Submitted:
15 September 2025
Posted:
16 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The Relevance of Iron in Cell Biology
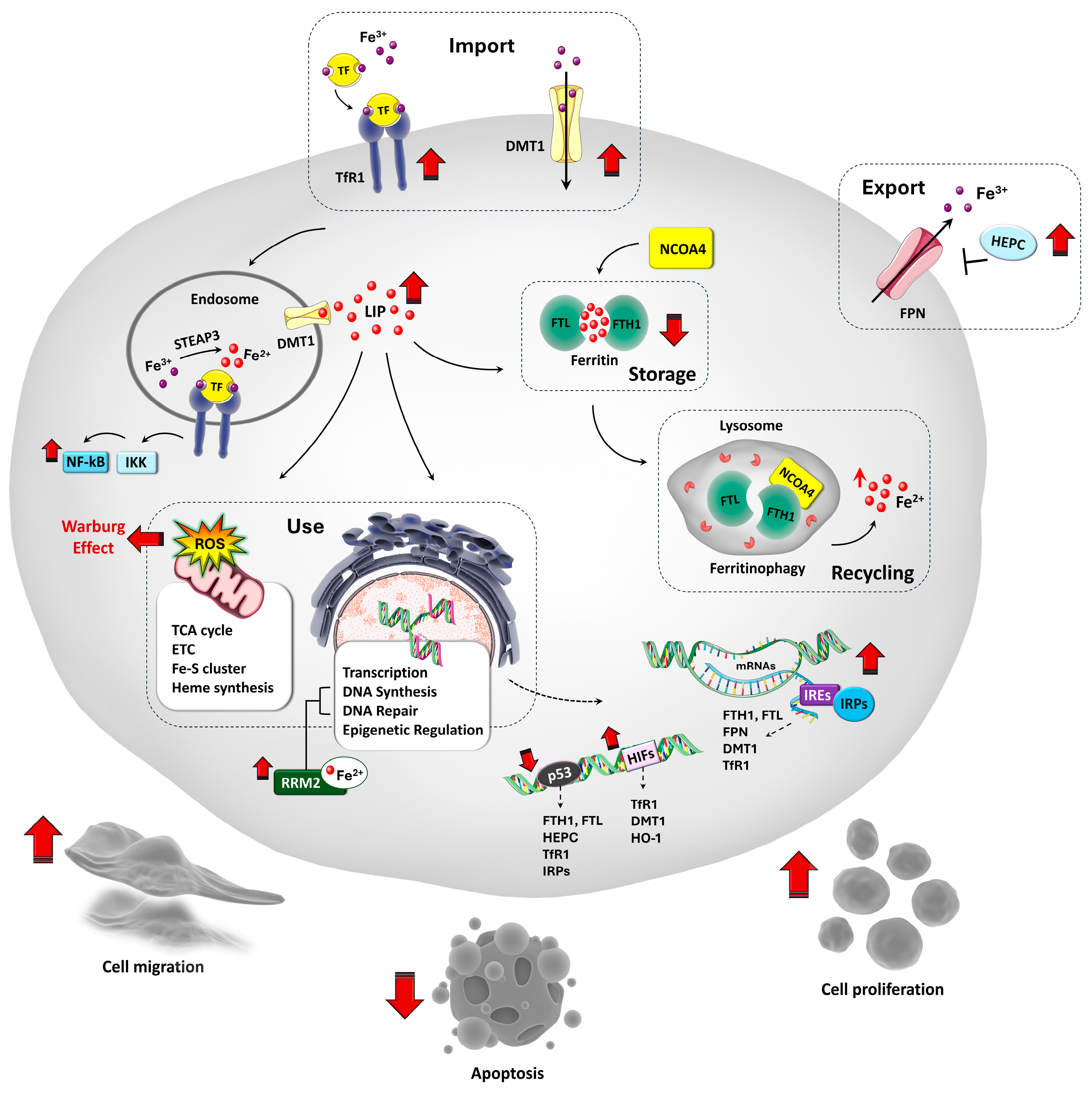
3. Iron Homeostasis and Its Regulation in Physiological States
4. Iron Metabolism Dysregulation in OS
5. Manipulating Iron Homeostasis in OS
5.1. Targeting TfR1 for OS Therapy
5.2. Iron Chelation for OS Therapy [14,48,60,65]
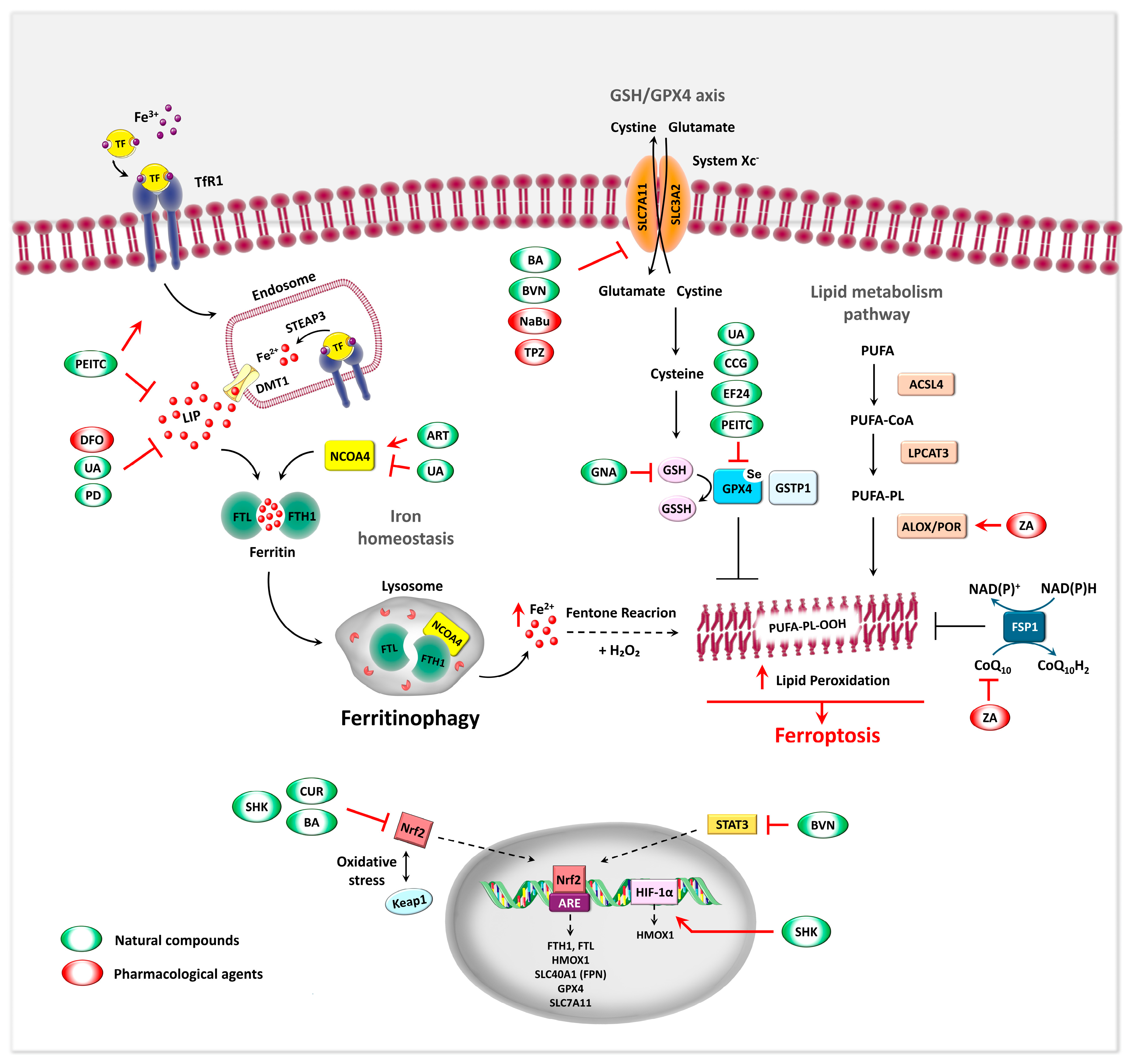
5.3. Ferroptosis as Iron-Related Metabolic Target in OS
5.3.1. Molecular Regulation and Morphological Features of Ferroptosis – General Aspects
5.3.2. Ferroptosis-Associated Markers in the Regulation of OS
5.3.3. Ferroptosis Inducers in OS
5.3.3.1. Natural Products Inducing Ferroptosis
5.3.3.2. Pharmacological Agents Inducing Ferroptosis
5.3.4. Nanomedicine Strategies to Promote Ferroptosis in OS
5.3.5. Genetic and RNA Biomarkers of Ferroptosis in the Regulation of OS
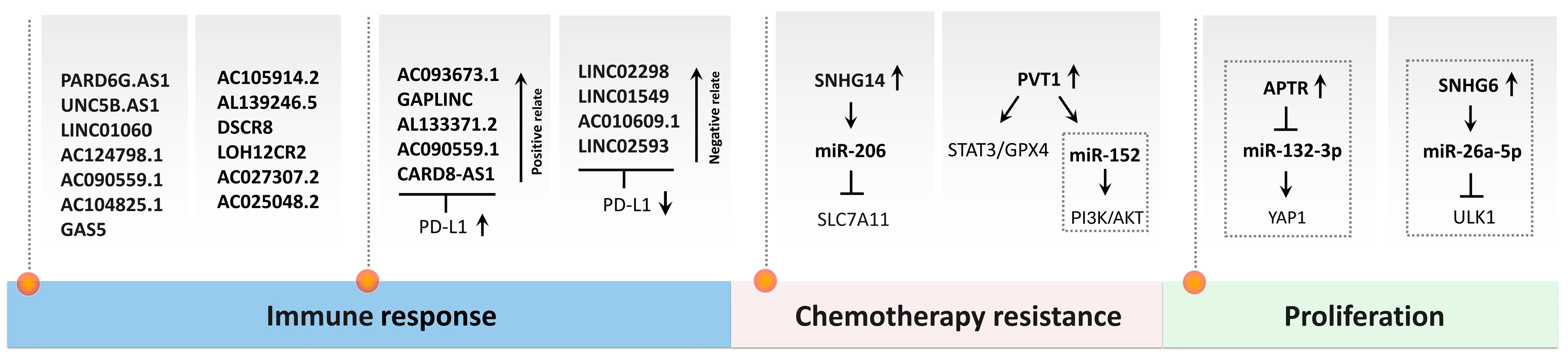
5.3.5.1. Ferroptosis-Related Genes in OS
5.3.5.2. Ferroptosis-Related ncRNA Networks in OS
5.4. Ferritinophagy: A Novel Target for OS Therapy
5.4.1. Ferritinophagy regulatory mechanism
5.4.2. Modulation of Ferritinophagy in OS: Emerging Therapeutic Approaches
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- M. U. Muckenthaler, S. Rivella, M. W. Hentze, and B. Galy, ‘A Red Carpet for Iron Metabolism’, Cell, vol. 168, no. 3, pp. 344–361, Jan. 2017. [CrossRef]
- S. Recalcati, E. Gammella, P. Buratti, and G. Cairo, ‘Molecular regulation of cellular iron balance’, IUBMB Life, vol. 69, no. 6, pp. 389–398, June 2017. [CrossRef]
- X. Bu and L. Wang, ‘Iron metabolism and the tumor microenvironment: A new perspective on cancer intervention and therapy (Review)’, Int. J. Mol. Med., vol. 55, no. 3, p. 39, Dec. 2024. [CrossRef]
- Y. Wang, L. Yu, J. Ding, and Y. Chen, ‘Iron Metabolism in Cancer’, Int. J. Mol. Sci., vol. 20, no. 1, p. 95, Dec. 2018. [CrossRef]
- R. Rodriguez, S. L. Schreiber, and M. Conrad, ‘Persister cancer cells: Iron addiction and vulnerability to ferroptosis’, Mol. Cell, vol. 82, no. 4, pp. 728–740, Feb. 2022. [CrossRef]
- W. Liang and N. Ferrara, ‘Iron Metabolism in the Tumor Microenvironment: Contributions of Innate Immune Cells’, Front. Immunol., vol. 11, Feb. 2021. [CrossRef]
- S. Cole, D. M. Gianferante, B. Zhu, and L. Mirabello, ‘Osteosarcoma: A Surveillance, Epidemiology, and End Results program-based analysis from 1975 to 2017’, Cancer, vol. 128, no. 11, pp. 2107–2118, June 2022. [CrossRef]
- M. A. Harris and C. J. Hawkins, ‘Recent and Ongoing Research into Metastatic Osteosarcoma Treatments’, Int. J. Mol. Sci., vol. 23, no. 7, p. 3817, Mar. 2022. [CrossRef]
- B. Zhang, Y. Zhang, R. Li, J. Li, X. Lu, and Y. Zhang, ‘The efficacy and safety comparison of first-line chemotherapeutic agents (high-dose methotrexate, doxorubicin, cisplatin, and ifosfamide) for osteosarcoma: a network meta-analysis’, J. Orthop. Surg., vol. 15, no. 1, p. 51, Feb. 2020. [CrossRef]
- C. Chen, L. Xie, T. Ren, Y. Huang, J. Xu, and W. Guo, ‘Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs’, Cancer Lett., vol. 500, pp. 1–10, Mar. 2021. [CrossRef]
- I. Lilienthal and N. Herold, ‘Targeting Molecular Mechanisms Underlying Treatment Efficacy and Resistance in Osteosarcoma: A Review of Current and Future Strategies’, Int. J. Mol. Sci., vol. 21, no. 18, p. 6885, Sept. 2020. [CrossRef]
- O. Badran, I. Cohen, and G. Bar-Sela, ‘The Impact of Iron on Cancer-Related Immune Functions in Oncology: Molecular Mechanisms and Clinical Evidence’, Cancers, vol. 16, no. 24, p. 4156, Dec. 2024. [CrossRef]
- Y. Xue et al., ‘Iron Chelator Induces Apoptosis in Osteosarcoma Cells by Disrupting Intracellular Iron Homeostasis and Activating the MAPK Pathway’, Int. J. Mol. Sci., vol. 22, no. 13, p. 7168, Jan. 2021. [CrossRef]
- P. V. Candelaria, L. S. Leoh, M. L. Penichet, and T. R. Daniels-Wells, ‘Antibodies Targeting the Transferrin Receptor 1 (TfR1) as Direct Anti-cancer Agents’, Front. Immunol., vol. 12, p. 607692, 2021. [CrossRef]
- G. De Vico, M. Martano, P. Maiolino, F. Carella, and L. Leonardi, ‘Expression of transferrin receptor-1 (TFR-1) in canine osteosarcomas’, Vet. Med. Sci., vol. 6, no. 3, pp. 272–276, Aug. 2020. [CrossRef]
- Y. Ma, L. Cong, W. Shen, C. Yang, and K. Ye, ‘Ferroptosis defense mechanisms: The future and hope for treating osteosarcoma’, Cell Biochem. Funct., vol. 42, no. 4, p. e4080, June 2024. [CrossRef]
- R. Huang, R. Xu, J. Shi, Z. Yang, J. Zheng, and D. Wei, ‘Artesunate induces ferroptosis in osteosarcoma through NCOA4-mediated ferritinophagy’, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol., vol. 39, no. 7, p. e70488, Apr. 2025. [CrossRef]
- K. Power, R. Leandri, G. Federico, G. De Vico, and L. Leonardi, ‘Ferritinophagy: a possible new iron-related metabolic target in canine osteoblastic osteosarcoma’, Front. Vet. Sci., vol. 12, p. 1546872, 2025. [CrossRef]
- C. Andreini, V. Putignano, A. Rosato, and L. Banci, ‘The human iron-proteome’, Met. Integr. Biometal Sci., vol. 10, no. 9, pp. 1223–1231, Sept. 2018. [CrossRef]
- S. J. F. Cronin, C. J. Woolf, G. Weiss, and J. M. Penninger, ‘The Role of Iron Regulation in Immunometabolism and Immune-Related Disease’, Front. Mol. Biosci., vol. 6, Nov. 2019. [CrossRef]
- M. R. Teh, A. E. Armitage, and H. Drakesmith, ‘Why cells need iron: a compendium of iron utilisation’, Trends Endocrinol. Metab., vol. 35, no. 12, pp. 1026–1049, Dec. 2024. [CrossRef]
- T. Ganz, ‘Systemic iron homeostasis’, Physiol. Rev., vol. 93, no. 4, pp. 1721–1741, Oct. 2013. [CrossRef]
- T. S. Koskenkorva-Frank, G. Weiss, W. H. Koppenol, and S. Burckhardt, ‘The complex interplay of iron metabolism, reactive oxygen species, and reactive nitrogen species: insights into the potential of various iron therapies to induce oxidative and nitrosative stress’, Free Radic. Biol. Med., vol. 65, pp. 1174–1194, Dec. 2013. [CrossRef]
- K. Pantopoulos, S. K. Porwal, A. Tartakoff, and L. Devireddy, ‘Mechanisms of mammalian iron homeostasis’, Biochemistry, vol. 51, no. 29, pp. 5705–5724, July 2012. [CrossRef]
- C. Gardi, B. Arezzini, V. Fortino, and M. Comporti, ‘Effect of free iron on collagen synthesis, cell proliferation and MMP-2 expression in rat hepatic stellate cells’, Biochem. Pharmacol., vol. 64, no. 7, pp. 1139–1145, Oct. 2002. [CrossRef]
- S. Dutt, I. Hamza, and T. B. Bartnikas, ‘Molecular Mechanisms of Iron and Heme Metabolism’, Annu. Rev. Nutr., vol. 42, pp. 311–335, Aug. 2022. [CrossRef]
- D.-L. Zhang, M. C. Ghosh, and T. A. Rouault, ‘The physiological functions of iron regulatory proteins in iron homeostasis - an update’, Front. Pharmacol., vol. 5, June 2014. [CrossRef]
- N. Koleini, J. S. Shapiro, J. Geier, and H. Ardehali, ‘Ironing out mechanisms of iron homeostasis and disorders of iron deficiency’, J. Clin. Invest., vol. 131, no. 11, p. e148671, June 2021. [CrossRef]
- C. P. Anderson, M. Shen, R. S. Eisenstein, and E. A. Leibold, ‘Mammalian iron metabolism and its control by iron regulatory proteins’, Biochim. Biophys. Acta BBA - Mol. Cell Res., vol. 1823, no. 9, pp. 1468–1483, Sept. 2012. [CrossRef]
- N. K. Kotla, P. Dutta, S. Parimi, and N. K. Das, ‘The Role of Ferritin in Health and Disease: Recent Advances and Understandings’, Metabolites, vol. 12, no. 7, p. 609, June 2022. [CrossRef]
- F. Hoelzgen et al., ‘Structural basis for the intracellular regulation of ferritin degradation’, Nat. Commun., vol. 15, no. 1, p. 3802, May 2024. [CrossRef]
- G. P. Holmes-Hampton, M. C. Ghosh, and T. A. Rouault, ‘Methods for Studying Iron Regulatory Protein 1: An Important Protein in Human Iron Metabolism’, Methods Enzymol., vol. 599, pp. 139–155, 2018. [CrossRef]
- C. Camaschella, A. Nai, and L. Silvestri, ‘Iron metabolism and iron disorders revisited in the hepcidin era’, Haematologica, vol. 105, no. 2, pp. 260–272, 2020. [CrossRef]
- F. Du, Z. Qian, Q. Gong, Z. J. Zhu, L. Lu, and Y. Ke, ‘The iron regulatory hormone hepcidin inhibits expression of iron release as well as iron uptake proteins in J774 cells’, J. Nutr. Biochem., vol. 23, no. 12, pp. 1694–1700, Dec. 2012. [CrossRef]
- G. N. Masoud and W. Li, ‘HIF-1α pathway: role, regulation and intervention for cancer therapy’, Acta Pharm. Sin. B, vol. 5, no. 5, pp. 378–389, Sept. 2015. [CrossRef]
- Y. M. Shah and L. Xie, ‘Hypoxia-inducible factors link iron homeostasis and erythropoiesis’, Gastroenterology, vol. 146, no. 3, pp. 630–642, Mar. 2014. [CrossRef]
- C. Peyssonnaux et al., ‘Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs)’, J. Clin. Invest., vol. 117, no. 7, pp. 1926–1932, July 2007. [CrossRef]
- R. M. Slominski, C. Raman, J. Y. Chen, and A. T. Slominski, ‘How cancer hijacks the body’s homeostasis through the neuroendocrine system’, Trends Neurosci., vol. 46, no. 4, pp. 263–275, Apr. 2023. [CrossRef]
- K. V. Kowdley, E. M. Gochanour, V. Sundaram, R. A. Shah, and P. Handa, ‘Hepcidin Signaling in Health and Disease: Ironing Out the Details’, Hepatol. Commun., vol. 5, no. 5, pp. 723–735, May 2021. [CrossRef]
- Z. Wu, W. Yang, J. Liu, and F. Zhang, ‘Interleukin-6 upregulates SOX18 expression in osteosarcoma’, OncoTargets Ther., vol. 10, pp. 5329–5336, 2017. [CrossRef]
- M. Y. Hsu, E. Mina, A. Roetto, and P. E. Porporato, ‘Iron: An Essential Element of Cancer Metabolism’, Cells, vol. 9, no. 12, p. 2591, Dec. 2020. [CrossRef]
- B. Li et al., ‘Increased hepcidin in hemorrhagic plaques correlates with iron-stimulated IL-6/STAT3 pathway activation in macrophages’, Biochem. Biophys. Res. Commun., vol. 515, no. 2, pp. 394–400, July 2019. [CrossRef]
- G. Ren, J. Zhou, Y. Su, Q. Yang, and J. Li, ‘TFRC promotes the proliferation, migration, and invasion of osteosarcoma cells by increasing the intracellular iron content and RRM2 expression’, Front. Oncol., vol. 15, May 2025. [CrossRef]
- H. Wu, J. Zhang, R. Dai, J. Xu, and H. Feng, ‘Transferrin receptor-1 and VEGF are prognostic factors for osteosarcoma’, J. Orthop. Surg., vol. 14, no. 1, p. 296, Sept. 2019. [CrossRef]
- N. S. Kenneth, S. Mudie, S. Naron, and S. Rocha, ‘TfR1 interacts with the IKK complex and is involved in IKK-NF-κB signalling’, Biochem. J., vol. 449, no. 1, pp. 275–284, Jan. 2013. [CrossRef]
- Y. Kuang et al., ‘Iron-dependent CDK1 activity promotes lung carcinogenesis via activation of the GP130/STAT3 signaling pathway’, Cell Death Dis., vol. 10, no. 4, p. 297, Apr. 2019. [CrossRef]
- S. Ni, Y. Kuang, Y. Yuan, and B. Yu, ‘Mitochondrion-mediated iron accumulation promotes carcinogenesis and Warburg effect through reactive oxygen species in osteosarcoma’, Cancer Cell Int., vol. 20, p. 399, 2020. [CrossRef]
- M. Morales and X. Xue, ‘Targeting iron metabolism in cancer therapy’, Theranostics, vol. 11, no. 17, pp. 8412–8429, July 2021. [CrossRef]
- M. Gao, A. Zheng, L. Chen, F. Dang, X. Liu, and J. Gao, ‘Benzo(a)pyrene affects proliferation with reference to metabolic genes and ROS/HIF-1α/HO-1 signaling in A549 and MCF-7 cancer cells’, Drug Chem. Toxicol., vol. 45, no. 2, pp. 741–749, Mar. 2022. [CrossRef]
- M. Liu, D. Wang, and N. Li, ‘MicroRNA-20b Downregulates HIF-1α and Inhibits the Proliferation and Invasion of Osteosarcoma Cells’, Oncol. Res., vol. 23, no. 5, pp. 257–266, 2016. [CrossRef]
- C. Renassia and C. Peyssonnaux, ‘New insights into the links between hypoxia and iron homeostasis’, Curr. Opin. Hematol., vol. 26, no. 3, pp. 125–130, May 2019. [CrossRef]
- A. M. Czarnecka et al., ‘Molecular Biology of Osteosarcoma’, Cancers, vol. 12, no. 8, p. 2130, July 2020. [CrossRef]
- S. Pollino et al., ‘CXCR4 in human osteosarcoma malignant progression. The response of osteosarcoma cell lines to the fully human CXCR4 antibody MDX1338’, J. Bone Oncol., vol. 17, p. 100239, Aug. 2019. [CrossRef]
- Y. Zhang, X. Feng, J. Zhang, M. Chen, E. Huang, and X. Chen, ‘Iron regulatory protein 2 is a suppressor of mutant p53 in tumorigenesis’, Oncogene, vol. 38, no. 35, pp. 6256–6269, Aug. 2019. [CrossRef]
- F. Zhang, W. Wang, Y. Tsuji, S. V. Torti, and F. M. Torti, ‘Post-transcriptional modulation of iron homeostasis during p53-dependent growth arrest’, J. Biol. Chem., vol. 283, no. 49, pp. 33911–33918, Dec. 2008. [CrossRef]
- G.-H. Yu, L. Fu, J. Chen, F. Wei, and W.-X. Shi, ‘Decreased expression of ferritin light chain in osteosarcoma and its correlation with epithelial-mesenchymal transition’, Eur. Rev. Med. Pharmacol. Sci., vol. 22, no. 9, pp. 2580–2587, May 2018. [CrossRef]
- Z. Chen, W. Wang, S. R. Abdul Razak, T. Han, N. H. Ahmad, and X. Li, ‘Ferroptosis as a potential target for cancer therapy’, Cell Death Dis., vol. 14, no. 7, p. 460, July 2023. [CrossRef]
- R. Ding et al., ‘Downregulation of ferroptosis-related Genes can regulate the invasion and migration of osteosarcoma cells’, Sci. Rep., vol. 15, no. 1, p. 17582, May 2025. [CrossRef]
- X. Ma, J. Zhao, and H. Feng, ‘Targeting iron metabolism in osteosarcoma’, Discov. Oncol., vol. 14, no. 1, p. 31, Mar. 2023. [CrossRef]
- S. Tortorella and T. C. Karagiannis, ‘Transferrin Receptor-Mediated Endocytosis: A Useful Target for Cancer Therapy’, J. Membr. Biol., vol. 247, no. 4, pp. 291–307, Apr. 2014. [CrossRef]
- X. Cheng et al., ‘TfR1 binding with H-ferritin nanocarrier achieves prognostic diagnosis and enhances the therapeutic efficacy in clinical gastric cancer’, Cell Death Dis., vol. 11, no. 2, p. 92, Feb. 2020. [CrossRef]
- B. Chiou and J. R. Connor, ‘Emerging and Dynamic Biomedical Uses of Ferritin’, Pharm. Basel Switz., vol. 11, no. 4, p. 124, Nov. 2018. [CrossRef]
- T. R. Daniels et al., ‘The transferrin receptor and the targeted delivery of therapeutic agents against cancer’, Biochim. Biophys. Acta, vol. 1820, no. 3, pp. 291–317, Mar. 2012. [CrossRef]
- M. Songbo, H. Lang, C. Xinyong, X. Bin, Z. Ping, and S. Liang, ‘Oxidative stress injury in doxorubicin-induced cardiotoxicity’, Toxicol. Lett., vol. 307, pp. 41–48, June 2019. [CrossRef]
- J. C. Almagro, T. R. Daniels-Wells, S. M. Perez-Tapia, and M. L. Penichet, ‘Progress and Challenges in the Design and Clinical Development of Antibodies for Cancer Therapy’, Front. Immunol., vol. 8, p. 1751, 2017. [CrossRef]
- P. P. Ng et al., ‘Molecular events contributing to cell death in malignant human hematopoietic cells elicited by an IgG3-avidin fusion protein targeting the transferrin receptor’, Blood, vol. 108, no. 8, pp. 2745–2754, Oct. 2006. [CrossRef]
- T. R. Daniels et al., ‘An Antibody-based Multifaceted Approach Targeting the Human Transferrin Receptor for the Treatment of B-cell Malignancies’, J. Immunother., vol. 34, no. 6, pp. 500–508, July 2011. [CrossRef]
- T. R. Daniels-Wells, D. P. Widney, L. S. Leoh, O. Martínez-Maza, and M. L. Penichet, ‘Efficacy of an Anti-transferrin Receptor 1 Antibody Against AIDS-related Non-Hodgkin Lymphoma: A Brief Communication’, J. Immunother., vol. 38, no. 8, pp. 307–310, Oct. 2015. [CrossRef]
- M. Neiveyans et al., ‘A recycling anti-transferrin receptor-1 monoclonal antibody as an efficient therapy for erythroleukemia through target up-regulation and antibody-dependent cytotoxic effector functions’, mAbs, vol. 11, no. 3, pp. 593–605, Apr. 2019. [CrossRef]
- M. Nakase, M. Inui, K. Okumura, T. Kamei, S. Nakamura, and T. Tagawa, ‘p53 gene therapy of human osteosarcoma using a transferrin-modified cationic liposome’, Mol. Cancer Ther., vol. 4, no. 4, pp. 625–631, Apr. 2005. [CrossRef]
- M. R. Bedford, S. J. Ford, R. D. Horniblow, T. H. Iqbal, and C. Tselepis, ‘Iron chelation in the treatment of cancer: a new role for deferasirox?’, J. Clin. Pharmacol., vol. 53, no. 9, pp. 885–891, Sept. 2013. [CrossRef]
- S. Tury et al., ‘The iron chelator deferasirox synergises with chemotherapy to treat triple-negative breast cancers’, J. Pathol., vol. 246, no. 1, pp. 103–114, Sept. 2018. [CrossRef]
- J.-H. Moon, J.-K. Jeong, and S.-Y. Park, ‘Deferoxamine inhibits TRAIL-mediated apoptosis via regulation of autophagy in human colon cancer cells’, Oncol. Rep., vol. 33, no. 3, pp. 1171–1176, Mar. 2015. [CrossRef]
- J. L. Kim et al., ‘Iron chelator-induced apoptosis via the ER stress pathway in gastric cancer cells’, Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med., vol. 37, no. 7, pp. 9709–9719, July 2016. [CrossRef]
- P. Li et al., ‘The iron chelator Dp44mT suppresses osteosarcoma’s proliferation, invasion and migration: in vitro and in vivo’, Am. J. Transl. Res., vol. 8, no. 12, pp. 5370–5385, 2016.
- M. Argenziano et al., ‘Effects of Iron Chelation in Osteosarcoma’, Curr. Cancer Drug Targets, vol. 21, no. 5, pp. 443–455, 2021. [CrossRef]
- S. J. Dixon et al., ‘Ferroptosis: an iron-dependent form of nonapoptotic cell death’, Cell, vol. 149, no. 5, pp. 1060–1072, May 2012. [CrossRef]
- G. Lei, L. Zhuang, and B. Gan, ‘Targeting ferroptosis as a vulnerability in cancer’, Nat. Rev. Cancer, vol. 22, no. 7, pp. 381–396, July 2022. [CrossRef]
- F. Peng et al., ‘Regulated cell death (RCD) in cancer: key pathways and targeted therapies’, Signal Transduct. Target. Ther., vol. 7, no. 1, p. 286, Aug. 2022. [CrossRef]
- C. Zhang, X. Liu, S. Jin, Y. Chen, and R. Guo, ‘Ferroptosis in cancer therapy: a novel approach to reversing drug resistance’, Mol. Cancer, vol. 21, no. 1, p. 47, Feb. 2022. [CrossRef]
- E. Dai et al., ‘A guideline on the molecular ecosystem regulating ferroptosis’, Nat. Cell Biol., vol. 26, no. 9, pp. 1447–1457, Sept. 2024. [CrossRef]
- D. Coradduzza, A. Congiargiu, Z. Chen, A. Zinellu, C. Carru, and S. Medici, ‘Ferroptosis and Senescence: A Systematic Review’, Int. J. Mol. Sci., vol. 24, no. 4, p. 3658, Feb. 2023. [CrossRef]
- D. Wang et al., ‘Regulatory pathways and drugs associated with ferroptosis in tumors’, Cell Death Dis., vol. 13, no. 6, p. 544, June 2022. [CrossRef]
- J. Van Loenhout, M. Peeters, A. Bogaerts, E. Smits, and C. Deben, ‘Oxidative Stress-Inducing Anticancer Therapies: Taking a Closer Look at Their Immunomodulating Effects’, Antioxid. Basel Switz., vol. 9, no. 12, p. 1188, Nov. 2020. [CrossRef]
- Z. Yin, G. Shen, M. Fan, and P. Zheng, ‘Lipid metabolic reprogramming and associated ferroptosis in osteosarcoma: From molecular mechanisms to potential targets’, J. Bone Oncol., vol. 51, p. 100660, Apr. 2025. [CrossRef]
- G. Fanelli, G. Alloisio, V. Lelli, S. Marini, S. Rinalducci, and M. Gioia, ‘Mechano-induced cell metabolism disrupts the oxidative stress homeostasis of SAOS-2 osteosarcoma cells’, Front. Mol. Biosci., vol. 10, p. 1297826, 2023. [CrossRef]
- Z. Wang et al., ‘Ferroptosis and its implications in bone-related diseases’, PeerJ, vol. 12, p. e18626, Nov. 2024. [CrossRef]
- C. Yan et al., ‘Research progress on the role of lncRNA, circular RNA, and microRNA networks in regulating ferroptosis in osteosarcoma’, Biomed. Pharmacother., vol. 176, p. 116924, July 2024. [CrossRef]
- M. Yang et al., ‘Ferroptosis-related lncRNAs guiding osteosarcoma prognosis and immune microenvironment’, J. Orthop. Surg., vol. 18, no. 1, p. 787, Oct. 2023. [CrossRef]
- W. Huang, N. Aabed, and Y. M. Shah, ‘Reactive Oxygen Species and Ferroptosis at the Nexus of Inflammation and Colon Cancer’, Antioxid. Redox Signal., vol. 39, no. 7–9, pp. 551–568, Sept. 2023. [CrossRef]
- D. Tang, X. Chen, and G. Kroemer, ‘Cuproptosis: a copper-triggered modality of mitochondrial cell death’, Cell Res., vol. 32, no. 5, pp. 417–418, May 2022. [CrossRef]
- B. R. Stockwell and X. Jiang, ‘The Chemistry and Biology of Ferroptosis’, Cell Chem. Biol., vol. 27, no. 4, pp. 365–375, Apr. 2020. [CrossRef]
- F. Ursini and M. Maiorino, ‘Lipid peroxidation and ferroptosis: The role of GSH and GPx4’, Free Radic. Biol. Med., vol. 152, pp. 175–185, May 2020. [CrossRef]
- Y. Xie, R. Kang, D. J. Klionsky, and D. Tang, ‘GPX4 in cell death, autophagy, and disease’, Autophagy, vol. 19, no. 10, pp. 2621–2638, Oct. 2023. [CrossRef]
- S. J. Dixon et al., ‘Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis’, eLife, vol. 3, p. e02523, May 2014. [CrossRef]
- M.-R. Liu, W.-T. Zhu, and D.-S. Pei, ‘System Xc-: a key regulatory target of ferroptosis in cancer’, Invest. New Drugs, vol. 39, no. 4, pp. 1123–1131, Aug. 2021. [CrossRef]
- P. Liu et al., ‘Ferroptosis: A New Regulatory Mechanism in Osteoporosis’, Oxid. Med. Cell. Longev., vol. 2022, p. 2634431, 2022. [CrossRef]
- A. Anandhan, M. Dodson, C. J. Schmidlin, P. Liu, and D. D. Zhang, ‘Breakdown of an Ironclad Defense System: The Critical Role of NRF2 in Mediating Ferroptosis’, Cell Chem. Biol., vol. 27, no. 4, pp. 436–447, Apr. 2020. [CrossRef]
- I. Bellezza, I. Giambanco, A. Minelli, and R. Donato, ‘Nrf2-Keap1 signaling in oxidative and reductive stress’, Biochim. Biophys. Acta Mol. Cell Res., vol. 1865, no. 5, pp. 721–733, May 2018. [CrossRef]
- Q. Liu and K. Wang, ‘The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin’, Cell Biol. Int., vol. 43, no. 11, pp. 1245–1256, Nov. 2019. [CrossRef]
- D. Li and Y. Li, ‘The interaction between ferroptosis and lipid metabolism in cancer’, Signal Transduct. Target. Ther., vol. 5, no. 1, p. 108, June 2020. [CrossRef]
- X. Lang et al., ‘Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11’, Cancer Discov., vol. 9, no. 12, pp. 1673–1685, Dec. 2019. [CrossRef]
- Y. Yang et al., ‘ACSL3 and ACSL4, Distinct Roles in Ferroptosis and Cancers’, Cancers, vol. 14, no. 23, p. 5896, Nov. 2022. [CrossRef]
- F. Kuang, J. Liu, D. Tang, and R. Kang, ‘Oxidative Damage and Antioxidant Defense in Ferroptosis’, Front. Cell Dev. Biol., vol. 8, p. 586578, 2020. [CrossRef]
- D. Lin, M. Zhang, C. Luo, P. Wei, K. Cui, and Z. Chen, ‘Targeting Ferroptosis Attenuates Inflammation, Fibrosis, and Mast Cell Activation in Chronic Prostatitis’, J. Immunol. Res., vol. 2022, p. 6833867, 2022. [CrossRef]
- W. Zhang et al., ‘SMURF2 predisposes cancer cell toward ferroptosis in GPX4-independent manners by promoting GSTP1 degradation’, Mol. Cell, vol. 83, no. 23, pp. 4352-4369.e8, Dec. 2023. [CrossRef]
- Y. Zhu et al., ‘Glutathione S-transferase-Pi 1 protects cells from irradiation-induced death by inhibiting ferroptosis in pancreatic cancer’, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol., vol. 38, no. 17, p. e70033, Sept. 2024. [CrossRef]
- F. Pu, F. Chen, S. Chen, B. Wang, J. Liu, and Z. Shao, ‘Association between GSTP1 polymorphisms and prognosis of osteosarcoma in patients treated with chemotherapy: a meta-analysis’, OncoTargets Ther., vol. 8, pp. 1835–1842, 2015. [CrossRef]
- Y. D. Smirnova et al., ‘Osteosarcoma Cells and Undifferentiated Human Mesenchymal Stromal Cells Are More Susceptible to Ferroptosis than Differentiated Human Mesenchymal Stromal Cells’, Antioxidants, vol. 14, no. 2, p. 189, Feb. 2025. [CrossRef]
- Y. Yang et al., ‘Mitochondria and Mitochondrial ROS in Cancer: Novel Targets for Anticancer Therapy’, J. Cell. Physiol., vol. 231, no. 12, pp. 2570–2581, Dec. 2016. [CrossRef]
- P. Koppula et al., ‘A targetable CoQ-FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers’, Nat. Commun., vol. 13, no. 1, p. 2206, Apr. 2022. [CrossRef]
- E. Panczyszyn et al., ‘FSP1 is a predictive biomarker of osteosarcoma cells’ susceptibility to ferroptotic cell death and a potential therapeutic target’, Cell Death Discov., vol. 10, no. 1, p. 87, Feb. 2024. [CrossRef]
- X. Chen, P. B. Comish, D. Tang, and R. Kang, ‘Characteristics and Biomarkers of Ferroptosis’, Front. Cell Dev. Biol., vol. 9, p. 637162, 2021. [CrossRef]
- D. Tang, X. Chen, R. Kang, and G. Kroemer, ‘Ferroptosis: molecular mechanisms and health implications’, Cell Res., vol. 31, no. 2, pp. 107–125, Feb. 2021. [CrossRef]
- C. Qiu, T. Liu, D. Luo, D. Luan, L. Cheng, and S. Wang, ‘Novel Therapeutic Savior for Osteosarcoma: The Endorsement of Ferroptosis’, Front. Oncol., vol. 12, p. 746030, 2022. [CrossRef]
- G. Isani et al., ‘Cytotoxic Effects of Artemisia annua L. and Pure Artemisinin on the D-17 Canine Osteosarcoma Cell Line’, Oxid. Med. Cell. Longev., vol. 2019, p. 1615758, 2019. [CrossRef]
- A. C. Guanizo, C. D. Fernando, D. J. Garama, and D. J. Gough, ‘STAT3: a multifaceted oncoprotein’, Growth Factors Chur Switz., vol. 36, no. 1–2, pp. 1–14, Apr. 2018.
- Y. Liu et al., ‘STAT3 and its targeting inhibitors in osteosarcoma’, Cell Prolif., vol. 54, no. 2, p. e12974, Feb. 2021. [CrossRef]
- D. Shin, E. H. Kim, J. Lee, and J.-L. Roh, ‘Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer’, Free Radic. Biol. Med., vol. 129, pp. 454–462, Dec. 2018. [CrossRef]
- J. Fu et al., ‘Activatable nanomedicine for overcoming hypoxia-induced resistance to chemotherapy and inhibiting tumor growth by inducing collaborative apoptosis and ferroptosis in solid tumors’, Biomaterials, vol. 268, p. 120537, Jan. 2021. [CrossRef]
- H.-H. Lv, C.-X. Zhen, J.-Y. Liu, and P. Shang, ‘PEITC triggers multiple forms of cell death by GSH-iron-ROS regulation in K7M2 murine osteosarcoma cells’, Acta Pharmacol. Sin., vol. 41, no. 8, pp. 1119–1132, Aug. 2020. [CrossRef]
- M. Moghimi et al., ‘Association of GSTM1, GSTT1, GSTM3, and GSTP1 Genes Polymorphisms with Susceptibility to Osteosarcoma: a Case- Control Study and Meta-Analysis’, Asian Pac. J. Cancer Prev. APJCP, vol. 20, no. 3, pp. 675–682, Mar. 2019. [CrossRef]
- G. Zhu, Y. Xia, Z. Zhao, A. Li, H. Li, and T. Xiao, ‘LncRNA XIST from the bone marrow mesenchymal stem cell derived exosome promotes osteosarcoma growth and metastasis through miR-655/ACLY signal’, Cancer Cell Int., vol. 22, no. 1, p. 330, Oct. 2022. [CrossRef]
- Z. Wu, T. Han, H. Su, J. Xuan, and X. Wang, ‘Comprehensive analysis of fatty acid and lactate metabolism-related genes for prognosis value, immune infiltration, and therapy in osteosarcoma patients’, Front. Oncol., vol. 12, p. 934080, 2022. [CrossRef]
- J. Nie et al., ‘Identification and experimental validation of Stearoyl-CoA desaturase is a new drug therapeutic target for osteosarcoma’, Eur. J. Pharmacol., vol. 963, p. 176249, Jan. 2024. [CrossRef]
- X. Du, Y. Ou, M. Zhang, K. Li, W. Huang, and D. Jiang, ‘The mevalonate pathway promotes the metastasis of osteosarcoma by regulating YAP1 activity via RhoA’, Genes Dis., vol. 9, no. 3, pp. 741–752, May 2022. [CrossRef]
- D. Fu et al., ‘iTRAQ-based proteomic analysis of the molecular mechanisms and downstream effects of fatty acid synthase in osteosarcoma cells’, J. Clin. Lab. Anal., vol. 35, no. 3, p. e23653, Mar. 2021. [CrossRef]
- H. Jin, Y. Ji, Y. Cui, L. Xu, H. Liu, and J. Wang, ‘Simvastatin-Incorporated Drug Delivery Systems for Bone Regeneration’, ACS Biomater. Sci. Eng., vol. 7, no. 6, pp. 2177–2191, June 2021. [CrossRef]
- X. Liao, R. Wei, J. Zhou, K. Wu, and J. Li, ‘Emerging roles of long non-coding RNAs in osteosarcoma’, Front. Mol. Biosci., vol. 11, p. 1327459, 2024. [CrossRef]
- M. Chen, Y. Jiang, and Y. Sun, ‘KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma’, Biochem. Biophys. Res. Commun., vol. 550, pp. 77–83, Apr. 2021. [CrossRef]
- E. Koren and Y. Fuchs, ‘Modes of Regulated Cell Death in Cancer’, Cancer Discov., vol. 11, no. 2, pp. 245–265, Feb. 2021. [CrossRef]
- F. Zhang et al., ‘Oridonin-induced ferroptosis and apoptosis: a dual approach to suppress the growth of osteosarcoma cells’, BMC Cancer, vol. 24, no. 1, p. 198, Feb. 2024. [CrossRef]
- A. Cimmino, M. Gioia, M. E. Clementi, I. Faraoni, S. Marini, and C. Ciaccio, ‘Polydatin-Induced Shift of Redox Balance and Its Anti-Cancer Impact on Human Osteosarcoma Cells’, Curr. Issues Mol. Biol., vol. 47, no. 1, p. 21, Dec. 2024. [CrossRef]
- M. Torrens-Mas and P. Roca, ‘Phytoestrogens for Cancer Prevention and Treatment’, Biology, vol. 9, no. 12, p. 427, Nov. 2020. [CrossRef]
- A. Cimmino, G. F. Fasciglione, M. Gioia, S. Marini, and C. Ciaccio, ‘Multi-Anticancer Activities of Phytoestrogens in Human Osteosarcoma’, Int. J. Mol. Sci., vol. 24, no. 17, p. 13344, Aug. 2023. [CrossRef]
- H. Lin et al., ‘EF24 induces ferroptosis in osteosarcoma cells through HMOX1’, Biomed. Pharmacother., vol. 136, p. 111202, Apr. 2021. [CrossRef]
- Y. Luo, X. Gao, L. Zou, M. Lei, J. Feng, and Z. Hu, ‘Bavachin Induces Ferroptosis through the STAT3/P53/SLC7A11 Axis in Osteosarcoma Cells’, Oxid. Med. Cell. Longev., vol. 2021, p. 1783485, 2021. [CrossRef]
- R. Wen et al., ‘Baicalin induces ferroptosis in osteosarcomas through a novel Nrf2/xCT/GPX4 regulatory axis’, Phytomedicine, vol. 116, p. 154881, July 2023. [CrossRef]
- P. Chen, H. Wang, F. Yang, H. Chen, W. He, and J. Wang, ‘Curcumin Promotes Osteosarcoma Cell Death by Activating miR-125a/ERRα Signal Pathway’, J. Cell. Biochem., vol. 118, no. 1, pp. 74–81, Jan. 2017. [CrossRef]
- M. Zhang, J. Zhang, J. Chen, Y. Zeng, Z. Zhu, and Y. Wan, ‘Fabrication of Curcumin-Modified TiO2 Nanoarrays via Cyclodextrin Based Polymer Functional Coatings for Osteosarcoma Therapy’, Adv. Healthc. Mater., vol. 8, no. 23, p. e1901031, Dec. 2019. [CrossRef]
- C. Yuan, R. Fan, K. Zhu, Y. Wang, W. Xie, and Y. Liang, ‘Curcumin induces ferroptosis and apoptosis in osteosarcoma cells by regulating Nrf2/GPX4 signaling pathway’, Exp. Biol. Med. Maywood NJ, vol. 248, no. 23, pp. 2183–2197, Dec. 2023. [CrossRef]
- Y. He, W. Li, G. Hu, H. Sun, and Q. Kong, ‘Bioactivities of EF24, a Novel Curcumin Analog: A Review’, Front. Oncol., vol. 8, p. 614, 2018. [CrossRef]
- J. Wu et al., ‘Design, synthesis, and evaluation of asymmetric EF24 analogues as potential anti-cancer agents for lung cancer’, Eur. J. Med. Chem., vol. 125, pp. 1321–1331, Jan. 2017. [CrossRef]
- Z. Liu et al., ‘Gambogenic acid induces cell death in human osteosarcoma through altering iron metabolism, disturbing the redox balance, and activating the P53 signaling pathway’, Chem. Biol. Interact., vol. 382, p. 110602, Sept. 2023. [CrossRef]
- Z. Tang et al., ‘The Synergistic Reducing Drug Resistance Effect of Cisplatin and Ursolic Acid on Osteosarcoma through a Multistep Mechanism Involving Ferritinophagy’, Oxid. Med. Cell. Longev., vol. 2021, p. 5192271, 2021. [CrossRef]
- C. Lu, Z. Zhang, Y. Fan, X. Wang, J. Qian, and Z. Bian, ‘Shikonin induces ferroptosis in osteosarcomas through the mitochondrial ROS-regulated HIF-1α/HO-1 axis’, Phytomedicine, vol. 135, p. 156139, Dec. 2024. [CrossRef]
- R. Huang, D. Chu, J. Shi, R. Xu, and K. Wang, ‘Shikonin suppresses proliferation of osteosarcoma cells by inducing ferroptosis through promoting Nrf2 ubiquitination and inhibiting the xCT/GPX4 regulatory axis’, Front. Pharmacol., vol. 15, p. 1490759, Dec. 2024. [CrossRef]
- Z. Ma et al., ‘Curculigoside exhibits multiple therapeutic efficacy to induce apoptosis and ferroptosis in osteosarcoma via modulation of ROS and tumor microenvironment’, Tissue Cell, vol. 93, p. 102745, Apr. 2025. [CrossRef]
- Y. Shi et al., ‘Tirapazamine suppress osteosarcoma cells in part through SLC7A11 mediated ferroptosis’, Biochem. Biophys. Res. Commun., vol. 567, pp. 118–124, Aug. 2021. [CrossRef]
- J. Nie et al., ‘Butyrate enhances erastin-induced ferroptosis of osteosarcoma cells via regulating ATF3/SLC7A11 pathway’, Eur. J. Pharmacol., vol. 957, p. 176009, Oct. 2023. [CrossRef]
- H. Jiacong, Y. Qirui, L. Haonan, S. Yichang, C. Yan, and C. Keng, ‘Zoledronic acid induces ferroptosis by upregulating POR in osteosarcoma’, Med. Oncol. Northwood Lond. Engl., vol. 40, no. 5, p. 141, Apr. 2023. [CrossRef]
- T. Ren et al., ‘Zoledronic acid induces ferroptosis by reducing ubiquinone and promoting HMOX1 expression in osteosarcoma cells’, Front. Pharmacol., vol. 13, p. 1071946, 2022. [CrossRef]
- D. L. Palliyaguru, J.-M. Yuan, T. W. Kensler, and J. W. Fahey, ‘Isothiocyanates: Translating the Power of Plants to People’, Mol. Nutr. Food Res., vol. 62, no. 18, p. e1700965, Sept. 2018. [CrossRef]
- T. Kasukabe, Y. Honma, J. Okabe-Kado, Y. Higuchi, N. Kato, and S. Kumakura, ‘Combined treatment with cotylenin A and phenethyl isothiocyanate induces strong antitumor activity mainly through the induction of ferroptotic cell death in human pancreatic cancer cells’, Oncol. Rep., vol. 36, no. 2, pp. 968–976, Aug. 2016. [CrossRef]
- H. Lv, C. Zhen, J. Liu, and P. Shang, ‘β -Phenethyl Isothiocyanate Induces Cell Death in Human Osteosarcoma through Altering Iron Metabolism, Disturbing the Redox Balance, and Activating the MAPK Signaling Pathway’, Oxid. Med. Cell. Longev., vol. 2020, pp. 1–23, Apr. 2020. [CrossRef]
- A. Bhaw-Luximon and D. Jhurry, ‘Artemisinin and its derivatives in cancer therapy: status of progress, mechanism of action, and future perspectives’, Cancer Chemother. Pharmacol., vol. 79, no. 3, pp. 451–466, Mar. 2017. [CrossRef]
- R. Salaroli et al., ‘Anticancer activity of an Artemisia annua L. hydroalcoholic extract on canine osteosarcoma cell lines’, Res. Vet. Sci., vol. 152, pp. 476–484, Dec. 2022. [CrossRef]
- Z. Li, X. Ding, H. Wu, and C. Liu, ‘Artemisinin inhibits angiogenesis by regulating p38 MAPK/CREB/TSP-1 signaling pathway in osteosarcoma’, J. Cell. Biochem., vol. 120, no. 7, pp. 11462–11470, July 2019. [CrossRef]
- C. Tang et al., ‘Effect and mechanism of dihydroartemisinin on proliferation, metastasis and apoptosis of human osteosarcoma cells’, J. Biol. Regul. Homeost. Agents, vol. 29, no. 4, pp. 881–887, 2015.
- Y. Zhang et al., ‘Chemotherapeutic drugs induce oxidative stress associated with DNA repair and metabolism modulation’, Life Sci., vol. 289, p. 120242, Jan. 2022. [CrossRef]
- X. Huang et al., ‘Combined Cancer Chemo-Photodynamic and Photothermal Therapy Based on ICG/PDA/TPZ-Loaded Nanoparticles’, Mol. Pharm., vol. 16, no. 5, pp. 2172–2183, May 2019. [CrossRef]
- S. B. Reddy and S. K. Williamson, ‘Tirapazamine: a novel agent targeting hypoxic tumor cells’, Expert Opin. Investig. Drugs, vol. 18, no. 1, pp. 77–87, Jan. 2009. [CrossRef]
- H. Liu, W. Jiang, Q. Wang, L. Hang, Y. Wang, and Y. Wang, ‘ROS-sensitive biomimetic nanocarriers modulate tumor hypoxia for synergistic photodynamic chemotherapy’, Biomater. Sci., vol. 7, no. 9, pp. 3706–3716, Aug. 2019. [CrossRef]
- Y. Sun et al., ‘Recent progress of hypoxia-modulated multifunctional nanomedicines to enhance photodynamic therapy: opportunities, challenges, and future development’, Acta Pharm. Sin. B, vol. 10, no. 8, pp. 1382–1396, Aug. 2020. [CrossRef]
- G. Chen et al., ‘Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model’, EBioMedicine, vol. 30, pp. 317–325, Apr. 2018. [CrossRef]
- R. Medzhitov and T. Horng, ‘Transcriptional control of the inflammatory response’, Nat. Rev. Immunol., vol. 9, no. 10, pp. 692–703, Oct. 2009. [CrossRef]
- H. T. Hatoum, S.-J. Lin, M. R. Smith, V. Barghout, and A. Lipton, ‘Zoledronic acid and skeletal complications in patients with solid tumors and bone metastases: analysis of a national medical claims database’, Cancer, vol. 113, no. 6, pp. 1438–1445, Sept. 2008. [CrossRef]
- N. Kohno et al., ‘Zoledronic acid significantly reduces skeletal complications compared with placebo in Japanese women with bone metastases from breast cancer: a randomized, placebo-controlled trial’, J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol., vol. 23, no. 15, pp. 3314–3321, May 2005. [CrossRef]
- A. Christou, N. Ferreira, and A. Sophocleous, ‘Effects of zoledronic acid on osteosarcoma progression and metastasis: systematic review and meta-analysis’, Clin. Exp. Med., vol. 23, no. 7, pp. 3041–3051, Nov. 2023. [CrossRef]
- E. H. Kim et al., ‘Carbon-Ion Beam Irradiation Alone or in Combination with Zoledronic acid Effectively Kills Osteosarcoma Cells’, Cancers, vol. 12, no. 3, p. 698, Mar. 2020. [CrossRef]
- H. Huang et al., ‘Employing machine learning using ferroptosis-related genes to construct a prognosis model for patients with osteosarcoma’, Front. Genet., vol. 14, p. 1099272, 2023. [CrossRef]
- N. Haider et al., ‘Nanomedicines in Diagnosis and Treatment of Cancer: An Update’, Curr. Pharm. Des., vol. 26, no. 11, pp. 1216–1231, 2020. [CrossRef]
- M. Pereira-Silva, C. Alvarez-Lorenzo, A. Concheiro, A. C. Santos, F. Veiga, and A. Figueiras, ‘Nanomedicine in osteosarcoma therapy: Micelleplexes for delivery of nucleic acids and drugs toward osteosarcoma-targeted therapies’, Eur. J. Pharm. Biopharm. Off. J. Arbeitsgemeinschaft Pharm. Verfahrenstechnik EV, vol. 148, pp. 88–106, Mar. 2020. [CrossRef]
- T. Xu et al., ‘A novel nanomedicine for osteosarcoma treatment: triggering ferroptosis through GSH depletion and inhibition for enhanced synergistic PDT/PTT therapy’, J. Nanobiotechnology, vol. 23, no. 1, p. 323, Apr. 2025. [CrossRef]
- J. Zheng, Z. Zuo, T. Zhang, Y. Wang, H. Liu, and W. Li, ‘Tumor Cell Membrane-Encapsulated Nanoparticles Composed of FeS2 and Radical-Initiator Nanoparticles for Osteosarcoma Cell Death’, ACS Appl. Nano Mater., vol. 8, no. 19, pp. 10138–10149, May 2025. [CrossRef]
- D. Wang et al., ‘Multiregion Sequencing Reveals the Genetic Heterogeneity and Evolutionary History of Osteosarcoma and Matched Pulmonary Metastases’, Cancer Res., vol. 79, no. 1, pp. 7–20, Jan. 2019. [CrossRef]
- W. Yu et al., ‘Autophagy inhibitor enhance ZnPc/BSA nanoparticle induced photodynamic therapy by suppressing PD-L1 expression in osteosarcoma immunotherapy’, Biomaterials, vol. 192, pp. 128–139, Feb. 2019. [CrossRef]
- F. He et al., ‘Enhanced up/down-conversion luminescence and heat: Simultaneously achieving in one single core-shell structure for multimodal imaging guided therapy’, Biomaterials, vol. 105, pp. 77–88, Oct. 2016. [CrossRef]
- Y. Wang et al., ‘Capsaicin Enhanced the Efficacy of Photodynamic Therapy Against Osteosarcoma via a Pro-Death Strategy by Inducing Ferroptosis and Alleviating Hypoxia’, Small, vol. 20, no. 26, p. 2306916, 2024. [CrossRef]
- A. Buglione et al., ‘A “Spicy” Mechanotransduction Switch: Capsaicin-Activated TRPV1 Receptor Modulates Osteosarcoma Cell Behavior and Drug Sensitivity’, Aug. 14, 2025, Biology and Life Sciences. [CrossRef]
- L. Lei et al., ‘Lonidamine liposomes to enhance photodynamic and photothermal therapy of hepatocellular carcinoma by inhibiting glycolysis’, J. Nanobiotechnology, vol. 21, no. 1, p. 482, Dec. 2023. [CrossRef]
- M. Li et al., ‘High-performance pyrite nano-catalyst driven photothermal/chemodynamic synergistic therapy for Osteosarcoma’, J. Nanobiotechnology, vol. 22, no. 1, p. 141, Apr. 2024. [CrossRef]
- Y. Zhang et al., ‘Iron-Based Nanovehicle Delivering Fin56 for Hyperthermia-Boosted Ferroptosis Therapy Against Osteosarcoma’, Int. J. Nanomedicine, vol. 19, pp. 91–107, 2024. [CrossRef]
- Z. Lin, Y. Li, Z. Wu, Q. Liu, X. Li, and W. Luo, ‘Eriodictyol-cisplatin coated nanomedicine synergistically promote osteosarcoma cells ferroptosis and chemosensitivity’, J. Nanobiotechnology, vol. 23, no. 1, p. 109, Feb. 2025. [CrossRef]
- K. Shimada et al., ‘Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis’, Nat. Chem. Biol., vol. 12, no. 7, pp. 497–503, July 2016. [CrossRef]
- S. Debnath et al., ‘Eriodictyol mediated selective targeting of the TNFR1/FADD/TRADD axis in cancer cells induce apoptosis and inhibit tumor progression and metastasis’, Transl. Oncol., vol. 21, p. 101433, July 2022. [CrossRef]
- H. Huang, Y. He, Y. Li, M. Gu, M. Wu, and L. Ji, ‘Eriodictyol suppresses the malignant progression of colorectal cancer by downregulating tissue specific transplantation antigen P35B (TSTA3) expression to restrain fucosylation’, Bioengineered, vol. 13, no. 3, pp. 5551–5563, Mar. 2022. [CrossRef]
- X. Liu, S. Du, S. Wang, and K. Ye, ‘Ferroptosis in osteosarcoma: A promising future’, Front. Oncol., vol. 12, p. 1031779, 2022. [CrossRef]
- G. Li, J. Lei, D. Xu, W. Yu, J. Bai, and G. Wu, ‘Integrative analyses of ferroptosis and immune related biomarkers and the osteosarcoma associated mechanisms’, Sci. Rep., vol. 13, no. 1, p. 5770, Apr. 2023. [CrossRef]
- L. Yang, J. Liu, and S. Liu, ‘Clinical significance and immune landscape of a novel ferroptosis-related prognosis signature in osteosarcoma’, BMC Cancer, vol. 23, no. 1, p. 229, Mar. 2023. [CrossRef]
- M. Rossi Sebastiano and G. Konstantinidou, ‘Targeting Long Chain Acyl-CoA Synthetases for Cancer Therapy’, Int. J. Mol. Sci., vol. 20, no. 15, p. 3624, July 2019. [CrossRef]
- H. Mo et al., ‘ATF4 regulated by MYC has an important function in anoikis resistance in human osteosarcoma cells’, Mol. Med. Rep., Dec. 2017. [CrossRef]
- A. Russo, A. Saide, R. Cagliani, M. Cantile, G. Botti, and G. Russo, ‘rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFκB upon 5-FU treatment’, Sci. Rep., vol. 6, no. 1, p. 38369, Dec. 2016. [CrossRef]
- N. Nishizawa et al., ‘Diagnostic potential of hypermethylation of the cysteine dioxygenase 1 gene ( CDO 1 ) promoter DNA in pancreatic cancer’, Cancer Sci., vol. 110, no. 9, pp. 2846–2855, Sept. 2019. [CrossRef]
- C. Piao et al., ‘Inhibition of stearoyl CoA desaturase-1 activity suppresses tumour progression and improves prognosis in human bladder cancer’, J. Cell. Mol. Med., vol. 23, no. 3, pp. 2064–2076, Mar. 2019. [CrossRef]
- L. H. Alfarsi et al., ‘Co-Expression Effect of SLC7A5/SLC3A2 to Predict Response to Endocrine Therapy in Oestrogen-Receptor-Positive Breast Cancer’, Int. J. Mol. Sci., vol. 21, no. 4, p. 1407, Feb. 2020. [CrossRef]
- J. Meng, H. Du, J. Lu, and H. Wang, ‘Construction and validation of a predictive nomogram for ferroptosis-related genes in osteosarcoma’, J. Cancer Res. Clin. Oncol., vol. 149, no. 15, pp. 14227–14239, Nov. 2023. [CrossRef]
- M. Si and J. Lang, ‘The roles of metallothioneins in carcinogenesis’, J. Hematol. Oncol.J Hematol Oncol, vol. 11, no. 1, p. 107, Dec. 2018. [CrossRef]
- C. Zhang, Z. Zhang, Y. Zhu, and S. Qin, ‘Glucose-6-phosphate Dehydrogenase: a Biomarker and Potential Therapeutic Target for Cancer’, Anticancer Agents Med. Chem., vol. 14, no. 2, pp. 280–289, Jan. 2014. [CrossRef]
- S. Mocellin, S. Tropea, C. Benna, and C. R. Rossi, ‘Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies’, BMC Med., vol. 16, no. 1, p. 20, Dec. 2018. [CrossRef]
- F. Ye, H. Wang, L. Zhang, Y. Zou, H. Han, and J. Huang, ‘Baicalein induces human osteosarcoma cell line MG-63 apoptosis via ROS-induced BNIP3 expression’, Tumor Biol., vol. 36, no. 6, pp. 4731–4740, June 2015. [CrossRef]
- Y. Qin et al., ‘A Novel Long Non-Coding RNA lnc030 Maintains Breast Cancer Stem Cell Stemness by Stabilizing SQLE mRNA and Increasing Cholesterol Synthesis’, Adv. Sci., vol. 8, no. 2, p. 2002232, Jan. 2021. [CrossRef]
- J. Ma, Z. Guo, X. Yang, and Y. Zhu, ‘Exploration of various roles of hypoxia genes in osteosarcoma’, Sci. Rep., vol. 12, no. 1, p. 18293, Oct. 2022. [CrossRef]
- F. J. Slack and A. M. Chinnaiyan, ‘The Role of Non-coding RNAs in Oncology’, Cell, vol. 179, no. 5, pp. 1033–1055, Nov. 2019. [CrossRef]
- S. Hong-bin, Y. Wan-jun, D. Chen-hui, Y. Xiao-jie, L. Shen-song, and Z. Peng, ‘Identification of an Iron Metabolism-Related lncRNA Signature for Predicting Osteosarcoma Survival and Immune Landscape’, Front. Genet., vol. 13, Mar. 2022. [CrossRef]
- Y. Liu, W. Ding, J. Wang, X. Ao, and J. Xue, ‘Non-coding RNAs in lung cancer: molecular mechanisms and clinical applications’, Front. Oncol., vol. 13, Sept. 2023. [CrossRef]
- S. Qin, Y. Wang, C. Ma, and Q. Lv, ‘Competitive endogenous network of circRNA, lncRNA, and miRNA in osteosarcoma chemoresistance’, Eur. J. Med. Res., vol. 28, no. 1, p. 354, Sept. 2023. [CrossRef]
- Y. Zhi, L. Gao, B. Wang, W. Ren, K. X. Liang, and K. Zhi, ‘Ferroptosis Holds Novel Promise in Treatment of Cancer Mediated by Non-coding RNAs’, Front. Cell Dev. Biol., vol. 9, June 2021. [CrossRef]
- L. Li et al., ‘LncSNHG14 promotes nutlin3a resistance by inhibiting ferroptosis via the miR-206 /SLC7A11 axis in osteosarcoma cells’, Cancer Gene Ther., vol. 30, no. 5, pp. 704–715, May 2023. [CrossRef]
- J. Shao et al., ‘A novel ferroptosis-related microRNA signature with prognostic value in osteosarcoma’, Acta Biochim. Biophys. Sin., vol. 55, no. 11, pp. 1758–1769, Nov. 2023. [CrossRef]
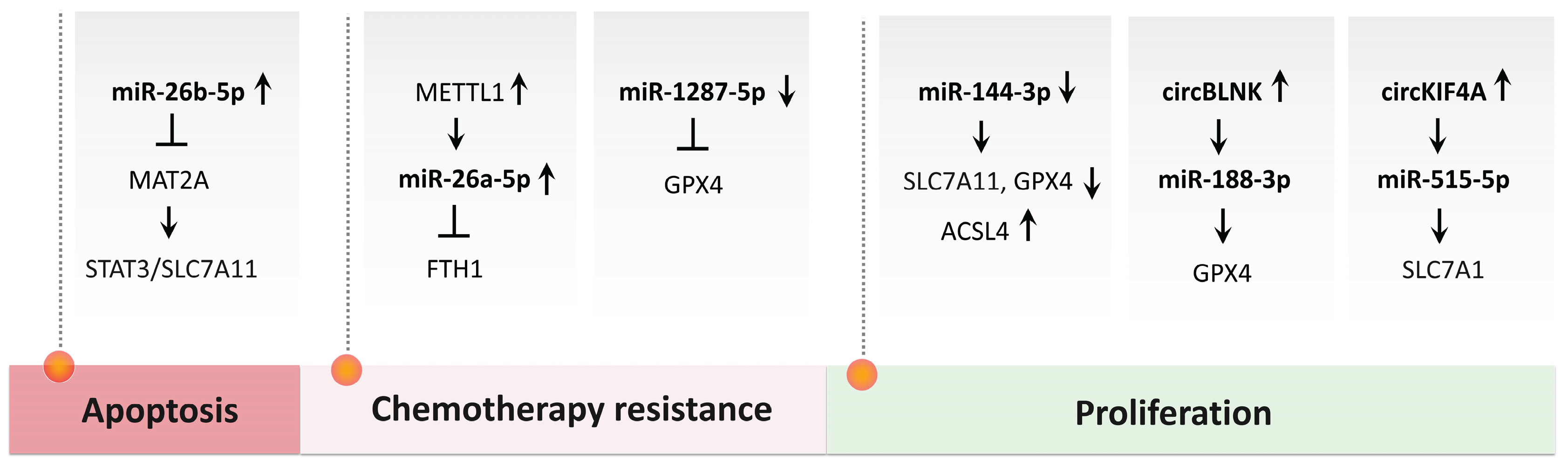
- Z. Li, Y. Luo, C. Wang, D. Han, and W. Sun, ‘Circular RNA circBLNK promotes osteosarcoma progression and inhibits ferroptosis in osteosarcoma cells by sponging miR-188-3p and regulating GPX4 expression’, Oncol. Rep., vol. 50, no. 5, p. 192, Nov. 2023. [CrossRef]
- A. Gupta, J. L. Andresen, R. S. Manan, and R. Langer, ‘Nucleic acid delivery for therapeutic applications’, Adv. Drug Deliv. Rev., vol. 178, p. 113834, Nov. 2021. [CrossRef]
- C. Wang et al., ‘Delivery of miRNAs Using Nanoparticles for the Treatment of Osteosarcoma’, Int. J. Nanomedicine, vol. 19, pp. 8641–8660, Aug. 2024. [CrossRef]
- M. Jiang et al., ‘Exosome-mediated miR-144-3p promotes ferroptosis to inhibit osteosarcoma proliferation, migration, and invasion through regulating ZEB1’, Mol. Cancer, vol. 22, no. 1, p. 113, July 2023. [CrossRef]
- S. Xia, Y. Liang, Y. Shen, W. Zhong, and Y. Ma, ‘MAT2A inhibits the ferroptosis in osteosarcoma progression regulated by miR-26b-5p’, J. Bone Oncol., vol. 41, p. 100490, Aug. 2023. [CrossRef]
- M. He et al., ‘M7G modification of FTH1 and pri-miR-26a regulates ferroptosis and chemotherapy resistance in osteosarcoma’, Oncogene, vol. 43, no. 5, pp. 341–353, Jan. 2024. [CrossRef]
- Z. Xu, L. Chen, C. Wang, L. Zhang, and W. Xu, ‘MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4’, Free Radic. Res., vol. 55, no. 11–12, pp. 1119–1129, Dec. 2021. [CrossRef]
- G. Li, J. Feng, S. Huang, and Q. Li, ‘LncRNA-PVT1 Inhibits Ferroptosis through Activating STAT3/GPX4 Axis to Promote Osteosarcoma Progression’, Front. Biosci. Landmark Ed., vol. 29, no. 6, p. 207, May 2024. [CrossRef]
- Z.-Y. Sun, Y.-K. Jian, H.-Y. Zhu, and B. Li, ‘lncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to gemcitabine through activating c-MET/PI3K/AKT pathway’, Pathol. Res. Pract., vol. 215, no. 3, pp. 555–563, Mar. 2019. [CrossRef]
- X. Zhu, G. Yang, J. Xu, and C. Zhang, ‘Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma’, Cancer Cell Int., vol. 19, no. 1, p. 82, Apr. 2019. [CrossRef]
- Y. Zhang et al., ‘Comprehensive Analysis of a Ferroptosis-Related lncRNA Signature for Predicting Prognosis and Immune Landscape in Osteosarcoma’, Front. Oncol., vol. 12, p. 880459, 2022. [CrossRef]
- H. Guan et al., ‘Long noncoding RNA APTR contributes to osteosarcoma progression through repression of miR-132-3p and upregulation of yes-associated protein 1’, J. Cell. Physiol., vol. 234, no. 6, pp. 8998–9007, June 2019. [CrossRef]
- P. He et al., ‘CircKIF4A enhances osteosarcoma proliferation and metastasis by sponging MiR-515-5p and upregulating SLC7A11’, Mol. Biol. Rep., vol. 49, no. 6, pp. 4525–4535, June 2022. [CrossRef]
- S. Huang et al., ‘LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1’, Cancer Biol. Ther., vol. 21, no. 4, pp. 379–387, Apr. 2020. [CrossRef]
- R. Li, X. Wang, C. Zhu, and K. Wang, ‘lncRNA PVT1: a novel oncogene in multiple cancers’, Cell. Mol. Biol. Lett., vol. 27, no. 1, p. 84, Oct. 2022. [CrossRef]
- K. Low, P. Foulkes, F. Hills, H. C. Roberts, and B. Stordal, ‘The efficacy of gemcitabine and docetaxel chemotherapy for the treatment of relapsed and refractory osteosarcoma: A systematic review and pre-clinical study’, Cancer Med., vol. 13, no. 18, p. e70248, Sept. 2024. [CrossRef]
- G. Li, H. Kawashima, T. Sasaki, T. Ariizumi, N. Oike, and A. Ogose, ‘Heterogeneous c-Met Activation in Osteosarcoma Dictates Synergistic Vulnerability to Combined c-Met Inhibition and Methotrexate Therapy’, Anticancer Res., vol. 45, no. 7, pp. 2791–2806, July 2025. [CrossRef]
- K. Wang, Y. Zhuang, C. Liu, and Y. Li, ‘Inhibition of c-Met activation sensitizes osteosarcoma cells to cisplatin via suppression of the PI3K–Akt signaling’, Arch. Biochem. Biophys., vol. 526, no. 1, pp. 38–43, Oct. 2012. [CrossRef]
- J. Song et al., ‘Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma’, Biochem. Biophys. Res. Commun., vol. 490, no. 2, pp. 217–224, Aug. 2017. [CrossRef]
- B. Wang, L. Fang, H. Zhao, T. Xiang, and D. Wang, ‘MDM2 inhibitor Nutlin-3a suppresses proliferation and promotes apoptosis in osteosarcoma cells’, Acta Biochim. Biophys. Sin., vol. 44, no. 8, pp. 685–691, Aug. 2012. [CrossRef]
- L. S. Kristensen, M. S. Andersen, L. V. W. Stagsted, K. K. Ebbesen, T. B. Hansen, and J. Kjems, ‘The biogenesis, biology and characterization of circular RNAs’, Nat. Rev. Genet., vol. 20, no. 11, pp. 675–691, Nov. 2019. [CrossRef]
- Y. Zhang, J. Li, Y. Wang, J. Jing, and J. Li, ‘The Roles of Circular RNAs in Osteosarcoma’, Med. Sci. Monit., vol. 25, pp. 6378–6382, Aug. 2019. [CrossRef]
- Z. Q. Li, ‘CircRNA_103801 accelerates proliferation of osteosarcoma cells by sponging miR-338-3p and regulating HIF-1/Rap1 /PI3K-Akt pathway’, J. Biol. Regul. Homeost. AGENTS, vol. 35, no. 3, June 2021. [CrossRef]
- G. Gong, Z. Han, W. Wang, Q. Xu, and J. Zhang, ‘Silencing hsa_circRNA_0008035 exerted repressive function on osteosarcoma cell growth and migration by upregulating microRNA-375’, Cell Cycle, vol. 19, no. 17, pp. 2139–2147, Sept. 2020. [CrossRef]
- X. Jin et al., ‘Ferritinophagy in the etiopathogenic mechanism of related diseases’, J. Nutr. Biochem., vol. 117, p. 109339, July 2023. [CrossRef]
- J. D. Mancias, X. Wang, S. P. Gygi, J. W. Harper, and A. C. Kimmelman, ‘Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy’, Nature, vol. 509, no. 7498, pp. 105–109, May 2014. [CrossRef]
- L. Rochette, G. Dogon, E. Rigal, M. Zeller, Y. Cottin, and C. Vergely, ‘Lipid Peroxidation and Iron Metabolism: Two Corner Stones in the Homeostasis Control of Ferroptosis’, Int. J. Mol. Sci., vol. 24, no. 1, p. 449, Dec. 2022. [CrossRef]
- Y. Le, Q. Liu, Y. Yang, and J. Wu, ‘The emerging role of nuclear receptor coactivator 4 in health and disease: a novel bridge between iron metabolism and immunity’, Cell Death Discov., vol. 10, no. 1, p. 312, July 2024. [CrossRef]
- N. Santana-Codina et al., ‘NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins’, Cancer Discov., vol. 12, no. 9, pp. 2180–2197, Sept. 2022. [CrossRef]
- K. Sun et al., ‘Ferritinophagy, a form of autophagic ferroptosis: New insights into cancer treatment’, Front. Pharmacol., vol. 13, p. 1043344, 2022. [CrossRef]
- J. Wang et al., ‘Ferritinophagy: research advance and clinical significance in cancers’, Cell Death Discov., vol. 9, no. 1, p. 463, Dec. 2023. [CrossRef]
- J. Lee and D.-H. Hyun, ‘The Interplay between Intracellular Iron Homeostasis and Neuroinflammation in Neurodegenerative Diseases’, Antioxid. Basel Switz., vol. 12, no. 4, p. 918, Apr. 2023. [CrossRef]
- N. Santana-Codina, A. Gikandi, and J. D. Mancias, ‘The Role of NCOA4-Mediated Ferritinophagy in Ferroptosis’, in Ferroptosis: Mechanism and Diseases, vol. 1301, A. F. Florez and H. Alborzinia, Eds, in Advances in Experimental Medicine and Biology, vol. 1301. , Cham: Springer International Publishing, 2021, pp. 41–57. [CrossRef]
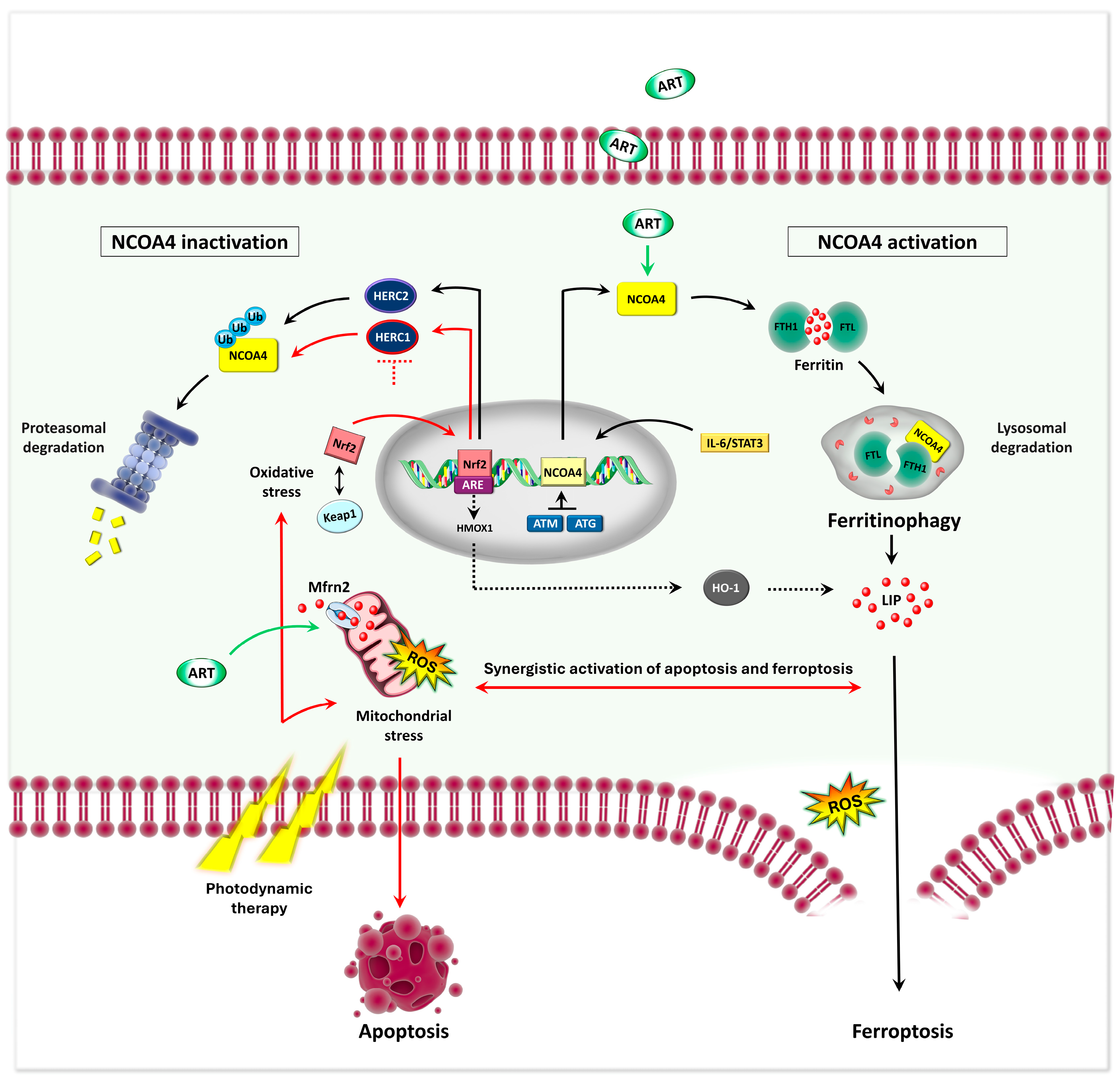
- Y. Zhang et al., ‘Synergistic induction of ferroptosis by targeting HERC1-NCOA4 axis to enhance the photodynamic sensitivity of osteosarcoma’, Redox Biol., vol. 76, p. 103328, Oct. 2024. [CrossRef]
- X. Li et al., ‘NCOA4 is Regulated by HIF and Mediates Mobilization of Murine Hepatic Iron Stores After Blood Loss’, Blood, p. blood.2020006321, July 2020. [CrossRef]
- S. Kuno, H. Fujita, Y. Tanaka, Y. Ogra, and K. Iwai, ‘Iron-induced NCOA4 condensation regulates ferritin fate and iron homeostasis’, EMBO Rep., vol. 23, no. 5, p. e54278, May 2022. [CrossRef]
- B. Galy, M. Conrad, and M. Muckenthaler, ‘Mechanisms controlling cellular and systemic iron homeostasis’, Nat. Rev. Mol. Cell Biol., vol. 25, no. 2, pp. 133–155, Feb. 2024. [CrossRef]
- Y. Fang, X. Chen, Q. Tan, H. Zhou, J. Xu, and Q. Gu, ‘Inhibiting Ferroptosis through Disrupting the NCOA4–FTH1 Interaction: A New Mechanism of Action’, ACS Cent. Sci., vol. 7, no. 6, pp. 980–989, June 2021. [CrossRef]
- J. Kaur and J. Debnath, ‘Autophagy at the crossroads of catabolism and anabolism’, Nat. Rev. Mol. Cell Biol., vol. 16, no. 8, pp. 461–472, Aug. 2015. [CrossRef]
- J. Ito et al., ‘Iron derived from autophagy-mediated ferritin degradation induces cardiomyocyte death and heart failure in mice’, eLife, vol. 10, p. e62174, Feb. 2021. [CrossRef]
- W. Hou et al., ‘Autophagy promotes ferroptosis by degradation of ferritin’, Autophagy, vol. 12, no. 8, pp. 1425–1428, Aug. 2016. [CrossRef]
- H. Wu, Q. Liu, X. Shan, W. Gao, and Q. Chen, ‘ATM orchestrates ferritinophagy and ferroptosis by phosphorylating NCOA4’, Autophagy, vol. 19, no. 7, pp. 2062–2077, July 2023. [CrossRef]
- J. Wang, Q. Zhu, R. Li, J. Zhang, X. Ye, and X. Li, ‘YAP1 protects against septic liver injury via ferroptosis resistance’, Cell Biosci., vol. 12, no. 1, p. 163, Oct. 2022. [CrossRef]
- M. Zhu et al., ‘STAT3 signaling promotes cardiac injury by upregulating NCOA4-mediated ferritinophagy and ferroptosis in high-fat-diet fed mice’, Free Radic. Biol. Med., vol. 201, pp. 111–125, May 2023. [CrossRef]
- M. M. Facchinetti, ‘Heme-Oxygenase-1’, Antioxid. Redox Signal., vol. 32, no. 17, pp. 1239–1242, June 2020. [CrossRef]
- J. D. Mancias et al., ‘Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis’, eLife, vol. 4, p. e10308, Oct. 2015. [CrossRef]
- J. Li et al., ‘d-Borneol enhances cisplatin sensitivity via autophagy dependent EMT signaling and NCOA4-mediated ferritinophagy’, Phytomedicine, vol. 106, p. 154411, Nov. 2022. [CrossRef]
- A. Nai et al., ‘NCOA4-mediated ferritinophagy in macrophages is crucial to sustain erythropoiesis in mice’, Haematologica, vol. 106, no. 3, pp. 795–805, Mar. 2021. [CrossRef]
- J. Lee, J. H. You, and J.-L. Roh, ‘Poly(rC)-binding protein 1 represses ferritinophagy-mediated ferroptosis in head and neck cancer’, Redox Biol., vol. 51, p. 102276, May 2022. [CrossRef]
- H. Man et al., ‘Super-Resolution Imaging of Autophagy by a Preferred Pair of Self-Labeling Protein Tags and Fluorescent Ligands’, Anal. Chem., vol. 94, no. 43, pp. 15057–15066, Nov. 2022. [CrossRef]
- S. Masaldan et al., ‘Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis’, Redox Biol., vol. 14, pp. 100–115, Apr. 2018. [CrossRef]
- O. Marques et al., ‘Expression of iron-related proteins in feline and canine mammary gland reveals unexpected accumulation of iron’, Biotech. Histochem., vol. 92, no. 8, pp. 584–594, Nov. 2017. [CrossRef]
- S. Simpson, M. D. Dunning, S. De Brot, L. Grau-Roma, N. P. Mongan, and C. S. Rutland, ‘Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics’, Acta Vet. Scand., vol. 59, no. 1, p. 71, Dec. 2017. [CrossRef]
- Z. He et al., ‘Oxygen-boosted biomimetic nanoplatform for synergetic phototherapy/ferroptosis activation and reversal of immune-suppressed tumor microenvironment’, Biomaterials, vol. 290, p. 121832, Nov. 2022. [CrossRef]
- Z. Jiang, H. Wang, G. Qi, C. Jiang, K. Chen, and Z. Yan, ‘Iron overload-induced ferroptosis of osteoblasts inhibits osteogenesis and promotes osteoporosis: An in vitro and in vivo study’, IUBMB Life, vol. 74, no. 11, pp. 1052–1069, 2022. [CrossRef]
- Q. Ru et al., ‘Fighting age-related orthopedic diseases: focusing on ferroptosis’, Bone Res., vol. 11, no. 1, p. 12, Mar. 2023. [CrossRef]
- F. Wang, N. Naowarojna, and Y. Zou, ‘Stratifying ferroptosis sensitivity in cells and mouse tissues by photochemical activation of lipid peroxidation and fluorescent imaging’, STAR Protoc., vol. 3, no. 2, p. 101189, June 2022. [CrossRef]
| Compound | Cell line/in vivo Model | Concentrations | Combined treatment | Molecular Mechanism | Observed Effects | References |
|---|---|---|---|---|---|---|
| Bavachin | MG63, HOS | 5 - 80 μM | DFO, Fer-1, Lip-1, Vit E | ↓ SLC7A11 via STAT3 inhibition and P53 upregulation | ↑ Fe²⁺, ROS, LPO, ferroptosis | [137] |
| Baicalin | MG63, 143B/ xenograft | 60 - 120 μg/ml 200 mg/kg/day |
Fer-1 | ↓ Nrf2 → ↓ xCT GPX4 axis | ↑ ferroptosis, ↓tumor growth | [138] |
| Curcumin | MG63, MNNG/HOS/ xenograft | 22.5 μM | Lip-1, BM | ↓ Nrf2 → ↓ GPX4 | ↑ ROS, ferroptosis, apoptosis ↓tumor growth |
[141] |
| EF24 | U2OS, SaOS-2 | 0.75 - 1.5 μM | Fer-1 | ↑ HMOX1 → ↓ GPX4 | ↑ Fe²⁺, ROS, LPO ferroptosis | [136] |
| Gambogenic Acid | HOS, 143B/ xenograft | 0.25 - 8 μM 30 mg/kg/day |
Fer-1, Eastin, NAC | ↑ P53, ↓SLC7A11, GPX4 | ↓ GSH, ↑ ROS, Mito dysfunction, ferroptosis and apoptosis ↓tumor growth |
[144] |
| β-Phenethyl isothiocyanate | MNNG/HOS, U-2OS, MG-63, 143B/ xenograft | 30 μM 30 mg/kg/day |
z-VAD-FMK, Ner-1, Fer-1, Lip-1, Baf-A1, 3-MA, NAC | ↑ TfR1, ↓ FTH1/FPN/DMT1 via MAPK, ↓ GPX4 | ↑ ferroptosis, apoptosis, autophagy ↓tumor growth |
[121] |
| Artesunate | MG63, 143B/ xenograft | 5 - 100 μM/ 200 mg/kg/day |
Fer-1, DFO, 3-MA, Nec-1, NAC |
↑TFR, DMT1, NCOA4, Mfrn2 ↓GPX4, xCT |
↓ GSH, ↑ Fe²⁺, LPO ferroptosis, ferritinophagy, apoptosis ↓tumor growth |
[17] |
| Ursolic Acid | HOS, 143B/ xenograft | 35 μmol/L | DFO, CIS (20 μmol/L) | ↑TFR, ↓NCOA4, GPX4 | ↑ Fe²⁺, LPO ferroptosis, ferritinophagy, ↓ drug resistance ↓tumor growth |
[145] |
| Shikonin | MG63, HOS/ xenograft |
1 or 4 μM/ 2 mg/kg/day |
Fer-1, DFO | ↑HIF-1α/HO-1 → mito ROS | ↑ Fe²⁺, ROS, LPO, ferroptosis ↓tumor growth | [146] |
| MG63, 143B | 0.25 - 10 μM | Fer-1 | ↓ Nrf2 → ↓ xCT GPX4 axis | ↑ Fe²⁺, ROS, LPO, ferroptosis | [147] | |
| Polydatin | SAOS-2, U2OS | 25 - 200 μM | Fer-1, NAC, DOX, CIS (10 - 20 µM) | ¯ | ↑ Fe²⁺, ROS, LPO, ↓ GSH, drug resistance | [133] |
| Curculigoside | HOS, U2OS, sjsa1, 143b/ xenograft and mini-PDX | 50 and 75 μM/mL | Fer-1, DFO |
↑ TFR, ↓ GPX4, ↑ NF-κB, iNOS in macrophages |
↑ Fe²⁺, ROS, LPO, ferroptosis, apoptosis, maturation of RAW264.7 cells | [148] |
| Tirapazamine | 143B, MNNG/HOS, U2OS | 5 - 20 μM | Fer-1 | ↓ SLC7A11, GPX4 | ↑ Fe²⁺, ROS, ferroptosis under hypoxia | [149] |
| Sodium Butyrate | MNNG/HOS, U-2OS/xenograft |
0.5 – 2.5 mM | erastin | ↑ ATF3 → ↓ SLC7A11 | ↑ LPO, erastin-induced ferroptosis, ↓ tumor growth | [150] |
| Zoledronic Acid | MG63, 143B/ xenograft | 2 - 16 μM/ 100 μg/kg | Lip-1, Z-VAD-FMK, Ner-1 |
↑ POR | ↑ ferroptosis ↓tumor growth |
[151] |
| U2OS, MNNG/HOS | 1 - 80 μM |
Fer-1 | ↑ HMOX1, ↓ CoQ10 | ↑ LPO, ROS, ferroptosis | [152] |
| Nanoplatform | Cell line/in vivo Model | Combined Treatment | Molecular Mechanism | Observed Effects | References |
|---|---|---|---|---|---|
|
CI@HSA NPs (capsaicin + IR780 in albumin NPs) |
143B, HOS xenograft |
capsaicin + PDT | ↑ TRPV1/Ca²⁺, ↓ Nrf2/GPX4, HIF-1α; ↑ MAPK, ↓ PI3K/AKT |
↑ ferroptosis, ↓ tumor hypoxia, ↓ tumor growth, good biosafety | [179] |
| CSIR (SRF@CuSO4·5H2O@IR780) | K7M2 xenograft |
PDT + PTT | ↓ xCT | ↓ GSH, ↑ ROS, ↑ ferroptosis, ↓ tumor growth | [174] |
| FeS2@CP NPs | U2OS, MNNG/HOS xenograft |
PTT + CDT | ↓ GPX4 | ↑ Fe2+, ROS, LPO, ↓ GSH, ↑ ferroptosis, apoptosis, ↓ tumor growth, minimal side effects | [182] |
|
FAM (FeS2-AIPH@Membrane) |
MNNG/HOS xenograft | PTT + CDT + TDT | __ | ROS-induced tumor ablation, low systemic toxicity | [175] |
|
SR-Fin56 (Fe₃O₄@SiO₂-RGD loaded with Fin56) |
MNNG/HOS xenograft | PTT + CDT | ↓ GPX4 | ↓ GSH , ↑ Fe2+, ROS, LPO, ↑ ferroptosis, safe and effective in vivo | [183] |
| HMPB@Cisplatin@Eriodictyol | U2OS, MG63, 293T, xenograft | cisplatin + eriodictyol | ↓ GPX4 |
↓ GSH, ↑ Fe2+, ROS, LPO, ↑ ferroptosis, ↑ cisplatin sensitivity; no organ toxicity | [184] |
| FR-miRNAs | ||||
|---|---|---|---|---|
| Expression Trend in OS | Functional Effects | Model Type | References | |
| miR-593 | upregulated | regulates FRGs; promotes proliferation and migration; associated with poor prognosis. | HOS, in silico | [210] |
| miR-635 | downregulated | regulates FRGs; suppresses proliferation and migration; high expression correlates with favorable prognosis. | HOS, in silico | [210] |
| miR-206 | upregulated | promotes ferroptosis via ↑ PTGS2, KEAP1 and ↓ GPX4, SLC7A11, Nrf2, HO-1; increases ROS, Fe², LPO; negatively regulated by lncRNA SNHG14 |
SJSA1 | [209] |
| miR-188-3p | downregulated | promotes ferroptosis via ↓ GPX4T; tumor-suppressive; negatively regulated by circBLNK | HOS, SJSA-1, MG63, U2OS, tissues | [211] |
| miR-144-3p | downregulated | promotes ferroptosis via ↓ ZEB, ↑ ACSL4, ↓ GPX4, SLC7A11; affects iron homeostasis and metabolism, inhibits proliferation, migration, and invasion of OS cells. | 143B, SW1353, MG-63, SaOS-2, U2OS, nude mice | [214] |
| miR-26b-5p | upregulated | ↓ MAT2A → promotes ferroptosis via ↓ STAT3/SLC7A11 | MNNG/HOS, U2OS, nude mice |
[215] |
| miR-26a-5p | upregulated | promotes ferroptosis via ↓ FTH1; increases sensitivity to cisplatin/doxorubicin; regulated by METTL1, which enhances miR-26a/FTH1 interaction |
U2OS, 143B xenograft |
[216] |
| miR-1287-5p | downregulated | promotes ferroptosis via ↓ GPX4; ↑ cisplatin sensitivity | SaOS2, U2OS | [217] |
| FR-lncRNAs | ||||
| PVT1 | upregulated | inhibits ferroptosis via ↑ STAT3/GPX4; ↑ metastasis |
MG63 | [218] |
| upregulated | regulates glycolysis and chemoresistance through ↑ c-MET/PI3K/AKT | MG63 | [219] | |
| SNHG14 | upregulated | sponges miR-206 → inhibits ferroptosis via ↑ SLC7A11; increases nutlin-3a resistance | nutlin3a-resistant NR-SJSA1 | [209] |
| SNHG6 | upregulated | promotes autophagy via miR-26a-5p/ULK1 axis | MG63 |
[220] |
| upregulated | possibly regulates ferroptosis indirectly, correlates to OS prognosis and immunity |
in silico | [221] | |
| APTR | upregulated | suppresses miR-132-3p → ↑ YAP1 → promotes proliferation and invasion | MG63, 143B, Saos-2, HOS | [222] |
| upregulated | included in high-risk ferroptosis-related signatures | in silico |
[221] |
|
| GAS5 | — | included in iron metabolism–related prognostic models; correlates with OS progression and immunotherapy response. | in silico | [205] |
| UNC5B-AS1 | ||||
| LINC01060 | ||||
| AC124798.1 | ||||
| AC104825.1 | ||||
| LINC02298 | downregulated | negatively correlated with PD-L1 → reduce immune suppression | in silico | [221] |
| LINC01549 | ||||
| AC010609.1 | ||||
| LINC02593 | ||||
| AC093673.1 | upregulated | positively correlated with PD-L1 → promote immune evasion | in silico | [221] |
| GAPLINC | ||||
| AL133371.2 | ||||
| CARD8-AS1 | ||||
| DSCR8 | downregulated | correlates with favorable prognosis; involved in immune cell infiltration | in silico | [89] |
| LOH12CR2 | ||||
| AC027307.2 | ||||
| AC025048.2 | ||||
| FR-circRNAs | ||||
| circBLNK | upregulated | sponges miR-188-3p → inhibits ferroptosis via ↑ GPX4; promotes OS progression | HOS, SJSA-1, MG63 and U2OS, tissues | [211] |
| circKIF4A | upregulated | sponges miR-515-5p → inhibits ferroptosis via ↑ SLC7A11, ↑ metastasis and proliferation | SOSP-9607, HOS, U2OS, SW1353, Saos-2, tissues | [223] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





