNT4 | 10 | 617 | 296 |
| WNT2B - ABC-C5 | 2213 | 1685 | 840 | WNT4 - ABC-C5 | 1383 | 2119 | 215 |
| WNT2B - ABC-C13 | 1149 | 1191 | 2175 | WNT4 - ABC-C13 | 1649 | 1814 | 542 |
| WNT2B - ABC-D1 | 1119 | 177 | 2163 | ABC-D1 - WNT4 | 1041 | 1171 | 1740 |
| WNT2B - ABC-G1 | 1068 | 1583 | 214 | ABC-G1 - WNT4 | 1020 | 1146 | 2025 |
| WNT2B - ABC-G2 | 1500 | 1533 | 172 | ABC-G2 - WNT4 | 784 | 1431 | 435 |
| Ranking of ABC family w.r.t WNT-7B | Ranking of ABC family w.r.t WNT-9A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - WNT7B | 1550 | 516 | 995 | ABC-A5 - WNT9A | 735 | 349 | 1479 |
| ABC-B11 - WNT7B | 968 | 599 | 324 | ABC-B11 - WNT9A | 843 | 1647 | 689 |
| ABC-C3 - WNT7B | 694 | 1668 | 695 | ABC-C3 - WNT9A | 1590 | 359 | 2136 |
| WNT7B - ABC-C5 | 979 | 1715 | 2268 | ABC-C5 - WNT9A | 1295 | 368 | 2265 |
| WNT7B - ABC-C13 | 950 | 2245 | 2298 | ABC-C13 - WNT9A | 1394 | 2294 | 1134 |
| ABC-D1 - WNT7B | 252 | 850 | 1215 | ABC-D1 - WNT9A | 910 | 2367 | 675 |
| ABC-G1 - WNT7B | 269 | 733 | 1160 | ABC-G1 - WNT9A | 426 | 2457 | 1074 |
| ABC-G2 - WNT7B | 1717 | 224 | 264 | ABC-G2 - WNT9A | 1108 | 2350 | 960 |
| Ranking WNT family w.r.t ABC family | |||||||
| Ranking of WNT family w.r.t ABC-A5 | Ranking of WNT family w.r.t ABC-B11 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - WNT2B | 1549 | 2018 | 2132 | WNT2B - ABC-B11 | 1083 | 703 | 1887 |
| ABC-A5 - WNT4 | 1375 | 2436 | 2449 | WNT4 - ABC-B11 | 156 | 298 | 1517 |
| ABC-A5 - WNT7B | 2420 | 1527 | 460 | WNT7B - ABC-B11 | 1134 | 204 | 2323 |
| ABC-A5 - WNT9A | 1989 | 2209 | 2365 | WNT9A - ABC-B11 | 226 | 2134 | 1480 |
| Ranking of WNT family w.r.t ABC-C3 | Ranking of WNT family w.r.t ABC-C5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-C3 - WNT2B | 1127 | 1482 | 1905 | WNT2B - ABC-C5 | 1970 | 2309 | 2248 |
| ABC-C3 - WNT4 | 897 | 1454 | 489 | WNT4 - ABC-C5 | 2129 | 229 | 230 |
| ABC-C3 - WNT7B | 656 | 2080 | 772 | WNT7B - ABC-C5 | 1539 | 756 | 1258 |
| ABC-C3 - WNT9A | 2339 | 1616 | 814 | ABC-C5 - WNT9A | 213 | 2183 | 2480 |
| Ranking of WNT family w.r.t ABC-C13 | Ranking of WNT family w.r.t ABC-D1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - ABC-C13 | 950 | 2150 | 2048 | WNT2B - ABC-D1 | 1751 | 1370 | 1174 |
| WNT4 - ABC-C13 | 538 | 326 | 2242 | WNT4 - ABC-D1 | 45 | 1784 | 101 |
| WNT7B - ABC-C13 | 2508 | 1830 | 1219 | WNT7B - ABC-D1 | 2238 | 2021 | 1121 |
| WNT9A - ABC-C13 | 738 | 2501 | 634 | WNT9A - ABC-D1 | 732 | 1526 | 1759 |
| Ranking of WNT family w.r.t ABC-G1 | Ranking of WNT family w.r.t ABC-G2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - ABC-G1 | 318 | 775 | 2040 | WNT2B - ABC-G2 | 1342 | 1987 | 1230 |
| WNT4 - ABC-G1 | 2169 | 157 | 39 | WNT4 - ABC-G2 | 862 | 1352 | 1985 |
| WNT7B - ABC-G1 | 587 | 1808 | 1866 | WNT7B - ABC-G2 | 2334 | 2145 | 1526 |
| WNT9A - ABC-G1 | 856 | 2350 | 920 | WNT9A - ABC-G2 | 1919 | 1284 | 2003 |
| Unexplored combinatorial hypotheses | |
| ABC w.r.t WNT | |
| WNT-2B | ABC-C3 |
| WNT-7B | ABC-C13 |
| WNT w.r.t ABC | |
| ABC-A5 | WNT-2B/4/9A |
| WNT-2B/9A | ABC-C5 |
| WNT-2B/7B | ABC-C13 |
| WNT-7B | ABC-D1 |
| WNT-7B | ABC-G1 |
| WNT-7B/9A | ABC-G2 |
| Ranking IL family VS WNT family | |||||||
| Ranking of IL family w.r.t WNT-2B | Ranking of WNT-2B w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - WNT2B | 6 | 2363 | 924 | IL1A - WNT2B | 2290 | 1360 | 2427 |
| IL1B - WNT2B | 1015 | 1278 | 794 | IL1B - WNT2B | 847 | 2168 | 1369 |
| IL1RAP - WNT2B | 1481 | 1391 | 799 | IL1RAP - WNT2B | 2488 | 35 | 1892 |
| IL1RN - WNT2B | 1229 | 1967 | 1582 | IL1RN - WNT2B | 1307 | 43 | 2514 |
| IL2RG - WNT2B | 1434 | 1100 | 2335 | IL2RG - WNT2B | 1384 | 1255 | 1283 |
| IL6ST - WNT2B | 1157 | 1797 | 2088 | IL6ST - WNT2B | 776 | 242 | 1481 |
| IL8 - WNT2B | 2107 | 1817 | 2251 | IL8 - WNT2B | 2157 | 2025 | 593 |
| IL10RB - WNT2B | 961 | 2494 | 512 | IL10RB - WNT2B | 2419 | 856 | 1419 |
| IL15 - WNT2B | 1008 | 1214 | 1714 | IL15 - WNT2B | 1171 | 625 | 1215 |
| IL15RA - WNT2B | 728 | 1782 | 1382 | IL15RA - WNT2B | 2262 | 1021 | 657 |
| IL17C - WNT2B | 477 | 2357 | 1483 | IL17C - WNT2B | 1947 | 1304 | 1331 |
| IL17REL - WNT2B | 1824 | 12 | 2241 | IL17REL - WNT2B | 1980 | 919 | 1617 |
| Ranking of IL family w.r.t WNT-4 | Ranking of WNT-4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - WNT4 | 2500 | 1346 | 955 | IL1A - WNT4 | 507 | 221 | 91 |
| IL1B - WNT4 | 1867 | 1976 | 1682 | IL1B - WNT4 | 129 | 250 | 291 |
| IL1RAP - WNT4 | 2302 | 1826 | 803 | IL1RAP - WNT4 | 74 | 19 | 1553 |
| IL1RN - WNT4 | 1314 | 856 | 104 | IL1RN - WNT4 | 851 | 1218 | 2029 |
| IL2RG - WNT4 | 1289 | 590 | 319 | IL2RG - WNT4 | 520 | 920 | 424 |
| IL6ST - WNT4 | 1315 | 273 | 2422 | IL6ST - WNT4 | 991 | 1443 | 2454 |
| IL8 - WNT4 | 1722 | 549 | 11 | IL8 - WNT4 | 1980 | 2144 | 1267 |
| IL10RB - WNT4 | 1700 | 153 | 1055 | IL10RB - WNT4 | 1828 | 2259 | 1993 |
| IL15 - WNT4 | 1012 | 871 | 1658 | IL15 - WNT4 | 959 | 553 | 448 |
| IL15RA - WNT4 | 1987 | 2265 | 819 | IL15RA - WNT4 | 788 | 139 | 645 |
| IL17C - WNT4 | 2018 | 1639 | 1881 | IL17C - WNT4 | 406 | 276 | 232 |
| IL17REL - WNT4 | 1019 | 425 | 893 | IL17REL - WNT4 | 955 | 595 | 1689 |
| Ranking of IL family w.r.t WNT-7B | Ranking of WNT-7B w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - WNT7B | 662 | 950 | 149 | IL1A - WNT7B | 1058 | 2134 | 2312 |
| IL1B - WNT7B | 290 | 167 | 502 | IL1B - WNT7B | 1683 | 1871 | 1575 |
| IL1RAP - WNT7B | 872 | 1976 | 789 | IL1RAP - WNT7B | 381 | 1728 | 1517 |
| IL1RN - WNT7B | 1882 | 1796 | 503 | IL1RN - WNT7B | 1907 | 2162 | 1605 |
| IL2RG - WNT7B | 1381 | 446 | 482 | IL2RG - WNT7B | 1070 | 1695 | 2245 |
| IL6ST - WNT7B | 819 | 1284 | 1528 | IL6ST - WNT7B | 1268 | 1881 | 2020 |
| IL8 - WNT7B | 2232 | 220 | 701 | IL8 - WNT7B | 1551 | 58 | 2149 |
| IL10RB - WNT7B | 1318 | 1198 | 656 | IL10RB - WNT7B | 375 | 2145 | 803 |
| IL15 - WNT7B | 1000 | 290 | 245 | IL15 - WNT7B | 2307 | 1524 | 1687 |
| IL15RA - WNT7B | 1535 | 1054 | 2204 | IL15RA - WNT7B | 1575 | 191 | 1949 |
| IL17C - WNT7B | 515 | 263 | 113 | IL17C - WNT7B | 1956 | 2388 | 1982 |
| IL17REL - WNT7B | 2053 | 2445 | 2489 | IL17REL - WNT7B | 322 | 859 | 1631 |
| Ranking of IL family w.r.t WNT-9A | Ranking of WNT-9A w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - WNT9A | 199 | 2228 | 1270 | IL1A - WNT9A | 597 | 1322 | 469 |
| IL1B - WNT9A | 305 | 2266 | 466 | IL1B - WNT9A | 776 | 652 | 1010 |
| IL1RAP - WNT9A | 1773 | 2273 | 2159 | IL1RAP - WNT9A | 2003 | 2179 | 964 |
| IL1RN - WNT9A | 2479 | 1506 | 1503 | IL1RN - WNT9A | 1363 | 1829 | 1632 |
| IL2RG - WNT9A | 1489 | 598 | 865 | IL2RG - WNT9A | 186 | 260 | 1276 |
| IL6ST - WNT9A | 2229 | 761 | 1103 | IL6ST - WNT9A | 2099 | 1416 | 1674 |
| IL8 - WNT9A | 346 | 1103 | 1910 | IL8 - WNT9A | 589 | 1751 | 1529 |
| IL10RB - WNT9A | 1836 | 1556 | 1006 | IL10RB - WNT9A | 1021 | 2127 | 1534 |
| IL15 - WNT9A | 168 | 1445 | 855 | IL15 - WNT9A | 1357 | 1025 | 1709 |
| IL15RA - WNT9A | 1776 | 206 | 2380 | IL15RA - WNT9A | 2149 | 2362 | 737 |
| IL17C - WNT9A | 72 | 2442 | 569 | IL17C - WNT9A | 1532 | 2465 | 1607 |
| IL17REL - WNT9A | 2512 | 24 | 580 | IL17REL - WNT9A | 2101 | 1940 | 313 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t WNT | |
| IL-6ST/8/17REL | WNT-2B |
| IL-1B/1RAP/15RA/17C | WNT-4 |
| IL-1RN/17REL | WNT-7B |
| IL-1RAP/15RA | WNT-9A |
| WNT w.r.t IL | |
| IL-1A/1RAP/8 | WNT-2B |
| IL-8/10RB | WNT-4 |
| IL-1A/1RN/6ST/17C | WNT-7B |
| IL-1RAP/15RA/17REL | WNT-9A |
| Ranking UBE2 family VS WNT family | |||||||
| Ranking of UBE2-A w.r.t WNT family | Ranking of WNT family w.r.t UBE2-A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2A | 1608 | 203 | 181 | WNT2B - UBE2A | 1677 | 899 | 1671 |
| WNT4 - UBE2A | 1293 | 2314 | 2279 | WNT4 - UBE2A | 424 | 1062 | 545 |
| WNT7B - UBE2A | 1139 | 1217 | 1961 | WNT7B - UBE2A | 392 | 2345 | 2151 |
| WNT9A - UBE2A | 443 | 1705 | 287 | WNT9A - UBE2A | 806 | 1581 | 1098 |
| Ranking of UBE2-B w.r.t WNT family | Ranking of WNT family w.r.t UBE2-B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2B | 1473 | 2220 | 599 | WNT2B - UBE2B | 2020 | 553 | 73 |
| WNT4 - UBE2B | 2260 | 2008 | 2141 | WNT4 - UBE2B | 301 | 334 | 47 |
| WNT7B - UBE2B | 2116 | 2206 | 1454 | WNT7B - UBE2B | 1336 | 2052 | 1903 |
| WNT9A - UBE2B | 2291 | 79 | 1381 | WNT9A - UBE2B | 2300 | 2476 | 2326 |
| Ranking of UBE2-F w.r.t WNT family | Ranking of WNT family w.r.t UBE2-F | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2F | 1246 | 833 | 2387 | WNT2B - UBE2F | 1006 | 1917 | 49 |
| WNT4 - UBE2F | 2135 | 2505 | 1762 | WNT4 - UBE2F | 63 | 1109 | 664 |
| WNT7B - UBE2F | 2423 | 1673 | 2077 | WNT7B - UBE2F | 2236 | 1660 | 1751 |
| WNT9A - UBE2F | 2032 | 1165 | 128 | WNT9A - UBE2F | 1014 | 2251 | 2179 |
| Ranking of UBE2-H w.r.t WNT family | Ranking of WNT family w.r.t UBE2-H | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2H | 1841 | 351 | 2178 | WNT2B - UBE2H | 2015 | 1019 | 1331 |
| WNT4 - UBE2H | 1090 | 778 | 1224 | WNT4 - UBE2H | 218 | 2248 | 2155 |
| WNT7B - UBE2H | 1505 | 1215 | 527 | WNT7B - UBE2H | 2294 | 1209 | 1367 |
| WNT9A - UBE2H | 605 | 332 | 2479 | WNT9A - UBE2H | 437 | 1202 | 2379 |
| Ranking of UBE2-J1 w.r.t WNT family | Ranking of WNT family w.r.t UBE2-J1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2J1 | 1539 | 1251 | 1814 | WNT2B - UBE2J1 | 1500 | 1562 | 1255 |
| WNT4 - UBE2J1 | 1583 | 2478 | 1604 | WNT4 - UBE2J1 | 292 | 62 | 65 |
| WNT7B - UBE2J1 | 2349 | 1207 | 2183 | WNT7B - UBE2J1 | 552 | 1877 | 1846 |
| WNT9A - UBE2J1 | 1835 | 2053 | 1652 | WNT9A - UBE2J1 | 2471 | 2137 | 2469 |
| Ranking of UBE2-Z w.r.t WNT family | Ranking of WNT family w.r.t UBE2-Z | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| WNT2B - UBE2Z | 58 | 1756 | 1878 | WNT2B - UBE2Z | 1576 | 1171 | 1543 |
| WNT4 - UBE2Z | 2195 | 2468 | 938 | WNT4 - UBE2Z | 896 | 132 | 186 |
| WNT7B - UBE2Z | 2343 | 1973 | 723 | WNT7B - UBE2Z | 1972 | 1800 | 1399 |
| WNT9A - UBE2Z | 136 | 1986 | 4 | WNT9A - UBE2Z | 1149 | 865 | 813 |
| Ranking EXOSC family VS WNT10B | |||||||
| Ranking of WNT10B w.r.t EXOSC family | Ranking of EXOSC family w.r.t WNT10B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC2 - WNT10B | 221 | 433 | 699 | EXOSC2 - WNT10B | 1695 | 1077 | 992 |
| EXOSC3 - WNT10B | 906 | 1292 | 860 | EXOSC3 - WNT10B | 610 | 2496 | 2428 |
| EXOSC5 - WNT10B | 919 | 484 | 997 | EXOSC5 - WNT10B | 832 | 1445 | 1589 |
| EXOSC6 - WNT10B | 407 | 1195 | 1747 | EXOSC6 - WNT10B | 1319 | 1738 | 1689 |
| EXOSC7 - WNT10B | 2599 | 2571 | 2584 | EXOSC7 - WNT10B | 2710 | 13 | 4 |
| EXOSC8 - WNT10B | 336 | 1437 | 391 | EXOSC8 - WNT10B | 451 | 2284 | 2493 |
| EXOSC9 - WNT10B | 222 | 701 | 732 | EXOSC9 - WNT10B | 1378 | 1501 | 1651 |
| Unexplored combinatorial hypotheses | |
| EXOSC w.r.t WNT10B | |
| EXOSC-2/5/6/7/9 | WNT10B |
| WNT10B w.r.t EXOSC | |
| EXOSC-2/3/5/6/8/9 | WNT10B |
| Ranking TP53 family VS WNT | |||||||
| Ranking of TP53 family w.r.t WNT2B | Ranking of WNT2B w.r.t TP53 family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - WNT2B | 2286 | 234 | 1550 | TP53BP2 - WNT2B | 313 | 908 | 2457 |
| TP53I3 - WNT2B | 2056 | 1712 | 1461 | TP53I3 - WNT2B | 713 | 1223 | 1720 |
| TP53INP1 - WNT2B | 945 | 1805 | 2056 | TP53INP1 - WNT2B | 1853 | 2089 | 762 |
| TP53INP2 - WNT2B | 369 | 1277 | 453 | TP53INP2 - WNT2B | 754 | 1723 | 2335 |
| Ranking of TP53 family w.r.t WNT4 | Ranking of WNT4 w.r.t TP53 family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - WNT4 | 1034 | 315 | 1734 | TP53BP2 - WNT4 | 678 | 1464 | 2500 |
| TP53I3 - WNT4 | 1738 | 1631 | 232 | TP53I3 - WNT4 | 297 | 319 | 493 |
| TP53INP1 - WNT4 | 645 | 498 | 450 | TP53INP1 - WNT4 | 131 | 2414 | 2493 |
| TP53INP2 - WNT4 | 671 | 1440 | 405 | TP53INP2 - WNT4 | 529 | 467 | 154 |
| Ranking of TP53 family w.r.t WNT7B | Ranking of WNT7B w.r.t TP53 family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - WNT7B | 2333 | 1282 | 1673 | TP53BP2 - WNT7B | 1442 | 2217 | 1068 |
| TP53I3 - WNT7B | 324 | 712 | 284 | TP53I3 - WNT7B | 1712 | 1988 | 2393 |
| TP53INP1 - WNT7B | 1227 | 1585 | 1019 | TP53INP1 - WNT7B | 1226 | 1685 | 1497 |
| TP53INP2 - WNT7B | 845 | 1004 | 470 | TP53INP2 - WNT7B | 1017 | 1746 | 1925 |
| Ranking of TP53 family w.r.t WNT9A | Ranking of WNT9A w.r.t TP53 family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TP53BP2 - WNT9A | 908 | 2232 | 2143 | TP53BP2 - WNT9A | 1035 | 371 | 1218 |
| TP53I3 - WNT9A | 1707 | 2297 | 1018 | TP53I3 - WNT9A | 1351 | 1281 | 1695 |
| TP53INP1 - WNT9A | 447 | 243 | 1245 | TP53INP1 - WNT9A | 295 | 2045 | 2437 |
| TP53INP2 - WNT9A | 22 | 2497 | 1138 | TP53INP2 - WNT9A | 421 | 1765 | 1121 |
| Unexplored combinatorial hypotheses | |
| TP53 family w.r.t WNT | |
| TP53I3 | WNT2B |
| TP53INP1 | WNT2B |
| TP53BP2 | WNT9A |
| WNT family w.r.t TP53 | |
| TP53INP1 | WNT2B |
| TP53INP2 | WNT2B |
| TP53INP1 | WNT4 |
| TP53I3 | WNT7B |
| TP53INP1 | WNT9A |
| Unexplored combinatorial hypotheses | |
| MUC w.r.t RIKP family | |
| MUC1 | RIPK1 |
| MUC3A | RIPK3 |
| MUC12 | RIPK4 |
| MUC20 | RIPK3 |
| RIPK w.r.t MUC family | |
| MUC1 | RIPK1/RIPK2 |
| MUC4 | RIPK4 |
| MUC17 | RIPK4 |
| MUC20 | RIPK2 |
| Ranking TNF family w.r.t NFkB-2/I family | |||||||
| Ranking of TNF family w.r.t NFkB2 | Ranking of TNF family w.r.t NFkBI-A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB2 - TNF | 1620 | 615 | 1897 | NFkBI-A - TNF | 820 | 1495 | 1109 |
| NFkB2 - TNF-AIP1 | 324 | 649 | 1387 | NFkBI-A - TNF-AIP1 | 1779 | 1904 | 1400 |
| NFkB2 - TNF-AIP2 | 1437 | 715 | 1986 | NFkBI-A - TNF-AIP2 | 1247 | 217 | 766 |
| NFkB2 - TNF-AIP3 | 1272 | 1574 | 441 | NFkBI-A - TNF-AIP3 | 776 | 981 | 212 |
| NFkB2 - TNF-RSF1A | 30 | 2465 | 575 | NFkBI-A - TNF-RSF1A | 1580 | 1422 | 43 |
| NFkB2 - TNF-RSF10A | 2095 | 817 | 2509 | NFkBI-A - TNF-RSF10A | 2499 | 1438 | 2191 |
| NFkB2 - TNF-RSF10B | 37 | 1411 | 250 | NFkBI-A - TNF-RSF10B | 2075 | 1555 | 1401 |
| NFkB2 - TNF-RSF10D | 2473 | 12 | 1499 | NFkBI-A - TNF-RSF10D | 2498 | 2344 | 2501 |
| NFkB2 - TNF-RSF12A | 1813 | 824 | 1893 | NFkBI-A - TNF-RSF12A | 2337 | 1101 | 1491 |
| NFkB2 - TNF-RSF14 | 1799 | 834 | 302 | NFkBI-A - TNF-RSF14 | 1974 | 2045 | 1136 |
| NFkB2 - TNF-RSF21 | 332 | 1973 | 1719 | NFkBI-A - TNF-RSF21 | 1119 | 951 | 903 |
| NFkB2 - TNF-SF10 | 1627 | 1614 | 1299 | NFkBI-A - TNF-SF10 | 2185 | 499 | 2316 |
| NFkB2 - TNF-SF15 | 564 | 2437 | 1064 | NFkBI-A - TNF-SF15 | 564 | 1684 | 1473 |
| Ranking of TNF family w.r.t NFkBI-E | Ranking of TNF family w.r.t NFkBI-Z | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkBI-E - TNF | 2443 | 925 | 228 | NFkBI-Z - TNF | 851 | 776 | 850 |
| NFkBI-E - TNF-AIP1 | 1720 | 685 | 971 | NFkBI-Z - TNF-AIP1 | 153 | 397 | 621 |
| NFkBI-E - TNF-AIP2 | 2347 | 1863 | 964 | NFkBI-Z - TNF-AIP2 | 2188 | 432 | 566 |
| NFkBI-E - TNF-AIP3 | 559 | 1663 | 280 | NFkBI-Z - TNF-AIP3 | 775 | 10 | 2362 |
| NFkBI-E - TNF-RSF1A | 846 | 1624 | 176 | NFkBI-Z - TNF-RSF1A | 399 | 2006 | 93 |
| NFkBI-E - TNF-RSF10A | 840 | 359 | 952 | NFkBI-Z - TNF-RSF10A | 1380 | 2004 | 1540 |
| NFkBI-E - TNF-RSF10B | 835 | 2257 | 1294 | NFkBI-Z - TNF-RSF10B | 2204 | 1438 | 1991 |
| NFkBI-E - TNF-RSF10D | 2454 | 1018 | 1566 | NFkBI-Z - TNF-RSF10D | 2214 | 2033 | 2514 |
| NFkBI-E - TNF-RSF12A | 383 | 166 | 1464 | NFkBI-Z - TNF-RSF12A | 1638 | 2370 | 1841 |
| NFkBI-E - TNF-RSF14 | 1877 | 2282 | 1426 | NFkBI-Z - TNF-RSF14 | 1120 | 1505 | 1899 |
| NFkBI-E - TNF-RSF21 | 2129 | 1293 | 831 | NFkBI-Z - TNF-RSF21 | 207 | 804 | 344 |
| NFkBI-E - TNF-SF10 | 890 | 1096 | 1816 | NFkBI-Z - TNF-SF10 | 609 | 1088 | 1344 |
| NFkBI-E - TNF-SF15 | 523 | 1957 | 32 | NFkBI-Z - TNF-SF15 | 1237 | 1375 | 2196 |
| Ranking NFkB-2/I family w.r.t TNF family | |||||||
| Ranking of NFkB-2/I family w.r.t TNF | Ranking of NFkB-2/I family w.r.t TNF-AIP1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF | 1632 | 989 | 1453 | NFkB-2 - TNF-AIP1 | 2027 | 1807 | 1140 |
| NFkBI-A - TNF | 904 | 561 | 658 | NFkBI-A - TNF-AIP1 | 2072 | 349 | 1218 |
| NFkBI-E - TNF | 2116 | 1247 | 803 | NFkBI-E - TNF-AIP1 | 56 | 420 | 1551 |
| NFkBI-Z - TNF | 691 | 51 | 265 | NFkBI-Z - TNF-AIP1 | 499 | 1648 | 646 |
| Ranking of NFkB-2/I family w.r.t TNF-AIP2 | Ranking of NFkB-2/I family w.r.t TNF-AIP3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF-AIP2 | 2077 | 1027 | 2224 | NFkB-2 - TNF-AIP3 | 1042 | 2336 | 2130 |
| NFkBI-A - TNF-AIP2 | 499 | 22 | 1192 | NFkBI-A - TNF-AIP3 | 1452 | 411 | 637 |
| NFkBI-E - TNF-AIP2 | 526 | 1755 | 338 | NFkBI-E - TNF-AIP3 | 711 | 1686 | 2041 |
| NFkBI-Z - TNF-AIP2 | 452 | 988 | 1617 | NFkBI-Z - TNF-AIP3 | 1979 | 886 | 278 |
| Ranking of NFkB-2/I family w.r.t TNF-RSF1A | Ranking of NFkB-2/I family w.r.t TNF-RSF10A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF-RSF1A | 648 | 164 | 990 | NFkB-2 - TNF-RSF10A | 611 | 1007 | 454 |
| NFkBI-A - TNF-RSF1A | 435 | 1454 | 130 | NFkBI-A - TNF-RSF10A | 458 | 190 | 1412 |
| NFkBI-E - TNF-RSF1A | 431 | 980 | 1417 | NFkBI-E - TNF-RSF10A | 1719 | 263 | 374 |
| NFkBI-Z - TNF-RSF1A | 550 | 2213 | 1447 | NFkBI-Z - TNF-RSF10A | 342 | 742 | 732 |
| Ranking of NFkB-2/I w.r.t TNF-RSF10B | Ranking of NFkB-2/I w.r.t TNF-RSF10D | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF-RSF10B | 713 | 1408 | 2397 | NFkB-2 - TNF-RSF10D | 123 | 1939 | 543 |
| NFkBI-A - TNF-RSF10B | 1237 | 1054 | 562 | NFkBI-A - TNF-RSF10D | 371 | 948 | 584 |
| NFkBI-E - TNF-RSF10B | 1352 | 931 | 2142 | NFkBI-E - TNF-RSF10D | 2136 | 621 | 1811 |
| NFkBI-Z - TNF-RSF10B | 165 | 2407 | 361 | NFkBI-Z - TNF-RSF10D | 259 | 400 | 1341 |
| Ranking of NFkB-2/I family w.r.t TNF-RSF12A | Ranking of NFkB-2/I family w.r.t TNF-RSF14 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF-RSF12A | 250 | 341 | 1232 | NFkB-2 - TNF-RSF14 | 299 | 1253 | 543 |
| NFkBI-A - TNF-RSF12A | 689 | 2225 | 17 | NFkBI-A - TNF-RSF14 | 280 | 1126 | 277 |
| NFkBI-E - TNF-RSF12A | 1188 | 1133 | 765 | NFkBI-E - TNF-RSF14 | 278 | 2025 | 1557 |
| NFkBI-Z - TNF-RSF12A | 973 | 1590 | 2298 | NFkBI-Z - TNF-RSF14 | 131 | 893 | 1953 |
| Ranking of NFkB-2/I family w.r.t TNF-RSF21 | Ranking of NFkB-2/I family w.r.t TNF-SF10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| NFkB-2 - TNF-RSF21 | 250 | 341 | 1232 | NFkB-2 - TNF-SF10 | 1643 | 496 | 743 |
| NFkBI-A - TNF-RSF21 | 689 | 2225 | 17 | NFkBI-A - TNF-SF10 | 262 | 1238 | 1352 |
| NFkBI-E - TNF-RSF21 | 1188 | 1133 | 765 | NFkBI-E - TNF-SF10 | 985 | 1090 | 158 |
| NFkBI-Z - TNF-RSF21 | 973 | 1590 | 2298 | NFkBI-Z - TNF-SF10 | 537 | 1557 | 2104 |
| Ranking of NFkB-2/I family w.r.t TNF-SF15 | |||||||
| laplace | linear | rbf | |||||
| NFkB-2 - TNF-SF15 | 1521 | 786 | 1211 | ||||
| NFkBI-A - TNF-SF15 | 2367 | 325 | 1079 | ||||
| NFkBI-E - TNF-SF15 | 97 | 1868 | 1195 | ||||
| NFkBI-Z - TNF-SF15 | 774 | 407 | 372 | ||||
| Ranking STAT family vs IKBKE | |||||||
| Ranking of STAT family w.r.t IKBKE family | Ranking of IKBKE w.r.t STAT family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| STAT2 - IKBKE | 1267 | 2033 | 1892 | STAT2 - IKBKE | 1604 | 554 | 2108 |
| STAT3 - IKBKE | 1055 | 2144 | 1672 | STAT3 - IKBKE | 1442 | 2179 | 1976 |
| STAT5A - IKBKE | 178 | 1687 | 1183 | STAT5A - IKBKE | 2085 | 2409 | 2277 |
| Unexplored combinatorial hypotheses | |
| STAT w.r.t IKBKE | |
| STAT2 | IKBKE |
| IKBKE w.r.t STAT | |
| STAT3 | IKBKE |
| STAT5A | IKBKE |
| Ranking TRAF family vs IKBKE | |||||||
| Ranking of IKBKE w.r.t TRAF family | Ranking of TRAF family w.r.t IKBKE | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TRAF4 - IKBKE | 1235 | 2158 | 2416 | TRAF4 - IKBKE | 1606 | 461 | 1330 |
| TRAF6 - IKBKE | 1694 | 389 | 1554 | TRAF6 - IKBKE | 2105 | 1376 | 1819 |
| TRAFD1 - IKBKE | 1687 | 532 | 1793 | TRAFD1 - IKBKE | 866 | 733 | 496 |
| TRAF3IP2 - IKBKE | 1349 | 738 | 1987 | TRAF3IP2 - IKBKE | 924 | 1966 | 334 |
| Unexplored combinatorial hypotheses | |
| TRAF w.r.t IKBKE | |
| TRAF6 | IKBKE |
| IKBKE w.r.t TRAF | |
| TRAF4 | IKBKE |
| Unexplored combinatorial hypotheses | |
| NFkB-2/I family w.r.t REL-B | |
| NFkB2 | RELB |
| NFKBIZ | RELB |
| REL-A w.r.t NFkB-2/I family | |
| NFkB2 | RELA |
| NFKBIA | RELA |
| Unexplored combinatorial hypotheses | |
| TNF w.r.t WNT | |
| TNF-RSF1A/RSF10A/RSF10B/SF15 | WNT2B |
| TNF-RSF10A/RSF10D/RSF12A/SF10 | WNT4 |
| TNF-RSF12A/SF10 | WNT7B |
| TNF-RSF21 | WNT9A |
| WNT w.r.t TNF | |
| TNF-RSF10B/RSF10D/RSF14 | WNT2B |
| TNF-AIP3/RSF10B | WNT4 |
| TNF, TNF-RSF1A/RSF14 | WNT7B |
| TNF-AIP2/AIP3/RSF10A/RSF12A/SF10 | WNT9A |
| Ranking TNF family w.r.t MUC family | |||||||
| Ranking of TNF family w.r.t MUC1 | Ranking of TNF family w.r.t MUC3A | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - TNF | 112 | 72 | 88 | MUC3A - TNF | 1353 | 1659 | 1479 |
| MUC1 - TNFAIP1 | 1193 | 1603 | 997 | MUC3A - TNFAIP1 | 2178 | 1209 | 1347 |
| MUC1 - TNFAIP2 | 716 | 405 | 2340 | MUC3A - TNFAIP2 | 1075 | 1614 | 1158 |
| MUC1 - TNFAIP3 | 2115 | 1636 | 1882 | MUC3A - TNFAIP3 | 962 | 1020 | 2491 |
| MUC1 - TNFRSF1A | 1380 | 422 | 1390 | MUC3A - TNFRSF1A | 461 | 1708 | 189 |
| MUC1 - TNFRSF10A | 1009 | 2180 | 1095 | MUC3A - TNFRSF10A | 2237 | 1910 | 335 |
| MUC1 - TNFRSF10B | 1923 | 732 | 88 | MUC3A - TNFRSF10B | 450 | 1443 | 2040 |
| MUC1 - TNFRSF10D | 2303 | 2154 | 376 | MUC3A - TNFRSF10D | 1678 | 2049 | 102 |
| MUC1 - TNFRSF12A | 2019 | 2009 | 1700 | MUC3A - TNFRSF12A | 2349 | 1315 | 382 |
| MUC1 - TNFRSF14 | 1955 | 1899 | 1429 | MUC3A - TNFRSF14 | 956 | 1442 | 1953 |
| MUC1 - TNFRSF21 | 337 | 477 | 968 | MUC3A - TNFRSF21 | 1297 | 1492 | 1959 |
| MUC1 - TNFSF10 | 1111 | 1592 | 1198 | MUC3A - TNFSF10 | 891 | 257 | 798 |
| MUC1 - TNFSF15 | 936 | 986 | 2391 | MUC3A - TNFSF15 | 2285 | 795 | 1164 |
| Ranking of TNF family w.r.t MUC4 | Ranking of TNF family w.r.t MUC12 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC4 - TNF | 1896 | 231 | 1355 | MUC12 - TNF | 1862 | 102 | 135 |
| MUC4 - TNFAIP1 | 864 | 397 | 987 | MUC12 - TNFAIP1 | 1386 | 479 | 942 |
| MUC4 - TNFAIP2 | 73 | 1011 | 1087 | MUC12 - TNFAIP2 | 1056 | 303 | 1587 |
| MUC4 - TNFAIP3 | 1159 | 1751 | 179 | MUC12 - TNFAIP3 | 2493 | 1259 | 1330 |
| MUC4 - TNFRSF1A | 179 | 71 | 16 | MUC12 - TNFRSF1A | 1709 | 1440 | 837 |
| MUC4 - TNFRSF10A | 1668 | 1892 | 1652 | MUC12 - TNFRSF10A | 598 | 531 | 363 |
| MUC4 - TNFRSF10B | 2024 | 1396 | 331 | MUC12 - TNFRSF10B | 409 | 1572 | 1297 |
| MUC4 - TNFRSF10D | 2503 | 2403 | 2356 | MUC12 - TNFRSF10D | 30 | 102 | 149 |
| MUC4 - TNFRSF12A | 1684 | 700 | 745 | MUC12 - TNFRSF12A | 298 | 882 | 153 |
| MUC4 - TNFRSF14 | 1675 | 2029 | 1146 | MUC12 - TNFRSF14 | 1749 | 2237 | 135 |
| MUC4 - TNFRSF21 | 647 | 326 | 323 | MUC12 - TNFRSF21 | 1795 | 607 | 2438 |
| MUC4 - TNFSF10 | 936 | 2134 | 1957 | MUC12 - TNFSF10 | 801 | 1795 | 2435 |
| MUC4 - TNFSF15 | 1440 | 1180 | 1627 | MUC12 - TNFSF15 | 1741 | 889 | 1098 |
| Ranking of TNF family w.r.t MUC13 | Ranking of TNF family w.r.t MUC17 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC13 - TNF | 2282 | 220 | 127 | MUC17 - TNF | 683 | 362 | 515 |
| MUC13 - TNFAIP1 | 378 | 230 | 1935 | MUC17 - TNFAIP1 | 117 | 188 | 272 |
| MUC13 - TNFAIP2 | 2464 | 220 | 697 | MUC17 - TNFAIP2 | 1311 | 414 | 351 |
| MUC13 - TNFAIP3 | 2274 | 1233 | 1446 | MUC17 - TNFAIP3 | 1589 | 1547 | 1539 |
| MUC13 - TNFRSF1A | 274 | 2152 | 514 | MUC17 - TNFRSF1A | 428 | 205 | 329 |
| MUC13 - TNFRSF10A | 2500 | 938 | 1844 | MUC17 - TNFRSF10A | 2269 | 2364 | 2005 |
| MUC13 - TNFRSF10B | 1891 | 1497 | 225 | MUC17 - TNFRSF10B | 1199 | 1323 | 2120 |
| MUC13 - TNFRSF10D | 1191 | 2263 | 2294 | MUC17 - TNFRSF10D | 1798 | 1378 | 2302 |
| MUC13 - TNFRSF12A | 460 | 1753 | 1704 | MUC17 - TNFRSF12A | 2041 | 2303 | 1049 |
| MUC13 - TNFRSF14 | 2220 | 1602 | 1359 | MUC17 - TNFRSF14 | 2043 | 825 | 1700 |
| MUC13 - TNFRSF21 | 1612 | 1673 | 127 | MUC17 - TNFRSF21 | 2013 | 393 | 119 |
| MUC13 - TNFSF10 | 2236 | 1598 | 1495 | MUC17 - TNFSF10 | 280 | 1025 | 817 |
| MUC13 - TNFSF15 | 2423 | 1488 | 1292 | MUC17 - TNFSF15 | 833 | 967 | 950 |
| Ranking of TNF family w.r.t MUC20 | |||||||
| laplace | linear | rbf | |||||
| MUC20 - TNF | 2267 | 262 | 145 | ||||
| MUC20 - TNFAIP1 | 1273 | 2296 | 178 | ||||
| MUC20 - TNFAIP2 | 1062 | 598 | 339 | ||||
| MUC20 - TNFAIP3 | 2205 | 435 | 2136 | ||||
| MUC20 - TNFRSF1A | 483 | 2346 | 145 | ||||
| MUC20 - TNFRSF10A | 100 | 2305 | 917 | ||||
| MUC20 - TNFRSF10B | 775 | 1578 | 1556 | ||||
| MUC20 - TNFRSF10D | 200 | 1487 | 799 | ||||
| MUC20 - TNFRSF12A | 318 | 1607 | 2258 | ||||
| MUC20 - TNFRSF14 | 410 | 1832 | 745 | ||||
| MUC20 - TNFRSF21 | 1686 | 2259 | 164 | ||||
| MUC20 - TNFSF10 | 1005 | 2139 | 1548 | ||||
| MUC20 - TNFSF15 | 2493 | 387 | 2108 | ||||
| Ranking MUC family w.r.t TNF family | |||||||
| Ranking of MUC1 w.r.t TNF family | Ranking of MUC3A w.r.t TNF family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC1 - TNF | 368 | 142 | 21 | MUC3A - TNF | 1478 | 985 | 2373 |
| MUC1 - TNFAIP1 | 692 | 91 | 1591 | MUC3A - TNFAIP1 | 1485 | 536 | 1698 |
| MUC1 - TNFAIP2 | 2290 | 476 | 398 | MUC3A - TNFAIP2 | 1254 | 1265 | 75 |
| MUC1 - TNFAIP3 | 810 | 492 | 748 | MUC3A - TNFAIP3 | 1844 | 960 | 243 |
| MUC1 - TNFRSF1A | 1089 | 2344 | 2312 | MUC3A - TNFRSF1A | 496 | 574 | 792 |
| MUC1 - TNFRSF10A | 1263 | 351 | 826 | MUC3A - TNFRSF10A | 1315 | 1525 | 1815 |
| MUC1 - TNFRSF10B | 1630 | 1604 | 2103 | MUC3A - TNFRSF10B | 351 | 1920 | 1489 |
| MUC1 - TNFRSF10D | 975 | 1026 | 984 | MUC3A - TNFRSF10D | 596 | 950 | 1016 |
| MUC1 - TNFRSF12A | 1597 | 1811 | 1078 | MUC3A - TNFRSF12A | 436 | 595 | 2124 |
| MUC1 - TNFRSF14 | 739 | 2119 | 938 | MUC3A - TNFRSF14 | 1612 | 1383 | 329 |
| MUC1 - TNFRSF21 | 766 | 1495 | 2322 | MUC3A - TNFRSF21 | 1254 | 1357 | 1162 |
| MUC1 - TNFSF10 | 1360 | 1969 | 477 | MUC3A - TNFSF10 | 774 | 980 | 2053 |
| MUC1 - TNFSF15 | 424 | 1183 | 542 | MUC3A - TNFSF15 | 75 | 1261 | 624 |
| Ranking of MUC4 w.r.t TNF family | Ranking of MUC12 w.r.t TNF family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC4 - TNF | 1656 | 777 | 565 | MUC12 - TNF | 266 | 1223 | 628 |
| MUC4 - TNFAIP1 | 2483 | 895 | 390 | MUC12 - TNFAIP1 | 2321 | 668 | 2457 |
| MUC4 - TNFAIP2 | 1875 | 1792 | 180 | MUC12 - TNFAIP2 | 281 | 1829 | 1913 |
| MUC4 - TNFAIP3 | 54 | 498 | 464 | MUC12 - TNFAIP3 | 2353 | 153 | 576 |
| MUC4 - TNFRSF1A | 1074 | 753 | 68 | MUC12 - TNFRSF1A | 1481 | 1952 | 1406 |
| MUC4 - TNFRSF10A | 683 | 311 | 997 | MUC12 - TNFRSF10A | 445 | 337 | 888 |
| MUC4 - TNFRSF10B | 98 | 1413 | 704 | MUC12 - TNFRSF10B | 792 | 164 | 133 |
| MUC4 - TNFRSF10D | 1916 | 230 | 80 | MUC12 - TNFRSF10D | 167 | 193 | 521 |
| MUC4 - TNFRSF12A | 1321 | 2190 | 150 | MUC12 - TNFRSF12A | 216 | 2093 | 302 |
| MUC4 - TNFRSF14 | 606 | 704 | 1493 | MUC12 - TNFRSF14 | 105 | 59 | 69 |
| MUC4 - TNFRSF21 | 1225 | 1967 | 1093 | MUC12 - TNFRSF21 | 1471 | 1975 | 1769 |
| MUC4 - TNFSF10 | 815 | 1108 | 1906 | MUC12 - TNFSF10 | 662 | 2135 | 2255 |
| MUC4 - TNFSF15 | 1141 | 1841 | 920 | MUC12 - TNFSF15 | 1619 | 2204 | 1257 |
| Ranking of MUC13 w.r.t TNF family | Ranking of MUC17 w.r.t TNF family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| MUC13 - TNF | 623 | 292 | 295 | MUC17 - TNF | 203 | 811 | 57 |
| MUC13 - TNFAIP1 | 823 | 755 | 81 | MUC17 - TNFAIP1 | 381 | 193 | 118 |
| MUC13 - TNFAIP2 | 1118 | 2464 | 116 | MUC17 - TNFAIP2 | 1069 | 822 | 136 |
| MUC13 - TNFAIP3 | 1189 | 546 | 541 | MUC17 - TNFAIP3 | 2132 | 47 | 937 |
| MUC13 - TNFRSF1A | 978 | 1506 | 490 | MUC17 - TNFRSF1A | 120 | 497 | 864 |
| MUC13 - TNFRSF10A | 1180 | 540 | 1926 | MUC17 - TNFRSF10A | 852 | 218 | 346 |
| MUC13 - TNFRSF10B | 280 | 1105 | 190 | MUC17 - TNFRSF10B | 933 | 1667 | 1166 |
| MUC13 - TNFRSF10D | 655 | 725 | 1668 | MUC17 - TNFRSF10D | 546 | 133 | 304 |
| MUC13 - TNFRSF12A | 401 | 1242 | 999 | MUC17 - TNFRSF12A | 18 | 1675 | 86 |
| MUC13 - TNFRSF14 | 1324 | 374 | 389 | MUC17 - TNFRSF14 | 819 | 296 | 1014 |
| MUC13 - TNFRSF21 | 690 | 2337 | 107 | MUC17 - TNFRSF21 | 1659 | 814 | 889 |
| MUC13 - TNFSF10 | 1146 | 1208 | 2159 | MUC17 - TNFSF10 | 387 | 1542 | 156 |
| MUC13 - TNFSF15 | 1633 | 314 | 155 | MUC17 - TNFSF15 | 1207 | 1040 | 522 |
| Ranking of MUC20 w.r.t TNF family | |||||||
| laplace | linear | rbf | |||||
| MUC20 - TNF | 57 | 216 | 903 | ||||
| MUC20 - TNFAIP1 | 1265 | 2266 | 2057 | ||||
| MUC20 - TNFAIP2 | 241 | 2404 | 2157 | ||||
| MUC20 - TNFAIP3 | 484 | 1012 | 513 | ||||
| MUC20 - TNFRSF1A | 748 | 173 | 2193 | ||||
| MUC20 - TNFRSF10A | 620 | 427 | 1054 | ||||
| MUC20 - TNFRSF10B | 765 | 1563 | 790 | ||||
| MUC20 - TNFRSF10D | 509 | 2185 | 794 | ||||
| MUC20 - TNFRSF12A | 216 | 2093 | 302 | ||||
| MUC20 - TNFRSF14 | 2298 | 651 | 1368 | ||||
| MUC20 - TNFRSF21 | 2374 | 1611 | 1140 | ||||
| MUC20 - TNFSF10 | 1257 | 1088 | 1031 | ||||
| MUC20 - TNFSF15 | 142 | 2159 | 7 | ||||
| Unexplored combinatorial hypotheses | |
| TNF w.r.t MUC | |
| MUC1 | TNFAIP3/TNFRSF10D/TNFRSF12A/TNFRSF14 |
| MUC3A | TNFRSF10A/TNFRSF10D |
| MUC4 | TNFRSF10D/TNFSF10 |
| MUC12 | TNFRSF21/TNFSF10 |
| MUC13 | TNFRSF10A/TNFRSF10D |
| MUC17 | TNFRSF10A/TNFRSF10D/TNFRSF12A |
| MUC20 | TNFAIP3/TNFSF15 |
| MUC w.r.t TNF | |
| MUC1 | TNFRSF1A |
| MUC4 | TNFAIP2 |
| MUC12 | TNFAIP1/TNFAIP2/TNFRSF21/TNFSF10 |
| MUC13 | TNFAIP1/TNFAIP2 |
| Ranking TNF family vs STEAP4 family | |||||||
| Ranking of TNF family w.r.t STEAP4 | Ranking of STEAP4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - STEAP4 | 1579 | 1914 | 2130 | TNF - STEAP4 | 1116 | 1482 | 999 |
| TNFAIP1 - STEAP4 | 2189 | 1293 | 1910 | TNFAIP1 - STEAP4 | 691 | 611 | 1105 |
| TNFAIP2 - STEAP4 | 1172 | 2002 | 1840 | TNFAIP2 - STEAP4 | 228 | 1747 | 2463 |
| TNFAIP3 - STEAP4 | 1458 | 1882 | 2197 | TNFAIP3 - STEAP4 | 159 | 727 | 219 |
| TNFRSF1A - STEAP4 | 803 | 75 | 1086 | TNFRSF1A - STEAP4 | 1483 | 408 | 1966 |
| TNFRSF10A - STEAP4 | 239 | 1949 | 339 | TNFRSF10A - STEAP4 | 1512 | 1796 | 2026 |
| TNFRSF10B - STEAP4 | 2210 | 1717 | 1827 | TNFRSF10B - STEAP4 | 565 | 571 | 248 |
| TNFRSF10D - STEAP4 | 510 | 2192 | 1797 | TNFRSF10D - STEAP4 | 1018 | 2339 | 2405 |
| TNFRSF12A - STEAP4 | 757 | 338 | 1497 | TNFRSF12A - STEAP4 | 1495 | 1430 | 581 |
| TNFRSF14 - STEAP4 | 1323 | 1512 | 792 | TNFRSF14 - STEAP4 | 1363 | 1956 | 2256 |
| TNFRSF21 - STEAP4 | 1643 | 1920 | 165 | TNFRSF21 - STEAP4 | 1646 | 802 | 160 |
| TNFSF10 - STEAP4 | 2083 | 544 | 1773 | TNFSF10 - STEAP4 | 845 | 675 | 2468 |
| TNFSF15 - STEAP4 | 631 | 1296 | 1020 | TNFSF15 - STEAP4 | 1558 | 600 | 784 |
| Ranking TNF family vs BCL family | |||||||
| Ranking of BCL2L1 w.r.t TNF family | Ranking of TNF family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL2L1 | 174 | 14 | 235 | TNF - BCL2L1 | 56 | 1101 | 294 |
| TNFAIP1 - BCL2L1 | 1527 | 435 | 791 | TNFAIP1 - BCL2L1 | 1497 | 1150 | 74 |
| TNFAIP2 - BCL2L1 | 2142 | 1735 | 798 | TNFAIP2 - BCL2L1 | 1485 | 1735 | 400 |
| TNFAIP3 - BCL2L1 | 2467 | 842 | 867 | TNFAIP3 - BCL2L1 | 1109 | 1939 | 1553 |
| TNFRSF1A - BCL2L1 | 1004 | 1558 | 383 | TNFRSF1A - BCL2L1 | 492 | 376 | 1016 |
| TNFRSF10A - BCL2L1 | 1906 | 1270 | 1222 | TNFRSF10A - BCL2L1 | 2273 | 1928 | 508 |
| TNFRSF10B - BCL2L1 | 1506 | 2235 | 589 | TNFRSF10B - BCL2L1 | 1003 | 2252 | 2217 |
| TNFRSF10D - BCL2L1 | 1920 | 1555 | 1787 | TNFRSF10D - BCL2L1 | 1868 | 2420 | 2392 |
| TNFRSF12A - BCL2L1 | 1254 | 1388 | 1319 | TNFRSF12A - BCL2L1 | 1923 | 53 | 1936 |
| TNFRSF14 - BCL2L1 | 688 | 237 | 2009 | TNFRSF14 - BCL2L1 | 340 | 2350 | 2414 |
| TNFRSF21 - BCL2L1 | 1465 | 1269 | 100 | TNFRSF21 - BCL2L1 | 2139 | 718 | 289 |
| TNFSF10 - BCL2L1 | 532 | 560 | 2332 | TNFSF10 - BCL2L1 | 2115 | 2299 | 1307 |
| TNFSF15 - BCL2L1 | 1026 | 1551 | 1134 | TNFSF15 - BCL2L1 | 453 | 423 | 25 |
| Ranking of BCL2L2 w.r.t TNF family | Ranking of TNF family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL2L2 | 1822 | 1926 | 2359 | TNF - BCL2L2 | 2140 | 109 | 1062 |
| TNFAIP1 - BCL2L2 | 2266 | 2478 | 1847 | TNFAIP1 - BCL2L2 | 2235 | 1607 | 712 |
| TNFAIP2 - BCL2L2 | 823 | 535 | 1117 | TNFAIP2 - BCL2L2 | 109 | 1002 | 54 |
| TNFAIP3 - BCL2L2 | 1201 | 1103 | 1511 | TNFAIP3 - BCL2L2 | 1470 | 1696 | 1276 |
| TNFRSF1A - BCL2L2 | 1124 | 2311 | 1920 | TNFRSF1A - BCL2L2 | 1912 | 169 | 1531 |
| TNFRSF10A - BCL2L2 | 1063 | 1532 | 2458 | TNFRSF10A - BCL2L2 | 1643 | 1095 | 953 |
| TNFRSF10B - BCL2L2 | 2478 | 739 | 2239 | TNFRSF10B - BCL2L2 | 2153 | 1164 | 1983 |
| TNFRSF10D - BCL2L2 | 910 | 2278 | 2237 | TNFRSF10D - BCL2L2 | 35 | 1012 | 1905 |
| TNFRSF12A - BCL2L2 | 1945 | 240 | 2484 | TNFRSF12A - BCL2L2 | 1971 | 1633 | 975 |
| TNFRSF14 - BCL2L2 | 2358 | 1648 | 2310 | TNFRSF14 - BCL2L2 | 1027 | 825 | 1228 |
| TNFRSF21 - BCL2L2 | 2292 | 1850 | 1014 | TNFRSF21 - BCL2L2 | 1138 | 486 | 554 |
| TNFSF10 - BCL2L2 | 2438 | 547 | 2013 | TNFSF10 - BCL2L2 | 2212 | 902 | 169 |
| TNFSF15 - BCL2L2 | 1196 | 2443 | 2350 | TNFSF15 - BCL2L2 | 2285 | 165 | 1330 |
| Ranking of BCL2L13 w.r.t TNF family | Ranking of TNF family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL2L13 | 2437 | 2482 | 2482 | TNF - BCL2L13 | 1162 | 103 | 462 |
| TNFAIP1 - BCL2L13 | 1863 | 2386 | 989 | TNFAIP1 - BCL2L13 | 852 | 606 | 787 |
| TNFAIP2 - BCL2L13 | 793 | 293 | 1846 | TNFAIP2 - BCL2L13 | 438 | 479 | 742 |
| TNFAIP3 - BCL2L13 | 1350 | 1030 | 2129 | TNFAIP3 - BCL2L13 | 1804 | 879 | 626 |
| TNFRSF1A - BCL2L13 | 1173 | 1962 | 2489 | TNFRSF1A - BCL2L13 | 1577 | 1512 | 476 |
| TNFRSF10A - BCL2L13 | 737 | 2055 | 2499 | TNFRSF10A - BCL2L13 | 1534 | 2360 | 1105 |
| TNFRSF10B - BCL2L13 | 1992 | 885 | 906 | TNFRSF10B - BCL2L13 | 2177 | 960 | 1053 |
| TNFRSF10D - BCL2L13 | 2204 | 2159 | 2343 | TNFRSF10D - BCL2L13 | 171 | 1983 | 960 |
| TNFRSF12A - BCL2L13 | 2183 | 2509 | 241 | TNFRSF12A - BCL2L13 | 59 | 1706 | 2046 |
| TNFRSF14 - BCL2L13 | 1852 | 1974 | 2339 | TNFRSF14 - BCL2L13 | 2459 | 2381 | 1187 |
| TNFRSF21 - BCL2L13 | 2280 | 2424 | 2301 | TNFRSF21 - BCL2L13 | 52 | 1054 | 394 |
| TNFSF10 - BCL2L13 | 1088 | 2429 | 1803 | TNFSF10 - BCL2L13 | 1764 | 1186 | 1227 |
| TNFSF15 - BCL2L13 | 1286 | 2438 | 2252 | TNFSF15 - BCL2L13 | 638 | 1962 | 814 |
| Ranking TNF family vs BCL family | |||||||
| Ranking of BCL3 w.r.t TNF family | Ranking of TNF family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL3 | 652 | 370 | 642 | TNF - BCL3 | 598 | 311 | 2473 |
| TNFAIP1 - BCL3 | 168 | 723 | 124 | TNFAIP1 - BCL3 | 596 | 500 | 158 |
| TNFAIP2 - BCL3 | 642 | 856 | 1098 | TNFAIP2 - BCL3 | 59 | 776 | 323 |
| TNFAIP3 - BCL3 | 2377 | 534 | 567 | TNFAIP3 - BCL3 | 300 | 940 | 1527 |
| TNFRSF1A - BCL3 | 163 | 206 | 740 | TNFRSF1A - BCL3 | 83 | 476 | 1355 |
| TNFRSF10A - BCL3 | 799 | 865 | 1044 | TNFRSF10A - BCL3 | 2388 | 2493 | 88 |
| TNFRSF10B - BCL3 | 1632 | 2427 | 1868 | TNFRSF10B - BCL3 | 757 | 1508 | 1062 |
| TNFRSF10D - BCL3 | 1110 | 858 | 714 | TNFRSF10D - BCL3 | 2213 | 1091 | 1972 |
| TNFRSF12A - BCL3 | 273 | 931 | 623 | TNFRSF12A - BCL3 | 671 | 1869 | 1286 |
| TNFRSF14 - BCL3 | 232 | 85 | 1422 | TNFRSF14 - BCL3 | 2149 | 1311 | 1650 |
| TNFRSF21 - BCL3 | 340 | 1384 | 2474 | TNFRSF21 - BCL3 | 411 | 729 | 998 |
| TNFSF10 - BCL3 | 1537 | 1753 | 1638 | TNFSF10 - BCL3 | 1926 | 1523 | 2107 |
| TNFSF15 - BCL3 | 129 | 284 | 729 | TNFSF15 - BCL3 | 1649 | 1032 | 2122 |
| Ranking of BCL6 w.r.t TNF family | Ranking of TNF family w.r.t BCL6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL6 | 2271 | 2071 | 1810 | TNF - BCL6 | 806 | 437 | 1411 |
| TNFAIP1 - BCL6 | 2135 | 2158 | 1330 | TNFAIP1 - BCL6 | 1089 | 850 | 372 |
| TNFAIP2 - BCL6 | 2340 | 1428 | 1808 | TNFAIP2 - BCL6 | 152 | 334 | 703 |
| TNFAIP3 - BCL6 | 267 | 1336 | 1219 | TNFAIP3 - BCL6 | 1884 | 1935 | 855 |
| TNFRSF1A - BCL6 | 1598 | 1771 | 2503 | TNFRSF1A - BCL6 | 788 | 741 | 1130 |
| TNFRSF10A - BCL6 | 1327 | 1831 | 2096 | TNFRSF10A - BCL6 | 607 | 1249 | 2360 |
| TNFRSF10B - BCL6 | 1373 | 1873 | 1264 | TNFRSF10B - BCL6 | 1746 | 1282 | 361 |
| TNFRSF10D - BCL6 | 2213 | 2188 | 788 | TNFRSF10D - BCL6 | 1540 | 1301 | 2008 |
| TNFRSF12A - BCL6 | 1867 | 99 | 2261 | TNFRSF12A - BCL6 | 545 | 2200 | 1910 |
| TNFRSF14 - BCL6 | 1409 | 1337 | 2028 | TNFRSF14 - BCL6 | 731 | 1302 | 1902 |
| TNFRSF21 - BCL6 | 645 | 2071 | 2335 | TNFRSF21 - BCL6 | 40 | 1850 | 50 |
| TNFSF10 - BCL6 | 919 | 445 | 99 | TNFSF10 - BCL6 | 2119 | 1102 | 1626 |
| TNFSF15 - BCL6 | 2106 | 1692 | 1451 | TNFSF15 - BCL6 | 969 | 1475 | 226 |
| Ranking of BCL9L w.r.t TNF family | Ranking of TNF family w.r.t BCL9L | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL9L | 1964 | 1172 | 1478 | TNF - BCL9L | 2218 | 98 | 425 |
| TNFAIP1 - BCL9L | 439 | 1445 | 264 | TNFAIP1 - BCL9L | 766 | 2470 | 1802 |
| TNFAIP2 - BCL9L | 1250 | 1473 | 696 | TNFAIP2 - BCL9L | 646 | 567 | 85 |
| TNFAIP3 - BCL9L | 534 | 630 | 618 | TNFAIP3 - BCL9L | 1046 | 1223 | 2296 |
| TNFRSF1A - BCL9L | 2050 | 1096 | 978 | TNFRSF1A - BCL9L | 863 | 500 | 825 |
| TNFRSF10A - BCL9L | 212 | 1682 | 980 | TNFRSF10A - BCL9L | 2140 | 241 | 1547 |
| TNFRSF10B - BCL9L | 952 | 698 | 685 | TNFRSF10B - BCL9L | 286 | 414 | 2046 |
| TNFRSF10D - BCL9L | 1315 | 181 | 1423 | TNFRSF10D - BCL9L | 1956 | 112 | 990 |
| TNFRSF12A - BCL9L | 430 | 1167 | 1470 | TNFRSF12A - BCL9L | 1797 | 1280 | 1699 |
| TNFRSF14 - BCL9L | 1433 | 635 | 1497 | TNFRSF14 - BCL9L | 670 | 1055 | 1540 |
| TNFRSF21 - BCL9L | 495 | 2326 | 468 | TNFRSF21 - BCL9L | 1291 | 1378 | 246 |
| TNFSF10 - BCL9L | 1889 | 974 | 183 | TNFSF10 - BCL9L | 1812 | 1796 | 2095 |
| TNFSF15 - BCL9L | 878 | 2389 | 71 | TNFSF15 - BCL9L | 1939 | 2114 | 2405 |
| Ranking of BCL10 w.r.t TNF family | Ranking of TNF family w.r.t BCL10 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| TNF - BCL10 | 708 | 79 | 979 | TNF - BCL10 | 1931 | 114 | 1573 |
| TNFAIP1 - BCL10 | 1657 | 805 | 1298 | TNFAIP1 - BCL10 | 690 | 1941 | 7 |
| TNFAIP2 - BCL10 | 1101 | 2197 | 312 | TNFAIP2 - BCL10 | 523 | 2099 | 339 |
| TNFAIP3 - BCL10 | 985 | 813 | 767 | TNFAIP3 - BCL10 | 935 | 595 | 1870 |
| TNFRSF1A - BCL10 | 745 | 1191 | 1288 | TNFRSF1A - BCL10 | 362 | 173 | 448 |
| TNFRSF10A - BCL10 | 451 | 819 | 954 | TNFRSF10A - BCL10 | 1547 | 415 | 2426 |
| TNFRSF10B - BCL10 | 791 | 537 | 1446 | TNFRSF10B - BCL10 | 582 | 658 | 1464 |
| TNFRSF10D - BCL10 | 1831 | 1694 | 2040 | TNFRSF10D - BCL10 | 302 | 19 | 2497 |
| TNFRSF12A - BCL10 | 2015 | 1072 | 1883 | TNFRSF12A - BCL10 | 1865 | 1234 | 1540 |
| TNFRSF14 - BCL10 | 254 | 1400 | 847 | TNFRSF14 - BCL10 | 1175 | 1894 | 2227 |
| TNFRSF21 - BCL10 | 1912 | 571 | 958 | TNFRSF21 - BCL10 | 848 | 1943 | 418 |
| TNFSF10 - BCL10 | 1743 | 931 | 1657 | TNFSF10 - BCL10 | 2020 | 1522 | 1054 |
| TNFSF15 - BCL10 | 254 | 1469 | 577 | TNFSF15 - BCL10 | 1256 | 188 | 1074 |
| Ranking RAD family w.r.t XRCC family | |||||||
| Ranking of RAD family w.r.t XRCC1 | Ranking of RAD family w.r.t XRCC2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| RAD1 - XRCC1 | 1922 | 1658 | 1771 | XRCC2 - RAD1 | 1921 | 893 | 1774 |
| RAD18 - XRCC1 | 1027 | 456 | 1355 | XRCC2 - RAD18 | 1388 | 847 | 765 |
| XRCC1 - RAD50 | 2459 | 2254 | 2082 | XRCC2 - RAD50 | 1877 | 2185 | 2546 |
| RAD51 - XRCC1 | 282 | 365 | 1003 | XRCC2 - RAD51 | 1247 | 1033 | 629 |
| RAD51AP1 - XRCC1 | 753 | 5 | 275 | XRCC2 - RAD51AP1 | 302 | 247 | 42 |
| RAD51C - XRCC1 | 337 | 111 | 968 | XRCC2 - RAD51C | 1079 | 674 | 323 |
| RAD54B - XRCC1 | 175 | 224 | 782 | XRCC2 - RAD54B | 387 | 566 | 506 |
| RAD54L - XRCC1 | 327 | 889 | 709 | XRCC2 - RAD54L | 976 | 918 | 847 |
| Ranking of RAD family w.r.t XRCC6 | Ranking of RAD family w.r.t XRCC6BP1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| XRCC6 - RAD1 | 1929 | 2029 | 2627 | RAD1 - XRCC6BP1 | 1167 | 2417 | 308 |
| RAD18 - XRCC6 | 541 | 25 | 1068 | RAD18 - XRCC6BP1 | 656 | 1612 | 2271 |
| XRCC6 - RAD50 | 2434 | 2043 | 2603 | XRCC6BP1 - RAD50 | 1302 | 2263 | 328 |
| RAD51 - XRCC6 | 608 | 425 | 900 | RAD51 - XRCC6BP1 | 435 | 495 | 1275 |
| RAD51AP1 - XRCC6 | 216 | 67 | 83 | RAD51AP1 - XRCC6BP1 | 81 | 177 | 73 |
| RAD51C - XRCC6 | 426 | 865 | 503 | RAD51C - XRCC6BP1 | 645 | 1366 | 1414 |
| RAD54B - XRCC6 | 3 | 610 | 112 | RAD54B - XRCC6BP1 | 154 | 693 | 1398 |
| RAD54L - XRCC6 | 85 | 252 | 432 | RAD54L - XRCC6BP1 | 420 | 1060 | 2542 |
| Ranking XRN2 w.r.t RAD family | |||||||
| Ranking of RAD family w.r.t XRN2 | Ranking of XRN2 w.r.t RAD family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| XRN2 - RAD51AP1 | 340 | 545 | 290 | XRN2 - RAD51AP1 | 1905 | 1256 | 852 |
| XRN2 - RAD51 | 387 | 560 | 605 | XRN2 - RAD51 | 786 | 2647 | 1995 |
| XRN2 - RAD54L | 594 | 827 | 879 | XRN2 - RAD54L | 1541 | 1246 | 1819 |
| XRN2 - RAD51C | 639 | 1236 | 745 | XRN2 - RAD51C | 1037 | 1777 | 2228 |
| XRN2 - RAD18 | 794 | 688 | 804 | XRN2 - RAD18 | 904 | 2403 | 1801 |
| XRN2 - RAD1 | 898 | 1955 | 2506 | XRN2 - RAD1 | 255 | 122 | 2557 |
| XRN2 - RAD54B | 951 | 165 | 343 | XRN2 - RAD54B | 1818 | 2381 | 2603 |
| XRN2 - RAD50 | 1330 | 2312 | 2295 | XRN2 - RAD50 | 504 | 2100 | 1842 |
| Unexplored combinatorial hypotheses | |
| RAD w.r.t NKRF | |
| RAD51AP1 | NKRF |
| RAD51 | NKRF |
| RAD54L | NKRF |
| RAD51C | NKRF |
| RAD18 | NKRF |
| RAD1 | NKRF |
| RAD54B | NKRF |
| NKRF w.r.t RAD | |
| RAD51AP1 | NKRF |
| RAD51 | NKRF |
| RAD54L | NKRF |
| RAD51C | NKRF |
| RAD18 | NKRF |
| NKRF | RAD1 |
| NKRF | RAD50 |
| Unexplored combinatorial hypotheses | |
| RAD w.r.t BCL | |
| RAD-18/50/51/51AP1/51C/54B/54L | BCL-2L12 |
| RAD-18/51/51AP1/51C/54B/54L | BCL-6B |
| RAD-18/51/51AP1/51C/54B/54L | BCL-7A |
| RAD-18/50/51/51AP1/51C/54B/54L | BCL-9 |
| RAD-1/18/50/51/51AP1/51C/54B/54L | BCL-11A |
| RAD-1/50/51/51AP1/51C/54B/54L | BCL-11B |
| BCL w.r.t RAD | |
| RAD-1/18/50/51/51C/54B/54L | BCL-2L12 |
| RAD-1/18/50/51/51AP1/51C/54B/54L | BCL-6B |
| RAD-1/18/50/51/54L | BCL-7A |
| RAD-18/51/51C/54L | BCL-9 |
| RAD-1/18/50/51/51AP1/51C/54B | BCL-11A |
| RAD-50/51/51AP1/54B/54L | BCL-11B |
| Ranking RAD family VS EXOSC family | |||||||
| Ranking of EXOSC2 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC2 - RAD1 | 1033 | 1311 | 1207 | EXOSC2 - RAD1 | 2456 | 1368 | 2292 |
| EXOSC2 - RAD18 | 1210 | 995 | 1906 | EXOSC2 - RAD18 | 1115 | 979 | 654 |
| EXOSC2 - RAD50 | 1124 | 698 | 629 | EXOSC2 - RAD50 | 1647 | 2495 | 2375 |
| EXOSC2 - RAD51 | 1754 | 191 | 633 | EXOSC2 - RAD51 | 795 | 1332 | 441 |
| EXOSC2 - RAD51AP1 | 198 | 1462 | 2718 | EXOSC2 - RAD51AP1 | 2320 | 1316 | 2127 |
| EXOSC2 - RAD51C | 87 | 463 | 1130 | EXOSC2 - RAD51C | 636 | 564 | 152 |
| EXOSC2 - RAD54B | 351 | 135 | 142 | EXOSC2 - RAD54B | 278 | 132 | 282 |
| EXOSC2 - RAD54L | 1131 | 1652 | 320 | EXOSC2 - RAD54L | 125 | 888 | 545 |
| Ranking of EXOSC3 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC3 - RAD1 | 2492 | 1677 | 549 | EXOSC3 - RAD1 | 2200 | 1243 | 2711 |
| EXOSC3 - RAD18 | 1676 | 2516 | 184 | EXOSC3 - RAD18 | 2024 | 1468 | 767 |
| EXOSC3 - RAD50 | 2368 | 1892 | 2204 | EXOSC3 - RAD50 | 1062 | 596 | 2346 |
| EXOSC3 - RAD51 | 894 | 1066 | 2463 | EXOSC3 - RAD51 | 727 | 583 | 963 |
| EXOSC3 - RAD51AP1 | 1884 | 1037 | 804 | EXOSC3 - RAD51AP1 | 100 | 49 | 219 |
| EXOSC3 - RAD51C | 2499 | 2356 | 1248 | EXOSC3 - RAD51C | 663 | 869 | 887 |
| EXOSC3 - RAD54B | 2183 | 2518 | 2360 | EXOSC3 - RAD54B | 384 | 277 | 310 |
| EXOSC3 - RAD54L | 1735 | 469 | 736 | EXOSC3 - RAD54L | 546 | 1117 | 808 |
| Ranking of EXOSC5 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC5 - RAD1 | 568 | 1169 | 1699 | EXOSC5 - RAD1 | 2405 | 1716 | 1718 |
| EXOSC5 - RAD18 | 2481 | 219 | 1652 | EXOSC5 - RAD18 | 1026 | 550 | 253 |
| EXOSC5 - RAD50 | 447 | 195 | 475 | EXOSC5 - RAD50 | 1596 | 1952 | 2271 |
| EXOSC5 - RAD51 | 2548 | 431 | 1121 | EXOSC5 - RAD51 | 260 | 1095 | 137 |
| EXOSC5 - RAD51AP1 | 1290 | 487 | 430 | EXOSC5 - RAD51AP1 | 1555 | 1860 | 976 |
| EXOSC5 - RAD51C | 1284 | 1264 | 1790 | EXOSC5 - RAD51C | 233 | 1003 | 359 |
| EXOSC5 - RAD54B | 940 | 812 | 1036 | EXOSC5 - RAD54B | 834 | 1825 | 335 |
| EXOSC5 - RAD54L | 408 | 2539 | 1407 | EXOSC5 - RAD54L | 248 | 197 | 39 |
| Ranking of EXOSC6 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC6 - RAD1 | 2283 | 2490 | 1228 | EXOSC6 - RAD1 | 2405 | 142 | 639 |
| EXOSC6 - RAD18 | 1637 | 1599 | 2254 | EXOSC6 - RAD18 | 1118 | 1313 | 1549 |
| EXOSC6 - RAD50 | 2289 | 1969 | 1797 | EXOSC6 - RAD50 | 2309 | 1722 | 575 |
| EXOSC6 - RAD51 | 1056 | 1482 | 1007 | EXOSC6 - RAD51 | 998 | 2297 | 2219 |
| EXOSC6 - RAD51AP1 | 1854 | 2480 | 1827 | EXOSC6 - RAD51AP1 | 149 | 1060 | 2731 |
| EXOSC6 - RAD51C | 1996 | 940 | 1842 | EXOSC6 - RAD51C | 500 | 1628 | 2409 |
| EXOSC6 - RAD54B | 2289 | 2312 | 2005 | EXOSC6 - RAD54B | 262 | 2703 | 2465 |
| EXOSC6 - RAD54L | 987 | 2240 | 1642 | EXOSC6 - RAD54L | 885 | 271 | 1224 |
| Ranking of EXOSC7 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC7 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC7 - RAD1 | 2559 | 1735 | 1210 | EXOSC7 - RAD1 | 2079 | 2308 | 1604 |
| EXOSC7 - RAD18 | 490 | 1688 | 1331 | EXOSC7 - RAD18 | 441 | 385 | 1542 |
| EXOSC7 - RAD50 | 2661 | 1939 | 2021 | EXOSC7 - RAD50 | 1840 | 406 | 2100 |
| EXOSC7 - RAD51 | 842 | 1900 | 1876 | EXOSC7 - RAD51 | 376 | 1180 | 550 |
| EXOSC7 - RAD51AP1 | 2446 | 349 | 2374 | EXOSC7 - RAD51AP1 | 35 | 97 | 786 |
| EXOSC7 - RAD51C | 1113 | 1623 | 530 | EXOSC7 - RAD51C | 854 | 671 | 1459 |
| EXOSC7 - RAD54B | 2431 | 1612 | 1191 | EXOSC7 - RAD54B | 458 | 260 | 646 |
| EXOSC7 - RAD54L | 1550 | 1754 | 1728 | EXOSC7 - RAD54L | 464 | 528 | 790 |
| Ranking of EXOSC8 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC8 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC8 - RAD1 | 2380 | 2442 | 2630 | EXOSC8 - RAD1 | 1928 | 151 | 1563 |
| EXOSC8 - RAD18 | 805 | 2287 | 1564 | EXOSC8 - RAD18 | 764 | 523 | 29 |
| EXOSC8 - RAD50 | 1798 | 1830 | 1893 | EXOSC8 - RAD50 | 2103 | 2649 | 1822 |
| EXOSC8 - RAD51 | 404 | 1630 | 2092 | EXOSC8 - RAD51 | 98 | 1161 | 902 |
| EXOSC8 - RAD51AP1 | 1932 | 1567 | 1701 | EXOSC8 - RAD51AP1 | 408 | 1824 | 541 |
| EXOSC8 - RAD51C | 2439 | 1576 | 2554 | EXOSC8 - RAD51C | 906 | 738 | 1052 |
| EXOSC8 - RAD54B | 1562 | 2542 | 1736 | EXOSC8 - RAD54B | 23 | 1578 | 130 |
| EXOSC8 - RAD54L | 1248 | 622 | 239 | EXOSC8 - RAD54L | 651 | 1384 | 1047 |
| Ranking of EXOSC9 w.r.t RAD family | Ranking of RAD family w.r.t EXOSC9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC9 - RAD1 | 2240 | 175 | 1648 | EXOSC9 - RAD1 | 1335 | 1799 | 978 |
| EXOSC9 - RAD18 | 1533 | 774 | 1180 | EXOSC9 - RAD18 | 2529 | 54 | 540 |
| EXOSC9 - RAD50 | 545 | 183 | 467 | EXOSC9 - RAD50 | 211 | 2217 | 1377 |
| EXOSC9 - RAD51 | 866 | 106 | 99 | EXOSC9 - RAD51 | 807 | 74 | 429 |
| EXOSC9 - RAD51AP1 | 1570 | 1819 | 1807 | EXOSC9 - RAD51AP1 | 2480 | 103 | 1210 |
| EXOSC9 - RAD51C | 110 | 742 | 200 | EXOSC9 - RAD51C | 399 | 844 | 69 |
| EXOSC9 - RAD54B | 179 | 178 | 84 | EXOSC9 - RAD54B | 2385 | 466 | 1286 |
| EXOSC9 - RAD54L | 1113 | 2436 | 22 | EXOSC9 - RAD54L | 536 | 724 | 414 |
| Unexplored combinatorial hypotheses | |
| RAD w.r.t EXOSC | |
| EXOSC-2 | RAD-18/51/51C/54B/54L |
| EXOSC-3 | RAD-18/50/51/51AP1/51C/54B/54L |
| EXOSC-5 | RAD-1/18/51/51AP1/51C/54B/54L |
| EXOSC-6 | RAD-1/18/50/51AP1/51C/54L |
| EXOSC-7 | RAD-18/51/51AP1/51C/54B/54L |
| EXOSC-8 | RAD-1/18/51/51AP1/51C/54B/54L |
| EXOSC-9 | RAD-1/18/50/51/51AP1/51C/54B/54L |
| EXOSC w.r.t RAD | |
| EXOSC-2 | RAD-1/18/50/51/51AP1/51C/54B/54L |
| EXOSC-3 | RAD-1/18/51/51AP1/54L |
| EXOSC-5 | RAD-1/18/50/51/51AP1/51C/54B/54L |
| EXOSC-6 | RAD-18/51/54L |
| EXOSC-7 | RAD-1/18/51C/54B/54L |
| EXOSC-8 | RAD-18/51/51AP1/54B/54L |
| EXOSC-9 | RAD-1/18/50/51/51C/54B/54L |
| Unexplored combinatorial hypotheses | |
| RAD w.r.t FANC | |
| RAD-18/51/51AP1/51C/54B/54L | FANCB |
| RAD-18/51/51AP1/54B/54L | FANCD2 |
| RAD-1/18/50/51/51C/54B/54L | FANCD2OS |
| RAD-1/18/50/51/51AP1/51C/54B/54L | FANCF |
| RAD-1/18/50/51/51AP1/51C/54B/54L | FANCG |
| RAD-18/50/51/51C/54B/54L | FANCI |
| FANC w.r.t RAD | |
| FANCB | RAD-1/50/51/51AP1/51C/54B/54L |
| FANCD2 | RAD-1/50/51/51AP1/51C/54B/54L |
| FANCD2OS | RAD-1/18/5051C/54B |
| FANCF | RAD-1/18/50/51C/54B |
| FANCG | RAD-1/50/51/51AP1/51C/54B |
| FANCI | RAD-1/18/50/51/51AP1/51C/54B/54L |
| Ranking TRET vs ABC family | |||||||
| Ranking of ABC family w.r.t TRET | Ranking of TRET w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCF2 - TERT | 381 | 1047 | 316 | ABCF2 - TERT | 2069 | 238 | 2712 |
| ABCA2 - TERT | 1201 | 49 | 317 | ABCA2 - TERT | 1693 | 2739 | 1997 |
| ABCE1 - TERT | 1613 | 499 | 1217 | ABCE1 - TERT | 120 | 2736 | 294 |
| Unexplored combinatorial hypotheses | |
| ABC family w.r.t TRET | |
| TERT | ABCF2 |
| TERT | ABCA2 |
| TERT | ABCE1 |
| TRET w.r.t ABC family | |
| ABCE1 | TERT |
| Ranking ABC family w.r.t UBE2 family | |||||||
| Ranking of ABC family w.r.t UBE2-A | Ranking of ABC family w.r.t UBE2-B | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-A | 2101 | 185 | 382 | ABC-A5 - UBE2-B | 1223 | 1193 | 194 |
| ABC-B11 - UBE2-A | 129 | 2487 | 304 | ABC-B11 - UBE2-B | 125 | 103 | 571 |
| ABC-C3 - UBE2-A | 2137 | 2491 | 1023 | ABC-C3 - UBE2-B | 606 | 791 | 1411 |
| ABC-C5 - UBE2-A | 1630 | 490 | 2408 | ABC-C5 - UBE2-B | 1515 | 2317 | 2266 |
| ABC-C13 - UBE2-A | 742 | 1604 | 475 | ABC-C13 - UBE2-B | 2199 | 2254 | 2362 |
| ABC-D1 - UBE2-A | 316 | 620 | 596 | ABC-D1 - UBE2-B | 1082 | 374 | 1057 |
| ABC-G1 - UBE2-A | 46 | 819 | 533 | ABC-G1 - UBE2-B | 48 | 843 | 551 |
| ABC-G2 - UBE2-A | 398 | 259 | 261 | ABC-G2 - UBE2-B | 189 | 189 | 41 |
| Ranking of ABC family w.r.t UBE2-F | Ranking of ABC family w.r.t UBE2-H | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-F | 997 | 2408 | 1784 | ABC-A5 - UBE2-H | 1247 | 2068 | 2438 |
| ABC-B11 - UBE2-F | 141 | 1122 | 578 | ABC-B11 - UBE2-H | 932 | 429 | 409 |
| ABC-C3 - UBE2-F | 931 | 2420 | 681 | ABC-C3 - UBE2-H | 540 | 1962 | 563 |
| ABC-C5 - UBE2-F | 628 | 1373 | 217 | ABC-C5 - UBE2-H | 1551 | 865 | 1450 |
| ABC-C13 - UBE2-F | 403 | 2464 | 1307 | ABC-C13 - UBE2-H | 1192 | 2492 | 2051 |
| ABC-D1 - UBE2-F | 2069 | 1959 | 1235 | ABC-D1 - UBE2-H | 1094 | 1016 | 1474 |
| ABC-G1 - UBE2-F | 209 | 1216 | 1450 | ABC-G1 - UBE2-H | 683 | 173 | 18 |
| ABC-G2 - UBE2-F | 690 | 1995 | 2120 | ABC-G2 - UBE2-H | 1328 | 1374 | 78 |
| Ranking of ABC family w.r.t UBE2-J1 | Ranking of ABC family w.r.t UBE2-Z | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-J1 | 634 | 222 | 711 | ABC-A5 - UBE2-Z | 454 | 1059 | 1287 |
| ABC-B11 - UBE2-J1 | 1182 | 1075 | 403 | ABC-B11 - UBE2-Z | 134 | 503 | 436 |
| ABC-C3 - UBE2-J1 | 1232 | 719 | 1285 | ABC-C3 - UBE2-Z | 975 | 1722 | 2095 |
| ABC-C5 - UBE2-J1 | 964 | 1342 | 2373 | ABC-C5 - UBE2-Z | 2348 | 845 | 1859 |
| ABC-C13 - UBE2-J1 | 2095 | 2412 | 2360 | ABC-C13 - UBE2-Z | 1157 | 651 | 1335 |
| ABC-D1 - UBE2-J1 | 542 | 1198 | 704 | ABC-D1 - UBE2-Z | 392 | 1660 | 943 |
| ABC-G1 - UBE2-J1 | 306 | 97 | 122 | ABC-G1 - UBE2-Z | 545 | 142 | 354 |
| ABC-G2 - UBE2-J1 | 335 | 668 | 591 | ABC-G2 - UBE2-Z | 747 | 285 | 530 |
| Ranking UBE2 family w.r.t ABC family | |||||||
| Ranking of UBE2-A w.r.t ABC | Ranking of UBE2-B w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-A | 1037 | 253 | 2091 | ABC-A5 - UBE2-B | 1846 | 2038 | 936 |
| ABC-B11 - UBE2-A | 1491 | 1269 | 2179 | ABC-B11 - UBE2-B | 1623 | 1304 | 1995 |
| ABC-C3 - UBE2-A | 1726 | 1906 | 1390 | ABC-C3 - UBE2-B | 1999 | 832 | 2050 |
| ABC-C5 - UBE2-A | 880 | 2122 | 2297 | ABC-C5 - UBE2-B | 612 | 2276 | 1681 |
| ABC-C13 - UBE2-A | 412 | 234 | 670 | ABC-C13 - UBE2-B | 467 | 1863 | 2496 |
| ABC-D1 - UBE2-A | 2507 | 237 | 1319 | ABC-D1 - UBE2-B | 2322 | 1917 | 2426 |
| ABC-G1 - UBE2-A | 907 | 2291 | 1573 | ABC-G1 - UBE2-B | 1194 | 1592 | 1239 |
| ABC-G2 - UBE2-A | 2048 | 1829 | 1376 | ABC-G2 - UBE2-B | 1833 | 2445 | 2506 |
| Ranking of UNE2-F w.r.t ABC family | Ranking of UBE2-H w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-F | 2485 | 406 | 66 | ABC-A5 - UBE2-H | 508 | 2339 | 1110 |
| ABC-B11 - UBE2-F | 2003 | 1203 | 2422 | ABC-B11 - UBE2-H | 1950 | 1770 | 2461 |
| ABC-C3 - UBE2-F | 2132 | 2163 | 861 | ABC-C3 - UBE2-H | 2439 | 1972 | 2305 |
| ABC-C5 - UBE2-F | 406 | 1651 | 1838 | ABC-C5 - UBE2-H | 398 | 2473 | 2355 |
| ABC-C13 - UBE2-F | 821 | 959 | 1196 | ABC-C13 - UBE2-H | 2004 | 2317 | 1847 |
| ABC-D1 - UBE2-F | 2421 | 686 | 2176 | ABC-D1 - UBE2-H | 164 | 1641 | 648 |
| ABC-G1 - UBE2-F | 115 | 2202 | 1953 | ABC-G1 - UBE2-H | 201 | 1921 | 2288 |
| ABC-G2 - UBE2-F | 983 | 883 | 1012 | ABC-G2 - UBE2-H | 2063 | 1631 | 1354 |
| Ranking of UBE2-J1 w.r.t ABC family | Ranking of UBE2-Z w.r.t ABC family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - UBE2-J1 | 1740 | 1467 | 1244 | ABC-A5 - UBE2-Z | 2336 | 1710 | 35 |
| ABC-B11 - UBE2-J1 | 1806 | 991 | 1935 | ABC-B11 - UBE2-Z | 521 | 645 | 2168 |
| ABC-C3 - UBE2-J1 | 2073 | 2291 | 631 | ABC-C3 - UBE2-Z | 1978 | 1823 | 1859 |
| ABC-C5 - UBE2-J1 | 126 | 525 | 1409 | ABC-C5 - UBE2-Z | 1237 | 148 | 1928 |
| ABC-C13 - UBE2-J1 | 2329 | 2153 | 1951 | ABC-C13 - UBE2-Z | 1185 | 137 | 2475 |
| ABC-D1 - UBE2-J1 | 2263 | 1886 | 2249 | ABC-D1 - UBE2-Z | 2292 | 21 | 2381 |
| ABC-G1 - UBE2-J1 | 1262 | 2418 | 2277 | ABC-G1 - UBE2-Z | 426 | 2515 | 1858 |
| ABC-G2 - UBE2-J1 | 1558 | 2408 | 1304 | ABC-G2 - UBE2-Z | 2270 | 2080 | 2448 |
| Unexplored combinatorial hypotheses | |
| ABC w.r.t UBE2 | |
| ABC-C3 | UBE2-A |
| ABC-C5 | UBE2-B |
| ABC-A5/D1/G2 | UBE2-F |
| ABC-A5/C13 | UBE2-H |
| ABC-C13 | UBE2-J1 |
| ABC-C5 | UBE2-Z |
| UBE2 w.r.t ABC | |
| UBEA-2 | ABC-C5/G2 |
| UBE2-B | ABC-A5/C3/C13/D1/G2 |
| UBE2-F | ABC-B11/C3/D1/G1 |
| UBE2-H | ABC-B11/C3/C5/C13/G1 |
| UBE2-J1 | ABC-B11/C3/C13/D1/G1/G2 |
| UBE2-Z | ABC-C3/D1/G1/G2 |
| Ranking ABC family w.r.t ABC family | |||||||
| Ranking of ABC family w.r.t ABC-A5 | Ranking of ABC family w.r.t ABC-B11 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-B11 - ABC-A5 | 733 | 471 | 26 | ABC-A5 - ABC-B11 | 1148 | 1443 | 1782 |
| ABC-C3 - ABC-A5 | 111 | 493 | 2264 | ABC-C3 - ABC-B11 | 845 | 527 | 1257 |
| ABC-C5 - ABC-A5 | 1717 | 519 | 1921 | ABC-C5 - ABC-B11 | 2226 | 1644 | 2241 |
| ABC-C13 - ABC-A5 | 1243 | 1943 | 2151 | ABC-C13 - ABC-B11 | 1971 | 609 | 2150 |
| ABC-D1 - ABC-A5 | 1262 | 2387 | 1573 | ABC-D1 - ABC-B11 | 891 | 217 | 854 |
| ABC-G1 - ABC-A5 | 657 | 991 | 533 | ABC-G1 - ABC-B11 | 1957 | 1920 | 669 |
| ABC-G2 - ABC-A5 | 587 | 397 | 104 | ABC-G2 - ABC-B11 | 685 | 1978 | 226 |
| Ranking of ABC family w.r.t ABC-C3 | Ranking of ABC family w.r.t ABC-C5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - ABC-C3 | 163 | 861 | 1672 | ABC-A5 - ABC-C5 | 2086 | 411 | 1243 |
| ABC-B11 - ABC-C3 | 410 | 613 | 1501 | ABC-B11 - ABC-C5 | 2398 | 272 | 464 |
| ABC-C5 - ABC-C3 | 1591 | 2435 | 927 | ABC-C3 - ABC-C5 | 2084 | 2274 | 1758 |
| ABC-C13 - ABC-C3 | 405 | 880 | 1282 | ABC-C13 - ABC-C5 | 226 | 2476 | 2446 |
| ABC-D1 - ABC-C3 | 18 | 1145 | 2187 | ABC-D1 - ABC-C5 | 2010 | 891 | 1257 |
| ABC-G1 - ABC-C3 | 1858 | 173 | 842 | ABC-G1 - ABC-C5 | 2402 | 894 | 741 |
| ABC-G2 - ABC-C3 | 1462 | 275 | 1373 | ABC-G2 - ABC-C5 | 2463 | 736 | 661 |
| Ranking of ABC family w.r.t ABC-C13 | Ranking of ABC family w.r.t ABC-D1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - ABC-C13 | 2251 | 1219 | 1614 | ABC-A5 - ABC-D1 | 163 | 1068 | 291 |
| ABC-B11 - ABC-C13 | 1106 | 56 | 1171 | ABC-B11 - ABC-D1 | 1273 | 130 | 1655 |
| ABC-C3 - ABC-C13 | 2279 | 1431 | 365 | ABC-C3 - ABC-D1 | 568 | 251 | 149 |
| ABC-C5 - ABC-C13 | 1537 | 2178 | 690 | ABC-C5 - ABC-D1 | 2423 | 538 | 2388 |
| ABC-D1 - ABC-C13 | 2370 | 171 | 362 | ABC-C13 - ABC-D1 | 2383 | 2029 | 425 |
| ABC-G1 - ABC-C13 | 833 | 1544 | 1343 | ABC-G1 - ABC-D1 | 1462 | 1175 | 827 |
| ABC-G2 - ABC-C13 | 329 | 1323 | 1755 | ABC-G2 - ABC-D1 | 467 | 670 | 2491 |
| Ranking of ABC family w.r.t ABC-G1 | Ranking of ABC family w.r.t ABC-G2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABC-A5 - ABC-G1 | 2488 | 1776 | 1078 | ABC-A5 - ABC-G2 | 1011 | 1640 | 1705 |
| ABC-B11 - ABC-G1 | 2312 | 253 | 52 | ABC-B11 - ABC-G2 | 988 | 481 | 1849 |
| ABC-C3 - ABC-G1 | 273 | 1415 | 1139 | ABC-C3 - ABC-G2 | 1102 | 1082 | 1563 |
| ABC-C5 - ABC-G1 | 220 | 1988 | 437 | ABC-C5 - ABC-G2 | 2284 | 1904 | 1829 |
| ABC-C13 - ABC-G1 | 2389 | 427 | 1125 | ABC-C13 - ABC-G2 | 929 | 1238 | 222 |
| ABC-D1 - ABC-G1 | 1836 | 485 | 597 | ABC-D1 - ABC-G2 | 814 | 995 | 1152 |
| ABC-G2 - ABC-G1 | 2506 | 692 | 1143 | ABC-G1 - ABC-G2 | 596 | 460 | 848 |
| Unexplored combinatorial hypotheses | |
| ABC intra family | |
| ABC-C13 | ABC-A5 |
| ABC-C5/C13/G1 | ABC-B11 |
| ABC-C3/C13 | ABC-C5 |
| ABC-C5/C13 | ABC-D1 |
| ABC-A5 | ABC-G1 |
| ABC-C5 | ABC-G2 |
| Unexplored combinatorial hypotheses | |
| ABC w.r.t IL | |
| IL-1B/1RAP/10RB | ABCA5 |
| IL-10RB | ABCB11 |
| IL-1A/17REL | ABCC3 |
| IL-1A/1RAP/15/17C | ABCC5 |
| IL-1RAP/15RA | ABCC13 |
| IL-8/10RB | ABCD1 |
| IL-10RB | ABCG2 |
| IL w.r.t ABC | |
| IL-17REL | ABCA5 |
| IL-2RG/6ST/15/15RA | ABCB11 |
| IL-8/15RA | ABCC3 |
| IL-15RA/17REL | ABCC5 |
| IL-15RA/17REL | ABCC13 |
| IL-1A/1RAP/8/15RA | ABCD1 |
| IL-1RAP | ABCG1 |
| IL-1RAP/15RA | ABCG2 |
| Ranking BCL family VS ABC family | |||||||
| Ranking of BCL2L1 w.r.t ABC family | Ranking of ABC family w.r.t BCL2L1 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL2L1 | 18 | 560 | 1715 | ABCA5 - BCL2L1 | 1522 | 220 | 1818 |
| ABCB11 - BCL2L1 | 1124 | 2418 | 552 | ABCB11 - BCL2L1 | 2002 | 234 | 10 |
| ABCC3 - BCL2L1 | 564 | 394 | 64 | ABCC3 - BCL2L1 | 2085 | 2309 | 929 |
| ABCC5 - BCL2L1 | 2239 | 1845 | 823 | ABCC5 - BCL2L1 | 599 | 847 | 1282 |
| ABCC13 - BCL2L1 | 805 | 1590 | 2407 | ABCC13 - BCL2L1 | 744 | 616 | 614 |
| ABCD1 - BCL2L1 | 356 | 202 | 930 | ABCD1 - BCL2L1 | 839 | 352 | 195 |
| ABCG1 - BCL2L1 | 793 | 2005 | 885 | ABCG1 - BCL2L1 | 1249 | 265 | 1165 |
| ABCG2 - BCL2L1 | 199 | 99 | 906 | ABCG2 - BCL2L1 | 401 | 620 | 277 |
| Ranking of BCL2L2 w.r.t ABC family | Ranking of ABC family w.r.t BCL2L2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL2L2 | 1476 | 324 | 1792 | ABCA5 - BCL2L2 | 174 | 482 | 501 |
| ABCB11 - BCL2L2 | 2097 | 1311 | 2311 | ABCB11 - BCL2L2 | 148 | 380 | 1204 |
| ABCC3 - BCL2L2 | 1091 | 1569 | 259 | ABCC3 - BCL2L2 | 890 | 949 | 1398 |
| ABCC5 - BCL2L2 | 2195 | 2359 | 2322 | ABCC5 - BCL2L2 | 765 | 1875 | 736 |
| ABCC13 - BCL2L2 | 2438 | 2494 | 898 | ABCC13 - BCL2L2 | 2271 | 1436 | 1665 |
| ABCD1 - BCL2L2 | 2477 | 831 | 2156 | ABCD1 - BCL2L2 | 1432 | 1291 | 64 |
| ABCG1 - BCL2L2 | 352 | 1653 | 2234 | ABCG1 - BCL2L2 | 406 | 1206 | 966 |
| ABCG2 - BCL2L2 | 1515 | 2409 | 1496 | ABCG2 - BCL2L2 | 404 | 314 | 55 |
| Ranking of BCL2L13 w.r.t ABC family | Ranking of ABC family w.r.t BCL2L13 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL2L13 | 1398 | 202 | 2292 | ABCA5 - BCL2L13 | 655 | 951 | 1380 |
| ABCB11 - BCL2L13 | 2505 | 261 | 1855 | ABCB11 - BCL2L13 | 1579 | 53 | 224 |
| ABCC3 - BCL2L13 | 1642 | 1769 | 1334 | ABCC3 - BCL2L13 | 265 | 588 | 459 |
| ABCC5 - BCL2L13 | 1427 | 1835 | 2178 | ABCC5 - BCL2L13 | 1975 | 2421 | 927 |
| ABCC13 - BCL2L13 | 2484 | 2184 | 2410 | ABCC13 - BCL2L13 | 1894 | 2335 | 2475 |
| ABCD1 - BCL2L13 | 2472 | 1579 | 2201 | ABCD1 - BCL2L13 | 912 | 511 | 1041 |
| ABCG1 - BCL2L13 | 3 | 2276 | 2095 | ABCG1 - BCL2L13 | 957 | 649 | 488 |
| ABCG2 - BCL2L13 | 2172 | 1723 | 1502 | ABCG2 - BCL2L13 | 2142 | 392 | 1206 |
| Ranking of BCL3 w.r.t ABC family | Ranking of ABC family w.r.t BCL3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| ABCA5 - BCL3 | 940 | 45 | 777 | ABCA5 - BCL3 | 960 | 1354 | 2477 |
| ABCB11 - BCL3 | 260 | 1002 | 483 | ABCB11 - BCL3 | 731 | 1483 | 2028 |
| ABCC3 - BCL3 | 2101 | 214 | 304 | ABCC3 - BCL3 | 1782 | 2186 | 1251 |
| ABCC5 - BCL3 | 1155 | 775 | 1176 | ABCC5 - BCL3 | 2192 | 957 | 280 |
| ABCC13 - BCL3 | 270 | 1116 | 1619 | ABCC13 - BCL3 | 1725 | 1407 | 1747 |
| ABCD1 - BCL3 | 759 | 2194 | 2106 | ABCD1 - BCL3 | 836 | 811 | 1359 |
| ABCG1 - BCL3 | 2014 | 1559 | 2253 | ABCG1 - BCL3 | 550 | 247 | 247 |
| ABCG2 - BCL3 | 480 | 465 | 1949 | ABCG2 - BCL3 | 792 | 798 | 1418 |
| Unexplored combinatorial hypotheses | |
| BCL w.r.t ABC | |
| ABC-C5 | BCL2L1 |
| ABC-B11/C5/C13/D1 | BCL2L2 |
| ABC-B11/C5/C13/D1/G1 | BCL2L13 |
| ABC-D1/G1 | BCL3 |
| ABC-B11 | BCL6 |
| ABC-B11 | BCL10 |
| ABC w.r.t BCL | |
| ABC-C3 | BCL2L1 |
| ABC-C5/C13 | BCL2L13 |
| ABC-C3 | BCL3 |
| ABC-C5/C13 | BCL6 |
| ABC-C5/C13/D1 | BCL9L |
| ABC-A5/C5/C13/D1 | BCL10 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t NFkB-2/I | |
| IL15RA | NFkB2 |
| IL17C | NFkB2 |
| IL1RN | NFkBIA |
| IL6ST | NFkBIA |
| IL15RA | NFkBIA |
| IL1RAP | NFkBIE |
| IL6ST | NFkBIE |
| IL8 | NFkBIE |
| IL17REL | NFkBIE |
| IL1A | NFkBIZ |
| IL6ST | NFkBIZ |
| IL15 | NFkBIZ |
| NFkB-2/I w.r.t IL | |
| IL10RB | NFkB2 |
| IL10RB | NFKBIZ |
| IL17REL | NFkBIZ |
| Ranking Interleukin family vs KCN family | |||||||
| Ranking of IL family w.r.t KCND3 | Ranking of KCND3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCND3 | 1995 | 2255 | 718 | IL1A - KCND3 | 707 | 118 | 11 |
| IL1B - KCND3 | 2083 | 1897 | 691 | IL1B - KCND3 | 1064 | 411 | 133 |
| IL1RAP - KCND3 | 212 | 1086 | 1690 | IL1RAP - KCND3 | 2495 | 2390 | 114 |
| IL1RN - KCND3 | 1091 | 1875 | 1551 | IL1RN - KCND3 | 459 | 743 | 300 |
| IL2RG - KCND3 | 2027 | 1557 | 403 | IL2RG - KCND3 | 588 | 248 | 58 |
| IL6ST - KCND3 | 28 | 24 | 2501 | IL6ST - KCND3 | 1 | 1127 | 2482 |
| IL8 - KCND3 | 46 | 1098 | 1426 | IL8 - KCND3 | 1134 | 1639 | 890 |
| IL10RB - KCND3 | 1573 | 2172 | 1302 | IL10RB - KCND3 | 2048 | 2306 | 2197 |
| IL15 - KCND3 | 1905 | 1606 | 716 | IL15 - KCND3 | 296 | 68 | 240 |
| IL15RA - KCND3 | 2074 | 483 | 2495 | IL15RA - KCND3 | 2511 | 2517 | 1606 |
| IL17C - KCND3 | 1881 | 2139 | 368 | IL17C - KCND3 | 588 | 1383 | 277 |
| IL17REL - KCND3 | 1715 | 2242 | 359 | IL17REL - KCND3 | 1361 | 748 | 1905 |
| Ranking of IL family w.r.t KCNH2 | Ranking of KCNH2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCNH2 | 2103 | 1832 | 356 | IL1A - KCNH2 | 1897 | 2152 | 2179 |
| IL1B - KCNH2 | 2447 | 2068 | 930 | IL1B - KCNH2 | 1599 | 2025 | 653 |
| IL1RAP - KCNH2 | 423 | 1275 | 2487 | IL1RAP - KCNH2 | 2451 | 1805 | 2002 |
| IL1RN - KCNH2 | 1600 | 828 | 779 | IL1RN - KCNH2 | 233 | 2304 | 305 |
| IL2RG - KCNH2 | 1501 | 903 | 929 | IL2RG - KCNH2 | 823 | 701 | 1820 |
| IL6ST - KCNH2 | 1016 | 1565 | 1929 | IL6ST - KCNH2 | 435 | 1665 | 2142 |
| IL8 - KCNH2 | 863 | 258 | 1395 | IL8 - KCNH2 | 1103 | 1062 | 2255 |
| IL10RB - KCNH2 | 1238 | 1335 | 1441 | IL10RB - KCNH2 | 648 | 1445 | 1684 |
| IL15 - KCNH2 | 2295 | 1419 | 1038 | IL15 - KCNH2 | 389 | 1247 | 1033 |
| IL15RA - KCNH2 | 1738 | 2263 | 296 | IL15RA - KCNH2 | 515 | 1572 | 2265 |
| IL17C - KCNH2 | 2084 | 1399 | 49 | IL17C - KCNH2 | 1388 | 1021 | 1079 |
| IL17REL - KCNH2 | 90 | 1956 | 1491 | IL17REL - KCNH2 | 727 | 2338 | 524 |
| Ranking of IL family w.r.t KCNH8 | Ranking of KCNH8 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCNH8 | 2268 | 2507 | 1877 | IL1A - KCNH8 | 29 | 1939 | 1438 |
| IL1B - KCNH8 | 2223 | 2013 | 2204 | IL1B - KCNH8 | 2060 | 472 | 2177 |
| IL1RAP - KCNH8 | 1238 | 479 | 1717 | IL1RAP - KCNH8 | 1950 | 651 | 150 |
| IL1RN - KCNH8 | 1653 | 819 | 2040 | IL1RN - KCNH8 | 1094 | 329 | 988 |
| IL2RG - KCNH8 | 57 | 1530 | 651 | IL2RG - KCNH8 | 1853 | 1224 | 390 |
| IL6ST - KCNH8 | 2067 | 979 | 1640 | IL6ST - KCNH8 | 607 | 368 | 800 |
| IL8 - KCNH8 | 1558 | 439 | 1250 | IL8 - KCNH8 | 2484 | 269 | 1048 |
| IL10RB - KCNH8 | 937 | 448 | 416 | IL10RB - KCNH8 | 2381 | 2008 | 726 |
| IL15 - KCNH8 | 1575 | 1789 | 580 | IL15 - KCNH8 | 1365 | 1649 | 2187 |
| IL15RA - KCNH8 | 2082 | 1524 | 1550 | IL15RA - KCNH8 | 1667 | 638 | 1648 |
| IL17C - KCNH8 | 1847 | 1700 | 2354 | IL17C - KCNH8 | 1232 | 1825 | 1519 |
| IL17REL - KCNH8 | 1542 | 2 | 1803 | IL17REL - KCNH8 | 1120 | 681 | 2060 |
| Ranking Interleukin family vs KCN family | |||||||
| Ranking of IL family w.r.t KCNK1 | Ranking of KCNK1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCNK1 | 2290 | 2066 | 1071 | IL1A - KCNK1 | 1644 | 1818 | 2362 |
| IL1B - KCNK1 | 1941 | 2452 | 1905 | IL1B - KCNK1 | 813 | 966 | 1554 |
| IL1RAP - KCNK1 | 171 | 595 | 2490 | IL1RAP - KCNK1 | 1103 | 1318 | 1803 |
| IL1RN - KCNK1 | 2468 | 1897 | 391 | IL1RN - KCNK1 | 2130 | 73 | 1326 |
| IL2RG - KCNK1 | 2384 | 1028 | 755 | IL2RG - KCNK1 | 650 | 2324 | 1413 |
| IL6ST - KCNK1 | 862 | 131 | 807 | IL6ST - KCNK1 | 2226 | 688 | 2283 |
| IL8 - KCNK1 | 722 | 22 | 2147 | IL8 - KCNK1 | 1872 | 1978 | 1201 |
| IL10RB - KCNK1 | 1965 | 125 | 1204 | IL10RB - KCNK1 | 1087 | 1633 | 1021 |
| IL15 - KCNK1 | 2280 | 2009 | 502 | IL15 - KCNK1 | 1639 | 506 | 2369 |
| IL15RA - KCNK1 | 1567 | 1546 | 895 | IL15RA - KCNK1 | 1126 | 1499 | 784 |
| IL17C - KCNK1 | 2451 | 122 | 931 | IL17C - KCNK1 | 1331 | 2472 | 611 |
| IL17REL - KCNK1 | 1515 | 659 | 2391 | IL17REL - KCNK1 | 979 | 2329 | 329 |
| Ranking of IL family w.r.t KCNK5 | Ranking of KCNK5 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCNK5 | 609 | 608 | 796 | IL1A - KCNK5 | 1840 | 1512 | 576 |
| IL1B - KCNK5 | 1297 | 110 | 543 | IL1B - KCNK5 | 510 | 1379 | 347 |
| IL1RAP - KCNK5 | 1137 | 2237 | 1314 | IL1RAP - KCNK5 | 730 | 911 | 1003 |
| IL1RN - KCNK5 | 1583 | 1930 | 2136 | IL1RN - KCNK5 | 815 | 405 | 1431 |
| IL2RG - KCNK5 | 223 | 601 | 14 | IL2RG - KCNK5 | 1841 | 782 | 806 |
| IL6ST - KCNK5 | 1038 | 867 | 295 | IL6ST - KCNK5 | 455 | 848 | 1275 |
| IL8 - KCNK5 | 819 | 1737 | 105 | IL8 - KCNK5 | 272 | 479 | 1215 |
| IL10RB - KCNK5 | 1879 | 2298 | 1903 | IL10RB - KCNK5 | 457 | 1769 | 2206 |
| IL15 - KCNK5 | 981 | 1630 | 1669 | IL15 - KCNK5 | 1060 | 244 | 580 |
| IL15RA - KCNK5 | 791 | 124 | 555 | IL15RA - KCNK5 | 361 | 332 | 1662 |
| IL17C - KCNK5 | 2345 | 449 | 919 | IL17C - KCNK5 | 388 | 713 | 509 |
| IL17REL - KCNK5 | 18 | 2118 | 1873 | IL17REL - KCNK5 | 1377 | 2108 | 1634 |
| Ranking of IL family w.r.t KCNK6 | Ranking of KCNK6 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - KCNK6 | 140 | 180 | 615 | IL1A - KCNK6 | 1683 | 944 | 860 |
| IL1B - KCNK6 | 525 | 57 | 369 | IL1B - KCNK6 | 620 | 1047 | 903 |
| IL1RAP - KCNK6 | 1878 | 830 | 902 | IL1RAP - KCNK6 | 2386 | 1248 | 2053 |
| IL1RN - KCNK6 | 1834 | 1564 | 90 | IL1RN - KCNK6 | 2047 | 970 | 1311 |
| IL2RG - KCNK6 | 2181 | 974 | 7 | IL2RG - KCNK6 | 691 | 2454 | 1443 |
| IL6ST - KCNK6 | 728 | 1895 | 1270 | IL6ST - KCNK6 | 440 | 125 | 1682 |
| IL8 - KCNK6 | 2168 | 2442 | 869 | IL8 - KCNK6 | 774 | 98 | 654 |
| IL10RB - KCNK6 | 1821 | 560 | 85 | IL10RB - KCNK6 | 802 | 1903 | 2156 |
| IL15 - KCNK6 | 1589 | 304 | 2447 | IL15 - KCNK6 | 1944 | 130 | 2047 |
| IL15RA - KCNK6 | 87 | 436 | 2447 | IL15RA - KCNK6 | 1835 | 1531 | 1529 |
| IL17C - KCNK6 | 103 | 200 | 538 | IL17C - KCNK6 | 1056 | 1350 | 1408 |
| IL17REL - KCNK6 | 2066 | 2159 | 1735 | IL17REL - KCNK6 | 1044 | 1263 | 947 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t KCN | |
| IL-1A/1B/15RA/17C | KCND3 |
| IL-1A/1B | KCNH2 |
| IL-1A/1B/17C | KCNH8 |
| IL-1A/1B/1RN/15 | KCNK1 |
| IL-1RN/10RB/17REL | KCNK5 |
| IL-8/17REL | KCNK6 |
| KCN w.r.t IL family | |
| IL-1A/1B/15RA/17C | KCND3 |
| IL-1A/1RAP | KCNH2 |
| IL-1B/10RB | KCNH8 |
| IL-1A/6ST/8 | KCNK1 |
| IL-10RB | KCNK5 |
| IL-1RAP/10RB/15 | KCNK6 |
| Ranking Interleukin family vs TRAF family | |||||||
| Ranking of IL family w.r.t TRAF3IP2 | Ranking of TRAF3IP2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TRAF3IP2 | 2142 | 100 | 666 | IL1A - TRAF3IP2 | 1518 | 2265 | 1107 |
| IL1B - TRAF3IP2 | 1155 | 110 | 1193 | IL1B - TRAF3IP2 | 1953 | 1294 | 2359 |
| IL1RAP - TRAF3IP2 | 704 | 2482 | 2385 | IL1RAP - TRAF3IP2 | 913 | 2034 | 38 |
| IL1RN - TRAF3IP2 | 272 | 497 | 133 | IL1RN - TRAF3IP2 | 1044 | 538 | 1173 |
| IL2RG - TRAF3IP2 | 1948 | 1043 | 942 | IL2RG - TRAF3IP2 | 1767 | 2385 | 2059 |
| IL6ST - TRAF3IP2 | 49 | 1244 | 1098 | IL6ST - TRAF3IP2 | 257 | 1991 | 1871 |
| IL8 - TRAF3IP2 | 1165 | 598 | 344 | IL8 - TRAF3IP2 | 796 | 2192 | 2289 |
| IL10RB - TRAF3IP2 | 1252 | 1426 | 552 | IL10RB - TRAF3IP2 | 840 | 237 | 2096 |
| IL15 - TRAF3IP2 | 1550 | 433 | 163 | IL15 - TRAF3IP2 | 1428 | 1183 | 2219 |
| IL15RA - TRAF3IP2 | 2024 | 2162 | 1800 | IL15RA - TRAF3IP2 | 906 | 1995 | 1717 |
| IL17C - TRAF3IP2 | 2253 | 61 | 98 | IL17C - TRAF3IP2 | 1290 | 1587 | 1839 |
| IL17REL - TRAF3IP2 | 18 | 2515 | 2057 | IL17REL - TRAF3IP2 | 1836 | 2042 | 1568 |
| Ranking of IL family w.r.t TRAF4 | Ranking of TRAF4 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TRAF4 | 26 | 2316 | 707 | IL1A - TRAF4 | 1806 | 439 | 1465 |
| IL1B - TRAF4 | 582 | 2136 | 175 | IL1B - TRAF4 | 1026 | 746 | 378 |
| IL1RAP - TRAF4 | 1180 | 1714 | 961 | IL1RAP - TRAF4 | 909 | 2225 | 1546 |
| IL1RN - TRAF4 | 494 | 2347 | 590 | IL1RN - TRAF4 | 625 | 1031 | 1939 |
| IL2RG - TRAF4 | 1092 | 1860 | 275 | IL2RG - TRAF4 | 1130 | 339 | 826 |
| IL6ST - TRAF4 | 2333 | 344 | 1914 | IL6ST - TRAF4 | 676 | 1966 | 1556 |
| IL8 - TRAF4 | 749 | 604 | 950 | IL8 - TRAF4 | 406 | 450 | 1531 |
| IL10RB - TRAF4 | 580 | 2512 | 424 | IL10RB - TRAF4 | 2407 | 1781 | 1136 |
| IL15 - TRAF4 | 1131 | 2078 | 227 | IL15 - TRAF4 | 905 | 2408 | 1759 |
| IL15RA - TRAF4 | 551 | 1628 | 2237 | IL15RA - TRAF4 | 1197 | 2125 | 2073 |
| IL17C - TRAF4 | 236 | 2464 | 19 | IL17C - TRAF4 | 1538 | 914 | 1515 |
| IL17REL - TRAF4 | 2422 | 381 | 2487 | IL17REL - TRAF4 | 575 | 1394 | 320 |
| Ranking of IL family w.r.t TRAF6 | Ranking of TRAF6 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TRAF6 | 1 | 343 | 2237 | IL1A - TRAF6 | 1637 | 455 | 2334 |
| IL1B - TRAF6 | 224 | 143 | 2107 | IL1B - TRAF6 | 861 | 1386 | 1342 |
| IL1RAP - TRAF6 | 1875 | 1483 | 1433 | IL1RAP - TRAF6 | 2219 | 1984 | 1766 |
| IL1RN - TRAF6 | 107 | 706 | 988 | IL1RN - TRAF6 | 1334 | 1067 | 1301 |
| IL2RG - TRAF6 | 790 | 1706 | 1028 | IL2RG - TRAF6 | 695 | 1717 | 1986 |
| IL6ST - TRAF6 | 1508 | 928 | 930 | IL6ST - TRAF6 | 54 | 762 | 1130 |
| IL8 - TRAF6 | 2088 | 1883 | 2089 | IL8 - TRAF6 | 2457 | 2139 | 1218 |
| IL10RB - TRAF6 | 17 | 786 | 1211 | IL10RB - TRAF6 | 303 | 1825 | 1709 |
| IL15 - TRAF6 | 320 | 1692 | 2045 | IL15 - TRAF6 | 2071 | 2475 | 1500 |
| IL15RA - TRAF6 | 1560 | 303 | 2392 | IL15RA - TRAF6 | 1688 | 1189 | 1344 |
| IL17C - TRAF6 | 42 | 227 | 1457 | IL17C - TRAF6 | 2469 | 2309 | 1503 |
| IL17REL - TRAF6 | 2454 | 2517 | 412 | IL17REL - TRAF6 | 124 | 2067 | 823 |
| Ranking of IL family w.r.t TRAFD1 | Ranking of TRAFD1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - TRAFD1 | 2408 | 1040 | 1579 | IL1A - TRAFD1 | 2121 | 699 | 1587 |
| IL1B - TRAFD1 | 1478 | 2046 | 1321 | IL1B - TRAFD1 | 756 | 2435 | 571 |
| IL1RAP - TRAFD1 | 491 | 1639 | 447 | IL1RAP - TRAFD1 | 528 | 857 | 2043 |
| IL1RN - TRAFD1 | 895 | 1149 | 266 | IL1RN - TRAFD1 | 1033 | 848 | 1374 |
| IL2RG - TRAFD1 | 1025 | 1948 | 43 | IL2RG - TRAFD1 | 1243 | 492 | 1579 |
| IL6ST - TRAFD1 | 1835 | 1824 | 809 | IL6ST - TRAFD1 | 1064 | 868 | 699 |
| IL8 - TRAFD1 | 1318 | 896 | 663 | IL8 - TRAFD1 | 650 | 671 | 1088 |
| IL10RB - TRAFD1 | 329 | 2371 | 355 | IL10RB - TRAFD1 | 2403 | 556 | 800 |
| IL15 - TRAFD1 | 1165 | 1934 | 769 | IL15 - TRAFD1 | 339 | 623 | 634 |
| IL15RA - TRAFD1 | 351 | 260 | 2385 | IL15RA - TRAFD1 | 265 | 1369 | 386 |
| IL17C - TRAFD1 | 1191 | 1389 | 1486 | IL17C - TRAFD1 | 756 | 1068 | 1390 |
| IL17REL - TRAFD1 | 704 | 2222 | 788 | IL17REL - TRAFD1 | 370 | 640 | 137 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t TRAF | |
| IL-1RAP/15RA/17REL | TRAF3IP2 |
| IL-6ST/17REL | TRAF4 |
| IL-8/17REL | TRAF6 |
| IL-6ST | TRAFD1 |
| TRAF w.r.t IL | |
| IL-1B/2RG/6ST/8/17REL | TRAF3IP2 |
| IL-10RB/15/15RA | TRAF4 |
| IL-1RAP/8/15/17C | TRAF6 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t TNF | |
| IL-1RAP/6ST/15RA | TNF |
| IL-1B/2RG/15RA/17C | TNFAIP1 |
| IL-1RN/10RB | TNFAIP2 |
| IL-6ST/8/17REL | TNFAIP3 |
| IL-1RAP | TNFRSF1A |
| IL-1RAP/15RA/17REL | TNFRSF10A |
| IL-15RA | TNFRSF10B |
| IL-15RA | TNFRSF10D |
| IL-8/15RA/17REL | TNFRSF12A |
| IL-15RA | TNFRSF14 |
| IL-1B/1RAP/2RG | TNFRSF21 |
| IL-1B/15RA/17C | TNFSF10 |
| IL-17C | TNFSF15 |
| TNF w.r.t IL | |
| IL-6ST/10RB | TNF |
| IL-8/15RA | TNFAIP1 |
| IL-1B | TNFRSF1A |
| IL-1A/1B/1RN/2RG/6ST/15/15RA/17C | TNFRSF10A |
| IL-1RN | TNFRSF10B |
| IL-1A/1B/2RG/6ST/10RB/15/17C/17REL | TNFRSF10D |
| IL-6ST/17C | TNFRSF12A |
| IL-1A/1RN/2RG/6ST/8/15RA/17C/17REL | TNFRSF14 |
| IL-17REL | TNFRSF14 |
| IL10RB | TNFSF10 |
| IL15 | TNFSF15 |
| Ranking IL family vs BCL family | |||||||
| Ranking of IL family w.r.t BCL2L1 | Ranking of BCL2L1 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL2L1 | 2482 | 859 | 1834 | IL1A - BCL2L1 | 780 | 1156 | 1712 |
| IL1B - BCL2L1 | 2252 | 1920 | 1482 | IL1B - BCL2L1 | 1838 | 954 | 2132 |
| IL1RAP - BCL2L1 | 1128 | 815 | 1935 | IL1RAP - BCL2L1 | 870 | 1777 | 1262 |
| IL1RN - BCL2L1 | 648 | 2504 | 650 | IL1RN - BCL2L1 | 973 | 385 | 1297 |
| IL2RG - BCL2L1 | 1542 | 1439 | 700 | IL2RG - BCL2L1 | 2048 | 486 | 1949 |
| IL6ST - BCL2L1 | 663 | 553 | 1432 | IL6ST - BCL2L1 | 284 | 674 | 468 |
| IL8 - BCL2L1 | 260 | 202 | 2070 | IL8 - BCL2L1 | 1430 | 1343 | 1417 |
| IL10RB - BCL2L1 | 1867 | 347 | 17 | IL10RB - BCL2L1 | 1659 | 1965 | 2024 |
| IL15 - BCL2L1 | 1558 | 775 | 381 | IL15 - BCL2L1 | 690 | 542 | 1277 |
| IL15RA - BCL2L1 | 2136 | 1177 | 1533 | IL15RA - BCL2L1 | 581 | 1107 | 972 |
| IL17C - BCL2L1 | 2481 | 2410 | 2512 | IL17C - BCL2L1 | 695 | 1739 | 1775 |
| IL17REL - BCL2L1 | 815 | 657 | 374 | IL17REL - BCL2L1 | 981 | 1225 | 509 |
| Ranking of IL family w.r.t BCL2L2 | Ranking of BCL2L2 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL2L2 | 138 | 361 | 86 | IL1A - BCL2L2 | 2407 | 2362 | 2464 |
| IL1B - BCL2L2 | 165 | 389 | 108 | IL1B - BCL2L2 | 1807 | 2462 | 2344 |
| IL1RAP - BCL2L2 | 623 | 1523 | 861 | IL1RAP - BCL2L2 | 77 | 1897 | 1711 |
| IL1RN - BCL2L2 | 2324 | 530 | 984 | IL1RN - BCL2L2 | 2298 | 1620 | 2092 |
| IL2RG - BCL2L2 | 2137 | 285 | 347 | IL2RG - BCL2L2 | 2429 | 850 | 1744 |
| IL6ST - BCL2L2 | 2239 | 1927 | 2085 | IL6ST - BCL2L2 | 477 | 2046 | 1859 |
| IL8 - BCL2L2 | 894 | 1418 | 1346 | IL8 - BCL2L2 | 1803 | 1072 | 2024 |
| IL10RB - BCL2L2 | 2243 | 738 | 1020 | IL10RB - BCL2L2 | 1041 | 145 | 843 |
| IL15 - BCL2L2 | 110 | 650 | 1347 | IL15 - BCL2L2 | 2474 | 2142 | 2416 |
| IL15RA - BCL2L2 | 258 | 1715 | 361 | IL15RA - BCL2L2 | 1377 | 1211 | 2298 |
| IL17C - BCL2L2 | 554 | 12 | 147 | IL17C - BCL2L2 | 1168 | 2512 | 2447 |
| IL17REL - BCL2L2 | 2454 | 2510 | 2482 | IL17REL - BCL2L2 | 539 | 1875 | 1442 |
| Ranking of IL family w.r.t BCL2L13 | Ranking of BCL2L13 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL2L13 | 1572 | 458 | 174 | IL1A - BCL2L13 | 1456 | 811 | 2403 |
| IL1B - BCL2L13 | 927 | 227 | 424 | IL1B - BCL2L13 | 1286 | 1446 | 2348 |
| IL1RAP - BCL2L13 | 278 | 718 | 1941 | IL1RAP - BCL2L13 | 823 | 2450 | 2510 |
| IL1RN - BCL2L13 | 608 | 1277 | 881 | IL1RN - BCL2L13 | 2503 | 623 | 2378 |
| IL2RG - BCL2L13 | 507 | 1182 | 5 | IL2RG - BCL2L13 | 2483 | 1648 | 2248 |
| IL6ST - BCL2L13 | 1778 | 1403 | 246 | IL6ST - BCL2L13 | 1899 | 2473 | 2046 |
| IL8 - BCL2L13 | 178 | 468 | 1606 | IL8 - BCL2L13 | 2099 | 910 | 2294 |
| IL10RB - BCL2L13 | 991 | 1211 | 804 | IL10RB - BCL2L13 | 2120 | 1895 | 194 |
| IL15 - BCL2L13 | 1868 | 432 | 15 | IL15 - BCL2L13 | 2515 | 2160 | 2420 |
| IL15RA - BCL2L13 | 1629 | 2134 | 685 | IL15RA - BCL2L13 | 933 | 1844 | 2318 |
| IL17C - BCL2L13 | 995 | 84 | 20 | IL17C - BCL2L13 | 2004 | 2434 | 2500 |
| IL17REL - BCL2L13 | 2420 | 2419 | 2464 | IL17REL - BCL2L13 | 1490 | 760 | 442 |
| Ranking of IL family w.r.t BCL3 | Ranking of BCL3 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL3 | 880 | 2462 | 396 | IL1A - BCL3 | 474 | 436 | 1045 |
| IL1B - BCL3 | 975 | 1507 | 40 | IL1B - BCL3 | 799 | 303 | 926 |
| IL1RAP - BCL3 | 1425 | 821 | 1129 | IL1RAP - BCL3 | 44 | 164 | 1115 |
| IL1RN - BCL3 | 149 | 471 | 311 | IL1RN - BCL3 | 37 | 1784 | 477 |
| IL2RG - BCL3 | 454 | 365 | 505 | IL2RG - BCL3 | 524 | 2060 | 335 |
| IL6ST - BCL3 | 1928 | 755 | 2344 | IL6ST - BCL3 | 316 | 1457 | 607 |
| IL8 - BCL3 | 1052 | 743 | 2044 | IL8 - BCL3 | 2266 | 1236 | 1983 |
| IL10RB - BCL3 | 95 | 800 | 1625 | IL10RB - BCL3 | 2187 | 1600 | 2170 |
| IL15 - BCL3 | 1041 | 820 | 214 | IL15 - BCL3 | 17 | 966 | 182 |
| IL15RA - BCL3 | 2478 | 1820 | 2500 | IL15RA - BCL3 | 462 | 1476 | 1100 |
| IL17C - BCL3 | 737 | 1682 | 8 | IL17C - BCL3 | 1069 | 923 | 1926 |
| IL17REL - BCL3 | 218 | 424 | 2019 | IL17REL - BCL3 | 692 | 1897 | 1274 |
| Ranking IL family vs BCL family | |||||||
| Ranking of IL family w.r.t BCL6 | Ranking of BCL6 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL6 | 157 | 5 | 1029 | IL1A - BCL6 | 1034 | 2503 | 1669 |
| IL1B - BCL6 | 274 | 767 | 1904 | IL1B - BCL6 | 2298 | 2423 | 2294 |
| IL1RAP - BCL6 | 1021 | 2360 | 1813 | IL1RAP - BCL6 | 2403 | 1289 | 777 |
| IL1RN - BCL6 | 2015 | 366 | 506 | IL1RN - BCL6 | 1919 | 2301 | 1680 |
| IL2RG - BCL6 | 425 | 553 | 480 | IL2RG - BCL6 | 1389 | 2106 | 2478 |
| IL6ST - BCL6 | 2419 | 1589 | 1962 | IL6ST - BCL6 | 92 | 184 | 1752 |
| IL8 - BCL6 | 2363 | 2233 | 1343 | IL8 - BCL6 | 2123 | 2068 | 181 |
| IL10RB - BCL6 | 853 | 383 | 1983 | IL10RB - BCL6 | 847 | 1980 | 1186 |
| IL15 - BCL6 | 500 | 397 | 1767 | IL15 - BCL6 | 1297 | 1925 | 1014 |
| IL15RA - BCL6 | 1686 | 1432 | 2269 | IL15RA - BCL6 | 2084 | 1791 | 2203 |
| IL17C - BCL6 | 227 | 255 | 2412 | IL17C - BCL6 | 1349 | 1499 | 1321 |
| IL17REL - BCL6 | 2253 | 2396 | 63 | IL17REL - BCL6 | 38 | 1949 | 1930 |
| Ranking of IL family w.r.t BCL9L | Ranking of BCL9L w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL9L | 1932 | 1942 | 210 | IL1A - BCL9L | 1620 | 1559 | 986 |
| IL1B - BCL9L | 1966 | 88 | 79 | IL1B - BCL9L | 361 | 1449 | 2484 |
| IL1RAP - BCL9L | 218 | 957 | 881 | IL1RAP - BCL9L | 984 | 623 | 1689 |
| IL1RN - BCL9L | 1629 | 937 | 132 | IL1RN - BCL9L | 689 | 55 | 1593 |
| IL2RG - BCL9L | 415 | 104 | 92 | IL2RG - BCL9L | 2113 | 892 | 567 |
| IL6ST - BCL9L | 2249 | 1960 | 1142 | IL6ST - BCL9L | 1718 | 1210 | 737 |
| IL8 - BCL9L | 814 | 2197 | 2162 | IL8 - BCL9L | 1679 | 1920 | 933 |
| IL10RB - BCL9L | 743 | 632 | 660 | IL10RB - BCL9L | 1631 | 717 | 1236 |
| IL15 - BCL9L | 1343 | 279 | 280 | IL15 - BCL9L | 568 | 1068 | 1794 |
| IL15RA - BCL9L | 1714 | 111 | 1279 | IL15RA - BCL9L | 206 | 951 | 251 |
| IL17C - BCL9L | 2029 | 196 | 94 | IL17C - BCL9L | 1031 | 573 | 1870 |
| IL17REL - BCL9L | 128 | 2308 | 1926 | IL17REL - BCL9L | 1214 | 1341 | 839 |
| Ranking of IL family w.r.t BCL10 | Ranking of BCL10 w.r.t IL family | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| IL1A - BCL10 | 5 | 1720 | 506 | IL1A - BCL10 | 513 | 2405 | 1889 |
| IL1B - BCL10 | 201 | 2404 | 803 | IL1B - BCL10 | 2100 | 432 | 1251 |
| IL1RAP - BCL10 | 1597 | 598 | 1869 | IL1RAP - BCL10 | 1929 | 499 | 2112 |
| IL1RN - BCL10 | 107 | 724 | 126 | IL1RN - BCL10 | 1846 | 1823 | 209 |
| IL2RG - BCL10 | 232 | 665 | 650 | IL2RG - BCL10 | 1885 | 1803 | 1577 |
| IL6ST - BCL10 | 2008 | 1698 | 1816 | IL6ST - BCL10 | 451 | 71 | 337 |
| IL8 - BCL10 | 1614 | 719 | 1555 | IL8 - BCL10 | 204 | 1653 | 544 |
| IL10RB - BCL10 | 2009 | 466 | 1053 | IL10RB - BCL10 | 2244 | 2150 | 1578 |
| IL15 - BCL10 | 35 | 2072 | 580 | IL15 - BCL10 | 2174 | 1618 | 1375 |
| IL15RA - BCL10 | 1477 | 2064 | 1789 | IL15RA - BCL10 | 1810 | 1656 | 1835 |
| IL17C - BCL10 | 8 | 2009 | 1232 | IL17C - BCL10 | 705 | 1777 | 207 |
| IL17REL - BCL10 | 2397 | 89 | 550 | IL17REL - BCL10 | 839 | 1214 | 377 |
| Unexplored combinatorial hypotheses | |
| IL w.r.t BCL | |
| IL-1A/1B/17C | BCL2L1 |
| IL-6ST/17REL | BCL2L2 |
| IL-17REL | BCL2L13 |
| IL-6ST/15RA | BCL3 |
| IL-1RAP/6ST/8/17REL | BCL6 |
| IL-1A/6ST/8/17REL | BCL9L |
| IL-6ST/15RA | BCL10 |
| BCL w.r.t IL | |
| IL-1B/2RG/10RB | BCL2L1 |
| IL-1A/1B/1RN/6ST/8/15/17C | BCL2L2 |
| IL-1RAP/1RN/2RG/6ST/8/10RB/15/15RA/17C | BCL2L13 |
| IL-8/10RB | BCL3 |
| IL-1B/1RN/2RG/8/15RA/17REL | BCL6 |
| IL-1A/1RAP/1RN/2RG/10RB/15RA | BCL10 |
| Ranking EXOSC family vs BCL family | |||||||
| Ranking of EXOSC2 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC2 - BCL2L12 | 723 | 355 | 1211 | EXOSC2 - BCL2L12 | 1498 | 1889 | 1856 |
| EXOSC2 - BCL6B | 1092 | 1033 | 638 | EXOSC2 - BCL6B | 202 | 81 | 194 |
| EXOSC2 - BCL7A | 1633 | 1047 | 317 | EXOSC2 - BCL7A | 2403 | 2531 | 2405 |
| EXOSC2 - BCL9 | 699 | 559 | 425 | EXOSC2 - BCL9 | 2552 | 2230 | 1755 |
| EXOSC2 - BCL11A | 338 | 319 | 1598 | EXOSC2 - BCL11A | 574 | 834 | 1055 |
| EXOSC2 - BCL11B | 1285 | 1440 | 812 | EXOSC2 - BCL11B | 1067 | 1574 | 730 |
| Ranking of EXOSC3 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC3 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC3 - BCL2L12 | 2280 | 1640 | 1955 | EXOSC3 - BCL2L12 | 1976 | 1482 | 2399 |
| EXOSC3 - BCL6B | 2429 | 2273 | 2407 | EXOSC3 - BCL6B | 571 | 335 | 307 |
| EXOSC3 - BCL7A | 2100 | 1374 | 2674 | EXOSC3 - BCL7A | 1739 | 1882 | 1700 |
| EXOSC3 - BCL9 | 2437 | 2223 | 2245 | EXOSC3 - BCL9 | 2380 | 1912 | 2321 |
| EXOSC3 - BCL11A | 2212 | 2090 | 116 | EXOSC3 - BCL11A | 1018 | 1345 | 483 |
| EXOSC3 - BCL11B | 1677 | 199 | 267 | EXOSC3 - BCL11B | 2572 | 1876 | 2395 |
| Ranking of EXOSC5 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC5 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC5 - BCL2L12 | 498 | 1342 | 436 | EXOSC5 - BCL2L12 | 2174 | 1635 | 1824 |
| EXOSC5 - BCL6B | 786 | 1272 | 1194 | EXOSC5 - BCL6B | 330 | 193 | 107 |
| EXOSC5 - BCL7A | 374 | 1338 | 874 | EXOSC5 - BCL7A | 2582 | 2701 | 2415 |
| EXOSC5 - BCL9 | 613 | 946 | 772 | EXOSC5 - BCL9 | 1777 | 1511 | 2011 |
| EXOSC5 - BCL11A | 459 | 90 | 1034 | EXOSC5 - BCL11A | 756 | 389 | 1183 |
| EXOSC5 - BCL11B | 1404 | 2520 | 1558 | EXOSC5 - BCL11B | 1368 | 1353 | 1455 |
| Ranking of EXOSC6 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC6 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC6 - BCL2L12 | 1327 | 1857 | 2063 | EXOSC6 - BCL2L12 | 2268 | 1527 | 478 |
| EXOSC6 - BCL6B | 1965 | 2525 | 1825 | EXOSC6 - BCL6B | 18 | 2334 | 2512 |
| EXOSC6 - BCL7A | 1676 | 787 | 944 | EXOSC6 - BCL7A | 593 | 2653 | 2037 |
| EXOSC6 - BCL9 | 1838 | 1059 | 1091 | EXOSC6 - BCL9 | 1846 | 851 | 1564 |
| EXOSC6 - BCL11A | 1677 | 1573 | 2217 | EXOSC6 - BCL11A | 596 | 2307 | 2547 |
| EXOSC6 - BCL11B | 1897 | 1736 | 1126 | EXOSC6 - BCL11B | 2094 | 2223 | 81 |
| Ranking of EXOSC7 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC7 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC7 - BCL2L12 | 1899 | 1755 | 974 | EXOSC7 - BCL2L12 | 1721 | 1551 | 1099 |
| EXOSC7 - BCL6B | 666 | 98 | 743 | EXOSC7 - BCL6B | 2730 | 2690 | 2689 |
| EXOSC7 - BCL7A | 2290 | 1501 | 1513 | EXOSC7 - BCL7A | 1282 | 831 | 1218 |
| EXOSC7 - BCL9 | 2363 | 1134 | 2219 | EXOSC7 - BCL9 | 1845 | 1234 | 328 |
| EXOSC7 - BCL11A | 1477 | 2239 | 1217 | EXOSC7 - BCL11A | 520 | 117 | 686 |
| EXOSC7 - BCL11B | 2396 | 1524 | 2037 | EXOSC7 - BCL11B | 1529 | 2720 | 1418 |
| Ranking of EXOSC8 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC8 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC8 - BCL2L12 | 2042 | 2152 | 506 | EXOSC8 - BCL2L12 | 1967 | 1525 | 2275 |
| EXOSC8 - BCL6B | 2469 | 2134 | 2224 | EXOSC8 - BCL6B | 190 | 1630 | 472 |
| EXOSC8 - BCL7A | 1175 | 1504 | 1743 | EXOSC8 - BCL7A | 2065 | 2351 | 1069 |
| EXOSC8 - BCL9 | 1733 | 2452 | 1164 | EXOSC8 - BCL9 | 2640 | 1895 | 1747 |
| EXOSC8 - BCL11A | 1864 | 906 | 1130 | EXOSC8 - BCL11A | 944 | 2303 | 532 |
| EXOSC8 - BCL11B | 1547 | 605 | 374 | EXOSC8 - BCL11B | 2581 | 2728 | 2359 |
| Ranking of EXOSC9 w.r.t BCL family | Ranking of BCL family w.r.t EXOSC9 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| EXOSC9 - BCL2L12 | 1179 | 1018 | 687 | EXOSC9 - BCL2L12 | 2105 | 1762 | 1453 |
| EXOSC9 - BCL6B | 437 | 852 | 1358 | EXOSC9 - BCL6B | 634 | 304 | 146 |
| EXOSC9 - BCL7A | 821 | 346 | 727 | EXOSC9 - BCL7A | 985 | 2017 | 2207 |
| EXOSC9 - BCL9 | 1305 | 1849 | 299 | EXOSC9 - BCL9 | 1197 | 1279 | 2154 |
| EXOSC9 - BCL11A | 892 | 2426 | 2011 | EXOSC9 - BCL11A | 481 | 441 | 1372 |
| EXOSC9 - BCL11B | 1569 | 549 | 1456 | EXOSC9 - BCL11B | 2606 | 1454 | 133 |
| Unexplored combinatorial hypotheses | |
| EXOSC w.r.t BCL | |
| EXOSC2 | BCL-2L12/6B/7A/9/11A/11B |
| EXOSC3 | BCL-11B |
| EXOSC5 | BCL-2L12/6B/7A/9/11A/11B |
| EXOSC6 | BCL-7A/9/11A |
| EXOSC7 | BCL-6B/7A/11A |
| EXOSC8 | BCL-7A/11A/11B |
| EXOSC9 | BCL-2L12/6B/7A/9/11B |
| BCL w.r.t EXOSC | |
| EXOSC2 | BCL-6B/11A/11B |
| EXOSC3 | BCL-6B/7A/11A |
| EXOSC5 | BCL-6B/11A/11B |
| EXOSC6 | BCL-2L12/9 |
| EXOSC7 | BCL-2L12/7A/9/11A/11B |
| EXOSC8 | BCL-6B/11A |
| EXOSC9 | BCL-6B/9/11A/11B |
| Ranking ANTRX2 vs COL family | |||||||
| Ranking of ANTRX2 w.r.t COL family | Ranking of COL family w.r.t ANTXR2 | ||||||
| laplace | linear | rbf | laplace | linear | rbf | ||
| COL5A3-ANTXR2 | 2006 | 1473 | 2217 | COL5A3-ANTXR2 | 984 | 1782 | 933 |
| COL6A1-ANTXR2 | 1935 | 1061 | 1366 | COL6A1-ANTXR2 | 324 | 2211 | 398 |
| COL7A1-ANTXR2 | 1002 | 2119 | 1690 | COL7A1-ANTXR2 | 1722 | 956 | 2121 |
| COL9A2-ANTXR2 | 1498 | 552 | 1361 | COL9A2-ANTXR2 | 2391 | 80 | 135 |
| COL17A1-ANTXR2 | 1906 | 1086 | 780 | COL17A1-ANTXR2 | 576 | 2504 | 229 |
| COL28A1-ANTXR2 | 1409 | 2259 | 2296 | COL28A1-ANTXR2 | 478 | 1731 | 2362 |
| Unexplored combinatorial hypotheses | |
| ANTXR2 w.r.t COL | |
| COL5A3 | ANTXR2 |
| COL28A1 | ANTXR2 |
| COL w.r.t ANTXR2 | |
| COL7A1 | ANTXR2 |
| COL28A1 | ANTXR2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





