Submitted:
07 August 2025
Posted:
08 August 2025
You are already at the latest version
Abstract
Keywords:
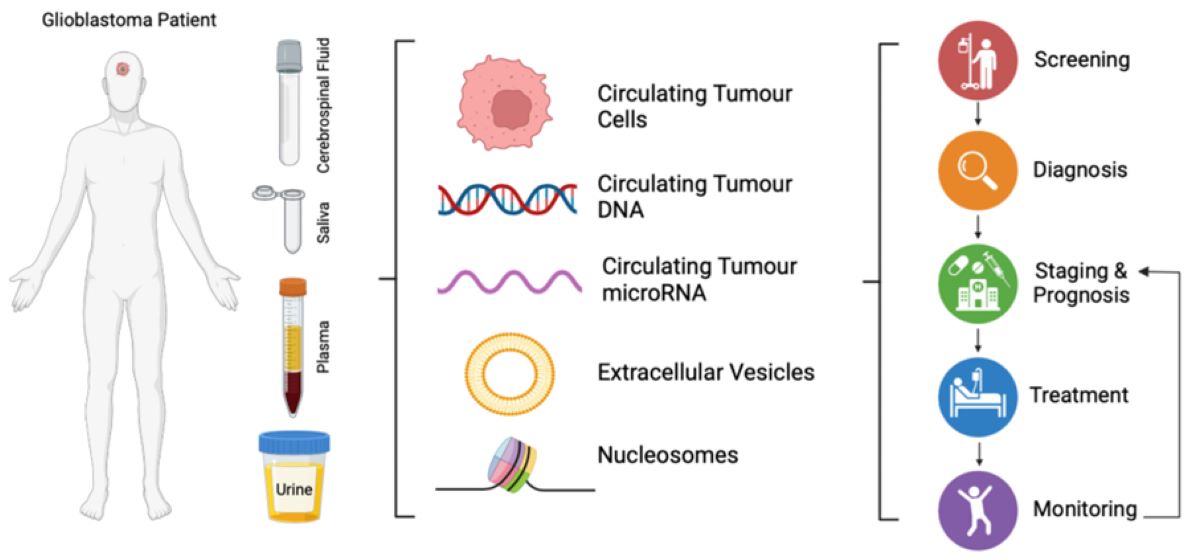
1. Introduction
2. Circulating Biomarkers of Glioblastoma from Liquid Biopsies
2.1. Circulating Tumour Cells
2.2. Circulating Tumour DNA
2.3. Nucleosomes
2.4. Circulating Tumour microRNA
2.5. Extracellular Vesicles
3. Application of Machine Learning and Artificial Intelligence
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lah, T. T.; Novak, M.; Breznik, B. Brain malignancies: Glioblastoma and brain metastases. Semin Cancer Biol 2020, 60, 262–273. [Google Scholar] [CrossRef]
- Ostrom, Q. T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J. S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro Oncol 2018, 20 (suppl_4), iv1–iv86. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M. E.; Mason, W. P.; van den Bent, M. J.; Taphoorn, M. J.; Janzer, R. C.; Ludwin, S. K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Tan, A. C.; Ashley, D. M.; López, G. Y.; Malinzak, M.; Friedman, H. S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 2015, 129, 829–848. [Google Scholar] [CrossRef]
- Clarke, J. L.; Chang, S. M. Neuroimaging: diagnosis and response assessment in glioblastoma. Cancer J 2012, 18, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Tanaka, R.; Takeda, N. Magnetic resonance imaging and histopathology of cerebral gliomas. Neuroradiology 1992, 34, 463–469. [Google Scholar] [CrossRef]
- Delgado-López, P. D.; Riñones-Mena, E.; Corrales-García, E. M. Treatment-related changes in glioblastoma: a review on the controversies in response assessment criteria and the concepts of true progression, pseudoprogression, pseudoresponse and radionecrosis. Clin Transl Oncol 2018, 20, 939–953. [Google Scholar] [CrossRef]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D'Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Saenz-Antoñanzas, A.; Auzmendi-Iriarte, J.; Carrasco-Garcia, E.; Moreno-Cugnon, L.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef]
- Sareen, H.; Garrett, C.; Lynch, D.; Powter, B.; Brungs, D.; Cooper, A.; Po, J.; Koh, E. S.; Vessey, J. Y.; McKechnie, S.; et al. The Role of Liquid Biopsies in Detecting Molecular Tumor Biomarkers in Brain Cancer Patients. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B. W.; Punyadeera, C. Circulating biomarkers in patients with glioblastoma. Br J Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef]
- Qazi, M. A.; Salim, S. K.; Brown, K. R.; Mikolajewicz, N.; Savage, N.; Han, H.; Subapanditha, M. K.; Bakhshinyan, D.; Nixon, A.; Vora, P.; et al. Characterization of the minimal residual disease state reveals distinct evolutionary trajectories of human glioblastoma. Cell Rep 2022, 40, 111420. [Google Scholar] [CrossRef]
- Strati, A.; Markou, A.; Kyriakopoulou, E.; Lianidou, E. Detection and Molecular Characterization of Circulating Tumour Cells: Challenges for the Clinical Setting. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Lion, T. Minimal residual disease. Curr Opin Hematol 1999, 6, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T. N. A.; Huang, P. S.; Chu, P. Y.; Hsieh, C. H.; Wu, M. H. Recent Progress in Enhanced Cancer Diagnosis, Prognosis, and Monitoring Using a Combined Analysis of the Number of Circulating Tumor Cells (CTCs) and Other Clinical Parameters. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M. M.; Ramani, V. C.; Jeffrey, S. S. Circulating tumor cell technologies. Mol Oncol 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Shankar, G. M.; Balaj, L.; Stott, S. L.; Nahed, B.; Carter, B. S. Liquid biopsy for brain tumors. Expert Rev Mol Diagn 2017, 17, 943–947. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Allard, W. J.; Matera, J.; Miller, M. C.; Repollet, M.; Connelly, M. C.; Rao, C.; Tibbe, A. G.; Uhr, J. W.; Terstappen, L. W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Lynch, D. , Powter, B., Po, J. W., Cooper, A., Garrett, C., Koh, E.-S., Sheridan, M., van Gelder, J., Darwish, B., Mckechnie, S., Bazina, R., Jaeger, M., Roberts, T. L., de Souza, P., & Becker, T. M.. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Applied Sciences 2020, 10. [Google Scholar]
- Sullivan, J. P.; Nahed, B. V.; Madden, M. W.; Oliveira, S. M.; Springer, S.; Bhere, D.; Chi, A. S.; Wakimoto, H.; Rothenberg, S. M.; Sequist, L. V.; et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov 2014, 4, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Peng, Z.; Lin, S. C.; Geri, M.; Li, S.; Li, P.; Chen, Y.; Dao, M.; Suresh, S.; Huang, T. J. Cell separation using tilted-angle standing surface acoustic waves. Proc Natl Acad Sci U S A 2014, 111, 12992–12997. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Wu, H.; Punyadeera, C.; Warkiani, M. E. The Use of Microfluidic Technology for Cancer Applications and Liquid Biopsy. Micromachines (Basel) 2018, 9. [Google Scholar] [CrossRef]
- Kolostova, K.; Pospisilova, E.; Pavlickova, V.; Bartos, R.; Sames, M.; Pawlak, I.; Bobek, V. Next generation sequencing of glioblastoma circulating tumor cells: non-invasive solution for disease monitoring. Am J Transl Res 2021, 13, 4489–4499. [Google Scholar]
- Valerius, A. R.; Webb, M. J.; Hammad, N.; Sener, U.; Malani, R. Cerebrospinal Fluid Liquid Biopsies in the Evaluation of Adult Gliomas. Curr Oncol Rep 2024, 26, 377–390. [Google Scholar] [CrossRef]
- Mathur, R.; Wang, Q.; Schupp, P. G.; Nikolic, A.; Hilz, S.; Hong, C.; Grishanina, N. R.; Kwok, D.; Stevers, N. O.; Jin, Q.; et al. Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective. Cell 2024, 187, 446–463.e416. [Google Scholar] [CrossRef]
- Berezovsky, A. D.; Poisson, L. M.; Cherba, D.; Webb, C. P.; Transou, A. D.; Lemke, N. W.; Hong, X.; Hasselbach, L. A.; Irtenkauf, S. M.; Mikkelsen, T.; et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia 2014, 16, 193–206, 206.e119-125. [Google Scholar] [CrossRef]
- Müller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M.; et al. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med 2014, 6, 247ra101. [Google Scholar] [CrossRef]
- Macarthur, K. M.; Kao, G. D.; Chandrasekaran, S.; Alonso-Basanta, M.; Chapman, C.; Lustig, R. A.; Wileyto, E. P.; Hahn, S. M.; Dorsey, J. F. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res 2014, 74, 2152–2159. [Google Scholar] [CrossRef]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef]
- Krol, I.; Castro-Giner, F.; Maurer, M.; Gkountela, S.; Szczerba, B. M.; Scherrer, R.; Coleman, N.; Carreira, S.; Bachmann, F.; Anderson, S.; et al. Detection of circulating tumour cell clusters in human glioblastoma. Br J Cancer 2018, 119, 487–491. [Google Scholar] [CrossRef]
- Müller Bark, J.; Kulasinghe, A.; Hartel, G.; Leo, P.; Warkiani, M. E.; Jeffree, R. L.; Chua, B.; Day, B. W.; Punyadeera, C. Isolation of Circulating Tumour Cells in Patients With Glioblastoma Using Spiral Microfluidic Technology - A Pilot Study. Front Oncol 2021, 11, 681130. [Google Scholar] [CrossRef]
- Gao, F.; Zhao, W.; Li, M.; Ren, X.; Jiang, H.; Cui, Y.; Lin, S. Role of circulating tumor cell detection in differentiating tumor recurrence from treatment necrosis of brain gliomas. Biosci Trends 2021, 15, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Parker, N. R.; Khong, P.; Parkinson, J. F.; Howell, V. M.; Wheeler, H. R. Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol 2015, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Menyailo, M. E.; Tretyakova, M. S.; Denisov, E. V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol Adv 2018, 36, 1063–1078. [Google Scholar] [CrossRef]
- Batth, I. S.; Mitra, A.; Rood, S.; Kopetz, S.; Menter, D.; Li, S. CTC analysis: an update on technological progress. Transl Res 2019, 212, 14–25. [Google Scholar] [CrossRef]
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of circulating tumor cells: opportunities and challenges. Biomark Res 2022, 10, 58. [Google Scholar] [CrossRef]
- Jan, Y. J.; Chen, J. F.; Zhu, Y.; Lu, Y. T.; Chen, S. H.; Chung, H.; Smalley, M.; Huang, Y. W.; Dong, J.; Chen, L. C.; et al. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv Drug Deliv Rev 2018, 125, 78–93. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F. O.; Hesch, R. D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001, 61, 1659–1665. [Google Scholar]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A. A. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncologist 2021, 26, 865–878. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S. J.; Tsui, D. W.; Gale, D.; Forshew, T.; Piskorz, A. M.; Parkinson, C.; Chin, S. F.; Kingsbury, Z.; Wong, A. S.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Leary, R. J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J. D.; Craig, D.; O'Shaughnessy, J.; Kinzler, K. W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012, 4, 162ra154. [Google Scholar] [CrossRef]
- Lebofsky, R.; Decraene, C.; Bernard, V.; Kamal, M.; Blin, A.; Leroy, Q.; Rio Frio, T.; Pierron, G.; Callens, C.; Bieche, I.; et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015, 9, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Sasmita, A. O.; Wong, Y. P.; Ling, A. P. K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol 2018, 14, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Birkó, Z.; Nagy, B.; Klekner, Á.; Virga, J. Novel Molecular Markers in Glioblastoma-Benefits of Liquid Biopsy. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Wadden, J.; Ravi, K.; John, V.; Babila, C. M.; Koschmann, C. Cell-Free Tumor DNA (cf-tDNA) Liquid Biopsy: Current Methods and Use in Brain Tumor Immunotherapy. Front Immunol 2022, 13, 882452. [Google Scholar] [CrossRef] [PubMed]
- Parker, A. L.; Kavallaris, M.; McCarroll, J. A. Microtubules and their role in cellular stress in cancer. Front Oncol 2014, 4, 153. [Google Scholar] [CrossRef]
- Schwaederle, M.; Husain, H.; Fanta, P. T.; Piccioni, D. E.; Kesari, S.; Schwab, R. B.; Banks, K. C.; Lanman, R. B.; Talasaz, A.; Parker, B. A.; et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016, 7, 9707–9717. [Google Scholar] [CrossRef]
- Piccioni, D. E.; Achrol, A. S.; Kiedrowski, L. A.; Banks, K. C.; Boucher, N.; Barkhoudarian, G.; Kelly, D. F.; Juarez, T.; Lanman, R. B.; Raymond, V. M.; et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol 2019, 8, Cns34. [Google Scholar] [CrossRef]
- Batool, S. M.; Escobedo, A. K.; Hsia, T.; Ekanayake, E.; Khanna, S. K.; Gamblin, A. S.; Zheng, H.; Skog, J.; Miller, J. J.; Stemmer-Rachamimov, A. O.; et al. Clinical utility of a blood based assay for the detection of IDH1.R132H-mutant gliomas. Nat Commun 2024, 15, 7074. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, G. S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D. L.; Petkova, L.; Ivanov, B. D.; Kaprelyan, A.; Ghenev, P. Reclassification of Glioblastoma Multiforme According to the 2021 World Health Organization Classification of Central Nervous System Tumors: A Single Institution Report and Practical Significance. Cureus 2022, 14, e21822. [Google Scholar] [CrossRef] [PubMed]
- Salkeni, M. A.; Zarzour, A.; Ansay, T. Y.; McPherson, C. M.; Warnick, R. E.; Rixe, O.; Bahassi el, M. Detection of EGFRvIII mutant DNA in the peripheral blood of brain tumor patients. J Neurooncol 2013, 115, 27–35. [Google Scholar] [CrossRef]
- Zill, O. A.; Banks, K. C.; Fairclough, S. R.; Mortimer, S. A.; Vowles, J. V.; Mokhtari, R.; Gandara, D. R.; Mack, P. C.; Odegaard, J. I.; Nagy, R. J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [PubMed]
- Miller, A. M.; Shah, R. H.; Pentsova, E. I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S. A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef]
- Liu, A. P. Y.; Smith, K. S.; Kumar, R.; Robinson, G. W.; Northcott, P. A. Low-coverage whole-genome sequencing of cerebrospinal-fluid-derived cell-free DNA in brain tumor patients. STAR Protoc 2022, 3, 101292. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R. J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B. R.; Wang, H.; Luber, B.; Alani, R. M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014, 6, 224ra224. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M. A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S. A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Yan, Y. Y.; Guo, Q. R.; Wang, F. H.; Adhikari, R.; Zhu, Z. Y.; Zhang, H. Y.; Zhou, W. M.; Yu, H.; Li, J. Q.; Zhang, J. Y. Cell-Free DNA: Hope and Potential Application in Cancer. Front Cell Dev Biol 2021, 9, 639233. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, W.; Wang, Y.; Guo, Y.; Cong, Z.; Du, F.; Song, B. MGMT promoter methylation in serum and cerebrospinal fluid as a tumor-specific biomarker of glioma. Biomed Rep 2015, 3, 543–548. [Google Scholar] [CrossRef]
- Juratli, T. A.; Stasik, S.; Zolal, A.; Schuster, C.; Richter, S.; Daubner, D.; Juratli, M. A.; Thowe, R.; Hennig, S.; Makina, M.; et al. TERT Promoter Mutation Detection in Cell-Free Tumor-Derived DNA in Patients with IDH Wild-Type Glioblastomas: A Pilot Prospective Study. Clin Cancer Res 2018, 24, 5282–5291. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Zhang, M.; McMahon, K. W.; Kinde, I.; Dobbyn, L.; Ptak, J.; Brem, H.; Chaichana, K.; Gallia, G. L.; et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A 2015, 112, 9704–9709. [Google Scholar] [CrossRef]
- Jones, J.; Nguyen, H.; Drummond, K.; Morokoff, A. Circulating Biomarkers for Glioma: A Review. Neurosurgery 2021, 88, E221–e230. [Google Scholar] [CrossRef]
- Mair, R.; Mouliere, F.; Smith, C. G.; Chandrananda, D.; Gale, D.; Marass, F.; Tsui, D. W. Y.; Massie, C. E.; Wright, A. J.; Watts, C.; et al. Measurement of Plasma Cell-Free Mitochondrial Tumor DNA Improves Detection of Glioblastoma in Patient-Derived Orthotopic Xenograft Models. Cancer Res 2019, 79, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Krauze, A. V.; Won, M.; Graves, C.; Corn, B. W.; Muanza, T. M.; Howard, S. P.; Mahadevan, A.; Schultz, C. J.; Haas, M. L.; Mehta, M. P.; et al. Predictive value of tumor recurrence using urinary vascular endothelial factor levels in patients receiving radiation therapy for Glioblastoma Multiforme (GBM). Biomark Res 2013, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Rieger, J.; Bähr, O.; Maurer, G. D.; Hattingen, E.; Franz, K.; Brucker, D.; Walenta, S.; Kämmerer, U.; Coy, J. F.; Weller, M.; et al. ERGO: a pilot study of ketogenic diet in recurrent glioblastoma. Int J Oncol 2014, 44, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Ishikawa, E.; Nakai, K.; Matsuda, M.; Masumoto, T.; Yamamoto, T.; Matsumura, A. Bevacizumab in Japanese patients with malignant glioma: from basic research to clinical trial. Onco Targets Ther 2014, 7, 1551–1562. [Google Scholar] [CrossRef]
- Constâncio, V.; Nunes, S. P.; Henrique, R.; Jerónimo, C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020, 9. [Google Scholar] [CrossRef]
- Lavon, I.; Refael, M.; Zelikovitch, B.; Shalom, E.; Siegal, T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncol 2010, 12, 173–180. [Google Scholar] [CrossRef]
- Watson, N.; Grieu, F.; Morris, M.; Harvey, J.; Stewart, C.; Schofield, L.; Goldblatt, J.; Iacopetta, B. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J Mol Diagn 2007, 9, 472–478. [Google Scholar] [CrossRef] [PubMed]
- McCord, M.; Steffens, A.; Javier, R.; Kam, K. L.; McCortney, K.; Horbinski, C. The efficacy of DNA mismatch repair enzyme immunohistochemistry as a screening test for hypermutated gliomas. Acta Neuropathol Commun 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and Circulating Biomarkers in Patients with Glioblastoma. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Mair, R.; Chandrananda, D.; Marass, F.; Smith, C. G.; Su, J.; Morris, J.; Watts, C.; Brindle, K. M.; Rosenfeld, N. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med 2018, 10. [Google Scholar] [CrossRef]
- Martínez-Ricarte, F.; Mayor, R.; Martínez-Sáez, E.; Rubio-Pérez, C.; Pineda, E.; Cordero, E.; Cicuéndez, M.; Poca, M. A.; López-Bigas, N.; Ramon, Y. C. S.; et al. Molecular Diagnosis of Diffuse Gliomas through Sequencing of Cell-Free Circulating Tumor DNA from Cerebrospinal Fluid. Clin Cancer Res 2018, 24, 2812–2819. [Google Scholar] [CrossRef]
- Wu, J.; Liu, Z.; Huang, T.; Wang, Y.; Song, M. M.; Song, T.; Long, G.; Zhang, X.; Li, X.; Zhang, L. Cerebrospinal fluid circulating tumor DNA depicts profiling of brain metastasis in NSCLC. Mol Oncol 2023, 17, 810–824. [Google Scholar] [CrossRef]
- Hickman, R. A.; Miller, A. M.; Holle, B. M.; Jee, J.; Liu, S. Y.; Ross, D.; Yu, H.; Riely, G. J.; Ombres, C.; Gewirtz, A. N.; et al. Real-world experience with circulating tumor DNA in cerebrospinal fluid from patients with central nervous system tumors. Acta Neuropathol Commun 2024, 12, 151. [Google Scholar] [CrossRef]
- Guo, W.; Jin, L.; Liang, J.; Lin, G.; Zheng, J.; Zhou, D.; Zhan, S.; Sun, H.; Jiang, X. Detection of mutation profiles and tumor mutation burden of cerebrospinal fluid circulating DNA by a cancer genomic panel sequencing in glioma patients. Clin Chim Acta 2022, 534, 81–92. [Google Scholar] [CrossRef]
- Bruzek, A. K.; Ravi, K.; Muruganand, A.; Wadden, J.; Babila, C. M.; Cantor, E.; Tunkle, L.; Wierzbicki, K.; Stallard, S.; Dickson, R. P.; et al. Electronic DNA Analysis of CSF Cell-free Tumor DNA to Quantify Multi-gene Molecular Response in Pediatric High-grade Glioma. Clin Cancer Res 2020, 26, 6266–6276. [Google Scholar] [CrossRef]
- Kornberg, R. D. Chromatin structure: a repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Cutter, A. R.; Hayes, J. J. A brief review of nucleosome structure. FEBS Lett 2015, 589, 2914–2922. [Google Scholar] [CrossRef]
- Peterson, C. L.; Laniel, M. A. Histones and histone modifications. Curr Biol 2004, 14, R546–551. [Google Scholar] [CrossRef]
- Kim, Y. Z. Altered histone modifications in gliomas. Brain Tumor Res Treat 2014, 2, 7–21. [Google Scholar] [CrossRef]
- Was, H.; Krol, S. K.; Rotili, D.; Mai, A.; Wojtas, B.; Kaminska, B.; Maleszewska, M. Histone deacetylase inhibitors exert anti-tumor effects on human adherent and stem-like glioma cells. Clin Epigenetics 2019, 11, 11. [Google Scholar] [CrossRef]
- McAnena, P.; Brown, J. A.; Kerin, M. J. Circulating Nucleosomes and Nucleosome Modifications as Biomarkers in Cancer. Cancers (Basel) 2017, 9. [Google Scholar] [CrossRef]
- Kunadis, E.; Lakiotaki, E.; Korkolopoulou, P.; Piperi, C. Targeting post-translational histone modifying enzymes in glioblastoma. Pharmacol Ther 2021, 220, 107721. [Google Scholar] [CrossRef]
- Holdenrieder, S.; Stieber, P.; Bodenmüller, H.; Fertig, G.; Fürst, H.; Schmeller, N.; Untch, M.; Seidel, D. Nucleosomes in serum as a marker for cell death. Clin Chem Lab Med 2001, 39, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Romani, M.; Pistillo, M. P.; Banelli, B. Epigenetic Targeting of Glioblastoma. Front Oncol 2018, 8, 448. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, B. S.; Xiong, Y. Y.; Yang, L. J.; Wu, L. X. 4,5-Dimethoxycanthin-6-one is a novel LSD1 inhibitor that inhibits proliferation of glioblastoma cells and induces apoptosis and pyroptosis. Cancer Cell Int 2022, 22, 32. [Google Scholar] [CrossRef] [PubMed]
- Lowe, B. R.; Maxham, L. A.; Hamey, J. J.; Wilkins, M. R.; Partridge, J. F. Histone H3 Mutations: An Updated View of Their Role in Chromatin Deregulation and Cancer. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef]
- Liu, B. L.; Cheng, J. X.; Zhang, X.; Wang, R.; Zhang, W.; Lin, H.; Xiao, X.; Cai, S.; Chen, X. Y.; Cheng, H. Global histone modification patterns as prognostic markers to classify glioma patients. Cancer Epidemiol Biomarkers Prev 2010, 19, 2888–2896. [Google Scholar] [CrossRef]
- Gahan, P. B. Circulating nucleic acids in plasma and serum: diagnosis and prognosis in cancer. Epma j 2010, 1, 503–512. [Google Scholar] [CrossRef]
- Holdenrieder, S. , Spuler, A., Tischinger, M., Nagel, D., Stieber, P. Presence of Nucleosomes in Cerebrospinal Fluid of Glioblastoma Patients – Potential for Therapy Monitoring. In Springer Nature, Gahan; 2010.
- Andreas Spuler, M. T. , Dorothea Nagel and Petra Stieber Presence of Nucleosomes in Cerebrospinal Fluid of Glioblastoma Patients – Potential for Therapy Monitoring; 2010.
- Erez, N.; Furth, N.; Fedyuk, V.; Wadden, J.; Aittaleb, R.; Schwark, K.; Niculcea, M.; Miclea, M.; Mody, R.; Franson, A.; et al. Single-molecule systems for detection and monitoring of plasma circulating nucleosomes and oncoproteins in Diffuse Midline Glioma. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Lee, R. C.; Feinbaum, R. L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236–240. [Google Scholar]
- Klekner, Á.; Szivos, L.; Virga, J.; Árkosy, P.; Bognár, L.; Birkó, Z.; Nagy, B. Significance of liquid biopsy in glioblastoma - A review. J Biotechnol 2019, 298, 82–87. [Google Scholar] [CrossRef]
- Ebrahimkhani, S.; Vafaee, F.; Hallal, S.; Wei, H.; Lee, M. Y. T.; Young, P. E.; Satgunaseelan, L.; Beadnall, H.; Barnett, M. H.; Shivalingam, B.; et al. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. NPJ Precis Oncol 2018, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Ebrahim Khani, S.; Wei, H.; Lee, M. Y. T.; Sim, H. W.; Sy, J.; Shivalingam, B.; Buckland, M. E.; Alexander-Kaufman, K. L. Deep Sequencing of Small RNAs from Neurosurgical Extracellular Vesicles Substantiates miR-486-3p as a Circulating Biomarker that Distinguishes Glioblastoma from Lower-Grade Astrocytoma Patients. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J Exp Clin Cancer Res 2012, 31, 97. [Google Scholar] [CrossRef]
- Sun, J.; Liao, K.; Wu, X.; Huang, J.; Zhang, S.; Lu, X. Serum microRNA-128 as a biomarker for diagnosis of glioma. Int J Clin Exp Med 2015, 8, 456–463. [Google Scholar]
- Lai, N. S.; Wu, D. G.; Fang, X. G.; Lin, Y. C.; Chen, S. S.; Li, Z. B.; Xu, S. S. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer 2015, 112, 1241–1246. [Google Scholar] [CrossRef]
- Shao, N.; Wang, L.; Xue, L.; Wang, R.; Lan, Q. Plasma miR-454-3p as a potential prognostic indicator in human glioma. Neurol Sci 2015, 36, 309–313. [Google Scholar] [CrossRef]
- Regazzo, G.; Terrenato, I.; Spagnuolo, M.; Carosi, M.; Cognetti, G.; Cicchillitti, L.; Sperati, F.; Villani, V.; Carapella, C.; Piaggio, G.; et al. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp Clin Cancer Res 2016, 35, 124. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, L.; Song, Z.; Guo, C.; Zhu, J.; Li, Z.; Zhu, S. Potential Diagnostic and Prognostic Value of Plasma Circulating MicroRNA-182 in Human Glioma. Med Sci Monit 2016, 22, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Lan, F.; Hu, M.; Pan, Q.; Wang, Q.; Wang, J. Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J Neurosurg 2016, 124, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Shao, N.; Wang, R.; Deng, D.; Xue, L.; Wang, Q.; Zhang, Y.; Shi, Y.; Xia, X.; Wang, S.; et al. Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro Oncol 2015, 17, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Chan, J. A.; Krichevsky, A. M.; Kosik, K. S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S. T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E. A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B. L.; Mak, R. H.; Ferrando, A. A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Johnson, S. M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K. L.; Brown, D.; Slack, F. J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Sampson, V. B.; Rong, N. H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N. J.; Dunn, S. P.; Krueger, L. J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Lee, S. T.; Chu, K.; Oh, H. J.; Im, W. S.; Lim, J. Y.; Kim, S. K.; Park, C. K.; Jung, K. H.; Lee, S. K.; Kim, M.; et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol 2011, 102, 19–24. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Friedmann-Morvinski, D.; Neveu, P.; Dugas, J. C.; Gill, R. M.; Huillard, E.; Liu, C.; Zong, H.; Rowitch, D. H.; Barres, B. A.; et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 2012, 31, 1884–1895. [Google Scholar] [CrossRef]
- Shi, L.; Wan, Y.; Sun, G.; Gu, X.; Qian, C.; Yan, W.; Zhang, S.; Pan, T.; Wang, Z.; You, Y. Functional differences of miR-125b on the invasion of primary glioblastoma CD133-negative cells and CD133-positive cells. Neuromolecular Med 2012, 14, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, D.; Lv, T.; Li, G.; Qu, S. Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol Neurobiol 2016, 53, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N. M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R. Y.; Saria, M. G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 2012, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; Ezz El Arab, L.; Al-Posttany, A. S.; S, B. S. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J Neurooncol 2019, 144, 545–551. [Google Scholar] [CrossRef]
- Morokoff, A.; Jones, J.; Nguyen, H.; Ma, C.; Lasocki, A.; Gaillard, F.; Bennett, I.; Luwor, R.; Stylli, S.; Paradiso, L.; et al. Serum microRNA is a biomarker for post-operative monitoring in glioma. J Neurooncol 2020, 149, 391–400. [Google Scholar] [CrossRef]
- Muhtadi, R.; Bernhardt, D.; Multhoff, G.; Hönikl, L.; Combs, S. E.; Krieg, S. M.; Gempt, J.; Meyer, B.; Barsegian, V.; Lindemann, M.; et al. Liquid Biopsy in Whole Blood for Identification of Gene Expression Patterns (mRNA and miRNA) Associated with Recurrence of Glioblastoma WHO CNS Grade 4. Cancers (Basel) 2024, 16. [Google Scholar] [CrossRef]
- Kitano, Y.; Aoki, K.; Ohka, F.; Yamazaki, S.; Motomura, K.; Tanahashi, K.; Hirano, M.; Naganawa, T.; Iida, M.; Shiraki, Y.; et al. Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds. ACS Appl Mater Interfaces 2021, 13, 17316–17329. [Google Scholar] [CrossRef]
- Akers, J. C.; Hua, W.; Li, H.; Ramakrishnan, V.; Yang, Z.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B. R.; et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 2017, 8, 68769–68779. [Google Scholar] [CrossRef]
- Drusco, A.; Bottoni, A.; Laganà, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M. P.; et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef]
- Cordonnier, M.; Chanteloup, G.; Isambert, N.; Seigneuric, R.; Fumoleau, P.; Garrido, C.; Gobbo, J. Exosomes in cancer theranostic: Diamonds in the rough. Cell Adh Migr 2017, 11, 151–163. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D'Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D. W.; Simpson, R. J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.; Kumari Singh, D.; Panda, S.; Shiras, A. Extracellular Vesicles As Modulators of Tumor Microenvironment and Disease Progression in Glioma. Front Oncol 2017, 7, 144. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, H.; Xiong, C.; Liu, Y. Hypoxic glioblastoma release exosomal VEGF-A induce the permeability of blood-brain barrier. Biochem Biophys Res Commun 2018, 502, 324–331. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Wu, X.; Shi, M.; Lian, Y.; Zhang, H. Exosomal circRNAs as promising liquid biopsy biomarkers for glioma. Front Immunol 2023, 14, 1039084. [Google Scholar] [CrossRef]
- Rosas-Alonso, R.; Colmenarejo-Fernández, J.; Pernía, O.; Burdiel, M.; Rodríguez-Antolín, C.; Losantos-García, I.; Rubio, T.; Moreno-Velasco, R.; Esteban-Rodríguez, I.; Martínez-Marín, V.; et al. Evaluation of the clinical use of MGMT methylation in extracellular vesicle-based liquid biopsy as a tool for glioblastoma patient management. Sci Rep 2024, 14, 11398. [Google Scholar] [CrossRef]
- Indira Chandran, V.; Gopala, S.; Venkat, E. H.; Kjolby, M.; Nejsum, P. Extracellular vesicles in glioblastoma: a challenge and an opportunity. NPJ Precis Oncol 2024, 8, 103. [Google Scholar] [CrossRef]
- Karami Fath, M.; Azami, J.; Masoudi, A.; Mosaddeghi Heris, R.; Rahmani, E.; Alavi, F.; Alagheband Bahrami, A.; Payandeh, Z.; Khalesi, B.; Dadkhah, M.; et al. Exosome-based strategies for diagnosis and therapy of glioma cancer. Cancer Cell Int 2022, 22, 262. [Google Scholar] [CrossRef]
- André-Grégoire, G.; Bidère, N.; Gavard, J. Temozolomide affects Extracellular Vesicles Released by Glioblastoma Cells. Biochimie 2018, 155, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.; Kushwaha, D.; Koay, D. C.; Reddy, H.; Mao, Y.; Zhou, L.; Ng, K.; Zinn, P.; Carter, B.; Chen, C. C. Post-transcriptional regulation of O(6)-methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark 2011, 10, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Garnier, D.; Meehan, B.; Kislinger, T.; Daniel, P.; Sinha, A.; Abdulkarim, B.; Nakano, I.; Rak, J. Divergent evolution of temozolomide resistance in glioblastoma stem cells is reflected in extracellular vesicles and coupled with radiosensitization. Neuro Oncol 2018, 20, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Akers, J. C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One 2013, 8, e78115. [Google Scholar] [CrossRef]
- Koch, C. J.; Lustig, R. A.; Yang, X. Y.; Jenkins, W. T.; Wolf, R. L.; Martinez-Lage, M.; Desai, A.; Williams, D.; Evans, S. M. Microvesicles as a Biomarker for Tumor Progression versus Treatment Effect in Radiation/Temozolomide-Treated Glioblastoma Patients. Transl Oncol 2014, 7, 752–758. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D. H.; Gainche, L.; Sena-Esteves, M.; Curry, W. T., Jr.; Carter, B. S.; Krichevsky, A. M.; Breakefield, X. O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Evans, S. M.; Putt, M.; Yang, X. Y.; Lustig, R. A.; Martinez-Lage, M.; Williams, D.; Desai, A.; Wolf, R.; Brem, S.; Koch, C. J. Initial evidence that blood-borne microvesicles are biomarkers for recurrence and survival in newly diagnosed glioblastoma patients. J Neurooncol 2016, 127, 391–400. [Google Scholar] [CrossRef]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G. V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin Cancer Res 2019, 25, 266–276. [Google Scholar] [CrossRef]
- Manda, S. V.; Kataria, Y.; Tatireddy, B. R.; Ramakrishnan, B.; Ratnam, B. G.; Lath, R.; Ranjan, A.; Ray, A. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J Neurosurg 2018, 128, 1091–1101. [Google Scholar] [CrossRef]
- Lan, F.; Qing, Q.; Pan, Q.; Hu, M.; Yu, H.; Yue, X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol (Dordr) 2018, 41, 25–33. [Google Scholar] [CrossRef]
- Manterola, L.; Guruceaga, E.; Gállego Pérez-Larraya, J.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol 2014, 16, 520–527. [Google Scholar] [CrossRef]
- Santangelo, A.; Imbrucè, P.; Gardenghi, B.; Belli, L.; Agushi, R.; Tamanini, A.; Munari, S.; Bossi, A. M.; Scambi, I.; Benati, D.; et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J Neurooncol 2018, 136, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Indira Chandran, V.; Welinder, C.; Månsson, A. S.; Offer, S.; Freyhult, E.; Pernemalm, M.; Lund, S. M.; Pedersen, S.; Lehtiö, J.; Marko-Varga, G.; et al. Ultrasensitive Immunoprofiling of Plasma Extracellular Vesicles Identifies Syndecan-1 as a Potential Tool for Minimally Invasive Diagnosis of Glioma. Clin Cancer Res 2019, 25, 3115–3127. [Google Scholar] [CrossRef]
- Tan, S. K.; Pastori, C.; Penas, C.; Komotar, R. J.; Ivan, M. E.; Wahlestedt, C.; Ayad, N. G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer 2018, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- de Mooij, T.; Peterson, T. E.; Evans, J.; McCutcheon, B.; Parney, I. F. Short non-coding RNA sequencing of glioblastoma extracellular vesicles. J Neurooncol 2020, 146, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ye, L.; Wang, L.; Quan, R.; Zhou, Y.; Li, X. Identification of miRNA signatures in serum exosomes as a potential biomarker after radiotherapy treatment in glioma patients. Ann Diagn Pathol 2020, 44, 151436. [Google Scholar] [CrossRef]
- Maire, C. L.; Fuh, M. M.; Kaulich, K.; Fita, K. D.; Stevic, I.; Heiland, D. H.; Welsh, J. A.; Jones, J. C.; Görgens, A.; Ricklefs, T.; et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol 2021, 23, 1087–1099. [Google Scholar] [CrossRef]
- Tzaridis, T.; Reiners, K. S.; Weller, J.; Bachurski, D.; Schäfer, N.; Schaub, C.; Hallek, M.; Scheffler, B.; Glas, M.; Herrlinger, U.; et al. Analysis of Serum miRNA in Glioblastoma Patients: CD44-Based Enrichment of Extracellular Vesicles Enhances Specificity for the Prognostic Signature. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Dobra, G.; Bukva, M.; Szabo, Z.; Bruszel, B.; Harmati, M.; Gyukity-Sebestyen, E.; Jenei, A.; Szucs, M.; Horvath, P.; Biro, T.; et al. Small Extracellular Vesicles Isolated from Serum May Serve as Signal-Enhancers for the Monitoring of CNS Tumors. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Chen, W. W.; Balaj, L.; Liau, L. M.; Samuels, M. L.; Kotsopoulos, S. K.; Maguire, C. A.; Loguidice, L.; Soto, H.; Garrett, M.; Zhu, L. D.; et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids 2013, 2, e109. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J. M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol 2017, 19, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F. L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A. B.; Speranza, M. C.; Nakashima, H.; Hayes, J. L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv 2018, 4, eaar2766. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D. D.; Carter, B. S.; Hochberg, F. H.; Breakefield, X. O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Müller Bark, J., Paul Leo7, Rosalind L. Jeffree, Benjamin Chua, Bryan W. Day, Chamindie Punyadeera. Isolation and characterization of exosomes from plasma and saliva of glioblastoma patients. Metro North Health QLD, 2020.
- Mikolajewicz, N.; Khan, S.; Trifoi, M.; Skakdoub, A.; Ignatchenko, V.; Mansouri, S.; Zuccatto, J.; Zacharia, B. E.; Glantz, M.; Zadeh, G.; et al. Leveraging the CSF proteome toward minimally-invasive diagnostics surveillance of brain malignancies. Neurooncol Adv 2022, 4, vdac161. [Google Scholar] [CrossRef]
- Buehler, M.; Yi, X.; Ge, W.; Blattmann, P.; Rushing, E.; Reifenberger, G.; Felsberg, J.; Yeh, C.; Corn, J. E.; Regli, L.; et al. Quantitative proteomic landscapes of primary and recurrent glioblastoma reveal a protumorigeneic role for FBXO2-dependent glioma-microenvironment interactions. Neuro Oncol 2023, 25, 290–302. [Google Scholar] [CrossRef]
- Soylemez, B.; Bulut, Z.; Şahin-Bölükbaşı, S. Investigating the Potential of Lipids for Use as Biomarkers for Glioblastoma via an Untargeted Lipidomics Approach. J Korean Neurosurg Soc 2022. [Google Scholar] [CrossRef]
- Tsvetkov, P. O.; Eyraud, R.; Ayache, S.; Bougaev, A. A.; Malesinski, S.; Benazha, H.; Gorokhova, S.; Buffat, C.; Dehais, C.; Sanson, M.; et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Eledkawy, A.; Hamza, T.; El-Metwally, S. Precision cancer classification using liquid biopsy and advanced machine learning techniques. Sci Rep 2024, 14, 5841. [Google Scholar] [CrossRef]
- Mouliere, F.; Smith, C. G.; Heider, K.; Su, J.; van der Pol, Y.; Thompson, M.; Morris, J.; Wan, J. C. M.; Chandrananda, D.; Hadfield, J.; et al. Fragmentation patterns and personalized sequencing of cell-free DNA in urine and plasma of glioma patients. EMBO Mol Med 2021, 13, e12881. [Google Scholar] [CrossRef]
- Sol, N.; In 't Veld, S.; Vancura, A.; Tjerkstra, M.; Leurs, C.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Zwaan, K.; et al. Tumor-Educated Platelet RNA for the Detection and (Pseudo)progression Monitoring of Glioblastoma. Cell Rep Med 2020, 1, 100101. [Google Scholar] [CrossRef] [PubMed]
- Herrgott, G. A.; Asmaro, K. P.; Wells, M.; Sabedot, T. S.; Malta, T. M.; Mosella, M. S.; Nelson, K.; Scarpace, L.; Barnholtz-Sloan, J. S.; Sloan, A. E.; et al. Detection of tumor-specific DNA methylation markers in the blood of patients with pituitary neuroendocrine tumors. Neuro Oncol 2022, 24, 1126–1139. [Google Scholar] [CrossRef]
- Menna, G.; Piaser Guerrato, G.; Bilgin, L.; Ceccarelli, G. M.; Olivi, A.; Della Pepa, G. M. Is There a Role for Machine Learning in Liquid Biopsy for Brain Tumors? A Systematic Review. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Theakstone, A. G.; Brennan, P. M.; Jenkinson, M. D.; Mills, S. J.; Syed, K.; Rinaldi, C.; Xu, Y.; Goodacre, R.; Butler, H. J.; Palmer, D. S.; et al. Rapid Spectroscopic Liquid Biopsy for the Universal Detection of Brain Tumours. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Bukva, M.; Dobra, G.; Gomez-Perez, J.; Koos, K.; Harmati, M.; Gyukity-Sebestyen, E.; Biro, T.; Jenei, A.; Kormondi, S.; Horvath, P.; et al. Raman Spectral Signatures of Serum-Derived Extracellular Vesicle-Enriched Isolates May Support the Diagnosis of CNS Tumors. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Aaes, T. L.; Vandenabeele, P. The intrinsic immunogenic properties of cancer cell lines, immunogenic cell death, and how these influence host antitumor immune responses. Cell Death Differ 2021, 28, 843–860. [Google Scholar] [CrossRef]
- Ginghina, O.; Hudita, A.; Zamfir, M.; Spanu, A.; Mardare, M.; Bondoc, I.; Buburuzan, L.; Georgescu, S. E.; Costache, M.; Negrei, C.; et al. Liquid Biopsy and Artificial Intelligence as Tools to Detect Signatures of Colorectal Malignancies: A Modern Approach in Patient's Stratification. Front Oncol 2022, 12, 856575. [Google Scholar] [CrossRef]
- Tiwari, A.; Mishra, S.; Kuo, T. R. Current AI technologies in cancer diagnostics and treatment. Mol Cancer 2025, 24, 159. [Google Scholar] [CrossRef]
- Foser, S.; Maiese, K.; Digumarthy, S. R.; Puig-Butille, J. A.; Rebhan, C. Looking to the Future of Early Detection in Cancer: Liquid Biopsies, Imaging, and Artificial Intelligence. Clin Chem 2024, 70, 27–32. [Google Scholar] [CrossRef]
- Parvin, N.; Joo, S. W.; Jung, J. H.; Mandal, T. K. Multimodal AI in Biomedicine: Pioneering the Future of Biomaterials, Diagnostics, and Personalized Healthcare. Nanomaterials (Basel) 2025, 15. [Google Scholar] [CrossRef]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer current: status, challenges and future prospects. Signal Transduct Target Ther 2024, 9, 336. [Google Scholar] [CrossRef]
- Cheung, H. M. C.; Rubin, D. Challenges and opportunities for artificial intelligence in oncological imaging. Clin Radiol 2021, 76, 728–736. [Google Scholar] [CrossRef]
- Bi, W. L.; Hosny, A.; Schabath, M. B.; Giger, M. L.; Birkbak, N. J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I. F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin 2019, 69, 127–157. [Google Scholar] [CrossRef]
| Isolation method | Patient numbers | Markers used to verify GBM origin | Findings | References and publication date |
|---|---|---|---|---|
| CTC-iChip microfluidic technology. Leukocyte depletion using magnetically-tagged anti-CD45 and anti-CD16 antibodies. | 33 | SOX-2, EGFR, c-MET, A2B5 tubulin β-3 | Isolated CTCs from peripheral blood of 39% (13 of 33) GBM patients. Greater CTC counts in patients with progressive disease relative to stable disease. CTCs have a mesenchymal phenotype. |
[22] (2014) |
| Density gradient centrifugation | 141 | Single-cell genomics for common GBM mutations, GFAP staining, tumour specific anomalies like amplification of EGFR gene and gains and losses in chromosomes 7 and 10 genomic regions by chromosomal and array CGH on whole genome amplification. | Isolated CTCs from 20.6% (29/141) of patients. A convenient diagnostic tool for identifying patients with extracranial tumour cell spread and indicates that CTCs could be used to monitor the progression of glioblastoma. | [29] (2014) |
| Density gradient centrifugation using the OncoQuick® system. | 11 | Telomerase-based test was used to identify CTCs. |
CTCs detected in 72% (8 of 11) of patients before radiotherapy but dropped to 8% (1 of 8) in post-radiotherapy patients. This suggests that CTCs may be useful to monitor the progression of cancers before and after therapies. | [30] (2014) |
| Immunoaffinity-based methods. CTC separation with a matrix and negative depletion of white blood cells using immunomagnetic beads. |
31 | Polyploidy chromosome-8-positive detection was employed as a positive measure for CTCs using subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH), in addition to GFAP-positive or GFAP-negative cells and CD45-negative cells grading to confirm cell origin. | CTCs were detected in 77% (24 of 31) of GBM patients. Monitoring treatment using CTCs was slightly better than MRI in distinguishing radionecrosis from recurrence of glioma. CTCs can dynamically monitor the microenvironment of gliomas which is a significant complement to radiographic imaging. |
[31] (2016) |
| Immunotargeted enrichment of MSP & MCAM expressing cells. | 13 | CTCs were isolated based on cell surface MSP and MCAM and identified by probing for GLAST and/or GFAP expression. | ≥ 1 CTCs were detected in 69% of patients (9/13), using the combination of 2 isolation and 2 identification markers increased CTC detection. | [21] (2020) |
| Immunopheno-typing. CTCs were isolated by size separation using a Parsortix® microfluidic cassette. | 13 | CTCs were isolated based on no expression of CD45, while expressing EGFR, Ki67, and EB1 microtubule associated protein. For confirmation, the GBM CTC clusters and a biopsy from the primary tumour of the patient were stained with GBM marker SOX-2. |
CTC clusters identified in 53.8% of 13 GBM patients. GBM markers validated that multicellular CTC clusters can be formed and pass the BBB in patients with GBM to reach peripheral circulation and be used for monitoring. |
[32] (2018) |
| Spiral microfluidic technology | 20 | CTCs exhibited characteristic molecular features of GBM, such as EGFR amplification and mutations in TP53 and IDH1. | The study found that CTCs could be isolated from both early- and late-stage GBM patients, highlighting the potential of this technique for non-invasive monitoring of tumour progression. | [33] (2021) |
| CTC Subtraction enrichment/depletion with magnetic immunoaffinity beads and immunostaining-FISH | 22 | Detection of CTCs to differentiate between treatment-induced necrosis and tumour recurrence in brain gliomas. CTC detection outperformed both DSC-MRP and MET-PET in diagnostic accuracy. Additionally, it showed potential for predicting recurrence in one patient. |
The mean CTC count was significantly higher in patients with tumour recurrence (6.10 ± 3.28) compared to those with treatment necrosis (1.08 ± 2.54). A threshold CTC count of 2 provided 100% sensitivity and 91.2% specificity (AUC = 0.933) for identifying tumour recurrence. CTC detection could be a valuable tool for distinguishing tumour recurrence from necrosis, warranting further validation in larger clinical studies. | [34] (2021) |
| Isolation method | Patient numbers | Markers used to verify GBM origin | Findings | References and publication date |
|---|---|---|---|---|
| Guardant360® and digital NGS | 171 | NGS targeting 54 cancer related genes, including assessments for copy number variants in EGFR, ERBB2, and MET. | Of the 33 patients diagnosed with GBM, 73% had unaltered ctDNA, 24% had one alteration and 3% had two or more alterations. | [50] (2016) |
| Guardant360® and digital NGS | 222 | Single nucleotide variants were detected in 61 genes, with amplifications detected in ERBB2, MET, EGFR. | ctDNA mutations were detected in blood samples from 55% of GBM patients. | [51] (2019) |
| Illustra triplePrep Kit (GE healthcare BioSciences Corp) and WGS | 13 | EGFRvIII mutation characterised by a deletion on exons 2 through 7. | EGFRvIII mutant DNA detected in the plasma of GBM patients, with its presence correlating with the mutation in tumour tissue. | [54] (2013) |
| DNA extraction and NGS | 107 | Tumour-specific mutations such as EGFRvIII | ctDNA detection rate was 51% Genomic alteration in the ctDNA of patients highlight the potential of guiding personalised cancer treatment. |
[55] (2018) |
| DNA extraction and methylation specific PCR assay | 19 | MGMT promoter methylation GBM patients serum and cerebrospinal fluid CSF samples for ctDNA detection using methylation specific PCR assay, |
Detected 37% ctDNA in serum and 61% ctDNA in CSF. MGMT promoter methylation was detected with higher sensitivity in CSF (72.0%) compared to serum (41.7%), suggesting CSF as a promising tool for early diagnosis, treatment monitoring, and recurrence detection. |
[61] (2015) |
| DNA extraction and nested PCR-based assays | 38 | ctDNA was analysed for TERT promoter mutations (C228T and C250T) and IDH hotspot mutations. | Detected 8% (3 of 38 patients) ctDNA in plasma and 92% (35 of 38 patients) ctDNA in CSF. | [62] (2018) |
| DNA extraction and WGS | 11 | Tumour-specific mutations in the ctDNA extracted from CSF. | 100% ctDNA detection rate in CSF. | [63] (2015) |
| DNA extraction and WGS | 13 | Copy number alterations in ctDNA. | 50% ctDNA detection rate in CSF. Identified copy number alterations in the ctDNA, which closely reflected tumour genetic profile. Fragmentation patterns in CSF-derived ctDNA Copy number alterations in ctDNA were consistent with tumour tissue. |
[74] (2018) |
| DNA extraction and NGS | 16 | IDH1/2 and 1p/19q codeletion. | ctDNA detected in 49.4% of CSF samples. Genetic alterations detected closely matched those found in tumour biopsies. |
[75] (2018) |
| DNA extraction and NGS | NA | IDH1 (R132H variant), TERT promoter (C228T mutation), TP53, ATRX, H3F3A and HIST1H3B. | CSF ctDNA more accurately reflected BM mutations, detecting all mutations in 83.33% of cases versus 27.78% for plasma ctDNA. CSF ctDNA more accurately reflected BM mutations, detecting all mutations in 83.33% of cases versus 27.78% for plasma ctDNA. Mutant allele frequency (MAF) in CSF ctDNA strongly correlated with BM tumour size (r = 0.95) and was higher than in plasma ctDNA (38.05% vs. 4.57%). MAF and tumour mutational burden in CSF ctDNA closely matched BM values (r = 0.96 and 0.97, respectively). CSF ctDNA exhibited superior concordance with BM (99.33%) compared to plasma ctDNA (67.44%), improving the identification of clinically relevant mutations. However, for multiple BM, plasma ctDNA performed well, achieving a 93.01% concordance, comparable to CSF ctDNA. |
[76] (2023) |
| DNA extraction and MSK-IMPACT™, a NGS assay | 711 | The distribution of clinically actionable somatic alterations was consistent with tumour-type specific alterations across the AACR GENIE cohort. | Genetic alterations were detected in 53% (489/922) of CSF samples from patients with confirmed CNS tumours, while none of the 85 samples from patients without CNS tumours contained detectable ctDNA. The identified mutations aligned with tumour-type-specific alterations observed in the AACR GENIE cohort. Repeated ctDNA testing revealed clonal evolution and resistance mechanisms, and the presence of ctDNA linked to reduced overall survival after CSF collection. |
[77] (2024) |
| QIAamp Circulating Nucleic Acids kit (QIAGEN), Western blot, histopathology, and immunochemistry, digital PCR, and shallow whole genome sequencing were utilised | 64 | Patient-derived orthotopically implanted xenograft models of GBM. | Analysis of fragment length profiles of host (rat) and tumour (human) ctDNA identified a peak at 145 bp in the human DNA fragments, indicating a difference in the origin or processing of the ctDNA. The concentration of ctDNA correlated with cell death only after treatment with TMZ and radiotherapy. ddPCR detection of plasma tumour mitochondrial DNA (tmtDNA), an alternative to detection of nuclear ctDNA, improved plasma DNA detection rate (82% versus 24%) and allowed detection in CSF and urine. The total amount of tmtDNA in the plasma (558 copies) is ~13 times higher than in the CSF (43 copies), showing therefore that the BBB does not prevent significant amounts of tumour DNA |
[65] (2019) |
| QIAamp DNA micro kit (Qiagen) and amplicon sequencing | 20 | DH1, IDH2, TP53, TERT, ATRX, H3F3A, and HIST1H3B gene mutations | The genomic analysis in CSF extracted ctDNA facilitates the diagnosis of diffuse gliomas into subtypes to support the surgical and clinical management of these patients. | [75] (2018) |
| QIAamp Circulating Nucleic Acid kit and NGS | 26 | Cancer genomic panel sequencing on the CSF-derived ctDNA. | A high detection rate of ctDNA (24/26, 92.3%) was observed in CSF. ctDNA mutations had high concordance rates with tumour DNA, especially in non-copy number variations and in GBM. CSF ctDNA TMB also exhibited a strong correlation with tumour DNA TMB (R2 = 0.879, P < 0.001), particularly in GBM (R2 = 0.992, P < 0.001). |
[78] (2022) |
| DNeasy Blood and Tissue Kit (Qiagen), and targeted DNA sequencing by Oxford Nanopore Technology MinION device. | 12 paediatric high-grade glioma patients and 6 controls | Analysis of ctDNA with a handheld platform (Oxford Nanopore MinION) to quantify patient-specific CSF ctDNA variant allele fraction (VAF) | Nanopore demonstrated 85% sensitivity and 100% specificity in CSF samples (n = 127 replicates) with 0.1 femtomole DNA limit of detection and 12 h results, all of which compared favourably with NGS. Multiplexed analysis provided concurrent analysis of H3.3A (H3F3A) and H3C2 (HIST1H3B) mutations in a non-biopsied patient and results were confirmed by ddPCR. Serial CSF ctDNA sequencing by Nanopore demonstrated correlation of radiological response on a clinical trial, with one patient showing dramatic multi-gene molecular response that predicted long-term clinical response. |
[79] (2020) |
| Isolation method | Patient numbers | Markers used to verify GBM origin | Findings | References |
|---|---|---|---|---|
| Cell Death Detection ELISA Plus kit | 10 | NA | Pre-therapeutic nucleosome levels in both serum and CSF did not significantly differ among GBM patients and control groups. Postoperative Increase: In GBM patients, nucleosome levels in serum and CSF increased moderately during the week following surgery and intracavitary chemotherapy. Cerebral Oedema Correlation: Three out of ten GBM patients developed cerebral oedema post-surgery. In these patients, CSF nucleosome levels increased almost 200-fold, peaking on day 3 postoperatively. In contrast, the seven patients without oedema exhibited only slight increases in nucleosome levels. Clinical Implication: Monitoring CSF nucleosome levels may serve as an indicator for postoperative complications such as cerebral oedema in GBM patients. |
[94] |
| Single-molecule technology to detect and monitor plasma-circulating nucleosomes | NA | H3-K27M mutation and mutant p53 | The single-molecule analysis revealed epigenetic patterns unique to diffuse midline glioma, enabling differentiation from healthy individuals and patients with other cancer types. This approach profiles multiple histone modifications on individual nucleosomes from less than 1 mL of plasma, revealing epigenetic patterns unique to glioma that significantly differentiate these patients from healthy individuals and those with other cancer types. The detection strategy demonstrated a correlation with MRI measurements and ddPCR assessments of ctDNA, highlighting its potential utility in non-invasive treatment monitoring. Suitable for paediatric patients and scenarios where sample volume is limited. |
[95] |
| Isolation method | Patient numbers | Markers used to verify GBM origin | Findings | Reference and publication date |
|---|---|---|---|---|
| miRNA extraction and qPCR | 20 | miR-221 and miR-222 | Both miR-221 and miR-222 were significantly upregulated in the plasma of GBM patients compared to healthy controls. miR-221 demonstrated 90% sensitivity and 100% specificity and miR-222 demonstrated 85% sensitivity and 100% specificity for GBM detection. Expression levels of miR-221 and miR-222 decreased following treatment |
[119] (2019) |
| miRNA profiling performed using the Nanostring® platform | 91 | miR-223 and miR-320e, IDH mutation status and1p/19q co-deletion. | Dynamic changes of miR-320e were linked to tumour volume of GBM patients. A 9-miRNA signature was established, distinguishing glioma patients from healthy controls with 99.8% accuracy. miRNA levels did not increase in cases of pseudo-progression. This supports their use in distinguishing true progression from treatment effects and in post-operative monitoring |
[120] (2020) |
| mirVana™ miRNA Isolation Kit and qRT-PCR | 50 | miR-21, miR-128, and miR-342-3p |
miR-21 was significantly upregulated, while miR-128 and miR-342-3p were markedly downregulated in glioma patients compared to healthy controls Notably, miR-21 demonstrated a high diagnostic performance with 90% sensitivity, and 100% specificity. |
[101] (2012) |
| RNA isolation and NGS, including mRNA-seq and small RNA-seq | 7 | mRNA and miRNA candidates | The study identified differentially expressed genes in individual patients, with up to 93 mRNA and 19 miRNA candidates linked to GBM recurrence. | [121] (2024) |
| Urinary microRNA-based diagnostic model for CNS tumours using nanowire scaffolds. | 119 | Differential miRNA expression profiles | The study reported high diagnostic performance, with sensitivity and specificity values of 100% and 97%, respectively, for detecting early-stage CNS tumours. Non-invasive method holds promise for early detection and monitoring of CNS tumours through urine-based liquid biopsy. |
[122] (2021) |
| miRCURY RNA Isolation Kit and qRT-PCR | 10 | miR-21, miR-218, miR-193b, miR- 331, and miR-374a, miR- 548c, miR-520f, miR-27b, and miR-130b | Sampling of CSF from the lumbar region to extract 9 signature miRNA for GBM. The overexpressed signatures were miR-21, miR-218, miR-193b, miR- 331, and miR-374a, while the down regulated were miR- 548c, miR-520f, miR-27b, and miR-130b in GBM CSF The study compared the diagnostic performance of miRNA detection between CSF obtained from the cisternal and lumbar regions. The cisternal CSF samples demonstrated a sensitivity of 80% and specificity of 67% for GBM detection, whereas the lumbar CSF samples showed a sensitivity of 28% and specificity of 95%. |
[123] (2017) |
| TaqMan Low Density Array platform for miRNA profiling and qRT-PCR | 16 GBM and 9 healthy patients | miR-451, miR-711, miR-935, miR-223 | Showed that miRNA from CSF can differentiate between tumour and non-tumour diseases states. Identified distinct miRNA signature in CSF that can distinguish CNS malignancies (GBM) from non-tumour controls. |
[124] (2015) |
| Isolation method | Patient numbers | Markers used to verify GBM origin | Findings | References and publication date |
|---|---|---|---|---|
| Differential centrifugation and flow cytometry | 11 | GFAP | Concentration and composition of circulating MVs in patient plasma correlated with tumour progression. Elevated levels of tumour-derived MVs were associated with true tumour progression, while lower levels were indicative of treatment-related changes or pseudoprogression. |
[140] (2014) |
| Differential centrifugation and ultracentrifugation, filtration and flotation density gradient centrifugation. | 25 | Tumour-specific EGFRvIII mRNA within the vesicles | EVs contain functional RNA and proteins that may influence the tumour microenvironment and serve as diagnostic tools GBM cells release MVs containing mRNA, miRNA, and angiogenic proteins. Detection of tumour-specific EGFRvIII mRNA in serum-derived MVs supports their potential as non-invasive biomarkers for GBM diagnosis and monitoring |
[141] (2008) |
| Serial centrifugation and flow cytometry | 16 | Annexin V, CD41, CD235 and Anti-EGFR | An increase in Annexin V-positive MV levels during chemoradiation therapy was associated with earlier tumour recurrence and shorter overall survival Patients with higher levels of MVs had > 4-fold increase in the hazard ratio for recurrence compared to those with lower levels The study provided initial evidence that monitoring blood-borne MV levels could serve as a non-invasive method to predict disease progression and patient outcomes in newly diagnosed GBM patients. |
[142] (2016) |
| Ultrafiltration and ultracentrifugation and NTA | 43 | GFAP |
Plasma EV concentration was higher in GBM compared with healthy controls (p=0.0099). Average size of GBM and healthy EVs were similar in discovery (p=0.548) and validation cohort (p=0.075). The amount of circulating EV was not affected by tumour size (p=0.318) However, the extent of necrosis influenced the degree of secretion (p=0.045): higher necrosis in GBM samples (grade 3) substantially reduced EV secretion. Elevated EV levels in GBM plasma decrease post-surgery and rose at recurrence. |
[143] (2019) |
| Precipitation using ThermoFisher kit and Semi quantitative RT-PCR | 96 | EGFRvIII mRNA within the vesicles | EGFRvIII prevalence in the data set was 39.58%. The sensitivity and specificity of serum EV analysis for EGFRvIII was 81.58% (95% CI 65.67%–92.96%) and 79.31% (95% CI 66.65%–88.83%), respectively |
[144] (2018) |
| Differential centrifugation and qRT-PCR | 60 | miR-301a | Serum exosomal miR-301a levels were significantly elevated in glioma patients compared to healthy controls. Higher miR-301a levels were associated with higher tumour grades and lower Karnofsky Performance Status (KPS) scores. Post-surgical samples showed a significant reduction in miR-301a levels, which increased again during tumour recurrence, suggesting its potential as a marker for disease monitoring. Kaplan-Meier survival analysis indicated that patients with higher serum exosomal miR-301a levels had shorter overall survival. |
[145] (2018) |
| Ultracentrifugation and TEM | 42 | miR-320, miR-574-3p and RNU6-1 | RNU6-1 identified in serum exosomes could effectively distinguish GBM patients from healthy individuals. The elevated levels of RNU6-1 in GBM patients' exosomes suggest its potential as a non-invasive diagnostic biomarker. |
[146] (2014) |
| Differential centrifugation and qPCR | 12 | miR-182-5p, miR-328-3p, miR-339-5p, miR-340-5p, miR-485-3p, miR-486-5p and miR-543 | The identified miRNA signature in serum exosomes could effectively distinguish GBM patients from healthy individuals and those with lower-grade gliomas. | [99] (2018) |
| Density gradient ultracentrifugation, using OptiPrep™ Density Gradient Medium and sequencing followed by differential expression analysis | 12 GBM (astrocytoma grade IV) and 5 astrocytoma grade II-III | Analysis of CUSA (cavitron ultrasonic surgical aspirate) EV and serum EV miRNA and piRNA | Seven miRNA species were determined to be the most stable classifiers for GBM (miR-182-5p, 382-3p, 339-5p, 340-5p, 485-3p, 486-3p and 543) with an overall predictive power of 91.7%; within multivariate models, six iterations of these markers were capable of distinguishing GBM patients from healthy controls with 100% accuracy. | [100] (2020) |
| Chemical precipitation using ExoQuick-TC and qPCR | 100 patients with glioma, 11 with metastatic brain tumours, 30 healthy patients | Expression of 3 miRNAs: miR-21, miR-222 and miR-124-3p, in serum exosomes. | Exosome miR-21, 222 and 124-3p were robust for discriminating patients with GBM from healthy controls, with an area under curve of 0.84 (95% CI 0.7538–0.9371, p < 0.001), 0.80 (95% CI 0.6967–0.8980, p < 0.001) and 0.78 (95% CI 0.6732–0.8904, p < 0.001), respectively. miR-21 alone appears to be the best predictor for distinguishing patients with High grade glioma from those with low grade glioma, displaying the highest AUC of 0.83 (95% CI 0.7395–0.9398, p < 0.001 |
[147] (2018) |
| Ultracentrifugation, NTA and TEM for plEV isolation with proximity extension assay–based ultrasensitive immunoprofiling. | 82 | Syndecan-1 |
SDC1 in plEVs could discriminate between GBM and low-grade glioma with a sensitivity of 71%, and specificity of 91%. The findings support the concept of circulating plEVs as a tool for non-invasive diagnosis and monitoring of gliomas. |
[148] (2019) |
| Total Exosome Isolation reagent followed by ultracentrifugation and qRT-PCR | 43 GBM, 23 other brain tumour patients and 40 healthy individuals | Serum analysed for presence of EVs and HOTAIR biomarker | HOTAIR can be used as a biomarker for Dx and progression of some brain tumours including GBM with sensitivity and specificity of HOTAIR 86.1% and 87.5% respectively. | [149] (2018) |
| Serial ultracentrifugation and short non-coding RNA sequencing using the OASIS-2.0 platform. | 5 | Isolate EV’s from human differentiated GBM cells in vitro and perform short, non-coding RNA sequencing to determine expression pattern | Small genome sequencing revealed a total of 712 non-coding RNA sequences most of which have not been associated with GBM EV’s previously, including the let-7 miRNA family, miR-3182, miR-4448, miR-100-5p and miR-27-3p. Furthermore, multiple non-microRNA short non-coding RNA species were identified including piRNA, snRNA, snoRNA, and yRNA. |
[150] (2020) |
| qPCR | 5 Glioma patients before and after radiotherapy | Expression signature of miRNAs in glioma patients before and after radiotherapy | Identified 18 up-regulated differentially expressed (DE) miRNAs and 16 down-regulated DE miRNAs and the target genes of DE miRNAs were predicted based on multiple miRNA-target databases. | [151] (2020) |
| Differential centrifugation and DNA analysis through methylation array analysis Proteome analysis with differential quantitative proteomics |
Unspecified number of glioma patients and non-tumorous temporal tissue from patients undergoing epilepsy surgery | DNA and protein analysis of Glioma and non-tumorous EVs | Tumour-specific mutations and copy number variations were detected in EV-DNA with high accuracy. Proteome analysis did not allow specific tumour identification or classification |
[152] (2021) |
| Size-exclusion chromatography (by using qEV columns from IZON®), followed by immunoprecipitation with CD44-conjugated beads and qRT-PCR | 55 | Suitability of novel EV isolation procedure and analysis of serum EVs for miRNA biomarkers and their correlation with prognosis. | Four serum biomarkers were identified to be predictive of prognosis, miR-15b-3p, miR-21-3p, and miR-328-3p exhibiting a negative correlation (high levels were associated with an inferior prognosis) and miR-106a-5p a positive correlation (high levels were associated with a better prognosis prediction of GBM prognosis |
[153] (2020) |
| Differential centrifugation and NTA | 96 total patients, 24 GBM, 24 meningioma, 24 BM from NSCLC and 24 controls from patients with benign disk herniation. | Proteomic analysis of serum and serum derived small EVs | Identified 10- and 17-membered protein panel for whole serum and sEV samples respectively. While none of these proteins appeared to be able to distinguish between the patient groups individually, their combination was found to reliably discriminate between the different patient groups suggesting that instead of a few candidates, a specific protein panel is required for a perfect differentiation between various tumour types. | [154] (2020) |
| Ultracentrifugation and RNA extraction from EVs, ddPCR and flow cytometry for rare mutation detection. | 14 glioma patients (4 with GBM) | Mutant IDH1 G395A | CSF is a viable bio fluid to examine contents of EVs. Mutant IDH1 G395A identified in CSF EVs with a sensitivity of 63% and specificity of 100%. CSF EVs generally contained higher levels of mutant mRNA than serum EVs. EVs carry tumour-specific RNA signatures that can be detected non-invasively, supporting the potential of EVs as liquid biopsy tools for glioma diagnosis and monitoring. |
[155] (2013) |
| Ultracentrifugation and purification with sucrose cushion for EV isolation. qRT-PCR for mRNA transcript detection. | 25 GBM, 5 low-grade glioma and 4 healthy patients | EGFRvIII mutant mRNA | EGFRvIII oncogene, in EV RNA can be accomplished with a sensitivity of 60% and 98% specificity in comparison to the gold standard qPCR of EGFRvIII transcript from brain tumour tissue. | [156] (2017) |
| Ultracentrifugation and. TEM for EV isolation. Western blotting for exosomal markers. qRT-PCR for mRNA transcript quantification. |
9 GBM and 5 healthy patients | Exosomal markers (CD63 and TSG101) miR-21 |
Showed that miR-21 from CSF EVs can differentiate between tumour and non-tumour diseases states. |
[139] (2013) |
| Ultracentrifugation and OptiPrep™ density gradient ultracentrifugation for further purification. NTA, flow cytometry and western blotting (to detect PD-L1 on EV surface). | 10 | Patient derived GBM stem cells. | PD-L1 expression on the surface of GBM derived EVs, can prevent T-cell activation and proliferation upon binding directly to PD-L1. This indicates PD-L1 expression on EVs can be an immune-escape mechanism for GBM |
[157] (2018) |
| Differential centrifugation of MVs and microfluidic chip-based immunomagnetic technique called μNMR (miniaturized nuclear magnetic resonance) for protein typing of EVs/MVs. Also used magnetic nanoparticles (MNPs) conjugated to antibodies to detect specific tumour-associated proteins on circulating MVs. | 15 | EGFRvIII mutant, PDPN and IDH1 |
Tumour-derived MVs carrying EGFR/EGFRvIII proteins were successfully detected in patient plasma. Showed four protein panels of EV surface proteins can be used to discriminate GBM patients from healthy controls using novel antibody capture method. The system enabled real-time monitoring of tumour progression and treatment response by tracking changes in circulating tumour-derived MV profiles. |
[158] (2012) |
| Ultracentrifugation for exosome isolation and characterisation by western blotting for expression protein markers, TEM for morphology and NTA for size and concentration. | 10 | CD63, CD81 and CD9 | Saliva and plasma were used as liquid biopsies to isolate exosomes before and after surgery. GBM patients exhibited smaller but more abundant exosomes compared to healthy donors. EVs from GBM plasma showed globally reduced levels of cytokines and co-stimulatory molecules, but PD-L1 presence was similar to healthy donors. |
[159] (2020) |
| Proteomics using mass spectrometry | 22 | MGMT and IDH statuses and GAP43 | CSF proteomics using mass spectrometry enables GBM biomarker discovery from small volumes (~30 μL). Mikolajewics et al., identified 755 unique proteins in 73 CSF samples (22 GBM), with MGMT and IDH statuses accurately detected at 94.1% and 33.3%, respectively. Single-cell RNA sequencing confirmed GAP43 as GBM-specific, while TFF3 and CACNA2D2 were specific to BM and CNS lymphoma. |
[160] (2022) |
| Proteomics with sequential window acquisition of all theoretical fragment ion spectra mass spectrometry (SWATH-MS) and immunohistochemistry for validation. | 134 | BCAS1, INF1, and FBXO2 | Identified overexpressed proteins CSF proteomics in recurrent GBM, to quantify the proteomes of newly diagnosed and recurrent GBM patients and validated the markers using immunohistochemistry. | [161] (2023) |
| Lipidomics using Quadrupole time-of-flight liquid mass spectrometer Q-TOF LCMS/MS | 14 GBM and 14 healthy patients | NA; based on statistically significant differences in blood lipid species between GBM and controls | Lipidomics, also holds potential Identified differential lipid species including fatty acids, saccharolipid, sphingolipid, glycerolipid and sterol lipid from blood samples. |
[162] (2022) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





