Submitted:
03 September 2025
Posted:
04 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Overview of Acacetin
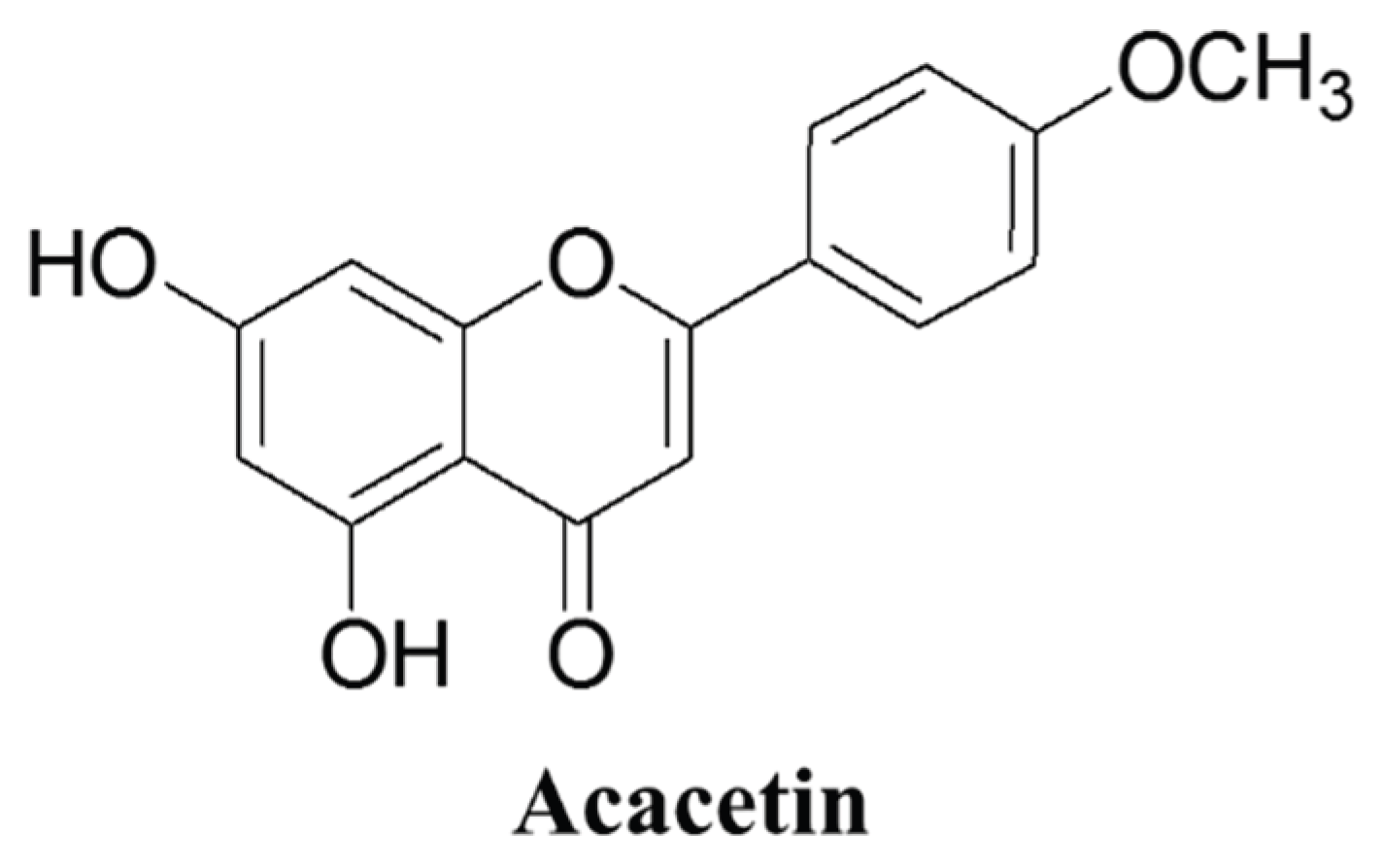
2.1. Chemical Structure and Sources
2.2. Pharmacological Activities of Acacetin
3. Pharmacological Evidence of Acacetin’s Anticancer Activity
| Interventions Cancer Type |
Cell Line(s) / Model | Up-Regulation | Down-Regulation | Mechanisms | Refs. |
|---|---|---|---|---|---|
| Breast | T-47D, MDA-MB-231, MCF-7, MDA-MB-468, |
DNA fragmentation, sub-G1 cells, Cyt c (cytosolic) AIF (cytosolic), p-SAPK/ JNK1/2, p-c-Jun, JNK1/2, ROS, cleaved caspase-7,-8 and -9, cleaved PARP, p-ERK, Chk1, Bax, Mitochondrial superoxide generation, RIP1, RIP3 p21, p27 |
Bcl-2, MMP, Cyt c (mitochondrial), AIF (mitochondrial) CDK1, CDK2, Cdc25C, Cyclin B1, Cyclin E, AKT, FADD, p-AKT, AKT, p38, ERK |
Apoptosis, Necroptosis, G2/M arrest, ERK activation Migration and EMT inhibition |
[41,42,43] |
| Colorectal |
HT-29, HCT 116, SW480 | ROS, MMP (mitochondrial), AIF | β-catenin, c-Myc, | Apoptosis, S and G2/M arrest |
[44,45] |
| Esophageal | TE-1, TE-10 | Bax | Ki-67, MMP-2 and -9, Bcl-2, p-JAK2, p-STAT3 | Apoptosis, JAK2/STAT3 inhibition, Migration suppression |
[26] |
| Gastric | MKN45, MKN45 xenograft, MGC803, AGS | E-cadherin, Bax, cleaved PARP, cleaved caspase-3, DNA fragmentation, Sub-G1 cells, ROS, Cyt c (cytosolic) caspase-3,-8, and -9 activity, Fas, FasL, cleaved Bid, p53 |
N-cadherin, MMP-2 and -9, Snail, p-PI3K, p-AKT, p-EGFR, Bcl-xL, p-STAT3, p-ERK, p-EGFR, PCNA, DFF-45, MMP | Apoptosis, EMT suppression, Anti-metastasis, PI3K/AKT/Snail inhibition |
[12,46,47] |
| Liver | HepG2, HepG2/RARγ xenografts | DNA fragmentation, cleaved PARP, p21, p53, FasL, mFasL, sFasL, caspase-8 activity, Bax, cleaved caspase-3, | p-STAT3, p-JAK1,p-JAK2, p-AKT, p-Src, Cyclin D1, Bcl-2, Bcl-xL, Mcl-1, Survivin, Mcl-2, VEGF, p-IκBα (cytosolic), p-p65 (cytosolic), pro-casapase-3, RARγ, p-GSK-3β, Ki67 | Apoptosis, STAT3 inhibition, Invasion inhibition, Angiogenesis inhibition, G1 arrest, Apoptosis via non-genomic RARγ-AKT-p53 pathway |
[48,49,50] |
| Lung | A549, H460, A549 xenografts | Bak, p53, miR-34a, IκBα(cytosolic), G1 phase, p21, Fas, FasL | Cyclin B1, , Cyclin D, Bcl-2, PD-L1, MMP-2 and -9, u-PA, p-p38α, p-MKK3, 6, p-MLK3, NF-κB, NF-κB p50 (nuclear), NF-κB p65 (nuclear) AP-1, c-Fos, c-Jun, p-IκBα (cytosolic) | Proliferation, invasion, and migration inhibition, G2/M arrest, Apoptosis |
[27,28,51,52] |
| Ovarian | SKOV3, OVCAR-3, A2780, A2780 (CAM assay in vivo model) | AKT | PCNA, MMP-2 and -9, IL-6 and -8, RAGE, p-PI3K, p-AKT, VEGF, HIF-1α | Cell proliferation and invasion inhibition, Apoptosis, RAGE-PI3K/AKT inhibition, Angiogenesis inhibition, AKT/HIF-1α inhibition |
[53,54] |
| Osteosarcoma | HOS | Bax, cleaved PARP, cleaved caspase-3, -8, and -9, Cyt c , ROS, p-c-Jun, c-Jun, p-JNK, | Colony-formation, Bcl-2, Survivin, MMP | Apoptosis, ROS/JNK activation | [55] |
| Prostate | DU145, DU145 xenograft, LNCaP | p53, IκBα, Bax, cleaved PARP, sub-G1 cells, ROS, p-JAK2, p-p38, p21 |
p-AKT, p-GSK-3β, p-NF-κB p65, p-IκBα, Bcl-2, XIAP, COX-2, p-STAT3 (Y705), Cyclin D1, Bcl-2, Bcl-xL, Mcl-1, Survivin, STAT3 activity, CDK2, CDK4, CDK6, Cdc25C, Cdc2, Cyclin B1 |
Apoptosis, STAT3 inhibition |
[25,56,57] |
| Skin (Melanoma) | SK-MEL-28, SK-MEL-28 xenograft | G1 arrest | p-AKT (Thr308), p-AKT (Ser473), p-GSK3β, Cyclin D1, Tumor volume | PI3K p110 binding, PI3K/AKT/p70S6K inhibition | [58] |
3.1. In Vitro Effects: Apoptosis, Proliferation, and Cell Cycle Arrest
3.2. Anti-Metastatic and Anti-Angiogenic Activities
3.3. In Vivo Efficacy and Safety Profiles
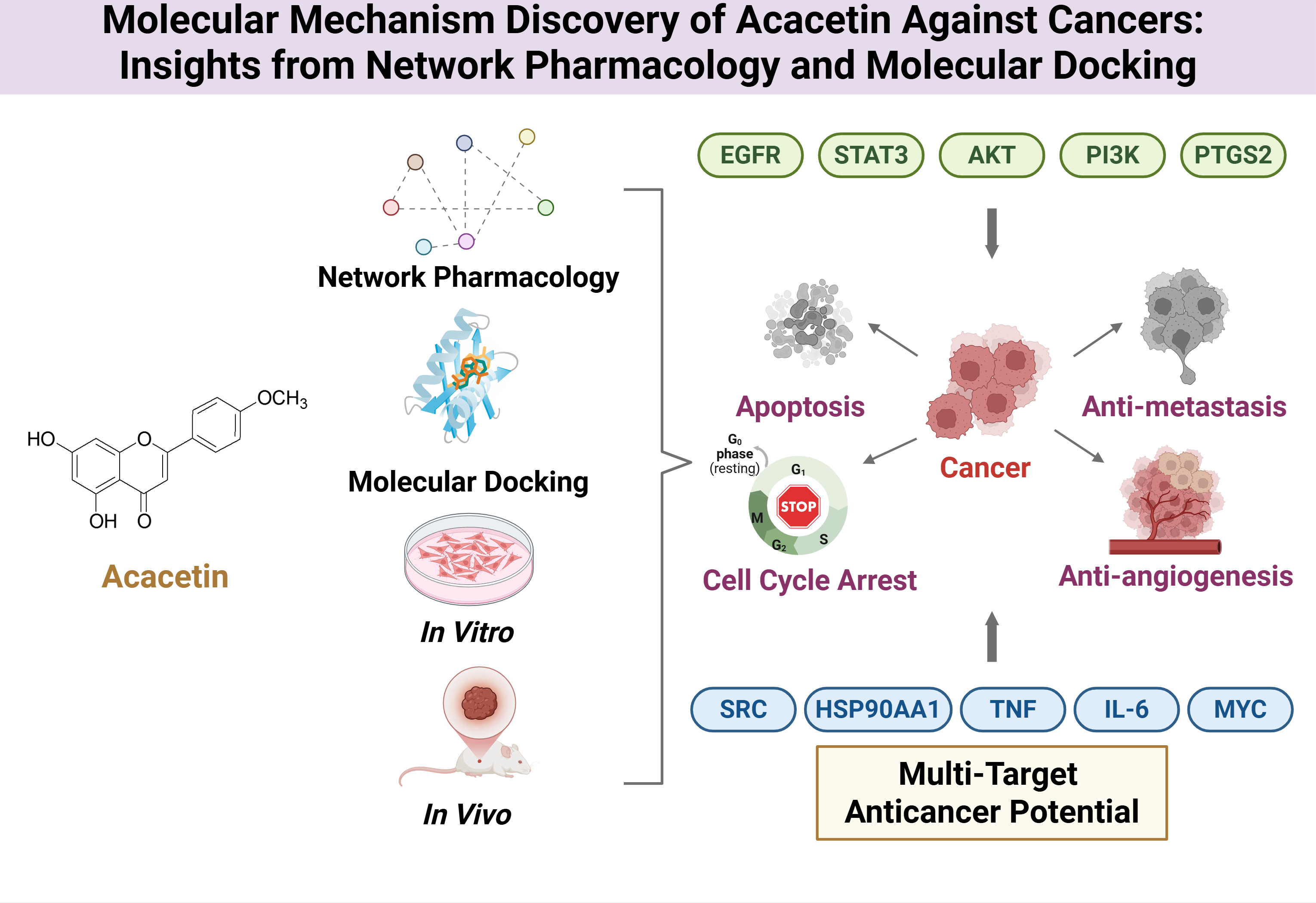
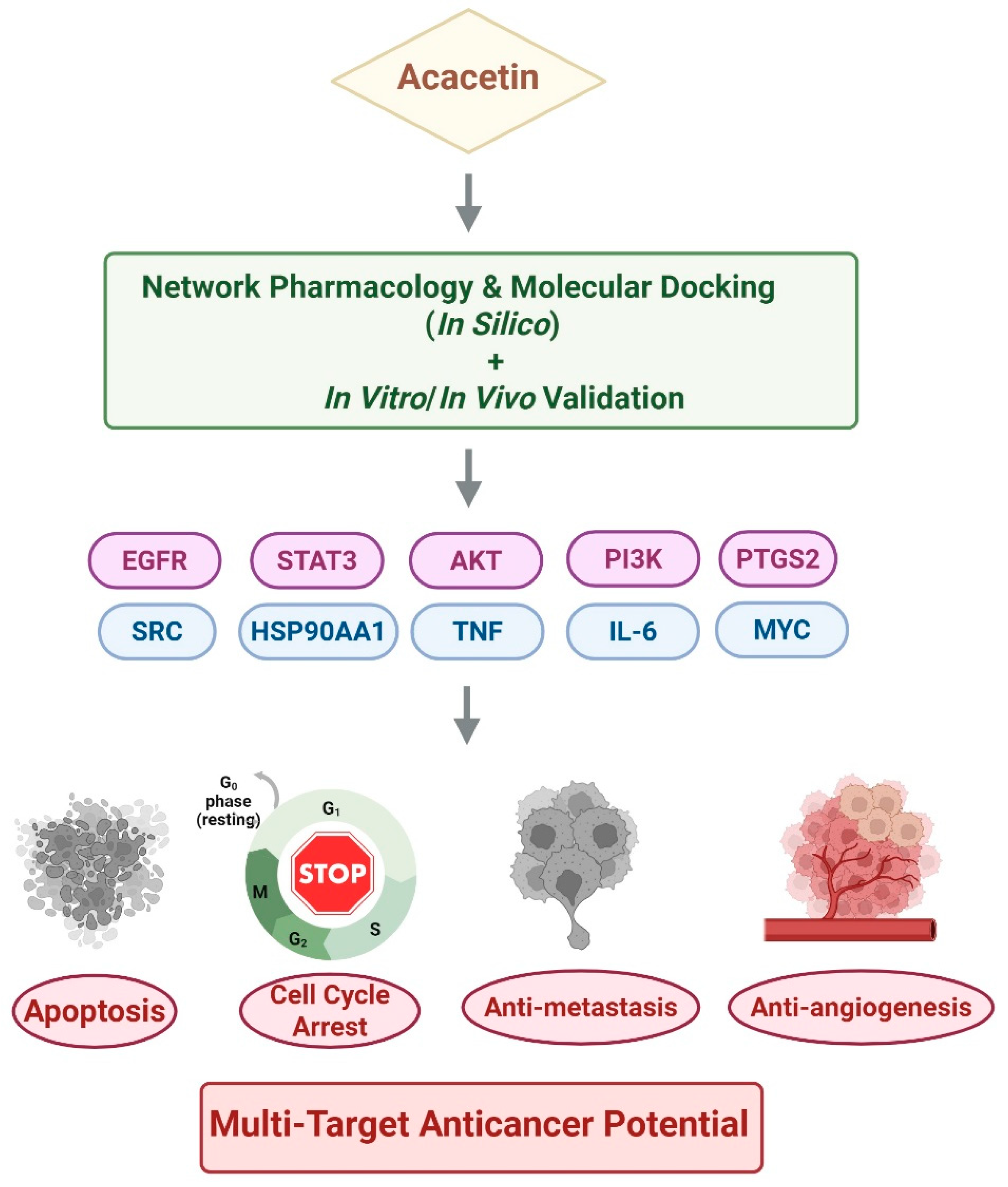
4. Network Pharmacology-Based Target Prediction of Acacetin
4.1. Methodological Pipeline and Identified Targets
5. Molecular Docking and Experimental Validation of Key Targets
5.1. Overview of Key Network-Derived Targets
5.2. EGFR
5.3. STAT3
5.4. AKT
5.5. Additional Network-Derived Targets
6. Challenges and Future Prospects
6.1. Bioavailability and Pharmacokinetics
6.2. Nanoparticle-Based Delivery and Structural Optimization
6.3. Synthetic Derivatives and Structural Analogs of Acacetin
6.4. Synergistic Drug Combinations
6.5. Need for Clinical Translation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADME | Absorption, distribution, metabolism, and excretion |
| AIF | Apoptosis-inducing factor |
| AKT | Serine/threonine kinase (also known as protein kinase B, PKB) |
| AMPK | AMP-activated protein kinase |
| Bak | Bcl-2 homologous antagonist/killer |
| Bax | Bcl-2–associated X protein |
| Bcl-xL | B-cell lymphoma-extra large |
| Bcl-2 | B-cell lymphoma 2 |
| Bid | BH3-interacting domain death agonist |
| CDK2 | Cyclin-dependent kinase 2 |
| CETSA | Cellular thermal shift assay |
| c-Myc | Cellular myelocytomatosis oncogene |
| DARTS | Drug affinity responsive target stability |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial–mesenchymal transition |
| ERK | Extracellular signal-regulated kinase |
| FYN | Proto-oncogene tyrosine-protein kinase Fyn |
| HIF-1α | Hypoxia-inducible factor-1 alpha |
| HRAS | Harvey rat sarcoma viral oncogene homolog |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 |
| IL-6 | Interleukin-6 |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| MDR1 | Multidrug resistance protein 1 |
| MAPK | Mitogen-activated protein kinase |
| mTOR | Mechanistic target of rapamycin |
| MYC | Myelocytomatosis oncogene |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| Nrf2 | Nuclear factor erythroid 2–related factor 2 |
| PAMPA | Parallel artificial membrane permeability assay |
| PD-L1 | Programmed death-ligand 1 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 |
| PI3K | Phosphoinositide 3-kinase |
| PPI | Protein–protein interaction |
| PTGS2 (COX-2) | Prostaglandin-endoperoxide synthase 2 (cyclooxygenase-2) |
| RHOA | Ras homolog family member A |
| RIP1 | Receptor-interacting protein kinase 1 |
| SIRT1 | Sirtuin 1 |
| STAT3 | Signal transducer and activator of transcription 3 |
| TNF | Tumor necrosis factor |
| VEGF | Vascular endothelial growth factor |
| Wnt | Wingless-related integration site |
References
- Zafar, A.; Khatoon, S.; Khan, M.J.; Abu, J.; Naeem, A. Advancements and limitations in traditional anti-cancer therapies: A comprehensive review of surgery, chemotherapy, radiation therapy, and hormonal therapy. Discov. Oncol. 2025, 16, 607. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, D.; Kim, N.D. Recent developments in combination chemotherapy for colorectal and breast cancers with topoisomerase inhibitors. Int. J. Mol. Sci. 2023, 24, 8457. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, D.; Im, E.; Kim, N.D. Etoposide as a key therapeutic agent in lung cancer: Mechanisms, efficacy, and emerging strategies. Int. J. Mol. Sci. 2025, 26, 796. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Cai, Q.; Deng, L.; Ouyang, Q.; Zhang, X.H.F.; Zheng, J. Invasion and metastasis in cancer: Molecular insights and therapeutic targets. Signal Transduct. Target. Ther. 2025, 10, 57. [Google Scholar] [CrossRef]
- Jang, J.Y.; Im, E.; Kim, N.D. Mechanism of resveratrol-induced programmed cell death and new drug discovery against cancer: A review. Int. J. Mol. Sci. 2022, 23, 13689. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhou, H.; Tan, L.; Siu, K.T.H.; Guan, X.-Y. Exploring treatment options in cancer: Tumor treatment strategies. Signal Transduct. Target. Ther. 2024, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, A.; Juchaniuk, P.; Oberda, J.; Wiśniewska, J.; Wojdan, W.; Szklener, K.; Mańdziuk, S. Phytochemicals in cancer treatment and cancer prevention—review on epidemiological data and clinical trials. Nutrients 2023, 15, 1896. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, T.; Qin, H.; Wang, X.; He, S.; Fan, Z.; Ye, Q.; Du, Y. Acacetin as a natural cardiovascular therapeutic: Mechanisms and preclinical evidence. Front. Pharmacol. 2025, 16, 1493981. [Google Scholar] [CrossRef]
- Zhang, G.; Dong, J.; Lu, L.; Liu, Y.; Hu, D.; Wu, Y.; Zhao, A.; Xu, H. Acacetin exerts antitumor effects on gastric cancer by targeting EGFR. Front. Pharmacol. 2023, 14, 1121643. [Google Scholar] [CrossRef]
- Noor, F.; Tahir Ul Qamar, M.; Ashfaq, U.A.; Albutti, A.; Alwashmi, A.S.S.; Aljasir, M.A. Network pharmacology approach for medicinal plants: Review and assessment. Pharmaceuticals 2022, 15, 572. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.R.; Jang, J.Y.; Kang, Y.J.; Oh, H.J.; Kang, M.K.; Yoon, D.; Kim, H.S.; Moon, H.R.; Chung, H.Y.; Kim, N.D. MHY446 induces apoptosis via reactive oxygen species-mediated endoplasmic reticulum stress in HCT116 human colorectal cancer cells. J. Chemother. 2024, 36, 483–500. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Jang, J.Y.; Kwon, Y.H.; Lee, J.H.; Lee, S.; Park, Y.; Jung, Y.-S.; Im, E.; Moon, H.R.; Chung, H.Y. MHY2245, a sirtuin inhibitor, induces cell cycle arrest and apoptosis in HCT116 human colorectal cancer cells. Int. J. Mol. Sci. 2022, 23, 1590. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Kang, Y.J.; Sung, B.; Kim, M.J.; Park, C.; Kang, D.; Moon, H.R.; Chung, H.Y.; Kim, N.D. MHY440, a novel topoisomerase Ι inhibitor, induces cell cycle arrest and apoptosis via a ROS-dependent DNA damage signaling pathway in AGS human gastric cancer cells. Molecules 2018, 24, 96. [Google Scholar] [CrossRef]
- Eom, S.H.; Liu, S.; Su, M.; Noh, T.H.; Hong, J.; Kim, N.D.; Chung, H.Y.; Yang, M.H.; Jung, J.H. Synthesis of phthalimide derivatives as potential PPAR-γ ligands. Mar. Drugs 2016, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yan, H.; Li, H.; Feng, Y.; Sun, W.; Ren, Y.; Ma, L.; Zeng, W.; Huang, F.; Jiang, Z.; et al. Use of network pharmacology and molecular docking to explore the mechanism of action of curcuma in the treatment of osteosarcoma. Sci. Rep. 2023, 13, 9569. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, Z.; Ye, X.; Zhu, M.; Li, Z.; Chen, Y.; Huang, S. Based on network pharmacology and molecular docking to predict the mechanism of Huangqi in the treatment of castration-resistant prostate cancer. PloS one 2022, 17, e0263291. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, L.; Li, L. The feasibility of using the compound kushen injection to treat cervical cancer based on network pharmacology and transcriptomics. Medicine 2023, 102, e35135. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C.; Elliott, P.C.; Kite, G.C.; Lewis, G.P. Flavonoid glycosides of the black locust tree, Robinia pseudoacacia (Leguminosae). Phytochemistry 2010, 71, 479–486. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, P.; Meena, A.; Luqman, S. Acacetin, a flavone with diverse therapeutic potential in cancer, inflammation, infections and other metabolic disorders. Food Chem. Toxicol. 2020, 145, 111708. [Google Scholar] [CrossRef]
- Alibi, S.; Crespo, D.; Navas, J. Plant-derivatives small molecules with antibacterial activity. Antibiotics 2021, 10, 231. [Google Scholar] [CrossRef]
- Kim, H.R.; Park, C.G.; Jung, J.Y. Acacetin (5, 7-dihydroxy-4-methoxyflavone) exhibits in vitro and in vivo anticancer activity through the suppression of NF-κB/Akt signaling in prostate cancer cells. Int. J. Mol. Med. 2014, 33, 317–324. [Google Scholar] [CrossRef]
- Wang, W.; Renquan, Z. Acacetin restrains the malignancy of esophageal squamous carcinoma cells via regulating JAK2/STAT3 pathway. Chem. Biol. Drug Des. 2023, 102, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Fong, Y.; Shen, K.H.; Chiang, T.A.; Shih, Y.W. Acacetin inhibits TPA-nduced MMP-2 and u-PA expressions of human lung cancer cells through inactivating JNK signaling pathway and reducing binding activities of NF-kappaB and AP-1. J. Food Sci. 2010, 75, H30–H38. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.-T.; Lin, S.-S.; Wang, C.-K.; Lee, Y.-B.; Chen, K.-S.; Fong, Y.; Shih, Y.-W. Acacetin inhibits the invasion and migration of human non-small cell lung cancer A549 cells by suppressing the p38α MAPK signaling pathway. Mol. Cell. Biochem. 2011, 350, 135–148. [Google Scholar] [CrossRef]
- Pan, M.-H.; Lai, C.-S.; Wang, Y.-J.; Ho, C.-T. Acacetin suppressed LPS-induced up-expression of iNOS and COX-2 in murine macrophages and TPA-induced tumor promotion in mice. Biochem. Pharmacol. 2006, 72, 1293–1303. [Google Scholar] [CrossRef]
- Wu, H.-J.; Sun, H.-Y.; Wu, W.; Zhang, Y.-H.; Qin, G.-W.; Li, G.-R. Properties and molecular determinants of the natural flavone acacetin for blocking hKv4. 3 channels. PloS one 2013, 8, e57864. [Google Scholar] [CrossRef]
- Li, G.-R.; Wang, H.-B.; Qin, G.-W.; Jin, M.-W.; Tang, Q.; Sun, H.-Y.; Du, X.-L.; Deng, X.-L.; Zhang, X.-H.; Chen, J.-B.; et al. Acacetin, a natural flavone, selectively inhibits human atrial repolarization potassium currents and prevents atrial fibrillation in dogs. Circulation 2008, 117, 2449–2457. [Google Scholar] [CrossRef]
- Wu, Y.; Song, F.; Li, Y.; Li, J.; Cui, Y.; Hong, Y.; Han, W.; Wu, W.; Lakhani, I.; Li, G. Acacetin exerts antioxidant potential against atherosclerosis through Nrf2 pathway in apoE−/− Mice. J. Cell. Mol. Med. 2021, 25, 521–534. [Google Scholar] [CrossRef]
- Simões, L.; Maciel, G.; Brandão, G.; Kroon, E.; Castilho, R.; Oliveira, A. Antiviral activity of Distictella elongata (Vahl) Urb.(Bignoniaceae), a potentially useful source of anti-dengue drugs from the state of Minas Gerais, Brazil. Lett. Appl. Microbiol. 2011, 53, 602–607. [Google Scholar] [CrossRef]
- Critchfield, J.W.; Butera, S.T.; Folks, T.M. Inhibition of HIV activation in latently infected cells by flavonoid compounds. AIDS Res. Hum. Retroviruses 1996, 12, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-Q.; Chen, K.; Shi, Q.; Kilkuskie, R.E.; Cheng, Y.-C.; Lee, K.-H. Anti-AIDS agents, 10. acacetin-7-O-β-D-galactopyranoside, an anti-HIV principle from chrysanthemum morifolium and a structure-activity correlation with some related flavonoids. J. Nat. Prod. 1994, 57, 42–51. [Google Scholar] [CrossRef]
- Wu, Q.; Yu, C.; Yan, Y.; Chen, J.; Zhang, C.; Wen, X. Antiviral flavonoids from Mosla scabra. Fitoterapia 2010, 81, 429–433. [Google Scholar] [CrossRef]
- Cha, J.-D.; Choi, S.-M.; Park, J.H. Combination of acacetin with antibiotics against methicillin resistant Staphylococcus aureus isolated from clinical specimens. Adv. Biosci. Biotechnol. 2014, 5, 398–408. [Google Scholar] [CrossRef]
- Komape, N.P.M.; Aderogba, M.; Bagla, V.P.; Masoko, P.; Eloff, J.N. Anti-bacterial and anti-oxidant activities of leaf extracts of Combretum vendee (combretecacea) and the isolation of an anti-bacterial compound. Afr. J. Tradit. Complement. Altern. Med. 2014, 11, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Dong, X.; Zhong, X.; Cai, H.; Wang, D.; Wang, L. Acacetin protects mice from Staphylococcus aureus bloodstream infection by inhibiting the activity of sortase A. Molecules 2016, 21, 1285. [Google Scholar] [CrossRef]
- Liou, C.-J.; Wu, S.-J.; Chen, L.-C.; Yeh, K.-W.; Chen, C.-Y.; Huang, W.-C. Acacetin from traditionally used Saussurea involucrata Kar. et Kir. suppressed adipogenesis in 3T3-L1 adipocytes and attenuated lipid accumulation in obese mice. Front. Pharmacol. 2017, 8, 589. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.-Y.; Park, J.-H.; Paik, H.-D.; Nah, S.-Y.; Kim, D.S.; Han, Y.S.J.M.; cells. Acacetin-induced apoptosis of human breast cancer MCF-7 cells involves caspase cascade, mitochondria-mediated death signaling and SAPK/JNK1/2-c-Jun activation. 2007, 24, 95-104.
- Kandhari, K.; Mishra, J.P.N.; Agarwal, R.; Singh, R.P. Acacetin induces sustained ERK1/2 activation and RIP1-dependent necroptotic death in breast cancer cells. Toxicol. Appl. Pharmacol. 2023, 462, 116409. [Google Scholar] [CrossRef] [PubMed]
- Kandhari, K.; Mishra, J.; Singh, R. Acacetin inhibits cell proliferation, survival, and migration in human breast cancer cells. Int. J. Pharm. Biol. Sci. 2019, 9, 443–452. [Google Scholar] [CrossRef]
- Aslan, B.T.; Ertugrul, B.; Iplik, E.S.; Cakmakoglu, B. Apoptotic effects of acacetin in human colon cancer HT-29 and HCT 116 cells. J. Cancer Res. Ther. 2021, 17, 1479–1482. [Google Scholar] [CrossRef]
- Prasad, N.; Sharma, J.R.; Yadav, U.C.S. Induction of growth cessation by acacetin via β-catenin pathway and apoptosis by apoptosis inducing factor activation in colorectal carcinoma cells. Mol. Biol. Rep. 2020, 47, 987–1001. [Google Scholar] [CrossRef]
- Zhang, G.; Li, Z.; Dong, J.; Zhou, W.; Zhang, Z.; Que, Z.; Zhu, X.; Xu, Y.; Cao, N.; Zhao, A. Acacetin inhibits invasion, migration and TGF-β1-induced EMT of gastric cancer cells through the PI3K/Akt/Snail pathway. BMC Complement. Med. Ther. 2022, 22, 10. [Google Scholar] [CrossRef]
- Pan, M.-H.; Lai, C.-S.; Hsu, P.-C.; Wang, Y.-J. Acacetin induces apoptosis in human gastric carcinoma cells accompanied by activation of caspase cascades and production of reactive oxygen species. J. Agric. Food Chem. 2005, 53, 620–630. [Google Scholar] [CrossRef]
- Alfwuaires, M.; Elsawy, H.; Sedky, A. Acacetin inhibits cell proliferation and induces apoptosis in human hepatocellular carcinoma cell lines. Molecules 2022, 27, 5361. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Kuo, P.-L.; Lin, C.-C. Acacetin inhibits the proliferation of Hep G2 by blocking cell cycle progression and inducing apoptosis. Biochem. Pharmacol. 2004, 67, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Zhang, C.; Cheng, H.; Wu, Y.-L.; Liu, J.; Chen, Z.; Huang, J.-g.; Ericksen, R.E.; Chen, L.; Zhang, H.; et al. Targeting to the non-genomic activity of retinoic acid receptor-gamma by acacetin in hepatocellular carcinoma. Sci. Rep. 2017, 7, 348. [Google Scholar] [CrossRef]
- Li, J.; Zhong, X.; Zhao, Y.; Shen, J.; Xiao, Z.; Pilapong, C. Acacetin inhibited non-small-cell lung cancer (NSCLC) cell growth via upregulating miR-34a in vitro and in vivo. Sci. Rep. 2024, 14, 2348. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-L.; Kuo, P.-L.; Liu, C.-F.; Lin, C.-C. Acacetin-induced cell cycle arrest and apoptosis in human non-small cell lung cancer A549 cells. Cancer letters 2004, 212, 53–60. [Google Scholar] [CrossRef]
- Tian, M.; Tang, Y.; Huang, T.; Liu, Y.; Pan, Y. Amelioration of human peritoneal mesothelial cell co-culture-evoked malignant potential of ovarian cancer cells by acacetin involves LPA release-activated RAGE-PI3K/AKT signaling. Cell. Mol. Biol. lett. 2021, 26, 51. [Google Scholar] [CrossRef]
- Liu, L.Z.; Jing, Y.; Jiang, L.L.; Jiang, X.E.; Jiang, Y.; Rojanasakul, Y.; Jiang, B.H. Acacetin inhibits VEGF expression, tumor angiogenesis and growth through AKT/HIF-1α pathway. Biochem. Biophys. Res. Commun. 2011, 413, 299–305. [Google Scholar] [CrossRef]
- Wang, S.; Lin, B.; Liu, W.; Wei, G.; Li, Z.; Yu, N.; Xue, X.; Ji, G. Acacetin induces apoptosis in human osteosarcoma cells by modulation of ROS/JNK activation. Drug Des. Devel. Ther. 2020, 5077–5085. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.; Lee, Y.-J.; Choi, J.; Kim, N.D.; Han, D.C.; Kwon, B.-M. Acacetin inhibits the growth of STAT3-activated DU145 prostate cancer cells by directly binding to signal transducer and activator of transcription 3 (STAT3). Molecules 2021, 26, 6204. [Google Scholar] [CrossRef]
- Singh, R.P.; Agrawal, P.; Yim, D.; Agarwal, C.; Agarwal, R. Acacetin inhibits cell growth and cell cycle progression, and induces apoptosis in human prostate cancer cells: Structure–activity relationship with linarin and linarin acetate. Carcinogenesis 2005, 26, 845–854. [Google Scholar] [CrossRef]
- Jung, S.K.; Kim, J.E.; Lee, S.Y.; Lee, M.H.; Byun, S.; Kim, Y.A.; Lim, T.G.; Reddy, K.; Huang, Z.; Bode, A.M.; et al. The P110 subunit of PI3-K is a therapeutic target of acacetin in skin cancer. Carcinogenesis 2014, 35, 123–130. [Google Scholar] [CrossRef]
- Jang, J.Y.; Sung, B.; Kim, N.D. Role of induced programmed cell death in the chemopreventive potential of apigenin. Int. J. Mol. Sci. 2022, 23, 3757. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Im, E.; Choi, Y.H.; Kim, N.D. Mechanism of bile acid-induced programmed cell death and drug discovery against cancer: A review. Int. J. Mol. Sci. 2022, 23, 7184. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Im, E.; Kim, N.D. Therapeutic potential of bioactive components from Scutellaria baicalensis Georgi in inflammatory bowel disease and colorectal cancer: A review. Int. J. Mol. Sci. 2023, 24, 1954. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-L.; Yang, B.-W.; Wang, X.-Y.; Chen, X.; Li, W.-D.; Zhai, H.-Y.; Wu, Y.; Cui, M.-Y.; Wu, J.-H.; Meng, Q.-H.J.W.J.o.G.O. Targeting colorectal cancer with Herba Patriniae and Coix seed: Network pharmacology, molecular docking, and in vitro validation. 2024, 16, 3539. [CrossRef]
- Yu, H.; Dong, Y.; Zou, G.-c.; Yang, Y.-j.; Liu, M.-z.; Han, C.-h. Exploring the molecular mechanism of Xiao Ji (Cirsium setosum) in treating bladder cancer using network pharmacology and molecular docking. Asian Biomed.(Res. Rev. News) 2025, 19, 94. [Google Scholar] [CrossRef]
- Guo, Z.; Yin, H.; Wu, T.; Wu, S.; Liu, L.; Zhang, L.; He, Y.; Zhang, R.; Liu, N. Study on the mechanism of Cortex Lycii on lung cancer based on network pharmacology combined with experimental validation. J. Ethnopharmacol. 2022, 293, 115280. [Google Scholar] [CrossRef]
- Zhang, H.-W.; Hu, J.-J.; Fu, R.-Q.; Liu, X.; Zhang, Y.-H.; Li, J.; Liu, L.; Li, Y.-N.; Deng, Q.; Luo, Q.-S. Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells. Sci. Rep. 2018, 8, 11255. [Google Scholar] [CrossRef]
- Huang, T.; Han, B.; Tao, Z.; Chen, X. Network pharmacology and experimental validation-based investigation of the underlying mechanism of Yi-Yi-Fu-Zi-Bai-Jiang-San of nasopharyngeal carcinoma. J. Cancer 2025, 16, 2212. [Google Scholar] [CrossRef]
- Lu, X.; Zheng, Y.; Wen, F.; Huang, W.; Chen, X.; Ruan, S.; Gu, S.; Hu, Y.; Teng, Y.; Shu, P. Study of the active ingredients and mechanism of Sparganii rhizoma in gastric cancer based on HPLC-Q-TOF–MS/MS and network pharmacology. Sci. Rep. 2021, 11, 1905. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, L.; Yin, Z.; Lin, J. Improving chemical similarity ensemble approach in target prediction. J. Cheminform. 2016, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 2016, 54, 1.30–1. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2014, 43, D789–D798. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef]
- Molina, D.M.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef]
- Han, D.G.; Cha, E.; Joo, J.; Hwang, J.S.; Kim, S.; Park, T.; Jeong, Y.S.; Maeng, H.J.; Kim, S.B.; Yoon, I.S. Investigation of the factors responsible for the poor oral bioavailability of acacetin in rats: Physicochemical and biopharmaceutical aspects. Pharmaceutics 2021, 13. [Google Scholar] [CrossRef]
- Fan, L.-h.; Li, X.; Chen, D.-y.; Zhang, N.; Wang, Y.; Shan, Y.; Hu, Y.; Xu, R.-a.; Jin, J.; Ge, R.-S. Determination of acacetin in rat plasma by UPLC-MS/MS and its application to a pharmacokinetic study. J. Chromatogr. B 2015, 986, 18–22. [Google Scholar] [CrossRef]
- Subhan, M.A.; Yalamarty, S.S.K.; Filipczak, N.; Parveen, F.; Torchilin, V.P. Recent advances in tumor targeting via EPR effect for cancer treatment. J. Pers. Med. 2021, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Xiang, J.; Zhou, Q.; Piao, Y.; Tang, J.; Shao, S.; Zhou, Z.; Bae, Y.H.; Shen, Y. The tumor EPR effect for cancer drug delivery: Current status, limitations, and alternatives. Adv. Drug Deliv. Rev. 2022, 191, 114614. [Google Scholar] [CrossRef]
- Ranjbar, S.; Emamjomeh, A.; Sharifi, F.; Zarepour, A.; Aghaabbasi, K.; Dehshahri, A.; Sepahvand, A.M.; Zarrabi, A.; Beyzaei, H.; Zahedi, M.M. Lipid-based delivery systems for flavonoids and flavonolignans: Liposomes, nanoemulsions, and solid lipid nanoparticles. Pharmaceutics 2023, 15, 1944. [Google Scholar] [CrossRef]
- Yan, S.; Na, J.; Liu, X.; Wu, P. Different targeting ligands-mediated drug delivery systems for tumor therapy. Pharmaceutics 2024, 16, 248. [Google Scholar] [CrossRef] [PubMed]
- Jurczyk, M.; Jelonek, K.; Musiał-Kulik, M.; Beberok, A.; Wrześniok, D.; Kasperczyk, J. Single-versus dual-targeted nanoparticles with folic acid and biotin for anticancer drug delivery. Pharmaceutics 2021, 13, 326. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tang, D.; Cui, J.; Jiang, H.; Yu, J.; Guo, Z. RGD-based self-assembling nanodrugs for improved tumor therapy. Front. Pharmacol. 2024, 15, 1477409. [Google Scholar] [CrossRef] [PubMed]
- Walle, T. Methylation of dietary flavones greatly improves their hepatic metabolic stability and intestinal absorption. Mol. Pharm. 2007, 4, 826–832. [Google Scholar] [CrossRef]
- Tang, S.; Wang, B.; Liu, X.; Xi, W.; Yue, Y.; Tan, X.; Bai, J.; Huang, L. Structural insights and biological activities of flavonoids: Implications for novel applications. Food Front. 2025, 6, 218–247. [Google Scholar] [CrossRef]
- Koirala, N.; Thuan, N.H.; Ghimire, G.P.; Thang, D.V.; Sohng, J.K. Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzyme Microb. Technol. 2016, 86, 103–116. [Google Scholar] [CrossRef]
- Gogineni, V.; Nael, M.A.; Chaurasiya, N.D.; Elokely, K.M.; McCurdy, C.R.; Rimoldi, J.M.; Cutler, S.J.; Tekwani, B.L.; León, F. Computationally assisted lead optimization of novel potent and selective MAO-B inhibitors. Biomedicines 2021, 9, 1304. [Google Scholar] [CrossRef]
- Zverev, Y.F.; Rykunova, A.Y. Modern nanocarriers as a factor in increasing the bioavailability and pharmacological activity of flavonoids. Appl. Biochem. Microbiol. 2022, 58, 1002–1020. [Google Scholar] [CrossRef] [PubMed]
- Stevens Barrón, J.C.; Chapa González, C.; Álvarez Parrilla, E.; De la Rosa, L.A. Nanoparticle-mediated delivery of flavonoids: Impact on proinflammatory cytokine production: A systematic review. Biomolecules 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.R.; Men, X.; Gao, X.H.; Liu, L.B.; Fan, H.Q.; Xia, X.H.; Wang, Q.A. Discovery of potent and selective acetylcholinesterase (AChE) inhibitors: acacetin 7-O-methyl ether Mannich base derivatives synthesised from easy access natural product naringin. Nat. Prod. Res. 2018, 32, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, L.; Cai, S.; Wang, Q. Semisynthesis of apigenin and acacetin-7-O-β-D-glycosides from naringin and their cytotoxic activities. Carbohydr. Res. 2012, 357, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Walle, T. Methylation of dietary flavones increases their metabolic stability and chemopreventive effects. Int. J. Mol. Sci. 2009, 10, 5002–5019. [Google Scholar] [CrossRef]
- Punia, R.; Raina, K.; Agarwal, R.; Singh, R.P. Acacetin enhances the therapeutic efficacy of doxorubicin in non-small-cell lung carcinoma cells. PLoS One 2017, 12, e0182870. [Google Scholar] [CrossRef]
- Chen, K.; Gao, Z. Acacetin, a natural flavone with potential in improving liver disease based on its anti-inflammation, anti-cancer, anti-infection and other effects. Molecules 2024, 29, 4872. [Google Scholar] [CrossRef]
- Fasolo, A.; Sessa, C. Translational research in phase I trials. Clini. Transl. Oncol. 2009, 11, 580–588. [Google Scholar] [CrossRef]
- Goldblatt, E.M.; Lee, W.H. From bench to bedside: the growing use of translational research in cancer medicine. Am. J. Transl. Res. 2010, 2, 1–18. [Google Scholar]
| Target | Cancer Type(s) | Function | Docking / Prediction / Validation Method | Validation Level | Functional Outcome | Refs. |
|---|---|---|---|---|---|---|
| EGFR | Gastric | EGFR-mediated signaling (STAT3, ERK) | Target prediction (SwissTargetPrediction, GeneCards, RNA-seq); network analysis (STRING, Cytoscape); docking (Schrödinger Maestro); validation (DARTS, CETSA, WB, TUNEL, colony formation, xenograft) | Confirmed (docking + in vitro + in vivo) | ↓: p-EGFR, p-STAT3, p-ERK (transient), Ki-67, PCNA, Bcl-xL, Colony formation, Tumor growth | [12] |
| ↑: Bax, cleaved caspase-3, cleaved PARP, Apoptosis, | ||||||
| Strong EGFR binding (docking) | ||||||
| Colorectal | - | Target prediction (SwissTargetPrediction, TCMSP, GeneCards, OMIM, DisGeNET); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -8.3 kcal/mol | [62] | |
| Bladder | EGFR-mediated signaling | Target prediction (TCMSP, GeneCards, OMIM); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -4.0 kcal/mol | [63] | |
| STAT3 | Prostate | STAT3-mediated signaling (proliferation, survival) | Docking (Glide, Schrödinger Maestro); binding validation (DARTS, CETSA, pull-down); functional assays (WB, Annexin V/PI, xenograft) | Confirmed (docking + in vitro + in vivo) | ↓: p-STAT3 (Tyr705), Cyclin D1, Bcl-2, Bcl-xL, Mcl-1, Survivin, Tumor volume, | [56] |
| ↑: Bax, cleaved PARP, cleaved caspase-3, Annexin V, p-JAK2, p-p38, ROS, Apoptosis | ||||||
| Strong STAT3 SH2 binding (3 H-bonds + cation–π; Glide) | ||||||
| AKT1 |
Colorectal | PI3K/AKT/p53 signaling | Target prediction (SwissTargetPrediction, TCMSP, GeneCards, OMIM, DisGeNET); network analysis (STRING, Cytoscape); docking (AutoDock Vina); validation (WB, CCK-8, Annexin V-FITC, scratch assay) | Confirmed (docking + multiple in vitro assays) | ↓: p-AKT, p-PI3K, Survivin, Migration, Proliferation | [62] |
| ↑: p53, cleaved caspase-3, Apoptosis, | ||||||
| Strong AKT1 binding (-9.2 kcal/mol; 1 H-bond, Thr211) | ||||||
| HSP90AB1 → AKT | Lung | Chaperone-mediated PI3K/AKT/mTOR activation | Network pharmacology; docking (AutoDock Tools); validation (WB, EMT, invasion assay ± terazosin) | Confirmed (docking + in vitro + in vivo) | ↓: p-AKT, p-mTOR, Migration, Invasion, EMT markers | [64] |
| ↑: E-Cadherin; effect reversed by HSP90 agonist terazosin | ||||||
| PI3Kγ | Breast | PI3K/AKT/mTOR/p70S6K/ULK signaling | Docking (SYBYL2.0); PI3Kγ kinase assay; validation (WB [PI3Kγ, p-AKT, p-mTOR, p-p70S6K, p-ULK1], Apoptosis assay, G2/M arrest, LC3 puncta) | Confirmed (docking + in vitro) | ↓: PI3Kγ, p-AKT, p-mTOR, p-p70S6K, p-ULK1 | [65] |
| ↑: Apoptosis, Autophagy, G2/M arrest | ||||||
| PI3Kγ ATP-site binding (H-bonds: Ser806, Ala885, Val882; hydrophobic: Lys833, Asp964) | ||||||
| SRC | Colorectal | Predicted PI3K/AKT signaling; proliferation/migration in CRC | Target prediction (SwissTargetPrediction, TCMSP, GeneCards, OMIM, DisGeNET); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -6.9 kcal/mol | [62] |
| HSP90AA1 | Colorectal | Predicted chaperone-mediated PI3K/AKT; cell survival in CRC | Target prediction (SwissTargetPrediction, TCMSP, GeneCards, OMIM, DisGeNET); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -7.1 kcal/mol | [62] |
| TNF | Colorectal | Predicted inflammatory/apoptotic signaling; CRC progression | Target prediction (SwissTargetPrediction, TCMSP, GeneCards, OMIM, DisGeNET); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -6.4 kcal/mol | [62] |
| IL-6 | Bladder | Inflammatory cytokine signaling | Target prediction (TCMSP, GeneCards, OMIM); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -4.12 kcal/mol | [63] |
| MYC | Bladder | Cell proliferation and transcription | Target prediction (TCMSP, GeneCards, OMIM); network analysis (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | Binding affinity -4.37 kcal/mol | [63] |
| PTGS2 | Nasopharyngeal carcinoma | Inflammation, Tumor proliferation and migration | Target prediction (TCMSP, SwissTargetPrediction, SEA, GeneCards, OMIM); network analysis (STRING, Cytoscape); docking (AutoDock Vina), MD simulation; validation (CCK-8, colony formation, migration, WB [PTGS2]) | Confirmed (docking + in vitro) | ↓: PTGS2 (docking: < -7 kcal/mol; Tyr385 contact), Proliferation, Migration | [66] |
| PI3K (p110 subunit) | Skin | PI3K-AKT-p70S6K signaling | Binding assays (pull-down, kinase); cell transformation (in vitro); docking (Glide/induced-fit, Maestro); xenograft | Confirmed (docking + in vitro + in vivo) | ↓: PI3K activity, p-AKT, p-p70S6K, Tumor growth, Colony formation, | [58] |
| ↑: G1 arrest; docking to PI3K-p110 ATP-binding site (H-bonds: Val828, Glu826, Asp911; hydrophobic: Trp760, Ile777, Tyr813) | ||||||
| STAT3, AKT1, MAPK1, HSP90AA1, HRAS, SRC, PIK3CA, PIK3R1, FYN, RHOA, |
Gastric | PI3K-AKT, MAPK, JAK/STAT3, and cytoskeleton regulation pathways | HPLC-Q-TOF–MS/MS identification; network pharmacology (STRING, Cytoscape); docking (AutoDock Vina) | Docking only | High binding affinity (mostly < -7.3 kcal/mol) |
[67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





