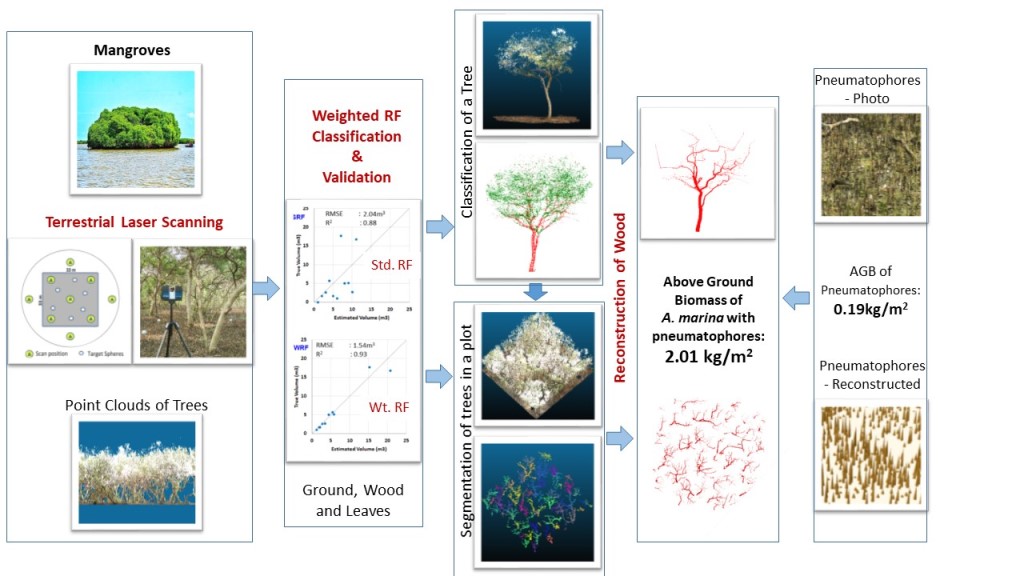
Accurately quantifying the above-ground volume (AGV) and thus above-ground biomass (AGB) of forest stands is an important aspect in the conservation of mangrove ecosystem owing to their ecological and economic benefits. However, the number of studies focusing on quantifying mangrove forests’ biomass has been relatively low due to their marshy terrain, making exploratory studies challenging. In recent times, the use of LiDAR technologies in forest inventory studies has become increasingly popular, due to the reliability of LiDAR as a highly accurate means of 3D spatial data acquisition. In this study, we propose an end-to-end methodology for estimating AGV of mangrove forest stands from terrestrial LiDAR data. Many of the recent studies on this topic effectively employ machine learning algorithms such as multi layer perceptron, random forests, etc. for filtering foliage in the point cloud data of single trees. This study further extends that approach by incorporating the impact of class imbalance of forest point cloud data in a weighted random forest classifier. For the task of segmentation of wood/foliage points in a single tree point cloud, this approach yielded an average increase of 2.737% in the balanced accuracy score, 0.007 in the Cohen’s kappa score, 2.745% in the ROC AUC score and 0.857% in the F1 score. For the task of AGV estimation of a single tree, this approach resulted in an average coefficient of determination of 0.93 with respect to the ground truth volumes. For the task of counting pneumatophores in a plot-level point cloud, the proposed breadth-first searching method yielded an average coefficient of determination of 0.9391. Also, the machine learning classifier and geometric features used in this study were invariant to tree species and hence could be generalised for the classification of point clouds of other tree species as well. Finally, a breadth-first graph-search segmentation based approach is also proposed as part of this pipeline to estimate the contribution of pneumatophores to the AGB of mangrove forest stands. Since pneumatophores are a special adaptation of mangrove forests for gaseous exchange in marshy environments, this study aims to incorporate the detection and AGB estimation of pneumatophores in the inventory of mangrove forest stands. Studying the contribution of pneumatophores to the AGB of mangrove forest plots could also aid future mangrove forest inventory studies in modeling the underlying root network and estimating the below-ground biomass of mangrove trees.