1. Introduction
Staphylococcus aureus is one of the main causative agents of mastitis in milk producing animals and the first for clinical bovine mastitis with ability to give origin to persistent intramammary infections [
1]. This bacterial species is also a major human pathogen capable of causing food poisoning for the production of multiple heat-stable enterotoxins (SE), localized soft tissue or skin infections and systemic infections triggered by virulence factors comprising staphylococcal superantigens (SAgs) [
2,
3,
4], cytotoxic proteins [
5] and factors that favor colonization and immune evasion [
6]. The emergence of antibiotic resistant (AR)
S. aureus strains, among which the methicillin-resistant
Staphylococcus aureus (MRSA) are listed by the World Health Organization among the pathogens of “high priority” against which new antibiotics are urgently needed, worsens the threat to human health posed by this bacterial species, since MRSA cause infections with high mortality rates [
7]. The transferable genetic element conferring the MRSA phenotype is the chromosomal cassette
mec (SCC
mec), that most often carry the
mecA or the
mecC gene, and sometimes other rare homologues, encoding for additional penicillin-binding proteins (PBP2a) with reduced affinity for β-lactams plus genes for site-specific recombinases [
8,
9].
Use of antibiotics in the animal farming sector to treat conditions such as mastitis can select for MRSA transmissible to humans through raw milk and derived products [
10]. Some risk factors have been identified for MRSA transmission in dairy farms, such as poor milking hygiene, while the role of antimicrobial usage has been little investigated, with the exception of one study reporting an increase in antibiotic minimum inhibitory concentration (MIC) values and in occurrence of AR genes
tetK,
tetM, and
blaZ after enrofloxacin treatment of persistent mastitis in goats, underlining the role of antimicrobial usage on the emergence of AR
S. aureus strains [
11]. Phylogenetic analysis based on multi-locus sequence typing (MLST) put in evidence that some
S. aureus lineages are found both in human and animal hosts, in particular strains from bovine mastitis, as a consequence of transfer from human to animal and
vice versa. Moreover, it was demonstrated that
S. aureus has the capacity to switch hosts [
12] so that animal isolates
S. aureus with resistance to antibiotics must be considered a threat to public health.
Investigating the trends of AR S. aureus prevalence can indicate if risk factors that favor their increase in farms are acting and allow to adopt measures to reduce the dissemination of the genetic determinants encoding resistance. Therefore, this study was undertaken to analyze the prevalence of AR S. aureus in farms by taking into account isolates from milk of animals affected by clinical mastitis requiring antibiotic treatment. The study was carried out in areas of the Abruzzo and Molise regions in Central Italy and aspects of mastitis management in the sampled farms were taken into account to explain the results of phenotypic and genotypic AR prevalence.
2. Materials and Methods
2.1. Bacterial strains and culture conditions
The bacterial strains used in this study were all isolates from mastitic milk samples analyzed upon request of veterinarians by the Istituto Zooprofilattico Sperimentale dell’Abruzzo e del Molise (IZSAM), Campobasso and Lanciano branches, for identification of the infectious agent and antibiogram execution. Strains phenotypically identified as S. aureus in routine analysis, 27 isolated in year 2021 and 27 in year 2022, were obtained after analysis of milk from 56 farms and 52 farms, respectively. These were propagated by streaking on blood agar (10/l g tryptose, 10 g/l meat extract, 5 g/l NaCl, 15 g/l agar, 100 ml of defibrinated sheep blood added aseptically after autoclaving and cooling of the base medium) incubated in aerobic conditions at 37°C for 24-48 h. Cell biomass from a colony isolated after two subsequent streaks on blood agar was used for each phenotypic or genotypic test. For long term storage the isolates were maintained in Microbank (Biolife Italiana, Milan, Italy) at -80°C.
2.2. Phenotypic AR testing
The antibiotics tested phenotypically were those of human usage belonging to the classes of antibiotics used for mastitis treatment and recommended for testing by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) to deduce resistance to the same antibiotic classes [
13], namely cefoxitin (FOX), clindamycin (CD), erythromycin (ERY), gentamicin (CN), kanamycin (KAN), norfloxacin (NOR), oxacillin (OXA) and tobramycin (TOB). The minimum inhibitory concentration (MIC) values were determined by using the Liofilchem® MIC Test Strips (Liofilchem, Roseto degli Abruzzi, TE, Italy) according to the instructions. The MIC values were assigned in accordance with EUCAST guidelines on antimicrobial suceptibility testing (AST) [
14]. For norfloxacin resistance was defined by using discs with 10 µg of the antibiotic (Liofilchem) as recommended [
13]. The reference to the epidemiological cut off (ECOFF) values [
13] was used to define the position of the new isolates in the range of observed MIC values for the species
S. aureus.
2.3. Quantitative PCR primer design
New qPCR tests for the transferable AR genes encoding resistance to the antibiotics tested phenotypically were designed after and defining the most frequently occurring genes in
S. aureus based on sequence database analysis. Oligonucleotides were designed by searching and aligning sequences in the NCBI databases (
https://www.ncbi.nlm.nih.gov/, accessed on 1 October 2022) and in the National Database of Antibiotic Resistant Organisms (NDARO,
https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/, accessed on 2 October 2022). For each gene a BLASTN analysis restricted to the
S. aureus taxon was carried out in order to consider different variants to be aligned such to design oligonucleotides targeting all of them. The primer/probe systems designed in this study are listed in
Table 1 with respective target genes and amplicon dimensions. These were synthetized by Eurofins Genomics (Ebersberg, Germany).
The gene regions comprised between each pair of oligonucleotides, ranging in size between 130 and 246 bp, were synthetized upon request by GenScript Biotech (Rijswijk, Netherlands) and delivered as pUC57 vector constructs to serve as positive controls in the qPCR runs.
2.4. DNA extraction
DNA was extracted from one loopful biomass resuspended in 200 µl of Macherey Nagel T1 buffer (Carlo Erba, Cornaredo, MI, Italy) containing 100 mg of sterile 200 µm diameter glass beads in safe lock Eppendorf tubes (Eppendorf). The suspension was bead beaten in a TissueLyser II (Qiagen) at 30 hz for 2 min). Then 200 µl of Macherey Nagel B3 buffer (Carlo Erba) were added and the extraction was continued according to the Macherey Nagel Nucleospin Tissue (Carlo Erba) protocol.
2.5. Quantitative PCR conditions
The qPCR reactions were carried out in a QuantStudio 5 thermal cycler (Thermo Fisher Scientific, Rodano, MI, Italy). Identification of isolates at the species level was carried out as described by Poli et al. [
15]. For AR gene detection a unique program suitable for all the primer/probe systems designed was used. This comprised initial denaturation at 94°C for 5 min and 40 cycles of denaturation at 94°C for 15 s and annealing at 51°C for 30 s. The qPCR reaction of 20 µl volume comprised 10 µl of Takara Premix Ex Taq (Probe qPCR) (Diatech, Jesi, AN, Italy), 0.2 µM primers and probe, TaqMan Exogenous Internal Positive Control Reagents (Thermo Fisher Scientific) in the recommended concentration, 2 µl of DNA sample and Nuclease Free water (Thermo Fisher Scientific) to the final volume. Four nanograms of synthetic positive control construct was used in the positive control reaction.
2.6. Veterinarian questionnaire
The 18 veterinarians who requested the bacteriological examinations and antibiograms for mastitis diagnosis in years 2021 and 2022 were interviewed to identify the antibiotic classes prescribed, the criteria adopted for antibiotic usage and different aspects of mastitis management in farms by delivering a questionnaire with closed ended questions.
2.7. Statistical analyses
MIC values plots, Student t test evaluation of distinctness of MIC data series obtained in 2021 and 2022 and correlation analyses were carried out by using PAST 4.03 free statistical software downloaded from
https://past.en.lo4d.com/windows (accessed on 23 December 2022). Data series were considered distinct for P<0.05.
3. Results and discussion
3.1. Rate of mastitis caused by S. aureus in 2021 and 2022
The number of farms with clinical mastitis caused by
S. aureus were 16 among 56 analyzed in 2021 and 13 among 52 analyzed in 2022, accounting for percentages of 28.5% and 25% of mastitis outbreaks, respectively. More than one isolate was obtained from the same sample if colonies of different dimension, appearance and hemolysis halo aspect were observed on blood agar. This led to obtaining 27 isolates for each year. All the isolates were received already phenotypically identified and identification was confirmed by qPCR targeted on the
nucA gene [
15]. The isolates are listed according to year and farm of isolation in
Table 2, section 3.3, reporting also the variable AR phenotypes and genotypes observed.
3.2. Phenotypic AR of S. aureus isolates
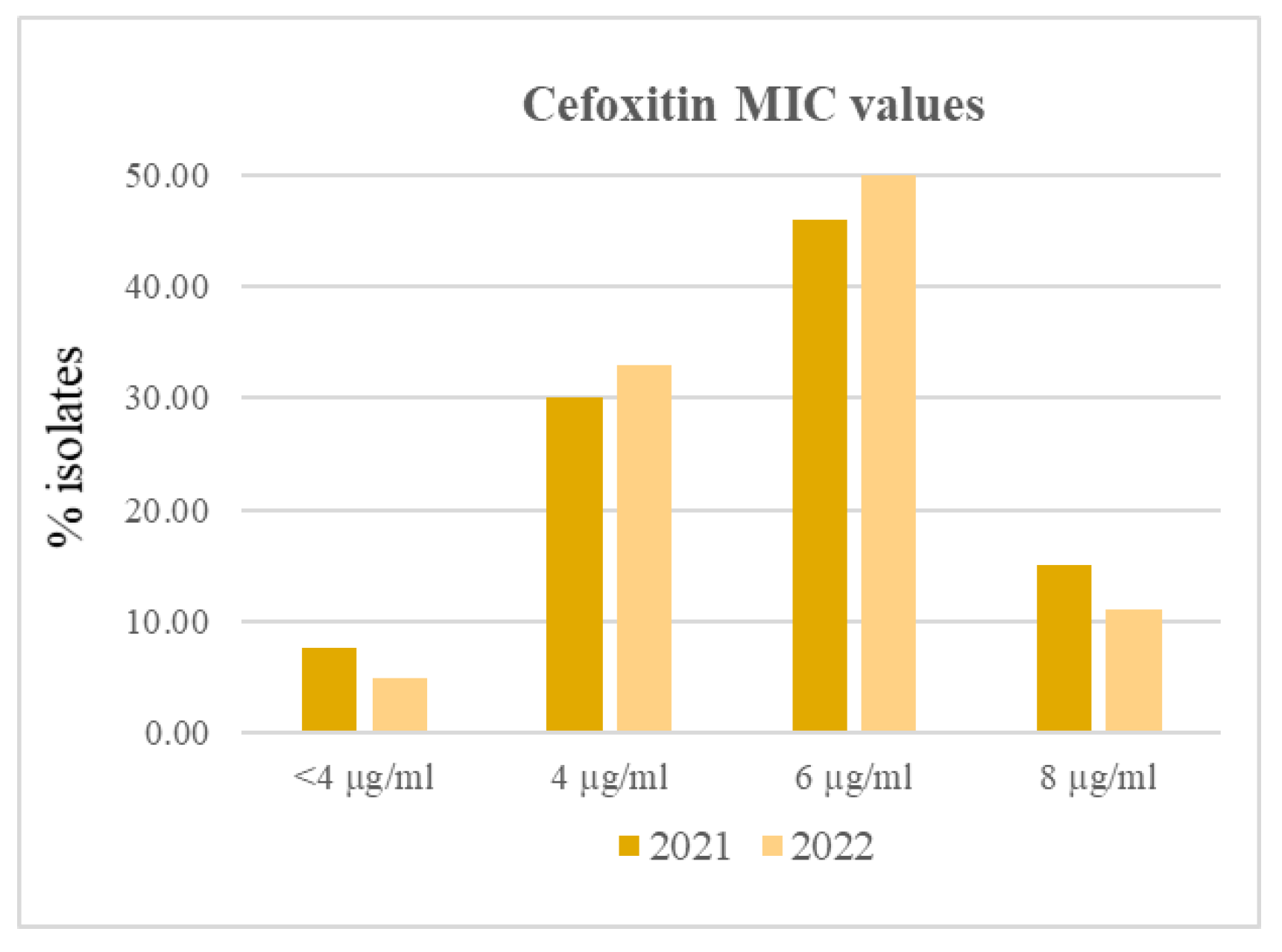
In this study isolates with cefoxitin MIC 6 – 8 µg/ml accounted for 59.2% isolates in both years. According to the EUCAST indications [
13] these isolates are resistant to this antibiotic, though at low levels, and must be considered methicillin resistant. However, all of them were sensitive to oxacillin and only one isolate from 2022 showed a MIC equal to the ECOFF for this antibiotic. The same isolate was also resistant to clindamycin (
S. aureus isolate 2022 6,
Table 2). The percentages of isolates assigned to groups with different cefoxitin MIC values in years 2021 and 2022 is shown in
Figure 1.
An high correlation with (r=0.99) was found between the numerosity of groups with different MIC values for cefoxitin in the two years, thus indicating that there was very little variation in the distribution of the isolates among different cefoxitin resistance levels in the investigation period.
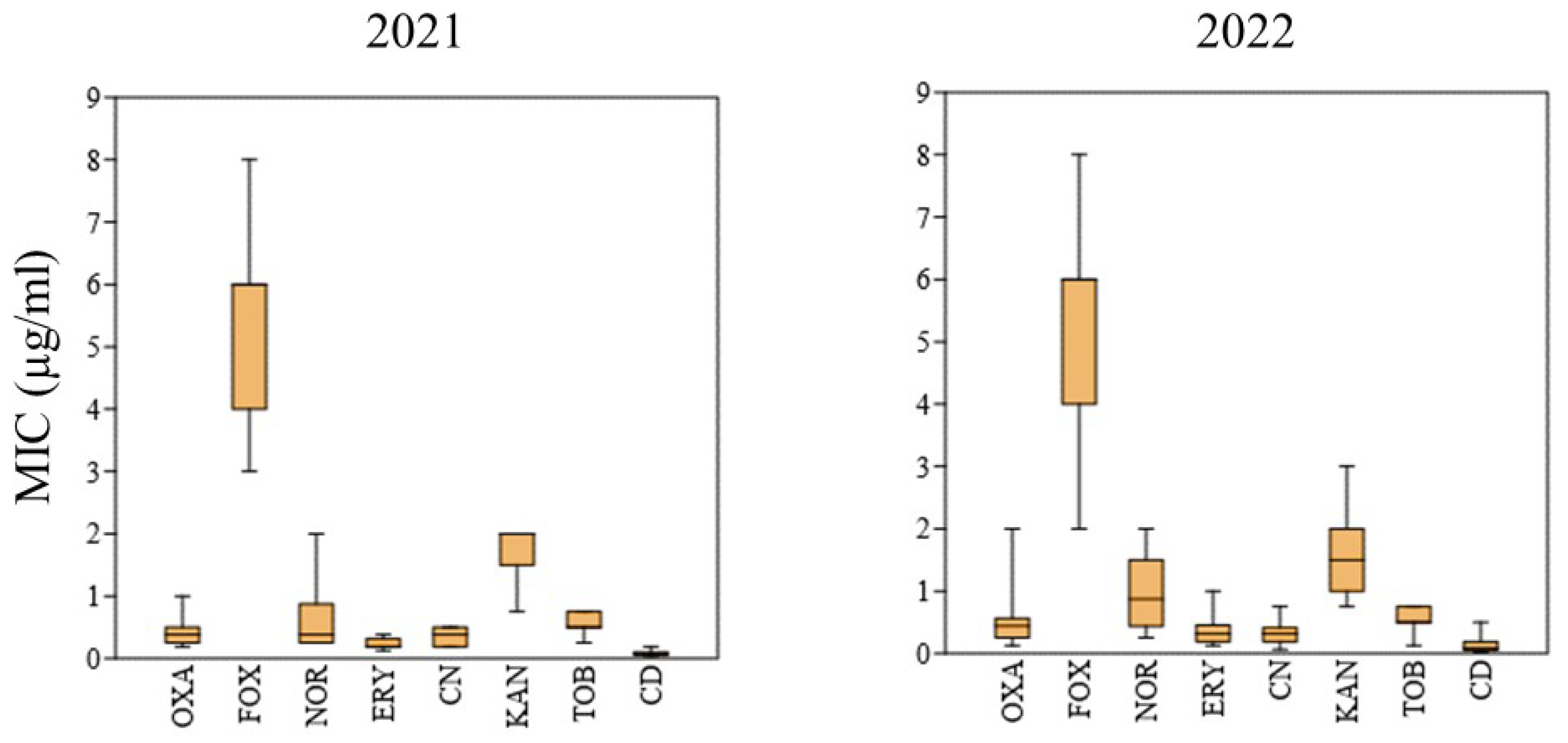
The distribution of MIC values for all antibiotics tested in the two years is shown in
Figure 2.
The MIC distributions for
S. aureus isolates between years 2021 and 2022 were not statistically distinct for any of the antibiotics considered. However, it can be noted that MICs for norfloxacin showed a shift to higher values in 2022, though resistance was not detected by the disc diffusion assay. Nevertheless, this trend could be indicative of an increase in resistance to quinolones that in
S. aureus is mediated by the core gene
norA encoding different variants of an efflux pump, and other efflux systems [
16]. These are increasingly expressed under antimicrobial pressure and can lead to the emergence of resistance phenotypes [
17]. Accordin to the MIC values, all the isolates were susceptible to oxacillin, norfloxacin, erythomycin, gentamicin, kanamycin, tobramycin and clindamycin, except isolate
S. aureus 2022 6 (
Table 2).
The percentage of isolates resistant to cefoxitin observed in this study was among the highest reported for European countries [
18]. On the other hand, an increase in AR to the other antimicrobials tested was not observed though it was reported to occur globally for
S. aureus strains causing bovine mastitis or isolated from milk and dairy products, specifically to clindamycin, erythromycin, gentamycin and oxacillin [
19,
20].
3.3. Occurrence of AR genes in the S. aureus isolates
The AR genes sought in this study were those encoding AR to the antibiotics of interest and found most frequently in
S. aureus, as deduced from consultation of the sequence databases and from a recent survey on identity and frequency of AR genes in 29,679 genomes of
S. aureus isolated worldwide [
21]. In addition, the
cfr gene was sought in this study since it codes for a 23S rRNA methyltransferase that confers resistance to different antibiotic classes, among which lincosamides, beyond phenicols [
22].
The AR gene most frequently detected in this study was
blaZ, present in 59.2% 2021 isolates and in 48.1% 2022 isolates (
Table 2).
Differences in the occurrence of this gene between the two years were not statistically significant according to the Student’s t test. In investigations carried out in different countries this gene was found at high frequencies with a maximum of 95.7% [
23].
Only a few other genetic determinants were identified in the isolates studied here. In particular, the
mecA gene was found only in one 2022 isolate, indicating a frequency lower than reported in other studies [
24,
25,
26,
27] but similar to that reported in Southern Italy for bulk tank milk of small ruminants [
28] indicating that its prevalence can vary on a local basis. The
mecA positive strain was not resistant to oxacillin, showing an MIC equal to the
S. aureus ECOFF for this antibiotic. The occurrence of
mecA positive and oxacillin sensitive strains was reported recently [
29].
According to the EUCAST AST guidelines
S. aureus strains resistant to cefoxitin have an MIC>4 µg/ml, a value that coincides with the ECOFF of the species for cefoxitin, and in most cases harbor a
mecA or
mecC gene [
12]. However, in this study cefoxitin resistance was not associated to the presence of the
mecA or the
mecC gene, resulting in an example of
mec-independent β-lactam resistance phenotype. The occurrence of cefoxitin resistant isolates without the
mec genetic determinants was described previously [
30,
31] and different genetic features were found to determine the cefoxitin resistance phenotype [
32,
33,
34] so further investigations should be devoted to definig the genetic basis of cefoxitin resistance in the isolates obtained in this study.
Other AR genes occurring in the
S. aureus isolates examined, namely
aph3’-III,
ant6-Ia,
ermB,
ermC/T and
mph, with the exception of
ermB found in a 2022 isolate harboring only this gene, were found mostly in association with other AR determinants (
Table 2). In particular, MDR genotypes
ant6-Ia-
aph3-III-
blaZ-
ermC/T and
aph3-III-
blaZ-
ermC/T were found each in one 2021 isolate. The occurrence of multiresistance encoding mobile genetic elements should be investigated in these isolates.
The gene mph for resistance to macrolides was found in the sole mecA positive strain. This strain was also resistant to clindamycin, possibly for the presence of a genetic determinant different from the genes lnuB and cfr, tested in this study. The finding that isolates harboring AR genes were susceptible to the antibiotics for which resistance was encoded suggests to carry out experiments to elucidate if those genes can be induced upon gradual exposure to antimicrobials.
A high prevalence of the
blaZ gene was observed in this study but none of the isolates overexpressed
blaZ to levels determining a borderline oxacillin resistant
S. aureus (BORSA) phenotype [
12].
3.4. Evaluation of antibiotic management by veterinarian interview
In order to understand if the results of AR screenings might be linked with a causal relation with the antibiotic usage practices adopted locally, the 18 veterinarians providing medical care to the sampled farms were interviewed by a questionnaire regarding antibiotics used, farm hygiene and the criteria adopted for antibiotic use decision in clinical mastitis. The results of the interviews are presented in
Table 3.
It is possible to observe that all the antibiotic classes allowed for mastitis treatment were used but β-lactams and fluoroquinolones prevailed. This could explain the high prevalence of strains harboring blaZ genes and the increase in norfloxacin MIC values. A further continuation of the antibiotic usage stated could enhance the trends observed.
Answers to the other aspects considered in the interview might indicate low usage of antibiotics since hygiene in the farm was considered good or acceptable in most cases and milking hygiene was found to be adequate in all instances. These conditions reduce the occurrence of infections and the need for antibiotic treatment. In addition, most farms were reported to adopt adequate mastitis prevention measures and protocols for antibiotic usage. Notably, half of the interviewed veterinarians declared to be committed to the reduction of antibiotic usage and all of them declared to use antibiotics based on the antibiogram outcomes for the specific pathogens.
However, according to the statements, strains causing mastitis were isolated and tested by antibiogram only in case of recidivating mastitis or in case of treatment failure and this could imply that initial treatments carried out without antibiogram execution can select for antibiotic resistant strains that become difficult to eradicate. Changes in this practice, together with improvements of mastitis management, could reduce prevalence of AR S. aureus in farms.
4. Conclusions
This study showed that the prevalence of both genotypic and phenotypic AR is currently low for non-β-lactam antibiotics and with no increasing trend in S. aureus isolates from the areas of Abruzzo and Molise considered. This is probably the consequence of overall good farm and milking hygiene, as reported by veterinarian professionals interviewed. However, strains harboring β-lactam resistance blaZ genes, already known to be widespread in the species S. aureus, occurred frequently, probably for the preferential use of β-lactams in mastitis therapy. Phenotypic resistance to cefoxitin in mecA/C negative isolates was frequent and its genetic basis needs to be identified. Moreover, the occurrence of one MRSA and two genotypically MDR isolates suggested to continue monitoring the presence of these AR profiles in dairy herds to understand if these genotypes tend to disseminate.
Author Contributions
Conceptualization, F.R. and I.D.; methodology, F.R., I.D. and M.A.S.; software, F.R.; validation, L.M. and L.R.; formal analysis, F.R., I.D. and M.A.S.; investigation, F.R.; data curation, F.R.; writing—original draft preparation, F.R.; writing—review and editing, L.R.; supervision, L.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data obtained in this study can made available by authors upon request.
Acknowledgments
The veterinarians participating to the interview are gratefully aknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zaatout, N.; Ayachi, A.; Kecha, M. Staphylococcus aureus persistence properties associated with bovine mastitis and alternative therapeutic modalities. J Appl Microbiol. 2020, 129, 1102–1119. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. Insights into the mechanism of superantigen. Nat Rev Microbiol. 2022, 20, 253. [Google Scholar] [CrossRef]
- Tuffs, S.W.; Goncheva, M.I.; Xu, S.X.; Craig, H.C.; Kasper, K.J.; Choi, J.; Flannagan, R.S.; Kerfoot, S.M.; Heinrichs, D.E.; McCormick, J.K. Superantigens promote Staphylococcus aureus bloodstream infection by eliciting pathogenic interferon-gamma production. Proc Natl Acad Sci U S A. 2022, 119, e2115987119. [Google Scholar] [CrossRef] [PubMed]
- Loffler, B.; Hussain, M.; Grundmeier, M.; Brück, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peters, G. Staphylococcus aureus Panton–Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010, 6, e1000715. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. 27 February 2017 News release, Geneva. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 December 2022).
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg Infect Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Schnitt, A.; Tenhagen, B.A. Risk Factors for the Occurrence of Methicillin-Resistant Staphylococcus aureus in Dairy Herds: An Update. Foodborne Pathog Dis. 2020, 17, 585–596. [Google Scholar] [CrossRef]
- Lima, M.C.; de Barros, M.; Scatamburlo, T.M.; Polveiro, R.C.; de Castro, L.K.; Guimarães, S.H.S.; da Costa, S.L.; da Costa, M.M.; Moreira, M.A.S. Profiles of Staphyloccocus aureus isolated from goat persistent mastitis before and after treatment with enrofloxacin. BMC Microbiol. 2020, 20, 127. [Google Scholar] [CrossRef]
- Shepheard, M.A.; Fleming, V.M.; Connor, T.R.; Corander, J.; Feil, E.J.; Fraser, C.; Hanage, W.P. Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS One 2013, 8, e62369. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 13.0, 2023. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 11 January 2023).
- Karatuna, O. Phenotypic Methods Used in Antimicrobial Susceptibility Testing. ICARS - ILRI Webinar Series. 1 December 2021. Available online: https://cgspace.cgiar.org/bitstream/handle/10568/119836/Phenotypic%20AST.pdf?sequence=1&isAllowed=y (accessed on 27 December 2022).
- Poli, A.; Guglielmini, E.; Sembeni, S.; Spiazzi, M.; Dellaglio, F.; Rossi, F.; Torriani, S. Detection of Staphylococcus aureus and enterotoxin genotype diversity in Monte Veronese, a Protected Designation of Origin Italian cheese. Lett Appl Microbiol. 2007, 45, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Santos Costa, S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 9, 710. [Google Scholar] [CrossRef]
- Santos Costa, S.; Viveiros, M.; Rosato, A.E.; Melo-Cristino, J.; Couto, I. Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus. BMC Microbiol. 2015, 15, 232. [Google Scholar]
- Naranjo-Lucena, A.; Slowey, R. Invited review: Antimicrobial resistance in bovine mastitis pathogens: A review of genetic determinants and prevalence of resistance in European countries. J Dairy Sci. 2023, 106, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Molineri, A.I.; Camussone, C.; Zbrun, M.V.; Suárez Archilla, G.; Cristiani, M.; Neder, V.; Calvinho, L.; Signorini, M. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: Systematic review and meta-analysis. Prev Vet Med. 2021, 188, 105261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Jin, J.; Li, X.; Zhang, H.; Shi, X.; Zhao, C. Prevalence, antibiotic resistance, and enterotoxin genes of Staphylococcus aureus isolated from milk and dairy products worldwide: A systematic review and meta-analysis. Food Res Int. 2022, 162 Pt A, 111969. [Google Scholar] [CrossRef] [PubMed]
- Pennone, V.; Prieto, M.; Álvarez-Ordóñez, A.; Cobo-Diaz, J.F. Antimicrobial Resistance Genes Analysis of Publicly Available Staphylococcus aureus Genomes. Antibiotics (Basel) 2022, 11, 1632. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Wang, J.; Wu, C.; Shen, Z.; Fu, X.; Yan, Y.; Zhang, Q.; Schwarz, S.; Shen, J. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother. 2012, 56, 1485–1490. [Google Scholar] [CrossRef]
- Gajewska, J.; Chajęcka-Wierzchowska, W., Zadernowska. Occurrence and Characteristics of Staphylococcus aureus Strains along the Production Chain of Raw Milk Cheeses in Poland. Molecules 2022, 4, 6569. [Google Scholar] [CrossRef]
- Titouche, Y.; Hakem, A.; Houali, K.; Meheut, T.; Vingadassalon, N.; Ruiz-Ripa, L.; Salmi, D.; Chergui, A.; Chenouf, N.; Hennekinne, J.A.; Torres, C.; Auvray, F. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) ST8 in raw milk and traditional dairy products in the Tizi Ouzou area of Algeria. J Dairy Sci. 2019, 102, 6876–6884. [Google Scholar] [CrossRef]
- Alghizzi, M.; Shami, A. The prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus in milk and dairy products in Riyadh, Saudi Arabia. Saudi J Biol Sci. 2021, 28, 7098–7104. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Bhattarai, R.K.; Luitel, H.; Karki, S.; Basnet, H.B. Prevalence of methicillin-resistant Staphylococcus aureus and pattern of antimicrobial resistance in mastitis milk of cattle in Chitwan, Nepal. BMC Vet Res. 2021, 17, 239. [Google Scholar] [CrossRef] [PubMed]
- Neelam; Jain, V.K.; Singh, M.; Joshi, V.G.; Chhabra, R.; Singh, K.; Rana, Y.S. Virulence and antimicrobial resistance gene profiles of Staphylococcus aureus associated with clinical mastitis in cattle. PLoS One 2022, 17, e0264762. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Latorre, L.; Santagada, G.; Fraccalvieri, R.; Miccolupo, A.; Sottili, R.; Palazzo, L.; Parisi, A. Methicillin-resistant Staphylococcus aureus (MRSA) in sheep and goat bulk tank milk from Southern Italy. Small Rumin. Res. 2016, 135, 26–31. [Google Scholar] [CrossRef]
- Pardo, L.; Giudice, G.; Mota, M.I.; Gutiérrez, C.; Varela, A.; Algorta, G.; Seija, V.; Galiana, A.; Aguerrebere, P. , Klein, M.; Varela, G. Phenotypic and genotypic characterization of oxacillin-susceptible and mecA positive Staphylococcus aureus strains isolated in Uruguay. Rev Argent Microbiol. 2022, 54, 293–298. [Google Scholar] [PubMed]
- Argudín, M.A.; Roisin, S.; Nienhaus, L.; Dodémont, M.; de Mendonça, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic Diversity among Staphylococcus aureus Isolates Showing Oxacillin and/or Cefoxitin Resistance Not Linked to the Presence of mec Genes. Antimicrob Agents Chemother. 2018, 62, e00091–18. [Google Scholar] [CrossRef] [PubMed]
- Velasco, V.; Mallea, A.; Bonilla, A.M.; Campos, J.; Rojas-García, P. Antibiotic-resistance profile of Staphylococcus aureus strains in the pork supply chain. Chil J Agric Anim Sc. 2022, 38, 234–240. [Google Scholar] [CrossRef]
- Ba, X.; Kalmar, L.; Hadjirin, N.F.; Kerschner, H.; Apfalter, P.; Morgan, F.J.; Paterson, G.K.; Girvan, S.L.; Zhou, R.; Harrison, E.M.; Holmes, M.A. Truncation of GdpP mediates β-lactam resistance in clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 2019, 74, 1182–1191. [Google Scholar] [CrossRef]
- Sommer, A.; Fuchs, S.; Layer, F.; Schaudinn, C.; Weber, R.E.; Richard, H.; Erdmann, M.B.; Laue, M.; Schuster, C.F.; Werner, G.; Strommenger, B. Mutations in the gdpP gene are a clinically relevant mechanism for β-lactam resistance in meticillin-resistant Staphylococcus aureus lacking mec determinants. Microb Genom. 2021, 7, 000623. [Google Scholar] [CrossRef]
- Sasaki, H.; Ishikawa, H.; Itoh, T.; Arano, M.; Hirata, K.; Ueshiba, H. Penicillin-Binding Proteins and Associated Protein Mutations Confer Oxacillin/Cefoxitin Tolerance in Borderline Oxacillin-Resistant Staphylococcus aureus. Microb Drug Resist. 2021, 27, 590–595. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).