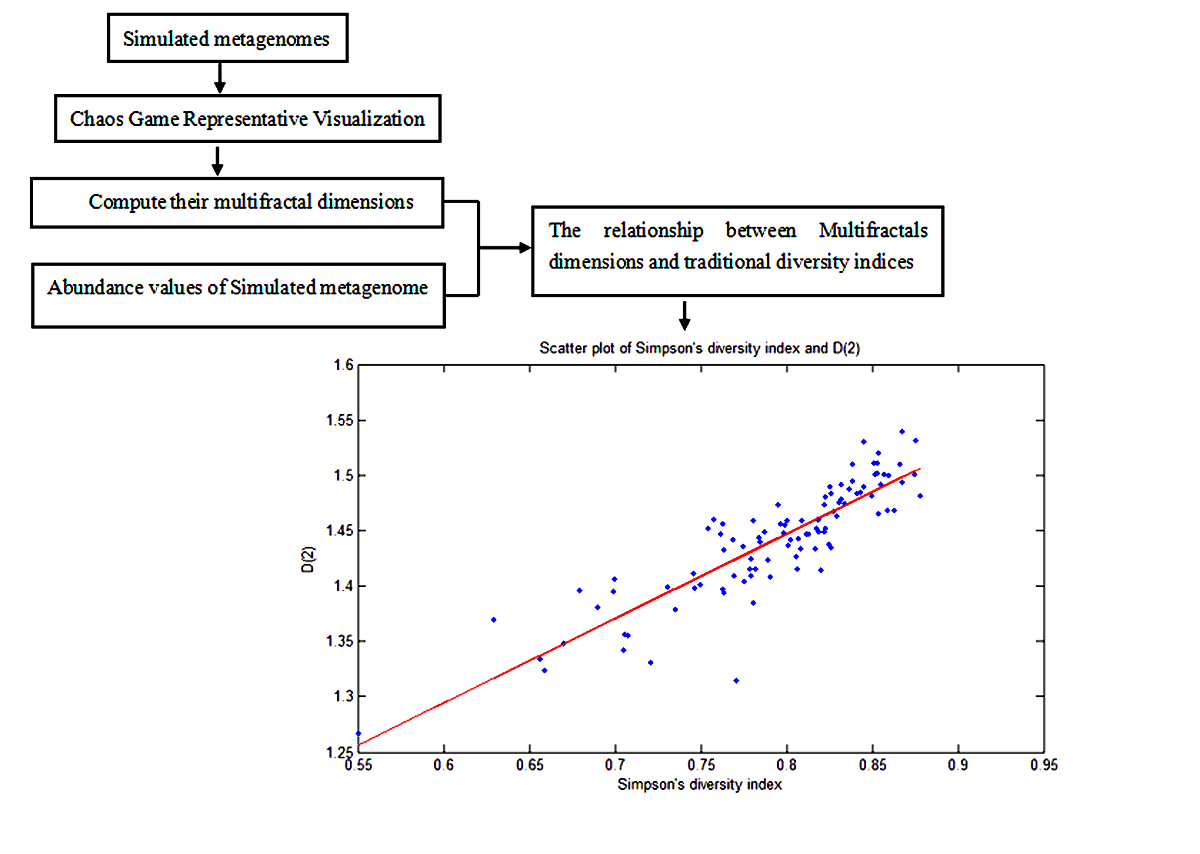
Species diversity in microbiome is a cutting-edge concept in metagenomic research. In this study, we propose a multifractal analysis for metagenomic research. From the chaos game representation (CGR) visualization of simulated and real metagenomes, we find that there exists self-similarity in the visualization of metagenomes. Then we compute the multifractal dimensions for simulated and real metagenomes. For simulated metagenomes, we also compute their diversity indices, such as species richness indices, Shannon’s diversity indices and Simpson’s diversity indices respectively for varying value of . Fom the Pearson correlation coefficients between their multifractal dimensions and traditional species diversity indices, we find that the correlation coefficients between the multifractal dimensions and species richness indices and Shannon diversity indices reach their maximums at respectively. The correlation coefficients between the multifractal dimensions and Simpson’s diversity indices reach their maximums at nearly. So the traditional diversity indices can be unified by the frame of multifractal analysis. These results coincided with the similar results in macrobial ecology. Finally, we apply our methods to real metagenomes of 100 infants’ gut microbiomes when they are newborn, 4 months and 12 months. Our results show that multifractal dimensions of infants’ gut microbiomes can discriminate the age difference.