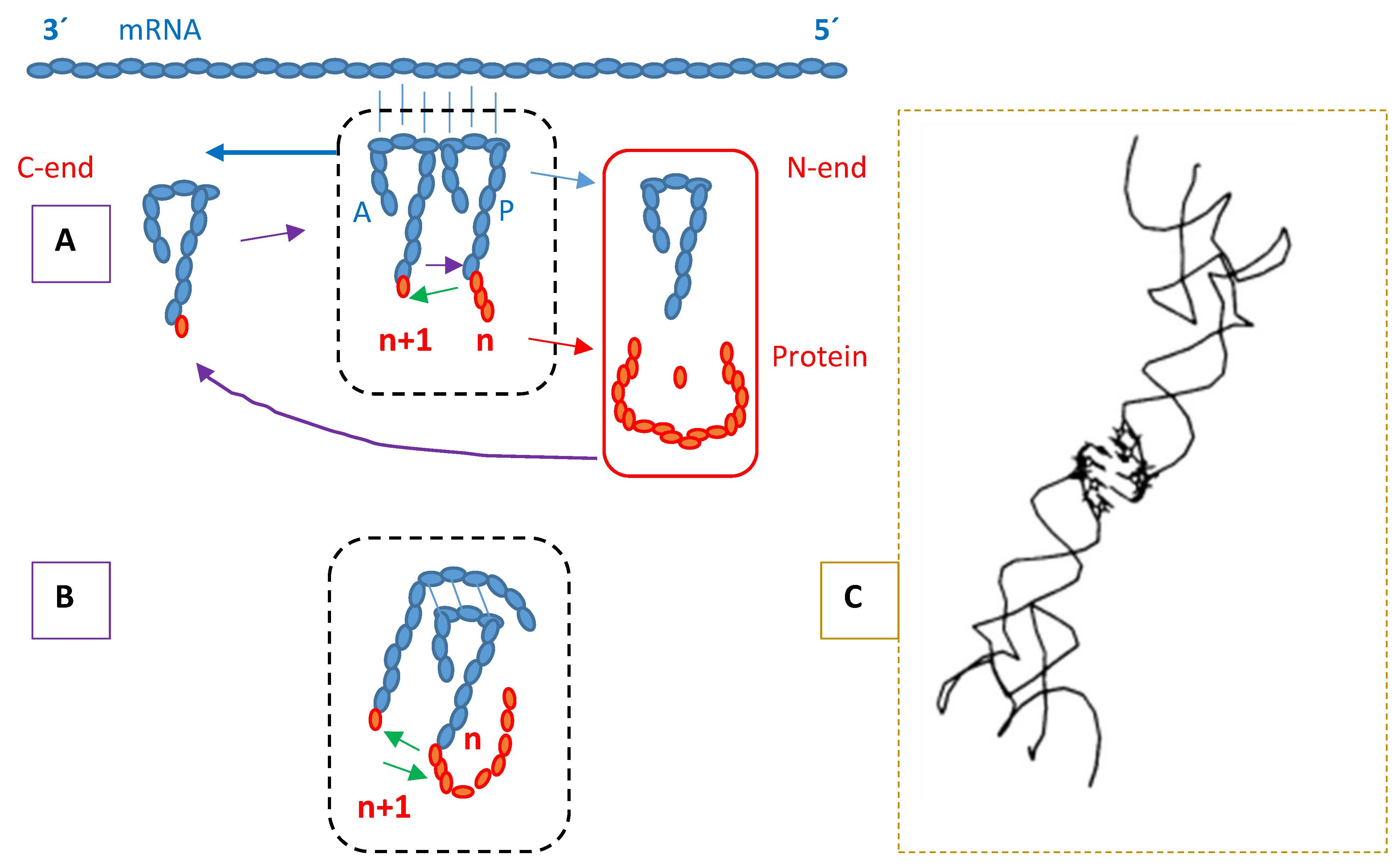
The proposal that the genetic code was formed on the basis of protein synthesis directed by tRNA dimers is reviewed and updated. The tRNAs paired through the anticodon loops are an indication on the process. Dimers are considered mimics of the ribosomes – structures that hold tRNAs together and facilitate the transferase reaction, and of the translation process – anticodons are at the same time codons for each other. The primitive protein synthesis system gets stabilized when the product peptides are able to bind the producers therewith establishing a self-stimulating production cycle. The chronology of amino acid encoding starts with Glycine and Serine, indicating the metabolic support of the Glycine-Serine C1-assimilation pathway, which is also consistent with evidence on origins of bioenergetics mechanisms. Since it is not possible to reach for substrates simpler than C1 and compounds in the identified pathway are apt for generating the other central metabolic routes, it is considered that protein synthesis is the beginning and center of a succession of sink-effective mechanisms that drive the formation and evolution of the metabolic flow system. Plasticity and diversification of proteins construct the cellular system following the orientation given by the flow and implementing it. Nucleic acid monomers participate in bioenergetics and the polymers are conservative memory systems for the synthesis of proteins. Protoplasmic fission is the final sink-effective mechanism, part of cell reproduction, guaranteeing that proteins don’t accumulate to saturation, which would trigger inhibition.