Noninvasive Preimplantation Genetic Testing in Recurrent Pregnancy Loss and Implantation Failure: Breakthrough or Overpromise?
Introduction
Recurrent pregnancy loss (RPL) and recurrent implantation failure (RIF) are representative of the significant challenges in reproductive medicine many face [
1,
2]. These two conditions foster significant clinical and emotional burdens [
2]. RPL, from a layman's point of view, refers to 2 or more consecutive pregnancy losses before 20 weeks [
1,
2]. On the other hand, RIF is hallmarked by multiple failed IVF cycles despite high-quality embryo transfers [
1,
2]. These two reproductive health complications frequently stem from multiple causes. However, embryonic aneuploidy is highlighted as the predominant contributor, more so in advanced maternal age and idiopathic cases [
2,
3].
Preimplantation genetic testing for aneuploidy (PGT-A) is the current gold standard via trophectoderm biopsy [
3,
4,
5,
6]. It central aim is to optimize embryo selection and reduce miscarriage risks [
7]. However, its invasive nature raises concerns about embryo integrity, diagnostic inaccuracies due to mosaicism, and ethical dilemmas [
6,
8]. These limitations have fostered and accelerated the development of noninvasive PGT (niPGT) [
9,
10,
11]. NiPGT analyzes cell-free DNA (cfDNA) in spent blastocyst culture media [
9]. Existing evidence in the form of early studies suggests that niPGT has the potential to mirror PGT-A in detecting aneuploidies [
9,
10,
11]. However, critical technical hurdles persist. These include contamination risks, inconsistent DNA yield, and insufficient validation in high-risk populations like RPL/RIF [
11].
This review critically evaluates niPGT's viability as a clinical tool for RPL/RIF management, arguing that despite its promise as a breakthrough, current evidence supports only cautious integration into IVF practice. Hence, thorough standardization and rigorous validation ought to be upheld before fully replacing invasive methods. This review begins by assessing the pathophysiology of RPL/RIF and aneuploidy's role and then advancing to the limitations of conventional PGT-A. NiPGT will then be addressed as the potential savior, specifically covering its methodological advances and reliability concerns, as well as the clinical evidence for niPGT in RPL/RIF cohorts. The review will then conclude by highlighting future directions integrating AI, epigenetics, and multicenter trials.
1. Understanding RPL and RIF: Clinical Definitions and Burden
1.1. Definitions and Incidence
Recurrent pregnancy loss (RPL) is clinically defined as an equivalent of 2 or more consecutive pregnancy losses before 20 weeks of gestation. However, some guidelines such as ESHRE distinctly identify the condition as 3 or more losses [
1,
2,
12]. Similarly, recurrent implantation failure (RIF) lacks universal definitive consensus. It typically refers to 3 or more failed IVF cycles despite high-quality embryo transfers [
13,
14]. RPL affects 1–5% of reproductive-aged women [
2,
15]. RIF as per existing statistics, impacts 10–15% of IVF patients [
13,
16] Both conditions show rising incidence rates, an occurrence that can be attributed to the significant demographic trends of delayed childbearing and increased ART utilization [
15,
17].
1.2. Common Etiologies
Common causes of RPL and RIF tend to be anatomic, endocrine, immunological, thrombophilic, and genetic. Anatomically, uterine anomalies such as septate uterus or adhesions account for 10–15% of RPL cases [
13,
18]. In the case of RIF, endometrial polyps and submucosal fibroids tend to disrupt implantation, contributing to its occurrence [
14,
15]. Regarding the endocrine contributors, thyroid dysfunction, luteal phase deficiency, and polycystic ovary syndrome (PCOS) are implicated. Thyroid dysfunction is evidenced by the levels of thyroid stimulating hormone being greater than 2.5 mIU/L. Alongside diabetes and prolactinemia, they contribute to 17% to 25% of RPL cases [
13,
19]. On the other hand, luteal phase deficiency and PCOS are key endocrine contributors to RIF [
20,
21].
On immunologic contributors, dysregulated uterine NK cells (uNKs) play a crucial role for both cases. Alongside aberrant cytokine profiles such as an increase IFN-γ and a decrease in IL-10, elevated uNK cytotoxicity is linked to both RPL and RIF [
19,
22,
23,
24]. Autoimmunity as a standalone factor, specifically through antiphospholipid syndrome (APS), causes 15% of RPL via thrombotic placental injury [
7,
25,
26]. Another significant immune-related contributor is alloimmune rejection. In this process, maternal-fetal human leukocyte antigen (HLA) compatibility such as shared paternal HLA-C alleles and regulatory T-cell (Treg) deficiencies disrupt tolerance [
18,
27,
28,
29]. There is also inflammation, hallmarked by the overexpression of pro-inflammatory cytokines, specifically TNF-α and IL-6 in the endometrium are associated with RIF [
14,
30].
Thrombophilic factors, specifically heritable thrombophilias also play a significant role. Aspects such as Factor V Leiden and prothrombin mutation increase miscarriage risk by 3 to 5 times [
26,
29]. Acquired thrombophilias such as anti-β2-glycoprotein antibodies are a major cause of placental infarction [
25,
26]. Genetically, parental karyotype abnormalities justify 2–5% of RPL [
31,
32]. On the other hand, embryonic aneuploidy drives more than 50% of miscarriages in RPL, escalating with maternal age [
31,
32]
1.3. The Challenge of Idiopathic Cases
Extensive workups have been fostered. However, 40–50% of RPL and approximately 30% of RIF cases remain idiopathic [
13,
14] This diagnostic gap can be attributed to various factors. One of them is undetectable molecular defects. This is evidenced by subtle endometrial transcriptomic dysregulation such as HOXA10 downregulation or sperm DNA fragmentation [
13,
32]. The second factor is microbiota imbalances, hallmarked by altered endometrial Lactobacillus dominance that correlates with RPL [
20,
33]. The third key aspect underlying idiopathy is the significant limitations in testing. Standard panels tend to often omit immune/metabolic biomarkers like ANXA5 or NK cell functional assays, resulting in the discrepancy [
26,
29].
1.4. Psychological and Financial Toll
RPL and RIF have significant effects on patients. Psychologically, patients with RPL/RIF experience 3 times higher rates of depression and anxiety compared to their fertile counterparts [
17,
21]. Further, many patients experience grief and stigma from their conditions and losses. As per existing evidence, 68% of those affected report strained relationships and social isolation [
12,
17]. The financial burden is also significant, with the most immediate concern being the high costs involved. As acknowledged in studies assessing the remedies, a single IVF cycle averages
$12,000–
$15,000. On the other hand, cumulative RIF treatments often exceed
$50,000 [
12,
16,
21]. Besides the cost, there is also lost productivity linked with both physical and emotional impacts of the 2 health conditions. RPL-associated absenteeism, for instance, is approximated to cost each patient
$6,000–10,000 annually in lost productivity [
17,
34].
2. The Role of Embryonic Aneuploidy in RPL and RIF
Embryonic aneuploidy is conceptualized as an abnormal number of chromosomes in a cell. It is a fundamental biological factor underlying both RPL and RIF. Understanding its prevalence, types, association to maternal factors, and implications for embryo selection is fundamental to efficiently managing the pertinent challenging reproductive conditions.
2.1. Understanding Aneuploidy: Definition and Types
Aneuploidy emanates from errors during meiosis (gamete formation) or mitosis (early embryonic cell division) [
4,
35]. These errors result in embryos with missing (monosomy) or extra (trisomy) chromosomes. There are several subtypes of aneuploidy. The first subtype is whole-chromosome aneuploidy, which refers to the gaining (trisomy, such as trisomy 16) or losing (monosomy, such as monosomy X) of chromosomes. It is responsible for over 90% of embryonic losses [
35,
36]. Linked to this the second subtype referred to as polyploidy. This refers to the extra full sets of chromosomes such as triploidy, an equivalent of 69 chromosomes. The third subtype is segmental aneuploidy. This refers to the partial deletions or duplications of chromosomes such as 22q11.2 microdeletion. It is implicated in 5–10% of miscarriages [
4,
37]. The fourth subtype is complex aneuploidy. This refers to multiple chromosomal errors that tend to be prevalent in advanced maternal age [
35,
38].
In addition to these subtypes is mosaicism. It refers to the presence of two or more cell lines. However, they have different chromosomal complements within the same embryo. The clinical impact of mosaicism on reproductive health outcomes varies depending on the type and proportion of abnormal cells [
4,
39,
40]. Overall, despite their slight variances, these abnormalities disrupt normal embryonic development. They often lead to implantation failure or early miscarriage. Further, as established, the risk of aneuploidy increases exponentially with advancing maternal age. This is due to age-related decline in oocyte quality, which particularly affects chromosomal segregation fidelity [
4,
35,
36,
38,
40,
41,
42,
43,
44,
45,
46]
2.2. Aneuploidy as a Primary Cause of Implantation Failure and Miscarriage
Regarding implantation failure, aneuploid embryos exhibit 3.7-fold lower implantation rates than euploid counterparts. This is due to aberrant gene expression affecting trophoblast invasion [
40,
42]. In RIF cohorts, 60–70% of transferred high-quality morphologic embryos exhibit aneuploidy [
43,
47,
48]. Hence, aneuploidy is the major contributor to RIF despite uterine factors, immunology, and endometrial receptivity being implicated and playing a substantial role to some extent. The failure of embryos with an abnormal chromosomal profile to implant successfully after transfer is the primary mechanism [
4,
40,
41,
43,
44,
47,
48,
49]. Studies consistently show that transferring euploid embryos, that is, those with a normal chromosomal profile, significantly improves implantation and live birth rates. At the same time, it reduces miscarriage rates in RIF patients compared to transferring untested embryos or those predicted to be aneuploidy [
4,
40,
42,
43,
44,
48,
49]. For instance, in their study, Wang et al. (2023) demonstrate that PGT-A helped RIF patients achieve live births with fewer transfer cycles [
40]. However, the absolute benefit of PGT-A in RIF, particularly in younger women, remains a hotly contested area. This is because some studies show no improvement in cumulative live birth rates (CLBR) [
41,
47].
When it comes to pregnancy loss, aneuploidy underlies 50–70% of RPL cases. Higher rates, that is, 80% and more, are in women above 40 years [
31,
35]. Even after successful implantation, aneuploidy causes 60% of first-trimester miscarriages in RPL [
45,
46]. Overall, a significant proportion of RPL, more so the unexplained RPL, is linked to embryonic aneuploidy. This is evidenced by studies comparing PGT-A outcomes between RPL patients and infertility controls that generally find similar overall aneuploidy rates [
35,
45]. However, in their study, Sato et al. (2019) reported a higher prevalence of aneuploidy embryos. This was specifically in patients with RPL due to documented prior embryonic aneuploidy compared to those with RIF [
35].
A key takeaway from this is that for some RPL patients, the likelihood to produce aneuploidy embryos is a key underlying factor. Miscarriages in RPL patients, as highlighted earlier by the 60% figure during first trimester, are frequently linked to chromosomal abnormalities [
4,
35,
36,
38,
41,
42,
46]. 2 significant mechanisms are implicated. One is placental insufficiency where trisomy 16 disrupts extravillous trophoblast differentiation [
35]. The second mechanism is cell cycle arrest where monosomies trigger p53-mediated apoptosis [
36].
2.3. Impact of Maternal Age and Diminished Ovarian Reserve (DOR)
Both maternal age and DOR are strong predictors of embryonic aneuploidy. Maternal age is implicated by the reality that aneuploidy rates rise exponentially with increasing age. At 30 years, aneuploidy rates are at 30%. At 40 years, the rates rise significantly to 80% [
4,
35,
36,
38,
40,
41,
42,
43,
44,
45,
46,
50]. This is because as oocytes age, the mechanism of ensuring accurate chromosome segregation during meiosis becomes less reliable. The outcome is a dramatic increase in the proportion of aneuploid embryos with advancing female age. Hence, women 35 years and above with RPL exhibit 2.5-fold higher aneuploidy risk compared to younger patients [
35,
40]. Consequently, the prevalence of RPL and RIF directly correlates with increasing maternal age as widely acknowledged in literature [
36,
38,
43,
44]. PGT-A is often considered more beneficial for older RPL/RIF patients where the aneuploidy burden is highest [
38,
42,
43,
44,
49].
For DOR, advanced maternal age also plays a role. However, it is also evident in younger women and it is defined by the reduced quality and quantity of oocytes. Low anti-Müllerian hormone (AMH) of less than 1.1 ng/mL correlates with 45% aneuploidy rate. This is independent of chronological age and it contributes to both RPL and RIF [
38,
40,
43,
44,
46]. It is also in contrast to the 28% in normal ovarian reserve [
41,
44]. With age not a strong predictor, younger women with DOR may paradoxically face aneuploidy risks similar to their older peers [
38]. Mechanistically, DOR-linked mitochondrial dysfunction elevates oxidative stress, which in turn increases meiotic errors [
38,
51].
2.4. Current Embryo Selection Strategies (Comparative Table)
Given the significant and determining role aneuploidy plays, selecting euploid embryos is fundamental to effectively managing RPL and RIF. There are several significant embryo selection strategies. One of them is morphokinetic grading. Under this is the traditional blastocyst morphology assessment using Gardner's criteria. This underlies the selection of euploid embryos with 65% accuracy [
4,
48]. A core part of the strategy's inferiority is that embryos considered to be high-grade morphologically can be aneuploid while those ranked as lower-grade, euploid [
4,
35,
40,
43,
44,
48,
49]. There is also time-lapse imaging such as pronuclei dynamics, which improves prediction [
43,
48]. However, it remains inferior to genetic testing due to the lack of sensitivity and specificity to predict euploidy reliably [
4,
48].
Another selection strategy is PGT-A where trophectoderm biopsy (TE) and genetic analysis are leveraged. TE entails the removal of 5-10 cells from the outer layer of the blastocyst at day 5 or 6. On the other hand, genetic analysis leverages Next-Generation Sequencing (NGS) to screen all 24 chromosomes for aneuploidy and significant mosaicism [
4,
35,
36,
37,
38,
40,
41,
42,
43,
44,
45,
46,
47,
48,
49,
50,
51,
52]. PGT-A is the gold standard for aneuploidy screening, and it increases live birth rates (LBR) per transfer by 20–30% in RIF/RPL [
49,
50,
52]. However, this strategy exhibits significant limitations, which include its invasive nature, the high possibility of harming the embryo's potential, and mosaicism is likely to yield false positives or negatives [
46,
49]. To counter these limitations is noninvasive PGT-A (niPGT-A), which uses spent culture medium cfDNA and shows 70–90% concordance with biopsy [
42,
53].
These selection strategies, despite their limitations, excel in identifying euploid embryos for transfer, aiming to bypass aneuploidy-related failures. The transfer then significantly reduces miscarriage rates in RPL and RIF populations [
4,
35,
38,
42,
46]. There is also improved efficiency in the form of reduced time to live birth by minimizing transfers of embryos destined to fail or miscarry. This is evident particularly in older women or those with previous failures [
38,
40,
42,
43,
44,
48,
49].
2.5. Controversies
PGT-A consistently shows benefits in reducing miscarriage per transfer and improving implantation rates in RPL/RIF. However, its impact on cumulative live birth rates (CLBR) per initiated cycle, more so in younger women aged between 35 and 37 with RIF, is less clear. Some studies show improvement with estimations being above 50% [
38,
40,
42,
48]. Others, particularly in younger unexplained RIF cohorts, show no significant CLBR benefit over transferring untested embryos [
41,
47,
51] Concerns regarding this embryo selection strategy include the invasiveness of biopsy, which is a key limiting factor when the embryo's integrity is considered. Other concerning factors include cost, potential embryo damage, diagnostic limitations (mosaicism), and the possibility of discarding embryos with developmental potential due to much emphasis being put on morphological underpinnings [
4,
41,
47,
49,
51,
52].
2.6. Evidence (Create an Image)
In their study, Kato et al. (2023) reported promising live birth rates using PGT-A in minimal stimulation cycles for RPL/RIF patients aged 35-42 [
38]. Similarly, Kim et al. (2023, 2024) highlighted that the efficacy of PGT-A in RIF/RPL may be predictable based on maternal age and embryo quality [
43,
44]. Mei et al. (2024), in their systematic review, concluded that PGT-A optimizes reproductive outcomes in recurrent reproductive failure [
48]. Mantravadi et al. (2020) specifically questioned its optimization role in idiopathic RPL [
49]. Findings by Kasaven et al. (2023) in their meta-analysis show that PGT-A at the blastocyst stage improves LBR [
52].
2.7. Summary
Embryonic aneuploidy is a predominant biological cause of both RPL and RIF. Its incident rate increases significantly with advanced maternal age and diminished ovarian reserve. Morphological assessment is foundational in IVF and has proven to be useful to a certain extent. However, its inability to accurately predict chromosomal status has driven the adoption of PGT-A via trophectoderm biopsy as the standard for selecting euploid embryos. PGT-A as confirmed by several key studies reduces miscarriage rates and improves per-transfer success rates in RPL and RIF. However, its impact on cumulative live birth rates, particularly in younger women with RIF, remains debatable. Further, the inherent limitations of its nature as an invasive biopsy procedure necessitates the need for continued refinement and the exploration of reliable non-invasive alternatives.
3. Invasive PGT-A: Current Standard and Its Limitations
PGT-A is a significant advancement in assisted reproductive technology (ART), more so in embryo selection. Its aim is to improve outcomes for patients experiencing RPL and RIF by selecting euploid embryos for transfer. However, its invasive nature and associated limitations warrant further critical evaluation.
3.1. Background – Purpose and Types (Images)
The primary objective of PGT-A is to identify embryos with the correct number of chromosomes (euploidy), screening out those with missing or extra chromosomes (aneuploidy). This selection increases implantation rates per embryo transfer, reduces the rate of clinical miscarriage, decreases the time to achieve a live birth by minimizing transfers of embryos with low developmental potential and it fosters single embryo transfer (SET), thus reducing the risk of multiple pregnancies [
3,
6,
39,
52,
54,
55,
56,
57].
PGT encompasses several types. These include PGT-M (Monogenic disorders). Its primary objective is to test for specific single-gene defects. There is also PGT-SR (Structural Rearrangements), which its main objective is to test for balanced chromosomal rearrangements in carriers such as translocations [
39,
54]. The third and significant type is PGT-A (Aneuploidy Screening). Its main focus is to screen for numerical chromosomal abnormalities across all 24 chromosomes [
6,
39,
54,
56,
57]. Of all the three, PGT-A is most commonly used. This is more particular in contexts like RPL, RIF, and advanced maternal age [
6,
39,
55,
56,
57].
3.2. Common Techniques: Trophectoderm Biopsy and Blastocyst Stage Analysis (Images)
The 2 significant techniques are Trophectoderm (TE) Biopsy and Blastocyst Stage Analysis. Despite their variances, they all begin with blastocyst culture. With this, embryos are cultured in vitro for 5-6 days post-fertilization. This is to enable them to reach the blastocyst stage, which is characterized by an inner cell mass (ICM) that becomes the fetus and TE that becomes the placenta. [
6,
39,
54,
55,
56]. TE biopsy, the current gold standard, begins with embryo preparation. This entails making a small opening in the zona pellucida, the outer shell, on day 3 or 5, often using laser ablation. The biopsy procedure is then performed on day 5 or 6 where approximately 5-10 cells are gently aspirated and removed from the TE layer. [
3,
6,
39,
54,
55,
56,
57,
58] This is usually achieved using micromanipulation techniques. The biopsied blastocyst is then cryopreserved while genetic analysis, that is, blastocyst stage analysis, is performed [
3,
6,
39,
55,
56]. During genetic analysis, the biopsied TE cells undergo DNA amplification through Whole Genome Amplification (WGA). This is then followed by chromosomal analysis, which is conducted by primarily using Next-Generation Sequencing (NGS), to detect aneuploidy and mosaicism [
6,
8,
39,
54,
55,
56,
58]. Quantitative PCR (qPCR) can also be leveraged for rapid aneuploidy screening [
39].
Compared to cleavage biopsy, TE biopsy has its superiority and benefits. It is preferred because it removes cells destined for the placenta. This theoretically minimizes the impact on fetal development. As per evidence, the lower embryo damage risk is 2-4% compared to the 15% in cleavage biopsy [
55]. Further, with TE, more cells are obtained hence higher DNA yield, which significantly improves diagnostic reliability, an attribute evidenced by the 99% amplification success. On top of these, blastocyst formation itself is a selection step for developmental efficiency [
6,
39,
54,
55,
56,
58]. TE biopsy also allows for cryopreservation of the biopsied embryo. This ensures that the biopsy procedure is separated from the transfer cycle [
3,
56].
3.3. Benefits: Improved Selection and Reduced Miscarriage Rates
TE biopsy-based PGT-A fosters several significant benefits. These include improved embryo selection. By identifying euploid embryos, PGT-A bypasses aneuploid embryos that are highly likely to fail or miscarry [
6,
39,
52,
55,
56,
57]. Evidence indicates that euploid embryos show 2.5 times higher implantation rates compared to the untested morphologically good embryos in RIF patients [
3,
59]. Secondly, PGT-A reduces miscarriage rates. Some studies show that in women 35 years and above with RPL, PGT-A reduces the miscarriage risk by 50% [
3,
56]. This is attributed to avoiding aneuploidy, a leading cause of miscarriage [
3,
52,
56,
57]. Further, in their systematic review and meta-analysis, Kasaven et al. (2023) found that that PGT-A is associated with a higher live birth rate per embryo transfer [
52]. Thirdly, PGT-A increases implantation rates (IR) per transfer. This is because transferring screened euploid embryos generally results in higher IR per transfer compared to when the embryos are transferred without screening [
52,
56]. Compared to conventional IVF where 3 or more cycles are required, a single euploid transfer achieves pregnancy in 1–2 cycles [
6,
56]. Fourthly, PGT-A underlies better prognosis and management of patient expectations [
56].
3.4. Limitations
Despite its benefits and status as the gold standard, invasive PGT-A exhibits significant limitations. Firstly, with its invasive nature, the embryo damage risk is high. This is because the biopsy procedure physically removes cells from the embryo. Concerns persist regarding potential short-term impacts such as the reduced developmental potential post-biopsy and long-term impacts, specifically subtle developmental effects. Regardless, robust evidence for major harm in live births is still limited [
6,
39,
54,
55,
56,
57,
58]. Further, due to the high technical expertise required, poor technique can damage the ICM or compromise embryo viability [
54,
55,
56]. Existing evidence indicates that double biopsies done for confirmation testing lower live birth rates by 39% [
55]. Still on embryo damage, double handling of combining biopsy and vitrification subjects the embryo to two potentially stressful procedures [
3,
55,
56,
58].
Besides embryo damage risk, another significant limitation is mosaicism and the high potential for false negatives or positives. Also referred to as biological limitation in layman's language, chromosomal mosaicism, that is, the presence of both euploid and aneuploid cells, is common in human blastocysts. TE biopsy operates by sampling only a few cells from one part of the trophectoderm. This is not fully representative of the chromosomal status of the entire embryo or the ICM [
8,
39,
54,
55,
56,
57]. The risk of false negatives is high and this is can be evidenced by a euploid TE biopsy result masking an aneuploid ICM hence the false negative effect. This falseness results in the transfer of an abnormal embryo that miscarries or results in an affected live birth [
8,
54,
55,
56,
57]. Conversely, in the case of a false positive, an aneuploid TE biopsy result tends to misrepresent a mosaic or euploid ICM. The outcome of this is the discarding of a viable embryo [
3,
8,
39,
54,
55,
56,
57]. Linked to these false outcomes is diagnostic uncertainty. In their systematic review and meta-analysis, Chen et al. (2025) note limitations in diagnostic accuracy [
8]. Further, Viville and Aboulghar (2025) posit that the inherent mosaicism and technical limitations insinuate that PGT-A results are not a definitive diagnosis of the embryo's true chromosomal profile [
57].
The third significant limitation is cost and logistical complexity. While IVF is already expensive, PGT-A adds substantial expenses to it in the form of biopsy procedure fees, genetic analysis costs, and embryo culture and cryopreservation-related costs. Based on existing literature, the cumulative costs for RIF patients exceed
$50,000 [
56]. This is because the procedure adds
$3,000–
$5,000 per cycle to IVF costs, totaling to about
$15,000–
$20,000 for most patients [
56,
57]. These fees create financial barriers, raising pertinent cost-effectiveness questions, especially in younger patients or those with few embryos [
6,
39,
54,
55,
56,
57]. Another cost issues is evidenced in the form of laboratory infrastructure. PGT-A requires specialized embryology skills, advanced genetic laboratory capabilities (NGS), and robust cryopreservation programs. All of these are costly, making the services exclusive only for the few that can afford.[
39,
55,
56]. Expenses are also incurred in terms of cycle management. The procedure requires a freeze-all strategy. This delays the transfer and increases the number of patient visits and overall cycle time, resulting in extra expenses for the patient [
6,
55,
56].
The fourth and last significant limitation is evident in the form of ethical and emotional concerns for patients. Firstly, there is a potential of discarding embryos based on biopsy results that may be inaccurate. More so for mosaic embryos. This raises significant ethical dilemmas for both patients and clinicians [
3,
55,
56,
57]. In their study, Morales (2024) highlights this as a major controversy [
56]. Secondly, the psychological burden is significant. The evident procedural complexity, cost, waiting periods for results, and the possible likelihood of having no transferable embryos can create significant emotional stress for patients. This is on top of them experiencing infertility or loss [
55,
56,
57]. It is indicated that 52% of patients report anxiety over no transferable embryos after PGT-A [
52,
56]. The third aspect is the issue of imperfect embryos. The possibility of mosaicism often places patients in difficult decision-making positions. This is regarding the transfer of embryos that experts have labeled as abnormal or intermediate, often with limited robust outcome data [
55,
56,
57]. Last is the aspect of overpromise and expectation management. Concerns exist that PGT-A may be oversold as a guarantee of success. This potentially underlies unrealistic expectations and significant disappointment if cycles fail despite euploid transfers [
56,
57]. To mitigate this, Kakourou et al. (2024) in their overview emphasize the importance of acknowledging the limitations of both biology and technology [
55].
3.5. Summary
Based on the above evidence, TE biopsy-based PGT-A is the established method for assessing embryonic aneuploidy. It offers potential benefits in selecting euploid embryos, reducing miscarriage rates per transfer, and improving implantation rates per transfer, particularly in RPL and AMA [
52,
56]. However, its invasive nature raises significant fundamental concerns. This include issues about embryo safety and diagnostic accuracy that is inherently challenged by embryonic mosaicism that underlies false results [
3,
8,
54,
55,
57]. It also imposes significant financial, logistical, and ethical burdens on patients and clinics [
39,
55,
56,
57]. These substantial limitations necessitate the need for continued refinement and rigorous counseling on realistic expectations. They also warrant the pursuit of less invasive and more reliable alternatives like noninvasive PGT (niPGT) [
58].
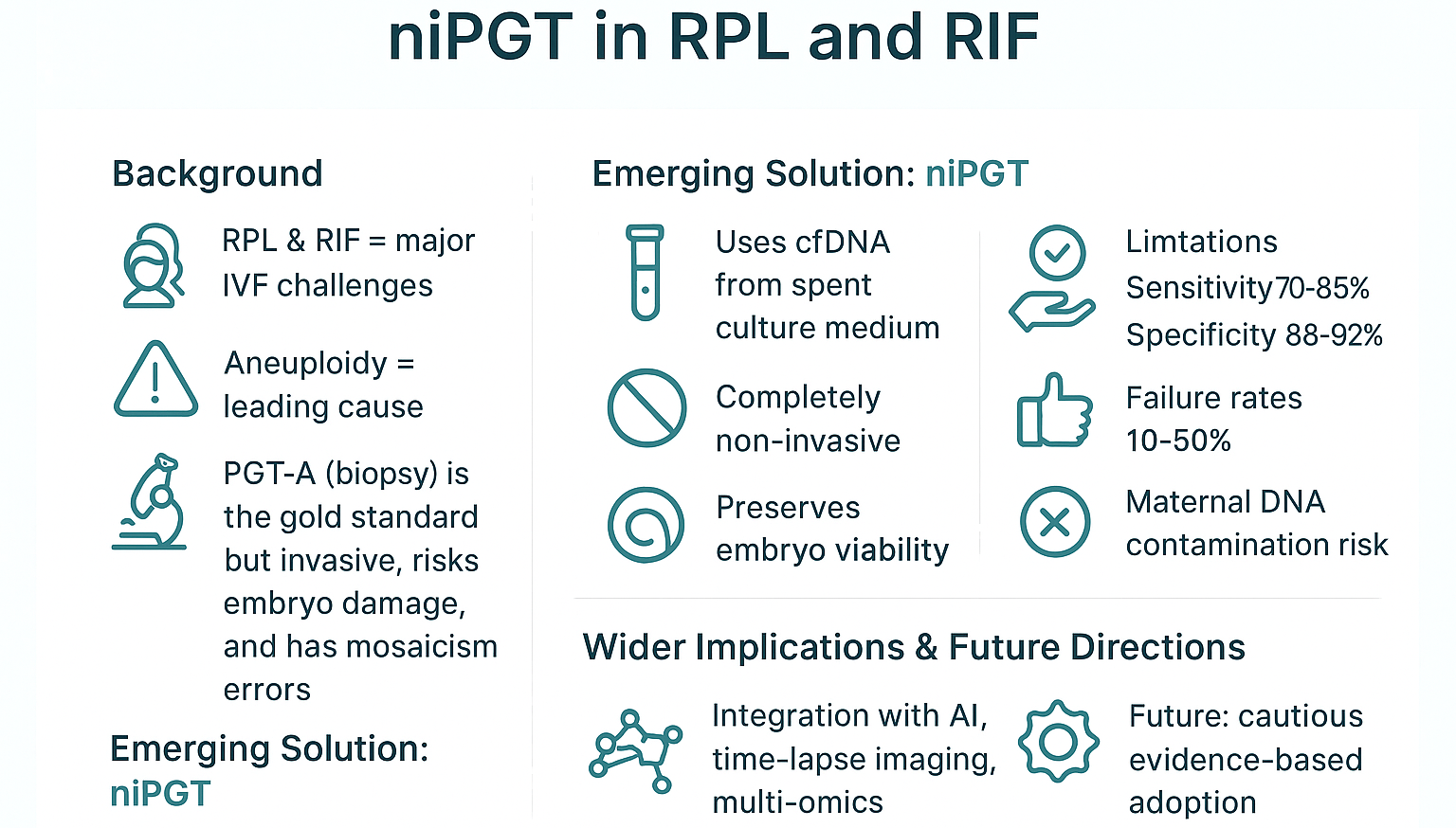
4. Noninvasive PGT (niPGT): Science and Methodologies (Image)
Noninvasive Preimplantation Genetic Testing (niPGT) is an evident paradigmal shift from the traditional invasive biopsy-based methods (PGT-A). It analyzes cell-free DNA (cfDNA) released by the embryo into its culture environment. Through this, niPGT's main objective is to assess embryonic chromosomal status without physically compromising the embryo. This segment of the article aims to understand its scientific basis and methodologies, aspects that are crucial for evaluating its potential and limitations.
4.1. Sources of Cell-Free DNA: Blastocoel Fluid vs. Spent Culture Medium (Differences Table)
The cfDNA analyzed in niPGT originates from embryonic cells undergoing apoptosis or other forms of cellular activity during in vitro undertakings. Balstocoel fluid (BF) and spent culture medium (SCM) are analyzed as the two primary sources.
BF refers to the fluid filling the blastocoel cavity of the blastocyst, usually at days 5-6. BF's DNA is presumed to be primarily embryonic in origin (90-95%), emanating from dying cells within the enclosed cavity [
9,
59,
60,
61,
62,
63]. BF's advantages include the potentially higher concentration of embryonic DNA. The fluid also has less direct exposure to external contaminants and is more representative of the inner cell mass (ICM) based on the underpinnings of the cavity formation dynamics [
59,
60,
61]. A significant disadvantage is that BF is challenging in terms of technicality, that is, aspirating without damaging the blastocyst. It also requires skilled micromanipulation similar to biopsy. Further, the aspiration in itself is a form of intervention, despite being less invasive than cell removal. On top of these, not all blastocysts have easily aspiratable fluid and the potential for DNA degradation is high if processing is not initiated as soon as possible [
59,
60,
61,
62].
SCM is the culture medium in which the embryo has been grown. It is typically collected after blastocyst culture, usually between days 5-6. cfDNA in SCM is released by the embryo's trophectoderm cells into the surrounding environment [
2,
11,
60,
64,
65,
66,
67]. SCM's key advantage is that it is a true non-invasive DNA collection method. Hence, it significantly limits any mechanical stress on the embryo. It is also simple to perform since only medium aspiration is required. With this, parallel testing of multiple embryos is possible and logistically, it integrates seamlessly into standard IVF lab workflows [
2,
9,
60,
61,
64,
67,
68,
69]. Its main weakness compared to BF is that the concentration of embryonic cfDNA is very low (10-40%) and the risk of contamination from maternal DNA or other external sources is fairly high [
2,
11,
62,
70]. Linked to these factors are the facts that the DNA is likely to be highly fragmented and due to the external influences, prone to degradation. Further, the dilution effect from the medium volume is substantial, underlying low DNA concentrations. Additionally, unlike BF, DNA may originate primarily from TE, thus less representative of the ICM [
2,
9,
11,
59,
60,
61,
63,
64,
65,
66,
68,
71].
Of the 2 primary sources, SCM is the predominant alternative for niPGT research and development. This is due to its practical advantages and truly noninvasive nature, albeit the significant weaknesses of low DNA yield and contamination [
2,
9,
60,
61,
64,
67,
68,
69,
70,
71].
4.2. Extraction, Amplification, and Sequencing Methods
The workflow for niPGT from SCM involves highly sensitive techniques. These are necessary to handle the minimal amount and already degraded cfDNA. The first step is sample collection and preparation. During this step, SCM is carefully aspirated after the embryo has been removed, all while ensuring minimal disturbance [
9,
11,
61,
64,
67,
68,
70]. Precautionary measures taken to minimize contamination include rigorously removing cumulus cells before fertilization and the use of sequential media that is changed at specific times such as before blastulation [
9,
11,
64,
68,
70]. To prevent DNA damage, immediate processing is fostered [
11,
60,
61,
65]. After the sample has been collected and prepared, cfDNA is then extracted. This step entails using specialized kits designed for lower concentration, fragmented cfDNA, which exhibits similarity to liquid biopsy techniques [
9,
11,
59,
60,
63,
65,
68]. The objective at this step is to maximize the recovery of the lowly concentrated embryonic DNA while minimizing contaminants. Efficiency is necessary at this step [
11,
60,
61,
65].
To maximize potential DNA yield, WGA is conducted. It is an essential step due to the extremely low quantity of embryonic cfDNA. However, while necessary, it presents key operational challenges. For instance, WGA introduces significant technical noise. It also exhibits amplification bias, allele dropout, and the introduction of aneuploidy signals [
59,
60,
61,
63,
65,
66]. Regardless, there is room for optimization. For instance, as noted by Hu et al. (2023), this can be achieved by comparing different WGA kits and protocols to improve fidelity and reduce artifacts [
63,
65]. Some protocols excel at amplifying specific chromosomes to reduce complexity [
72].
With the DNA extracted, the next step is genetic analysis, also described as sequencing or detection. This is achieved by next-generation sequencing (NGS), the dominant platform for niPGT-A. NGS excels in simultaneously sequencing millions of DNA fragments [
2,
9,
59,
60,
62,
63,
66,
67,
68,
69,
70,
71]. There are numerous ways it can achieve this. The first approach is low-pass whole genome sequencing (LP-WGS). This sequences the entire genome at low coverage. With that, it is sufficient to detect whole-chromosome aneuploidies, specifically by analyzing sequence read counts mapped to each chromosome. As per evidence, niPGT-A leverages this approach the most [
2,
59,
60,
61,
66,
68,
71]. There is also higher coverage WGS, also referred to as targeted NGS. This is less common compared to LP-WGS because it is costly and computationally complex. However, it still has its place, specifically being used to detect segmental imbalances or mosaicism [
66]. Besides NGS, there is quantitative polymerase chain reaction (qPCR). It operates by using fluorescent probes to quantify the number of copies of specific chromosomal regions [
72]. Compared to NGS, it is faster, less expensive, and easy to use due to the simple data analysis. Unlike NGS, it is limited to predefined chromosomal targets. It also has lower resolution and is less effective for detecting mosaicism or partial aneuploidies. Further, it is more susceptible to amplification bias and contamination artifacts [
66,
72].
The last step after DNA sequencing and detection is bioinformatics analysis. This step is a sophisticated computerized approach required to analyze NGS data [
2,
59,
60,
66,
68,
71]. Key tasks entail genome mapping, calculating chromosomal ratios, and aneuploidy identification through relevant statistical models [
2,
59,
60,
66,
68,
71].
4.3. Current Techniques
Several niPGT techniques are in use. However, niPGT for aneuploidy (niPGT-A) is the most common. It is the most developed application. Its primary focus is on detecting whole-chromosome aneuploidies using LP-WGS of cfDNA from SCM [
2,
62,
63,
66,
67,
68,
69,
71]. Within niPGT-A, NGS, specifically LP-WGS is the gold standard due to its genome-wide scope and scalability [
2,
60,
63,
66,
68,
70,
71]. There are also emerging applications, mainly niPGT for monogenic disorders (niPGT-M) and structural rearrangements (niPGT-SR). However, these still face significant operational limitations due to the complexities that must first be overcome, such as the need for structural variant detection from minute, fragmented DNA [
68,
72].
4.4. Challenges
Despite its promise, niPGT, just like the invasive PGT it is trying to replace, faces significant technical hurdles. The first key concern revolves around maternal DNA contamination. The contaminating DNA is primarily from maternal cumulus and granulosa cells surrounding the oocyte, residual sperm cells, and sometimes, even endometrial cells. It is often present in much higher quantities than embryonic cfDNA in SCM, thus fostering dilution [
2,
9,
11,
59,
60,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71]. The consequence of maternal DNA's presence is that it can completely mask the embryonic signal. This often results in false euploid calls or false aneuploid calls in cases where the contaminating cells are aneuploid [
9,
61,
64,
71]. A key mitigation measure for this challenge is rigorous oocyte denudation [
9,
11,
64]. Other relevant operational measures include the careful handling of media, changing medium after cumulus removal or early cleavage stages [
11,
64,
70], and adopting ways that can selectively digest maternal DNA to avoid pertinent contamination [
64].
The second key challenge linked to contamination issues is low cfDNA yield and quality. As significantly highlighted by relevant evidence, the amount of embryonic cfDNA released into SCM is extremely low. It is also highly variable between embryos [
11,
59,
60,
61,
62,
65,
66,
71]. Additionally, embryonic cfDNA tends to be highly fragmented. This further complicates analysis and amplification [
59,
60,
61]. The key consequence is that all these result in high rates of amplification failure or poor-quality WGA. The outcome is usually no result (NR) or inconclusive results [
11,
59,
60,
61,
65,
71]. Other consequences of the low yield and poor quality are the limitation in detecting mosaicism and segmental imbalances, as well as compromises on the robustness and reliability of niPGT [
59,
60,
66,
71]. Key ways to mitigate this challenge as per literature is to optimize culture conditions to potentially enhance DNA release [
11,
70]. Technicians can also leverage larger starting medium volumes [
11,
61] and enhance the amplification process [
72].
The third key challenge limiting this test's efficiency is inter-embryo variability. Biologically, embryos of similar morphology and development stage are highly likely to exhibit variances in the amount, timing, and pattern of cfDNA release. Key factors underlying this phenomenon include embryo quality, metabolic activity, and genetic status [
11,
59,
60,
61,
62,
71]. The key consequence of this challenge is that it significantly hampers standardization, making universal diagnostic thresholds hard to establish. The variability also underlies inconsistent performance, as well as discordance rates with TE biopsy [
59,
60,
61,
62,
66,
71]. To mitigate the variability, amplification protocols need to be continually refined, embryo-specific normalization factors need to be developed, and more studies conducted to fully conceptualize the biological determinants of release [
59,
60,
68].
4.5. Summary
niPGT primarily uses cfDNA from spent culture medium analyzed via NGS following WGA. It is a significant noninvasive alternative to biopsy. The core scientific premise banks on using naturally released embryonic DNA [
2,
60,
61,
68]. However, its technical efficiency is significantly limited by substantial maternal DNA contamination that masks the embryonic signal, the severely low and variable embryonic cfDNA that underlies high failure rates, and inter-embryo biological and technical variability that negatively impacts standardization [
9,
11,
59,
60,
61,
62,
63,
64,
65,
66,
71] Despite potential advances, these key limitations currently prevent niPGT-A from effectively replacing TE biopsy as the standard of care. As such, it is necessary that the results are cautiously interpreted and rigorously validated.
5. Clinical Evidence: Can niPGT Replace Biopsy?
The potential of niPGT fully replacing TE biopsy and becoming the standard method for aneuploidy screening fully depends on robust clinical validation. Current evidence, derived from key studies and meta-analyses comparing niPGT to TE biopsy-based PGT-A reveals significant promise. However, this promise is marred by substantial limitations, more so regarding reliability and consistency.
5.1. Key Studies and Meta-Analyses
Xu et al., 2016 provide seminal proof-of-concept. In their study, the scholars demonstrated that cfDNA in SCM has the potential to reflect embryonic ploidy status. This shows concordance with TE biopsy results in a small cohort, including detection of aneuploidy and mosaicism [
74]. This foundational study is heralded to have ignited the field. Part of this was acknowledging some of the key challenges such as low DNA quantity.
Huang et al., 2019 provide validation and optimism. This study reported high concordance of approximately 80% between niPGT-A (SCM) and TE biopsy. The scholars even went ahead to claim that niPGT-A might be more reliable than biopsy in some cases. This is down due to better representation of mosaic status [
53]. The assertions exceled in significantly boosting optimism about replacing biopsy.
In their pivotal prospective multicenter study, Rubio et al., 2020 explore the concepts of fertility and sterility. This is the first large-scale (n=533 blastocysts), prospective, multicenter trial using niPGT-A (SCM) for embryo selection. Pertinent findings indicated a high concordance rate with TE biopsy of approximately 85% for full chromosome aneuploidies. Further, the results also showed the promising ongoing pregnancy rates to be 50% after the intervention of euploid niPGT-A transfers [
75]. This study has been crucial in moving niPGT-A towards clinical application. However, it also acknowledged a significant rate of no-result (NR) and discordance. These are attributed to maternal contamination and biological variability.
Subsequent Validation and Discordance Studies:
Huang et al. 2019 validated a targeted NGS niPGT-A method. They showed high sensitivity and specificity. However, they also noted and acknowledged technical challenges [
76].
Shitara et al. 2021 found that cfDNA in SCM reflected chromosomal status. This was after an extended culture. Nevertheless, they also noted variability compared to TE biopsy [
61].
Sakkas et al. 2024 demonstrated that implementing a specific niPGT-A culture protocol can potentially impact viability and outcomes. This excels in highlighting the sensitivity of the method to procedural scrutiny [
70].
Studies Highlighting Limitations:
Hanson et al 2021 reported alarmingly high rates of DNA amplification failure at approximately 50%. They also indicated the poor correlation between niPGT-A (SCM) and TE biopsy results. These findings significantly challenge the reliability of niPGT-A, thus substantially countering the initial bout of optimism [
77].
Volovsky et al. 2024 critically assert that despite the promise of niPGT-A being real, current evidence shows otherwise. For instance, it has lower sensitivity and specificity than TE biopsy. niPGT-A also suffers from high failure rates and there significantly lacks validation for clinical use as a biopsy replacement [
71].
Meta-Analyses:
Huang et al. 2023 included 20 studies in their meta-analysis. Their findings supported the conclusion that niPGT-A demonstrates good diagnostic performance in research settings. However, they also emphasized significant heterogeneity and the influence of technical factors. Overall pooled sensitivity was indicated as 0.84 and specificity as 0.85 compared to TE biopsy [
78]. What these figures insinuate is the lack of near-perfect accuracy that is sometimes claimed.
Li Piani et al. 2025 primarily focus on biopsy harm. As such, they significantly highlight the need for a reliable non-invasive alternative. With this assertion, they indirectly insinuate that the current niPGT-A is yet to be proven to efficiently fulfill that role [
79].
5.2. Sensitivity and Specificity Compared to PGT-A
The metrics vary. However, they excel in indicating that niPGT-A is yet to be fully considered an equivalent. Regarding sensitivity, values range widely across studies, from approximately 50% to over 90% [
53,
61,
74,
75,
76,
77,
78]. Huang et al. 2023 indicate a pooled sensitivity of around 0.77-0.85 [
78]. The implication of these findings is that niPGT-A misses a significant proportion of aneuploid embryos, hence false negatives. Contributors to this limitation include low embryonic DNA concentration, contamination and dilution by maternal DNA, amplification failure, and biological sampling variances evident in SCM and TE cells [
61,
71,
77,
78].
Regarding specificity, findings also show variability. However, it is generally higher than sensitivity, often reported between 80-95%, but still slightly lower compared to TE biopsy [
53,
61,
75,
76,
77,
78]. Pooled estimates by Huang et al. (2023) are around 0.83-0.90 [
78]. The key contributors to the false positives include dilution by maternal DNA, WGA bias, and a significant emphasis on noisy data [
61,
71,
77,
78].
On the concordance rate, which tends to be reported instead of the traditional sensitivity and specificity, the value is indicated to be above 80% by high-impact studies [
53,
75]. However, real-world data and meta-analyses show substantial discordance of 15-30% or more. In this case, niPGT-A is performing worse than biopsy in head-to-head comparisons for diagnostic reliability [
52,
71,
77,
78]. Regarding the no-result (NR) rate, it is often reported to range from 10-50% [
70,
71,
75,
77]. The implications for this is that the embryo will have to be discarded, a rescue biopsy performed, thus defeating the whole purpose of noninvasiveness, or transfer the embryo blindly [
52,
71,
75].
5.3. Real-World Success Rates in RPL/RIF Subpopulations
Data on niPGT-A outcomes in RPL and RIF patients is significantly limited and less encouraging compared to the initial broad population studies highlighting its potential efficiency. Firstly, their lacks targeted RCTs comparing live birth rates after niPGT-A selection, TE biopsy selection, and morphology-based selection in RPL/RIF populations [
35,
52,
71]. Secondly, the high rate of low sensitivity is significantly concerning. This is because undetected aneuploid embryos can be transferred, increasing the risk of miscarriage [
35,
71,
78]. Thirdly, the high NR rate is a major issue for RIF/RPL patients, more so for those having fewer embryos. It is a major disadvantage to discard or blindly transfer embryos without genetic information due to test failure [
52,
71,
75]. In their analysis, Volovsky et al. (2024) argue the data from Rubio et al. actually showed lower ongoing pregnancy rates with niPGT-A selection compared to conventional PGT-A selection in their cohort [
71,
75]. However, it is worth noting that Rubio et al. (2020) included patients with previous failures and subgroup analysis specific to confirmed RIF/RPL wasn't the study's primary focus, thus limiting any significant conclusions [
75].
In a different analysis, Guan et al. (2024) focus on an RCT in young RIF patients that compared morphology alone against PGT-A (biopsy). The RCT did not directly test niPGT. However, it showed no CLBR benefit for PGT-A over morphology alone in the young RIF group [
42]. The findings raise valid concerns regarding the ability of any form of PGT-A, especially the less reliable niPGT-A, to offer a significant advantage for young RIF over standard care. This indirectly insinuates that niPGT-A might fail to meaningfully benefit patients. Overall, validation is inadequate in the high-risk groups to make definitive conclusions on niPGT-A's efficacy in RPL/RIF [
35,
71]. Hence, subgroup analysis is needed to effectively determine real-world success.
5.4. Cases Where niPGT Changed Clinical Management
Despite limitations, niPGT-A has shown potential utility in specific scenarios. One of them is rescue for biopsy failure or inconclusive results. In cases where TE biopsy fails by exhibiting outcomes such as no result or inconclusive diagnosis, niPGT-A on SCM has been indicated to have the potential to provide a genetic assessment. It offers information that would otherwise not be available, thus enabling the transfer of an embryo that would have been discarded [
9,
75,
80,
81]. In their study that provided this evidence, Li et al. 2021 explored this specifically for mosaic embryos [
80]. Another scenario where niPGT excels is detecting uniform aneuploidy. Despite confidence levels varying, niPGT-A has the potential to clearly identify uniform aneuploidy in cases of high confidence calls. Through such, it prevents the transfer of embryos with very low potential just like biopsy [
53,
74,
75]. niPGT also excels in avoiding biopsy risks. Granted that reliability is confirmed, it thrives on its non-invasiveness [
81]. Lastly, in the research context, niPGT-A allows repeated sampling of the same embryo's environment at different time points. This offers unique research insights into embryonic biology and DNA release dynamics that biopsy cannot provide [
61].
5.5. Summary
Current clinical evidence indicates that niPGT-A cannot yet reliably replace TE biopsy as the standard method for PGT-A. Landmark studies like Huang et al. (2019) and Rubio et al. (2020) demonstrate feasibility and potential. However, subsequent research and critical reviews reveal significant limitations. These include variable and often suboptimal sensitivity and specificity compared to biopsy, a significantly high rate of no-result, and susceptibility to errors from maternal contamination and low DNA yield. Further, evidence for its effectiveness in improving live birth rates for the target RPL and RIF subpopulations is lacking. Indirect data suggests that it is significantly limited [
35,
42,
71]. Overall, niPGT-A can be leveraged in rescuing biopsy failures, a back-up, and as a research tool [
9]. However, its inconsistency and diagnostic uncertainty prevent it from being the clinically validated substitute for TE biopsy, remaining an adjunct at best. To address the gaps, more evidence from large-scale studies and well-designed RCTs in RPL and RIF populations is needed.
6. Application in RPL/RIF: Hope or Hype?
The potential of niPGT presents a significant promise for patients and clinicians facing the harsh and heartbreaking realities of RPL and RIF. All the publicity emanates from its capability to revolutionize embryo selection noninvasively. However, translating this promise into reliable clinical practice, more so for RIF and RPL patients, remains a challenging course. The current application of niPGT in RPL/RIF sits precariously between hopeful innovation and premature hype.
6.1. Benefits: The Foundations of Hope
niPGT's key advantage and selling point is its non-invasiveness. It eliminates the need for physically removing cells. As such, this preserves embryo viability and developmental potential. This is mainly achieved by avoiding mechanical stress and potential damage to the ICM) [
56,
66,
82,
83]. Through non-invasiveness, niPGT addresses a core ethical and biological concern of invasive PGT-A. This is particularly important when dealing with precious embryos from patients with limited ovarian reserve, a common scenario in RPL/RIF [
25,
56,
66,
82]. Linked to non-invasiveness is the aspect of embryo preservation. By avoiding biopsy, niPGT eliminates the need for concurrent vitrification of the biopsied embryo. This eliminates the stress associated with double handling and allows embryos to be transferred fresh as required or vitrified intact [
10,
56,
66,
82] This is particularly more appealing in RIF cases where endometrial receptivity timing is crucial [
25,
84]. Thirdly, niPGT has high patient acceptability. Many patients prefer it due to reduced anxiety about biopsy-related embryo harm [
1,
56,
82]. Patients are also ethically comforted by the fact that this procedure avoids embryo manipulation [
56,
66,
82]. It is also a simplified process compared to invasive biopsy [
1,
82], thus improving the patient's experience in an already stressful journey [
1,
25,
84]. Additionally, it also has the potential for wide access. If proven reliable and cost-effective, the simpler workflow would make genetic testing accessible to more patients. This would include some RPL/RIF cases previously deemed unsuitable for PGT-A [
82].
6.2. Drawbacks: The Reality Check
Inconsistency and unreliability are the most significant issues hindering niPGTs clinical adoption, especially for RPL/RIF where diagnostic accuracy is paramount [
55,
56,
66,
83]. One key issue established in the earlier review of literature is the high rate of no-result (NR). Reported in 10-50% of the cases, amplification failure underlies niPGT's ability to provide a reliable genetic analysis [
10,
56,
83]. This is more devastating for RPL/RIF patients with few embryos as viable embryos can be discarded or blindly transferred [
56]. Another reliability issue is that niPGT's accuracy is variable. This is evidenced by several studies highlighting niPGT's suboptimal sensitivity and specificity compared to TE biopsy [
55,
66,
78,
83]. False negatives are more impactful for RPL patients given the high risk of miscarriage. Contrastingly, false positives are more detrimental for RIF patients with limited embryos [
56,
66,
83]. niPGT also exhibits high discordance; its results frequently disagree with TE biopsy. This undermines confidence in its ability to accurately reflect the embryo's true chromosomal status [
55,
66,
83].
Besides reliability and inconsistency, there is also regulatory skepticism. Major regulatory bodies such as FDA in the US and EMA in Europe currently classify niPGT as a research use only or investigational technology [
55,
56,
66]. The justification provided is the lack of sufficient clinical validation, standardized protocols, and concerns about reliability and potential misdiagnosis [
55,
56,
66] With such a regulatory stance, widespread clinical implementation and insurance coverage remain significantly limited [
55,
56].
Another key drawback is the lack of protocol standardization. For instance, the variance in culture conditions such as media type, volume, and exposure, significantly impact cfDNA quantity and quality [
10,
66]. Sample collection and handling, as well as DNA processing also vary between labs, directly contributing to inconsistent results [
55,
56,
66].
There is also the issue of unproven clinical utility in RPL/RIF. This is highlighted by the lack of robust evidence from well-designed RCTs demonstrating niPGT-A's efficiency over traditional methods in RPL or RIF populations [
48,
56,
83].
6.3. How IVF Centers Are Currently Using or Trialing niPGT
Clinical application is cautious and primarily experimental due to current limitations in evidence. niPGT is presently more leveraged in research and validation protocols [
10,
55,
56,
83]. For instance, Cheng et al. (2023) describe a protocol for a double-blinded RCT (NCT05622288) that seeks to compare niPGT-A to morphology alone [
10]. Such a trial is essential but the results are pending [
10]. It is also being leveraged as a back-up on specific scenarios. For instance it is being used as a rescue for biopsy failure, providing information where none was available [
66,
80]. It can be used when patients specifically prefer it. Individuals receive extensive counselling, more so about niPGT's limitations such as the risk of false negatives/positives and NR rates [
56,
66,
82]. It is also being used as adjunct to TE biopsy and morphokinetic assessment [
66]. Lastly, it is being leveraged for general infertility than a targeted first-line solution for RPL/RIF due to the higher risks involved [
56]. Despite these various applications, there lacks widespread adoption. Many IVF centers are cautious about niPGT's unresolved technical and validation issues, particularly when it comes to the welfare of RPL/RIF patients [
55,
56,
66].
6.4. Integration with Time-Lapse, Morphology, and AI
niPGT's efficiency and full potential lies in integration with other advanced embryo assessment tools. Amalgamated, they will create a multi-modal diagnostic approach. These tools include time-lapse imaging (TLI), which functions by providing continuous data on embryo morphokinetics [
85,
86]. Pertinent Integration aims to understand the dynamics of DNA shedding in relation to developmental events and embryo quality markers [
85]. It also targets to leverage pertinent morphokinetic parameters to predict which embryos are more likely to yield successful niPGT results [
85,
86]. In their overview, Oh et al. (2024) explore nucleolar dynamics as a potential marker [
85]. Additionally, integrating niPGT with TLI can enhance interpretation through holistic embryo assessment [
86]. With morphology, pertinent insights such as blastocyst grade alongside available niPGT results and TLI data can refine selection algorithms. This has the potential to mitigate the impact of niPGT's false results or NRs [
66,
86].
Integration with AI is crucial for several aspects. One of them is bioinformatics analysis. With effective AI algorithms, technicians can better distinguish true embryonic aneuploidy signals from background noise such as maternal DNA contamination in NGS data. This improves diagnostic accuracy [
66,
82,
83]. Further, AI integration underlies predictive modelling. There are numerous crucial datasets such as TLI, morphology, patient history, cfDNA characteristics, fragmentation patterns, and hormonal profiles. Integrating all of them via AI can predict embryo viability and ploidy status more accurately than any single method [
66,
82,
83,
86]. For instance, Stankewicz (2021) conceptualized optimizing IVF by controlling for both aneuploidy and receptivity using testing [
86]. This is a goal where integrated AI could play a role. AI can also be leveraged for complex scenarios. Managing and interpreting the vast, multi-dimensional data from various sources such as TLI and niPGT requires powerful AI [
66,
82].
6.5. Conclusion: Cautious Optimism Amid Significant Hurdles
niPGT offers a compelling vision for the future of embryo selection in RPL and RIF. It is truly noninvasive, embryo-preserving, and significantly more patient-centered [
1,
66,
82]. The theoretical benefits are substantial and have driven ongoing research [
10,
55] However, the current reality leans heavily towards its status as just hype. That is because it is not ready in any significant manner for routine clinical application in these challenging populations [
55,
56,
66,
83]. Key issues preventing it from reliably replacing invasive PGT-A include inconsistency, unreliability, lack of standardization, and regulatory skepticism. The most crucial of them all is the absence of proven clinical benefit in improving live birth rates for RPL/RIF [
48,
56,
83].
Some centers are already exploring its use in specific well-evaluated scenarios that include application in research protocols [
10,
56]. However, it is not the standard of care. The integration of niPGT with TLI, advanced morphology, and AI represents the most promising pathway forward. They have the potential to overcome current limitations by creating a more robust, multi-parameter assessment system [
66,
82,
83,
85,
86]. However, until these integrations become effective and rigorous evidence is available demonstrating clear efficacy and reliability in RIF/RPL, niPGT ought to be approached with caution and managed expectations. It presently just remains a promising tool under intense development, its application in managing RPL and RIF residing firmly in the realm of hope rather than established clinical practice [
55,
56,
66,
82,
83].
7. Future Perspectives and Clinical Recommendations
7.1. Need for Prospective, Multicenter RCTs
Current studies are largely retrospective or underpowered. Hence, there is a significant need for prospective, multicenter RCTs that compare LBR in niPGT-A versus TE-PGT-A or morphology-based selection. These trials must prioritize RPL/RIF subpopulations. This is where diagnostic accuracy impacts outcomes most significantly [
48,
56]. The focus of the pertinent RCTs should be on clinical utility such as the ability of niPGT-A reducing miscarriage rate, failure mitigation, and long-term outcomes, particularly on child health and development [
48,
56,
87].
7.2. Combining niPGT with Epigenetic and Metabolomic Profiling
The first key area is multi-omics integration. This entails epigenetic profiling where scholars can leverage cell-free DNA methylation patterns in SCM to assess embryonic epigenetic stability. This is relevant to implantation competence and miscarriage risk [
1,
87]. It also entails metabolomic signatures where the analysis of SCM metabolites such as amino acids may reflect embryo viability and mitochondrial function. In the case of amino acids, their flux such as glutamate increament is linked with euploidy. This offers complementary data to genetic aneuploidy screening [
1,
2]. The second key area is synergistic diagnostics which entails integrating niPGT-A with epigenomic or metabolomic biomarkers. This could enable epigenetic-metabolic-blastic readout for holistic embryo selection, especially in idiopathic RPL/RIF [
1,
48].
7.3. Standardization of Protocols and Reporting
Critical gaps in niPGT workflows highlight the urgent need for consensus as highlighted in
Table 1 below.
7.4. Tailored Approaches for Patient Subgroups
Risk-stratified niPGT applications are necessary. In AMA and DOR, technicians need to prioritize niPGT-A given the higher aneuploidy burden. However, they must validate against TE-biopsy due to the high risk of false-negatives [
48,
56]. In unexplained RPL, they should combine niPGT-A with endometrial immune profiling and thrombophilia panels. This is fundamental to addressing multifactorial etiology [
1,
88]. In young RIF patients, niPGT-A should be limited to research purposes only until more evidence is available confirming utility [
48]. Ethically, niPGT-A should be offered as a biopsy-free option after thorough counseling on its potential limitations such as the risk of false-negatives, which is critical for RPL [
56].
8. Conclusion: A Roadmap for Responsible Innovation
niPGT holds transformative potential. However, it requires rigorous validation before replacing TE biopsy. Until prospective RCTs confirm efficiency over TE-PGT-A, niPGT-A should remain investigational for RPL/RIF management. Its greatest value and potential lies in rescue testing for biopsy failures. It is also has the ability to thrive as a research tool in multi-parameter embryo selection models.
8.1. Reiterating the Promise and the Prerequisites
It is clear that niPGT’s potential is undeniable and compelling. Its core advantage is noninvasiveness, which addresses fundamental ethical and biological concerns associated with TE biopsy. This is through preserving embryo integrity and eliminating procedural risks. Through the strengths, it fosters enhanced patient acceptability, reducing potential negative psychological outcomes linked with embryo manipulation. Further, niPGT optimizes embryo preservation and allows flexibility in transfer timing. Research about its development is ongoing and this is premised on the core attributes of safety, efficiency, and patient-friendly genetic assessment.
8.2. Confronting the Current Reality: Barriers to Clinical Adoption
niPGT’s transition from promise to proven practice faces formidable hurdles. One key issue is the unresolved technical limitations. This is evidenced by the core issues of maternal DNA contamination, critically low and variable embryonic cfDNA yield, and significant inter-embryo biological variability. These aspects fundamentally compromise the reliability and consistency of results. They contribute to unacceptably high rates of test failure and diagnostic discordance compared to the traditional gold standard (TE biopsy). The second key issue is insufficient and inconclusive clinical evidence. Currently, there are no well-designed, large-scale RCTs demonstrating the efficiency of niPGT-A in improving LBRs or reducing miscarriage rates specifically in RPL or RIF patients compared to the conventional methods. The variable sensitivity and specificity fall short of the diagnostic accuracy required for routine clinical reliance in RIF/RPL. In fact, evidence suggests its performance in these specific populations may be suboptimal.
The third key issue is the lack of standardization and regulatory hurdles. This underlies inconsistent results across labs. It is based on this that reliability issues persist, informing regulatory skepticism that has seen niPGT being relegated to investigational purposes only. The last aspect is its unproven utility in complex etiologies.
8.3. Imperative Call for Cautious Integration and Further Research
The path forward demands cautious integration and intensified research. Firstly, rigorous RCTs comparing its outcomes with the current standards of TLI and TE PGT-A are needed. Secondly, standardization is needed to foster and guarantee consistency, comparability, and data quality. Thirdly, key stakeholders should explore multi-omic integration, as well as with TLI and AI-driven predictive modelling to enhance viability assessment. Fourthly, its application needs to be tailored and cautious, being leveraged only for certain selected scenarios, used as an adjunct, and implemented mostly in favorable cases and validity and reliability established.
Overall, as niPGT shines as a beacon of hope for a less invasive future in embryo selection, its light is currently too dim to effectively guide clinical decision-making for the complex and emotionally draining cases of RIF and RPL. Widespread adoption requires not just optimism. Instead, concrete evidence from rigorous research, standardized methodologies, and proven efficacy in the specific populations it aims to serve are needed. Hence, evidence-based integration, not haste or hope, need to define the next chapter of niPGT development.
Author Contributions
Conceptualization & Supervision: G.M.; Writing – Original Draft & Project Administration: A.M., J.G.; Literature Search & Data Curation: J.G., O.S., W.S., J.M., Y.S.; Writing – Review & Editing: G.M., A.M., L.K., W.G., P.B., A.B.; Visualization: L.K.; Correspondence: A.M.
Funding
This research received no external funding.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not applicable. No individual patient data are presented.
Data availability statement
Not Applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Abbreviation |
Full Form |
| ADO |
Allele Dropout |
| AI |
Artificial Intelligence |
| AMA |
Advanced Maternal Age |
| AMH |
Anti-Müllerian Hormone |
| ANXA5 |
Annexin A5 |
| APS |
Antiphospholipid Syndrome |
| ART |
Assisted Reproductive Technology |
| BF |
Blastocoel Fluid |
| cfDNA |
Cell-Free DNA |
| CLBR |
Cumulative Live Birth Rate |
| DOR |
Diminished Ovarian Reserve |
| EQC |
External Quality Controls |
| ESHRE |
European Society of Human Reproduction and Embryology |
| HLA |
Human Leukocyte Antigen |
| ICM |
Inner Cell Mass |
| IR |
Implantation Rate |
| ISO |
International Organization for Standardization |
| LBR |
Live Birth Rate |
| LP-WGS |
Low-Pass Whole Genome Sequencing |
| MDA |
Multiple Displacement Amplification |
| NGS |
Next-Generation Sequencing |
| niPGT |
Noninvasive Preimplantation Genetic Testing |
| niPGT-A |
Noninvasive Preimplantation Genetic Testing for Aneuploidy |
| niPGT-M |
Noninvasive Preimplantation Genetic Testing for Monogenic disorders |
| niPGT-SR |
Noninvasive Preimplantation Genetic Testing for Structural Rearrangements |
| NK cells (uNKs) |
Natural Killer cells (Uterine Natural Killer cells) |
| NR |
No Result |
| PCOS |
Polycystic Ovary Syndrome |
| PGT |
Preimplantation Genetic Testing |
| PGT-A |
Preimplantation Genetic Testing for Aneuploidy |
| PGT-M |
Preimplantation Genetic Testing for Monogenic disorders |
| PGT-SR |
Preimplantation Genetic Testing for Structural Rearrangements |
| PPV/NPV |
Positive Predictive Value / Negative Predictive Value |
| qPCR |
Quantitative Polymerase Chain Reaction |
| RCT |
Randomized Controlled Trial |
| RIF |
Recurrent Implantation Failure |
| RPL |
Recurrent Pregnancy Loss |
| SCM |
Spent Culture Medium |
| SET |
Single Embryo Transfer |
| STARD-PGT |
Standards for Reporting Diagnostic Accuracy Studies - PGT |
| STR |
Short Tandem Repeat |
| TE |
Trophectoderm |
| TLI |
Time-Lapse Imaging |
| Treg |
Regulatory T cell |
| TSH |
Thyroid-Stimulating Hormone |
| WGA |
Whole Genome Amplification |
| WGS |
Whole Genome Sequencing |
References
- Asghari, K.M.; Novinbahador, T.; Mehdizadeh, A.; Zolfaghari, M.; Yousefi, M. Revolutionized attitude toward recurrent pregnancy loss and recurrent implantation failure based on precision regenerative medicine. Heliyon 2024, 10. [Google Scholar]
- Cao, C.; Bai, S.; Zhang, J.; Sun, X.; Meng, A.; Chen, H. Understanding recurrent pregnancy loss: recent advances on its etiology, clinical diagnosis, and management. Medical Review 2023, 2, 570–589. [Google Scholar] [CrossRef] [PubMed]
- Barad, D.H.; Albertini, D.F.; Molinari, E.; Gleicher, N. IVF outcomes of embryos with abnormal PGT-A biopsy previously refused transfer: a prospective cohort study. Human Reproduction 2022, 37, 1194–1206. [Google Scholar] [CrossRef]
- Busnelli, A.; Somigliana, E.; Cirillo, F.; Baggiani, A.; Levi-Setti, P.E. Efficacy of therapies and interventions for repeated embryo implantation failure: a systematic review and meta-analysis. Scientific Reports 2021, 11, 1747. [Google Scholar] [CrossRef]
- Bakalova, D.N.; Navarro-Sánchez, L.; Rubio, C. (2025). Non-Invasive Preimplantation Genetic Testing.
- Capalbo, A.; Wells, D. The evolution of preimplantation genetic testing: where is the limit? Reproductive BioMedicine Online 2025, 50, 104845. [Google Scholar] [CrossRef]
- Cavalcante, M.B.; Sarno, M.; Barini, R. Immune biomarkers in cases of recurrent pregnancy loss and recurrent implantation failure. Minerva obstetrics and gynecology 2025, 77, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Hu, Z.; Lian, Y.; Han, Y.; Zhou, X.; Li, Y.; Xiang, L.; Jiang, W.; Li, M.; Zeng, P.; et al. The diagnostic accuracy of preimplantation genetic testing (PGT) in assessing the genetic status of embryos: a systematic review and meta-analysis. Reproductive Biology and Endocrinology 2025, 23, 39. [Google Scholar] [CrossRef]
- Chen, S.; Wang, L.; Hu, Y.; Yao, Y.; Gao, F.; Chang, C.; Zhang, L.; Huang, H.; Lu, D.; Xu, C. Noninvasive preimplantation genetic testing for aneuploidy using blastocyst spent culture medium may serve as a backup of trophectoderm biopsy in conventional preimplantation genetic testing. BMC Medical Genomics 2025, 18, 34. [Google Scholar] [CrossRef]
- Cheng, H.Y.H.; Chow, J.F.; Lam, K.K.; Lai, S.F.; Yeung, W.S.B.; Ng, E.H. Randomised double-blind controlled trial of non-invasive preimplantation genetic testing for aneuploidy in in vitro fertilisation: a protocol paper. BMJ open 2023, 13, e072557. [Google Scholar] [CrossRef]
- Chow, J.F.; Lam, K.K.; Cheng, H.H.; Lai, S.F.; Yeung, W.S.; Ng, E.H. Optimizing non-invasive preimplantation genetic testing: investigating culture conditions, sample collection, and IVF treatment for improved non-invasive PGT-A results. Journal of Assisted Reproduction and Genetics 2024, 41, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Nagori, C.; Panchal, S. (2022). Practical Guide to Recurrent Pregnancy Loss. Jaypee Brothers Medical Publishers.
- Ma, J.; Gao, W.; Li, D. Recurrent implantation failure: A comprehensive summary from etiology to treatment. Frontiers in Endocrinology 2023, 13, 1061766. [Google Scholar] [CrossRef]
- Sheikhansari, G.; Pourmoghadam, Z.; Danaii, S.; Mehdizadeh, A.; Yousefi, M. Etiology and management of recurrent implantation failure: a focus on intra-uterine PBMC-therapy for RIF. Journal of reproductive immunology 2020, 139, 103121. [Google Scholar] [CrossRef]
- Wang, C.; Lu, Y.; Ou, M.; Qian, L.; Zhang, Y.; Yang, Y.; Luo, L.; Wang, Q. Risk factors for recurrent implantation failure as defined by the European Society for Human Reproduction and Embryology. Human Reproduction 2025, deaf042. [Google Scholar] [CrossRef]
- Gonçalves, B.D.O. (2022). Aetiopathogenesis of Recurrent implantation failure after assisted reproductive techniques (Master's thesis).
- Linehan, L.; Hennessy, M.; O'Donoghue, K. Infertility and subsequent recurrent miscarriage: current state of the literature and future considerations for practice and research. HRB Open Research 2021, 4, 100. [Google Scholar] [CrossRef]
- Cuadrado-Torroglosa, I.; García-Velasco, J.A.; Alecsandru, D. Maternal--Fetal Compatibility in Recurrent Pregnancy Loss. Journal of Clinical Medicine 2024, 13, 2379. [Google Scholar] [CrossRef]
- Yang, X.; Tian, Y.; Zheng, L.; Luu, T.; Kwak-Kim, J. The update immune-regulatory role of pro-and anti-inflammatory cytokines in recurrent pregnancy losses. International journal of molecular sciences 2022, 24, 132. [Google Scholar] [CrossRef] [PubMed]
- Garmendia, J.V.; De Sanctis, C.V.; Hajdúch, M.; De Sanctis, J.B. Microbiota and recurrent pregnancy loss (RPL); more than a simple connection. Microorganisms 2024, 12, 1641. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; He, Z.; Ma, R.; Lin, N.; Li, L.; Li, Y.; Li, Y.; Ke, Y.; Meng, X.; Wu, Z. Narrative review of multifaceted approaches to managing recurrent implantation failure: insights and innovations. Clinical and Experimental Obstetrics Gynecology 2024, 51, 87. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Omidvar-Mehrabadi, A.; Shahbazi, M.; Mohammadnia-Afrouzi, M. Innate and adaptive immune dysregulation in women with recurrent implantation failure. Journal of Reproductive Immunology 2024, 164, 104262. [Google Scholar] [CrossRef] [PubMed]
- Lash, G.E.; Raghupathy, R. Immune involvement in recurrent pregnancy loss. Frontiers in Immunology 2023, 14, 1330701. [Google Scholar] [CrossRef]
- Sfakianoudis, K.; Rapani, A.; Grigoriadis, S.; Pantou, A.; Maziotis, E.; Kokkini, G.; Tsirligkani, C.; Bolaris, S.; Nikolettos, K.; Chronopoulou, M.; et al. The role of uterine natural killer cells on recurrent miscarriage and recurrent implantation failure: from pathophysiology to treatment. Biomedicines 2021, 9, 1425. [Google Scholar] [CrossRef] [PubMed]
- Garmendia, J.V.; De Sanctis, C.V.; Hajdúch, M.; De Sanctis, J.B. Exploring the Immunological Aspects and Treatments of Recurrent Pregnancy Loss and Recurrent Implantation Failure. International Journal of Molecular Sciences 2025, 26, 1295. [Google Scholar] [CrossRef]
- Mukherjee, N.; Sharma, R.; Modi, D. Immune alterations in recurrent implantation failure. American Journal of Reproductive Immunology 2023, 89, e13563. [Google Scholar] [CrossRef]
- George, J.S.; Mortimer, R.; Anchan, R.M. (2022). The role of reproductive immunology in recurrent pregnancy loss and repeated implantation failure. In Immunology of Recurrent Pregnancy Loss and Implantation Failure (pp. 223-240). Academic Press.
- Nakagawa, K.; Kuroda, K.; Sugiyama, R. Helper T cell pathology and repeated implantation failures. Immunology of Recurrent Pregnancy Loss and Implantation Failure 2022, 273–285. [Google Scholar]
- Uța, C.; Tîrziu, A.; Zimbru, E.L.; Zimbru, R.I.; Georgescu, M.; Haidar, L.; Panaitescu, C. Alloimmune Causes of Recurrent Pregnancy Loss: Cellular Mechanisms and Overview of Therapeutic Approaches. Medicina 2024, 60, 1896. [Google Scholar] [CrossRef]
- Montazeri, F.; Tajamolian, M.; Hosseini, E.S.; Hoseini, S.M. Immunologic Factors and Genomic Considerations in Recurrent Pregnancy Loss: A Review. International Journal of Medical Laboratory 2023. [Google Scholar] [CrossRef]
- Davalieva, K.; Kocarev, D.; Plaseska-Karanfilska, D. Decoding recurrent pregnancy loss: insights from comparative proteomics studies. Biology of Reproduction 2025, 112, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liaqat Ali Khan, N.; Nafee, T.; Shao, T.; Hart, A.R.; Elliott, S.; Ola, B.; Heath, P.R.; Fazeli, A. Dysregulation in multiple transcriptomic endometrial pathways is associated with recurrent implantation failure and recurrent early pregnancy loss. International Journal of Molecular Sciences 2022, 23, 16051. [Google Scholar] [CrossRef]
- Nørgaard-Pedersen, C. (2023). Recurrent Pregnancy Loss: immunological risk factors and immunomodulatory treatments.
- Zeng, Y.; Tu, W. Predicting Pregnancy Outcomes in the Recurrent Reproductive Failure. Immune Regulations in Reproductive Organs and Organ Transplant 2022, 18, 64216768. [Google Scholar]
- Sato, T.; Sugiura-Ogasawara, M.; Ozawa, F.; Yamamoto, T.; Kato, T.; Kurahashi, H.; Kuroda, T.; Aoyama, N.; Kato, K.; Kobayashi, R.; Fukuda, A.; Utsunomiya, T.; Kuwahara, A.; Saito, H.; Takeshita, T.; Irahara, M. Preimplantation genetic testing for aneuploidy: a comparison of live birth rates in patients with recurrent pregnancy loss due to embryonic aneuploidy or recurrent implantation failure. Human reproduction (Oxford, England) 2019, 34, 2340–2348. [Google Scholar] [CrossRef]
- Wang, X.; Chen, B.; Fang, L.; Wang, J.; Xu, A.; Xu, W.; Tong, X. Aneuploidy rates and clinical pregnancy outcomes after preimplantation genetic testing for aneuploidy using the progestin-primed ovarian stimulation protocol or the gonadotropin-releasing hormone antagonist protocol. Journal of Gynecology Obstetrics and Human Reproduction 2025, 54, 102883. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.X.; Ye, J.F.; Sui, Y.L.; Zhang, Y.P.; Sun, X.X. Retrospective cohort study of preimplantation genetic testing for aneuploidy with comprehensive chromosome screening versus nonpreimplantation genetic testing in normal karyotype, secondary infertility patients with recurrent pregnancy loss. Reproductive and Developmental Medicine 2019, 3, 205–212. [Google Scholar] [CrossRef]
- Kato, K.; Kuroda, T.; Yamadera-Egawa, R.; Ezoe, K.; Aoyama, N.; Usami, A.; Miki, T.; Yamamoto, T.; Takeshita, T. Preimplantation genetic testing for aneuploidy for recurrent pregnancy loss and recurrent implantation failure in minimal ovarian stimulation cycle for women aged 35--42 years: live birth rate, developmental follow-up of children, and embryo ranking. Reproductive Sciences 2023, 30, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Du, R.Q.; Zhao, D.D.; Kang, K.; Wang, F.; Xu, R.X.; Chi, C.L.; Kong, L.Y.; Liang, B. A review of pre-implantation genetic testing technologies and applications. Reproductive and Developmental Medicine 2023, 7, 20–31. [Google Scholar] [CrossRef]
- Wang, S.; Liu, L.; Ma, M.; Wang, H.; Han, Y.; Guo, X.; Yeung, W.S.; Cheng, Y.; Zhang, H.; Dong, F.; et al. Preimplantation genetic testing for aneuploidy helps to achieve a live birth with fewer transfer cycles for the blastocyst FET patients with unexplained recurrent implantation failure. Archives of Gynecology and Obstetrics 2023, 308, 599–610. [Google Scholar] [CrossRef]
- Du, Y.; Guan, Y.; Li, N.; Shi, C.; Zhang, Y.; Ren, B.; Liu, J.; Lou, H. Is it necessary for young patients with recurrent implantation failure to undergo preimplantation genetic testing for aneuploidy? Frontiers in Endocrinology 2023, 14, 1020055. [Google Scholar] [CrossRef]
- Guan, H.; He, Y.; Lu, Y.; Huang, J.; Wang, Y.; Zhu, Q.; Qi, J.; Lin, W.; Lindheim, S.R.; Wei, Z.; et al. Effect of Preimplantation Genetic Testing for Aneuploidy on Live Birth Rate in Young Women With Recurrent Implantation Failure: A Secondary Analysis of a Multicentre Randomised Trial. BJOG: An International Journal of Obstetrics Gynaecology 2024. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, S.Y.; Hur, C.Y.; Lim, J.H. P-507 Prediction of clinical efficacy of PGT-A based on maternal age and embryo quality in patients with recurrent implantation failure (RIF) or recurrent pregnancy loss (RPL). Human Reproduction 2024, 39(Supplement_1), deae108-847. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, S.Y.; Hur, C.Y.; Lim, J.H.; Park, C.K. Clinical outcomes of preimplantation genetic testing for aneuploidy in high-risk patients: a retrospective cohort study. Clinical and Experimental Reproductive Medicine 2023, 51, 75. [Google Scholar] [CrossRef]
- Kornfield, M.S.; Parker, P.; Rubin, E.; Garg, B.; O'Leary, T.; Amato, P.; Lee, D.; Wu, D.; Krieg, S. Patients with Recurrent Pregnancy Loss Have Similar Embryonic Preimplantation Genetic Testing Aneuploidy Rates and In Vitro Fertilization Outcomes to Infertility Patients. F&S Reports 2022, 3, 342–348. [Google Scholar] [CrossRef]
- Liang, Z.; Wen, Q.; Li, J.; Zeng, D.; Huang, P. A systematic review and meta-analysis: clinical outcomes of recurrent pregnancy failure resulting from preimplantation genetic testing for aneuploidy. Frontiers in Endocrinology 2023, 14, 1178294. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lan, X.; Lu, J.; Zhang, Q.; Zhou, T.; Ni, T.; Yan, J. Preimplantation genetic testing for aneuploidy could not improve cumulative live birth rate among 705 couples with unexplained recurrent implantation failure. The Application of Clinical Genetics 2024, 1–13. [Google Scholar] [CrossRef]
- Mei, Y.; Lin, Y.; Chen, Y.; Zheng, J.; Ke, X.; Liang, X.; Wang, F. Preimplantation genetic testing for aneuploidy optimizes reproductive outcomes in recurrent reproductive failure: a systematic review. Frontiers in Medicine 2024, 11, 1233962. [Google Scholar] [CrossRef] [PubMed]
- Mantravadi, K.; Mathew, S.; Soorve, S.; Rao, D.G.; Karunakaran, S. Does pre-implantation genetic testing for aneuploidy optimise the reproductive outcomes in women with idiopathic recurrent pregnancy loss? Fertility and Sterility 2020, 114, e437. [Google Scholar] [CrossRef]
- Sarkar, P.; Jindal, S.; New, E.P.; Sprague, R.G.; Tanner, J.; Imudia, A.N. The role of preimplantation genetic testing for aneuploidy in a good prognosis IVF population across different age groups. Systems Biology in Reproductive Medicine 2021, 67, 366–373. [Google Scholar] [CrossRef]
- Orvieto, R. Does preimplantation genetic testing for aneuploidy really improve IVF outcomes in advanced maternal age patients without compromising cumulative live-birth rate? Journal of Assisted Reproduction and Genetics 2020, 37, 159–159. [Google Scholar] [CrossRef]
- Kasaven, L.S.; Marcus, D.; Theodorou, E.; Jones, B.P.; Saso, S.; Naja, R.; Serhal, P.; Ben-Nagi, J. Systematic review and meta-analysis: does pre-implantation genetic testing for aneuploidy at the blastocyst stage improve live birth rate? Journal of Assisted Reproduction and Genetics 2023, 40, 2297–2316. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Bogale, B.; Tang, Y.; Lu, S.; Xie, X.S.; Racowsky, C. Noninvasive preimplantation genetic testing for aneuploidy in spent medium may be more reliable than trophectoderm biopsy. Proceedings of the National Academy of Sciences 2019, 116, 14105–14112. [Google Scholar] [CrossRef]
- Giuliano, R.; Maione, A.; Vallefuoco, A.; Sorrentino, U.; Zuccarello, D. Preimplantation genetic testing for genetic diseases: limits and review of current literature. Genes 2023, 14, 2095. [Google Scholar] [CrossRef]
- Kakourou, G.; Sofocleous, C.; Mamas, T.; Vrettou, C.; Traeger-Synodinos, J. The current clinical applications of preimplantation genetic testing (PGT): acknowledging the limitations of biology and technology. Expert Review of Molecular Diagnostics 2024, 24, 767–775. [Google Scholar] [CrossRef]
- Morales, C. Current applications and controversies in preimplantation genetic testing for aneuploidies (PGT-A) in in vitro fertilization. Reproductive Sciences 2024, 31, 66–80. [Google Scholar] [CrossRef]
- Viville, S.; Aboulghar, M. PGT-A: what's it for, what's wrong? Journal of Assisted Reproduction and Genetics 2025, 42, 63–69. [Google Scholar] [CrossRef]
- Phillips, K.R.; Kuzma-Hunt, A.G.; Neal, M.S.; Lisle, C.; Sribalachandran, H.; Carter, R.F.; Amin, S.; Karnis, M.F.; Faghih, M. Temporal Evaluation of a Minimally Invasive Method of Preimplantation Genetic Testing for Aneuploidy (mi-PGT-A) in Human Embryos. Reproductive Medicine 2024, 5. [Google Scholar] [CrossRef]
- He, F.; Yao, Y.X.; Wang, J.; Zhao, D.M.; Wan, A.Q.; Ren, J.; Lei, X. Non-invasive chromosome screening for embryo preimplantation using cell-free DNA. Reproductive and Developmental Medicine 2022, 6, 113–120. [Google Scholar] [CrossRef]
- Del Collado, M.; Andrade, G.M.; Gonçalves, N.J.N.; Fortini, S.; Perecin, F.; Carriero, M.M. The embryo non-invasive pre-implantation diagnosis era: how far are we? Animal Reproduction 2023, 20, e20230069. [Google Scholar] [CrossRef]
- Shitara, A.; Takahashi, K.; Goto, M.; Takahashi, H.; Iwasawa, T.; Onodera, Y.; Makino, K.; Miura, H.; Shirasawa, H.; Sato, W.; et al. Cell-free DNA in spent culture medium effectively reflects the chromosomal status of embryos following culturing beyond implantation compared to trophectoderm biopsy. PLoS One 2021, 16, e0246438. [Google Scholar] [CrossRef]
- Sialakouma, A.; Karakasiliotis, I.; Ntala, V.; Nikolettos, N.; Asimakopoulos, B. Embryonic cell-free DNA in spent culture medium: a non-invasive tool for aneuploidy screening of the corresponding embryos. in vivo 2021, 35, 3449–3457. [Google Scholar] [CrossRef]
- Xie, P.; Zhang, S.; Gu, Y.; Jiang, B.; Hu, L.; Tan, Y.Q.; Yao, Y.; Tang, Y.; Wan, A.; Cai, S.; et al. Non-invasive preimplantation genetic testing for conventional IVF blastocysts. Journal of Translational Medicine 2022, 20, 396. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, B.R. Great Challenges Remain for niPGT-A Reliability. Revista Brasileira de Ginecologia e Obstetrícia/RBGO Gynecology and Obstetrics 2022, 44, 721–722. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, Y.; Chen, K.; Li, M.; Tian, M.; Xiang, L.; Wu, X.; Zeng, P.; Li, M.; Shao, J.; et al. The comparison of two whole-genome amplification approaches for noninvasive preimplantation genetic testing (ni-PGT) and the application scenario of ni-PGT during the fresh cycle. The Journal of Molecular Diagnostics 2023, 25, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Karami, N.; Iravani, F.; Bavarsad, S.B.; Asadollahi, S.; Hoseini, S.M.; Montazeri, F.; Kalantar, S.M. Comparing the advantages, disadvantages and diagnostic power of different non-invasive pre-implantation genetic testing: A literature review. International Journal of Reproductive BioMedicine 2024, 22, 177. [Google Scholar] [CrossRef]
- Sousa, L.N.; Monteiro, P.B. Non-invasive preimplantation genetic testing: a literature review. JBRA Assisted Reproduction 2022, 26, 554. [Google Scholar] [CrossRef]
- Rubio, C.; Navarro-Sánchez, L.; García-Pascual, C.M. (2022). Embryo Cell-Free DNA in the Culture Medium and Its Potential for Non-Invasive Aneuploidy Testing. In Handbook of Genetic Diagnostic Technologies in Reproductive Medicin (pp. 160-172). CRC Press.
- Vendrell, X.; Escribà, M.J. Non-invasive PGT. Medicina Reproductiva y Embriología Clínica 2021, 8, 100101. [Google Scholar] [CrossRef]
- Sakkas, D.; Navarro-Sánchez, L.; Ardestani, G.; Barroso, G.; Bisioli, C.; Boynukalin, K.; Cimadomo, D.; Frantz, N.; Kopcow, L.; Andrade, G.M.; et al. The impact of implementing a non-invasive preimplantation genetic testing for aneuploidies (niPGT-A) embryo culture protocol on embryo viability and clinical outcomes. Human Reproduction 2024, 39, 1952–1959. [Google Scholar] [CrossRef]
- Volovsky, M.; Scott Jr, R.T.; Seli, E. Non-invasive preimplantation genetic testing for aneuploidy: is the promise real? Human Reproduction 2024, 39, 1899–1908. [Google Scholar] [CrossRef]
- Rogers, A.; Menezes, M.; Kane, S.C.; Zander-Fox, D.; Hardy, T. Preimplantation genetic testing for monogenic conditions: is cell-free DNA testing the next step? Molecular diagnosis therapy 2021, 25, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ai, Z.; Zhu, X.; Che, Z.; Pratikshya, A.; Tang, S.; Zhang, Q. Analysis of predictors of clinical pregnancy and live birth in patients with RIF treated with IVF-ET technology: A cohort study based on a propensity score approach. Frontiers in Medicine 2024, 11, 1348733. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; et al. Cell-free DNA in spent embryo culture medium reflects aneuploidy and mosaicism. Nature Communications 2016, 7, 11165. [Google Scholar]
- Rubio, C.; et al. Use of noninvasive preimplantation genetic testing (niPGT) to select euploid embryos in IVF: A prospective multicenter study. Fertility and Sterility 2020, 114, 538. [Google Scholar]
- Huang, L.; et al. Noninvasive preimplantation genetic testing for aneuploidy: Validation of a targeted next-generation sequencing-based method. Human Reproduction 2019, 34, 281. [Google Scholar]
- Hanson, B.M.; Tao, X.; Hong, K.H.; Comito, C.E.; Pangasnan, R.; Seli, E.; Jalas, C.; Scott, R.T., Jr. Noninvasive preimplantation genetic testing for aneuploidy exhibits high rates of deoxyribonucleic acid amplification failure and poor correlation with results obtained using trophectoderm biopsy. Fertility and Sterility 2021, 115, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Luo, X.; Wu, R.; Qiu, L.; Lin, S.; Huang, X.; Wu, J. Evaluation of non-invasive gene detection in preimplantation embryos: a systematic review and meta-analysis. Journal of Assisted Reproduction and Genetics 2023, 40, 1243–1253. [Google Scholar] [CrossRef]
- Li Piani, L.; Petrone, P.; Brutto, M.; De Vos, A.; Van Der Kelen, A.; Vaiarelli, A.; Rienzi, L.; Conforti, A.; Cimadomo, D.; Verpoest, W. A systematic review and meta-analysis of double trophectoderm biopsy and/or cryopreservation in PGT: balancing the need for a diagnosis against the risk of harm. Human Reproduction Update 2025, 31, 102–115. [Google Scholar] [CrossRef]
- Li, X.; Hao, Y.; Chen, D.; Ji, D.; Zhu, W.; Zhu, X.; Wei, Z.; Cao, Y.; Zhang, Z.; Zhou, P. Non-invasive preimplantation genetic testing for putative mosaic blastocysts: a pilot study. Human Reproduction 2021, 36, 2020–2034. [Google Scholar] [CrossRef]
- Rubio, C.; Racowsky, C.; Barad, D.H.; Scott, R.T.; Simon, C. Noninvasive preimplantation genetic testing for aneuploidy in spent culture medium as a substitute for trophectoderm biopsy. Fertility and Sterility 2021, 115, 841–849. [Google Scholar] [CrossRef]
- Leaver, M.; Wells, D. Non-invasive preimplantation genetic testing (niPGT): the next revolution in reproductive genetics? Human reproduction update 2020, 26, 16–42. [Google Scholar] [CrossRef] [PubMed]
- Tiegs, A.W.; et al. Assessment of noninvasive preimplantation genetic testing: what are we missing? Current Opinion in Obstetrics and Gynecology 2021, 33, 138–145. [Google Scholar]
- Girardi, G.; Bremer, A.A. Advancing research on recurrent pregnancy loss: Overcoming obstacles and opportunities for translation. American Journal of Reproductive Immunology 2022, 87, e13508. [Google Scholar] [CrossRef]
- Oh, H.S.; Jang, J.M.; Yoon, H.J.; Choo, C.W.; Lim, K.S.; Lim, J.H.; Cheon, Y.P. The kinetics of nucleolar precursor bodies clustering at the pronuclei interface: Positive correlations with the morphokinetic characteristics of cleaving embryos and euploidy in preimplantation genetic testing programs. Korean Journal of Fertility and Sterility 2024. [CrossRef]
- Stankewicz, T. (2021). Optimizing IVF by controlling for both embryonic aneuploidy and endometrial receptivity using genetic testing. University of Kent (United Kingdom).
- Ying, L.I. (2022). Characterizing the Spectrum of Genome-wide Chromosomal Abnormalities in Early Pregnancy Loss by Lowpass Genome Sequencing (Doctoral dissertation, The Chinese University of Hong Kong (Hong Kong)).
- Peng, L.; Yang, W.; Deng, X.; Bao, S. Research progress on ANXA5 in recurrent pregnancy loss. Journal of Reproductive Immunology 2022, 153, 103679. [Google Scholar] [CrossRef] [PubMed]
- Petch, S.; Crosby, D. Updates in preimplantation genetic testing (PGT). Best Practice Research Clinical Obstetrics Gynaecology 2024, 102526. [Google Scholar] [CrossRef]
- Rubio, C.; Simón, C. Noninvasive preimplantation genetic testing for aneuploidy: Is the glass half-empty or half-full? Fertility and Sterility 2021, 115, 1426–1427. [Google Scholar] [CrossRef]
- Kakourou, G.; Mamas, T.; Vrettou, C.; Traeger-Synodinos, J. An update on non-invasive approaches for genetic testing of the preimplantation embryo. Current Genomics 2022, 23, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Montazeri, F.; Kalantar, S.M. Comparing the advantages, disadvantages and diagnostic power of different non-invasive pre-implantation genetic testing: A literature.
- Li, J.; Liu, Y.; Qian, Y.; Zhang, D. Noninvasive preimplantation genetic testing in assisted reproductive technology: current state and future perspectives. Journal of Genetics and Genomics 2020, 47, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Wang, Y.; He, L.; Zheng, J.; Lin, Y.; Wang, F. Performance of preimplantation genetic testing for aneuploidy for patients with unexplained recurrent pregnancy loss and repeated implantation failure. Heliyon 2024, 10. [Google Scholar] [CrossRef] [PubMed]
Table 1.
niPGT’s standardization requirements for the technology’s improvement. For reporting frameworks, stakeholders should Implement STARD-NiPGT guidelines to ensure transparency in sensitivity and specificity metrics and NR rates [
56].
Table 1.
niPGT’s standardization requirements for the technology’s improvement. For reporting frameworks, stakeholders should Implement STARD-NiPGT guidelines to ensure transparency in sensitivity and specificity metrics and NR rates [
56].
| Domain |
Standardization requirements |
| Culture conditions |
Media type/volume, change intervals, incubation duration [2,56] |
| Sample handling |
SCM collection timing, storage, contamination controls [56] |
| Analytical methods |
WGA kits, sequencing depth, bioinformatics [2,56] |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).