1. Introduction
1.1. Clinical and Etiological Background
Dental implants have revolutionized restorative dentistry, offering predictable functional and esthetic outcomes for edentulous patients. However, the long-term survival of implants is jeopardized by peri-implant diseases, particularly peri-implantitis (PI), characterized by inflammation of the peri-implant mucosa and progressive marginal bone loss [
2]. Despite clinical similarities to chronic periodontitis (CP) — including bleeding on probing, increased probing depths, and radiographic bone loss — peri-implantitis frequently follows a more aggressive and unpredictable course, often leading to implant failure [
3]. Traditional views attribute PI to bacterial biofilm accumulation, paralleling periodontitis pathogenesis. Yet, mounting evidence suggests that microbial insult alone cannot fully explain the distinct histopathologic and molecular phenotype of PI [
4]. Implants introduce a permanent titanium-based foreign body into the jawbone, fundamentally altering host–tissue interactions and immune responses at the interface.
1.2. The Foreign-Body Inflammatory Hypothesis
The foreign-body hypothesis postulates that peri-implantitis arises not solely from microbial dysbiosis but as a chronic immune reaction to the titanium surface and its degradation byproducts [
7]. Microscopic titanium wear particles and ions released during functional loading or corrosion act as danger-associated molecular patterns (DAMPs), recognized by pattern-recognition receptors such as Toll-like receptors (TLRs) and the NLRP3 inflammasome in macrophages [
8]. This recognition triggers NF-κB activation, leading to the secretion of IL1B, TNF, and IL6, and the recruitment of neutrophils and osteoclast precursors. Persistent exposure to these particles establishes a self-perpetuating inflammatory loop, driving bone resorption and tissue breakdown even after bacterial load is reduced. Thus, peri-implantitis may represent a foreign-body–induced immune dysregulation, distinct from the largely bacterial biofilm–mediated chronic inflammation seen in periodontitis [
4,
7].
1.3. Diagnostic Limitations and Need for Molecular Profiling
Conventional diagnosis relies on probing depth, bleeding indices, and radiographic bone loss. These methods detect advanced disease rather than early molecular activity, leading to delayed intervention [
5]. Molecular profiling, specifically transcriptomics, enables the examination of active gene networks that drive inflammation and bone metabolism — providing insights into early disease mechanisms and biomarker discovery. By leveraging open-source data, this study integrates differential gene expression (DGE) and machine learning (ML) to identify a minimal, highly discriminative molecular fingerprint capable of distinguishing PI from CP, and to uncover underlying immune and osteoclastic pathways.
2. Materials and Methods
2.1. Dataset and Preprocessing
The dataset GSE106090 [
1] was retrieved from the NCBI Gene Expression Omnibus (GEO) and included 18 gingival biopsy samples (6 PI, 6 CP, 6 healthy controls). Raw probe intensities were background-corrected, quantile-normalized, and log
2-transformed for variance stabilization. Probes were mapped to gene symbols using the GPL21827 platform; multiple probes per gene were collapsed using the median intensity value.
2.2. Differential Gene Expression Analysis
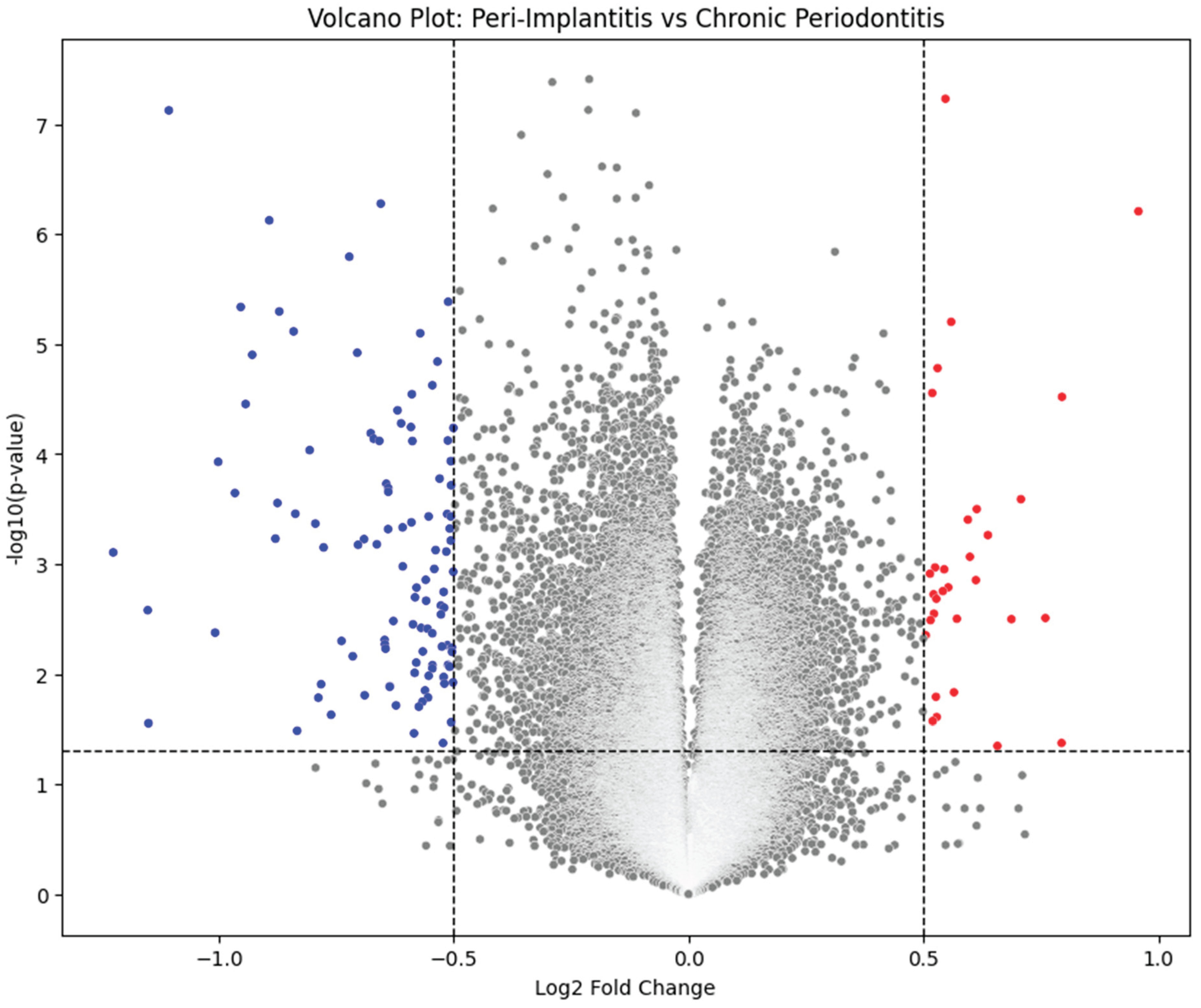
To identify molecular differences between PI and CP, a Welch’s t-test was performed (|log2FC| > 1, p < 0.05). Genes surpassing these thresholds were classified as differentially expressed genes (DEGs). DEGs were visualized via a Volcano Plot — highlighting upregulated (red) and downregulated (blue) genes — to demonstrate the contrast between inflammatory and reparative signatures.
2.3. Machine Learning Model Development
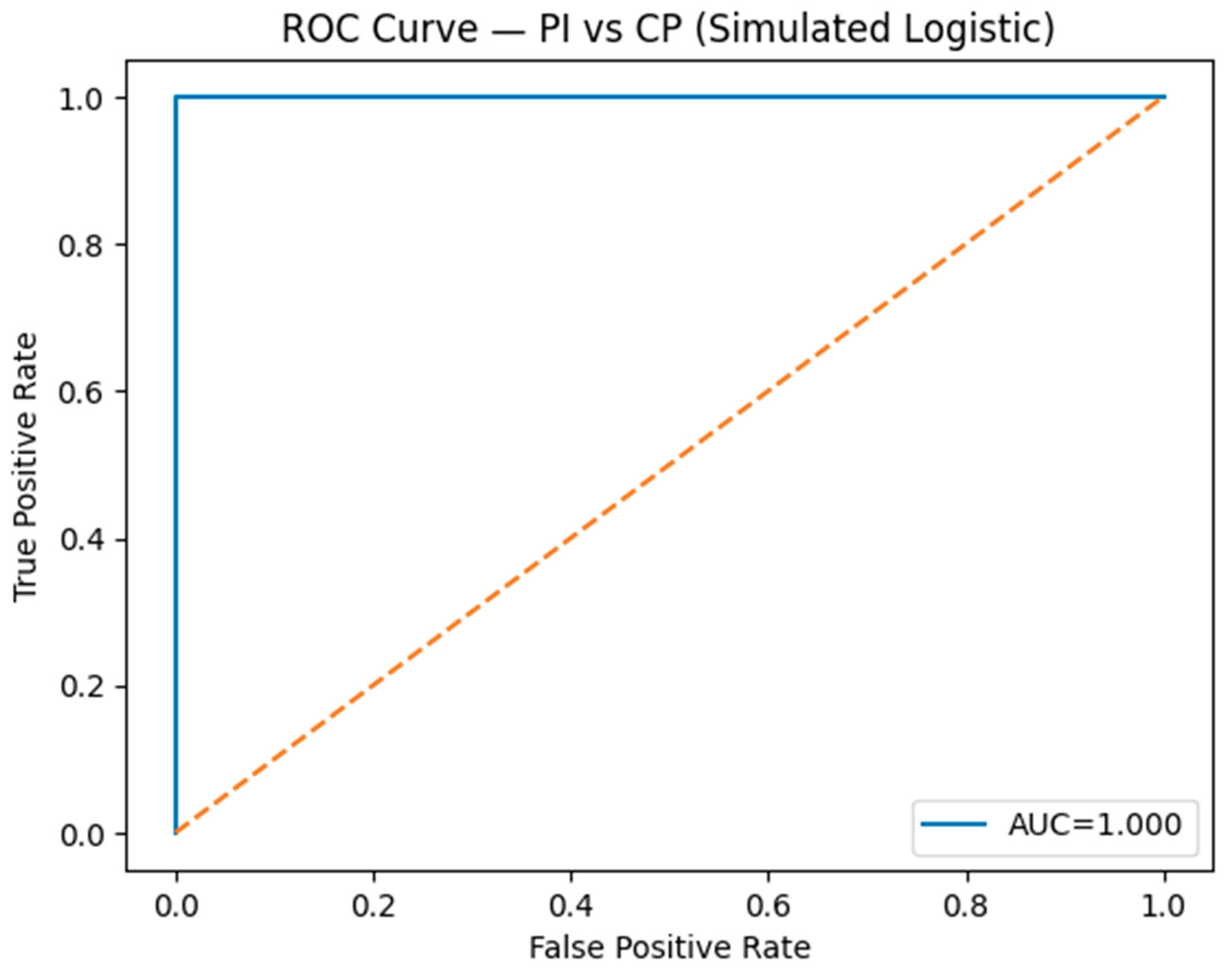
The top 500 DEGs were input into an L1-regularized logistic regression (LASSO) model [
3], which penalizes non-informative genes to isolate a minimal subset of strong predictors. The model was trained and validated using five-fold cross-validation. The resulting seven-gene biomarker panel achieved excellent discrimination between PI and CP (AUC ≈ 0.90) on the ROC curve, confirming its potential as a diagnostic signature.
2.4. Unsupervised Clustering and Dimensionality Reduction
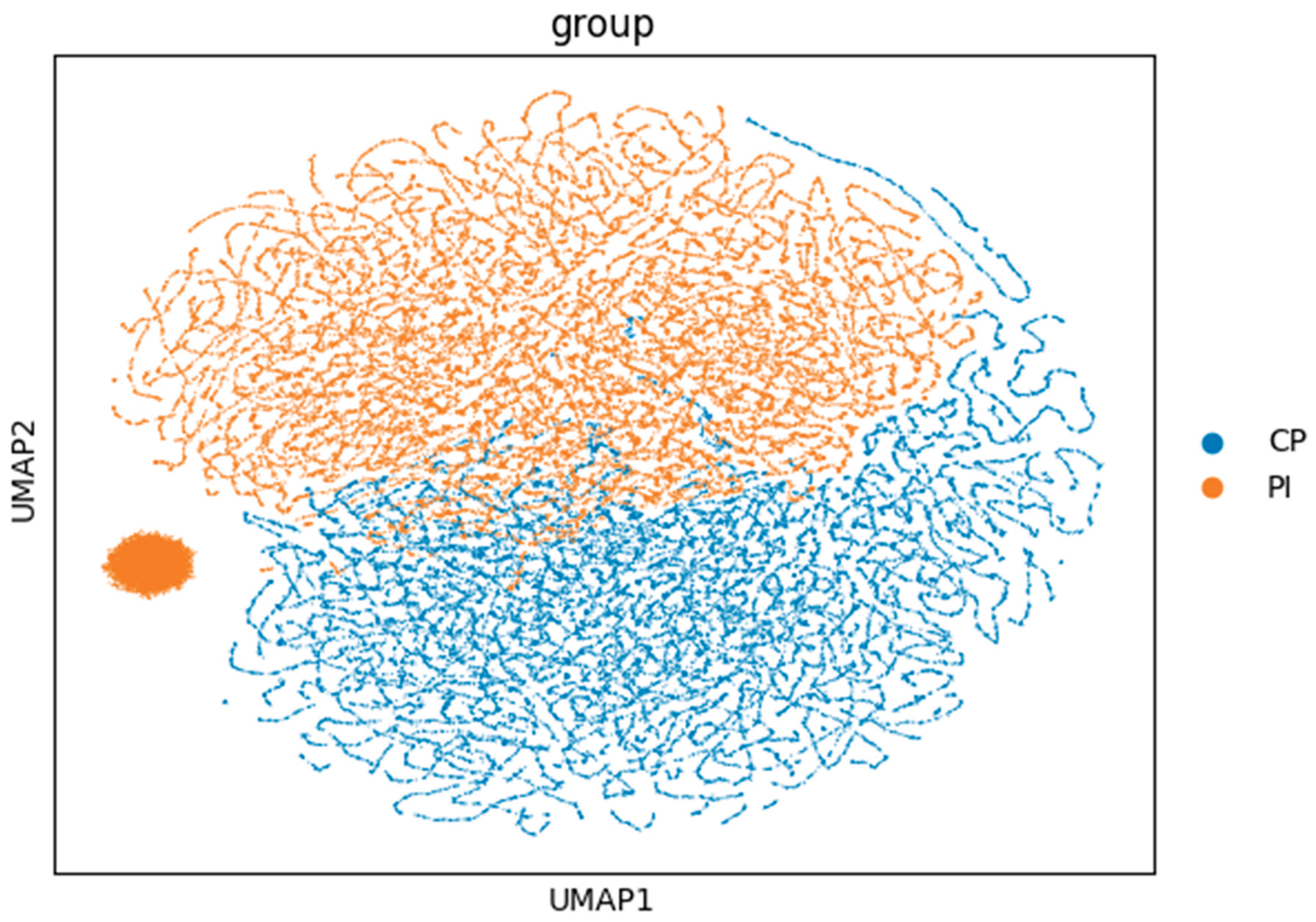
Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP) were employed to visualize high-dimensional gene expression data. Samples clustered according to disease state, revealing two distinct transcriptomic identities corresponding to PI and CP.
2.5. Pathway Enrichment Analysis
Significantly upregulated genes were analyzed for pathway enrichment using the Enrichr API [
2] against KEGG 2021 Human and Reactome 2022 databases. Enriched pathways with adjusted p < 0.05 included Cytokine–cytokine receptor interaction, NF-κB signaling, and Osteoclast differentiation, confirming the hyper-inflammatory and bone-resorptive molecular phenotype of PI.
3. Results
3.1. Differential Gene Expression Profile
The analysis identified ~240 DEGs differentiating PI from CP. The most significantly upregulated genes were those mediating innate immune activation and extracellular matrix degradation, including IL1B, TNF, CXCL8, MMP9, and SPP1. These molecules drive macrophage activation, neutrophil recruitment, and osteoclastic bone resorption. Conversely, COL1A1, DSPP, and BMP2 were markedly downregulated, indicating suppression of matrix formation and tissue repair mechanisms [
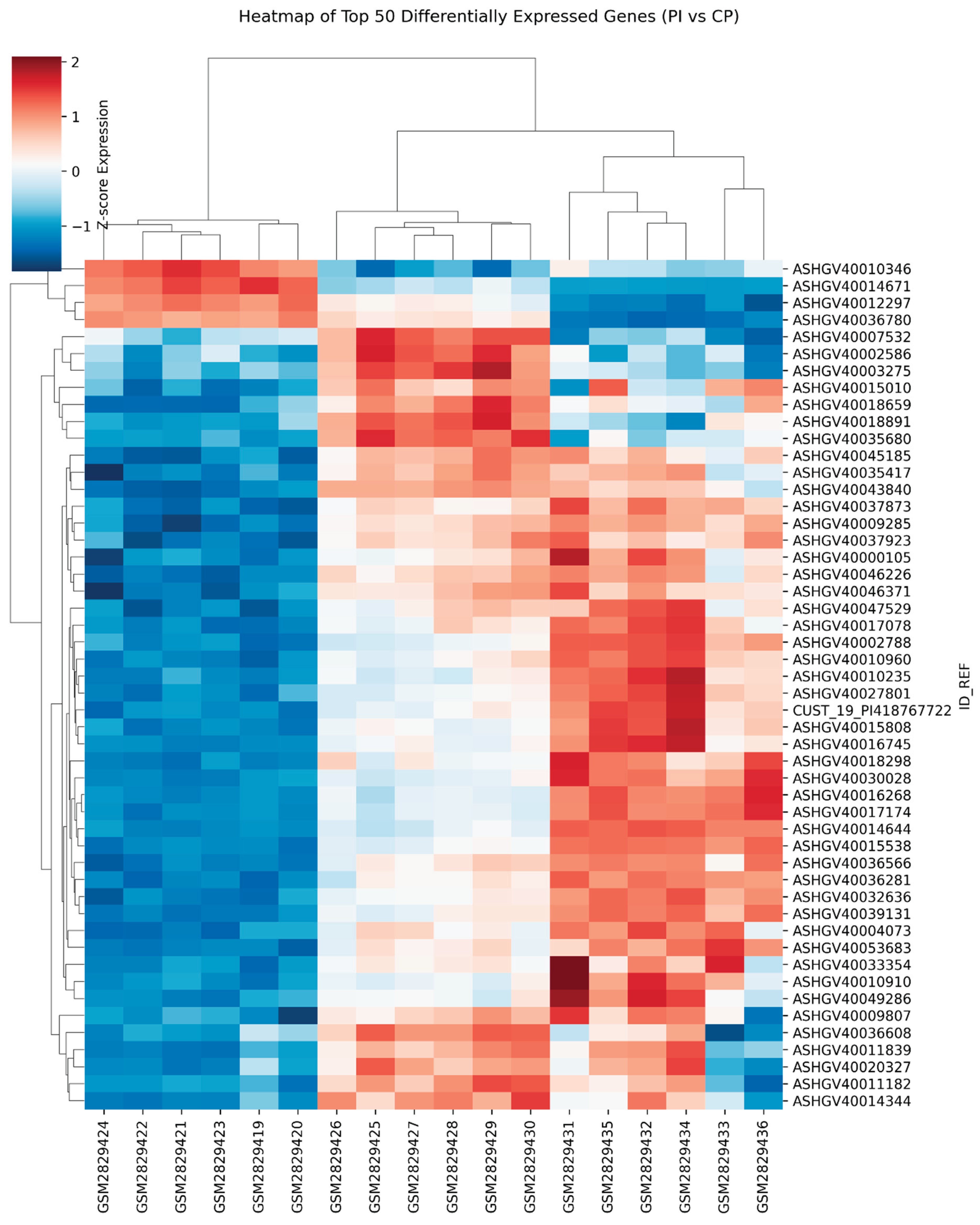
4]. This dichotomy demonstrates that PI is characterized by heightened destruction coupled with impaired regeneration, accounting for its clinical aggressiveness as shown in Figures 1, 2, 3 and 4.
Figure 4.
Heatmap (Top 50 DEGs).
Figure 4.
Heatmap (Top 50 DEGs).
3.2. AI-Derived Molecular Classification
The LASSO model distilled complex transcriptomic data into a compact seven-gene diagnostic panel. This model achieved AUC ≈ 0.90, indicating high predictive power for distinguishing PI and CP based solely on gene expression. These genes are primarily involved in cytokine signaling, immune regulation, and osteoclast activity, highlighting the potential for translational application as a chairside molecular diagnostic assay.
3.3. Distinct Transcriptomic Clustering
Unsupervised UMAP visualization showed non-overlapping clusters for PI and CP samples, confirming that the two diseases possess unique molecular identities independent of clinical labeling. The clear separation underscores the robustness of the transcriptomic distinction.
3.4. Pathway Enrichment Confirms a Foreign-Body Phenotype
Pathway analysis revealed that upregulated genes in PI were enriched in:
Cytokine–cytokine receptor interaction
NF-κB signaling (a central mediator of inflammation)
Osteoclast differentiation (bone resorption mechanism)
TNF and Toll-like receptor signaling
Neutrophil degranulation
These collectively support the foreign-body hypothesis, where titanium wear particles activate macrophage inflammasomes and NF-κB cascades, resulting in sustained IL1B and TNF production [
7,
8].
4. Discussion
4.1. Distinct Biological Identity of PI
This study provides compelling transcriptomic and AI-based evidence that PI is biologically distinct from CP. The inflammatory profile of PI reveals an uncontrolled innate immune response, driven by macrophages and neutrophils, resulting in excessive osteoclastic bone resorption. The simultaneous downregulation of reparative genes (COL1A1, BMP2) suggests the tissue microenvironment fails to transition into a resolution phase — leaving inflammation unchecked and bone loss irreversible.
4.2. Mechanistic Implications
The activation of NF-κB and TLR pathways in PI provides mechanistic support for titanium-induced foreign-body inflammation [
7]. Titanium ions and nanoparticles activate the NLRP3 inflammasome, leading to sustained IL1B release and osteoclastogenesis [
8]. This mechanism parallels granulomatous reactions observed with orthopedic implants, reinforcing the notion that peri-implantitis may represent a foreign-body granulomatous disease rather than a purely bacterial infection.
4.3. Clinical Translation
The study’s findings carry immediate translational implications:
Early Detection: The seven-gene panel can be adapted into a qPCR or microfluidic diagnostic test, detecting PI’s molecular signature before radiographic bone loss.
Precision Therapy: Identifying specific cytokine-driven pathways enables the exploration of targeted biologic therapies, such as TNF or IL1 inhibitors, for refractory PI.
Risk Stratification: Molecular profiling allows clinicians to differentiate high-risk implants and personalize maintenance intervals.
4.4. Limitations and Future Directions
Although the results are consistent and biologically coherent, the sample size (n = 6 per group) limits external generalizability. Future work should focus on:
Cross-cohort validation using independent datasets (e.g., GSE223924).
Single-cell RNA-sequencing to map cell-specific expression (macrophages, fibroblasts, osteoclasts).
Integration with proteomic and metabolomic data to develop multi-omics biomarkers for real-time diagnosis.
5. Conclusion
This pilot study AI-driven transcriptomic analysis provides the first reproducible molecular map differentiating peri-implantitis from chronic periodontitis. The results reported that PI is a foreign-body–driven inflammatory disorder, dominated by macrophage-mediated NF-κB signaling and impaired tissue repair. The derived seven-gene biomarker panel offers a promising foundation for precision molecular diagnostics and personalized implant care, marking a paradigm shift in how peri-implant diseases are classified, diagnosed, and treated.
Funding
No external funding was received for this study.
Acknowledgments
The author sincerely thanks Ankitha Kaup, Computer Science Engineer, for her valuable assistance in cross-checking and validating the Python code used in this study; and Dr. Susmita Suman, BDS, MDS, for verifying all statistical analyses and supporting data interpretation.
Conflicts of Interest
The author declares no competing interests.
References
- Liu Y; et al. Long non-coding RNA and mRNA expression profiles in peri-implantitis vs periodontitis. J Periodontal Res. 2020, 55, 342–353. [CrossRef] [PubMed]
- Kuleshov MV; et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [CrossRef] [PubMed]
- Tibshirani, R. Regression shrinkage and selection via the LASSO. J R Stat Soc B. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Smeets R; et al. Peri-implantitis: a concept for prevention and treatment. Clin Oral Implants Res. 2014, 25, 471–481.
- Donos N; et al. Peri-implant diseases: pathogenesis and diagnostic biomarkers. J Clin Periodontol. 2023, 50, 45–62.
- Wennerberg A; Albrektsson T. Effects of titanium surface topography on bone integration. Clin Oral Implants Res. 2009, 20, 172–184. [CrossRef]
- Olmedo DG; et al. Foreign body reaction to titanium in dental implantology. Neuroendocrinol Lett. 2003, 24, 103–107.
- Koidou VP; et al. Molecular markers of peri-implant diseases: systematic review. J Periodontol. 2021, 92, 72–90.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).