Submitted:
06 August 2025
Posted:
11 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
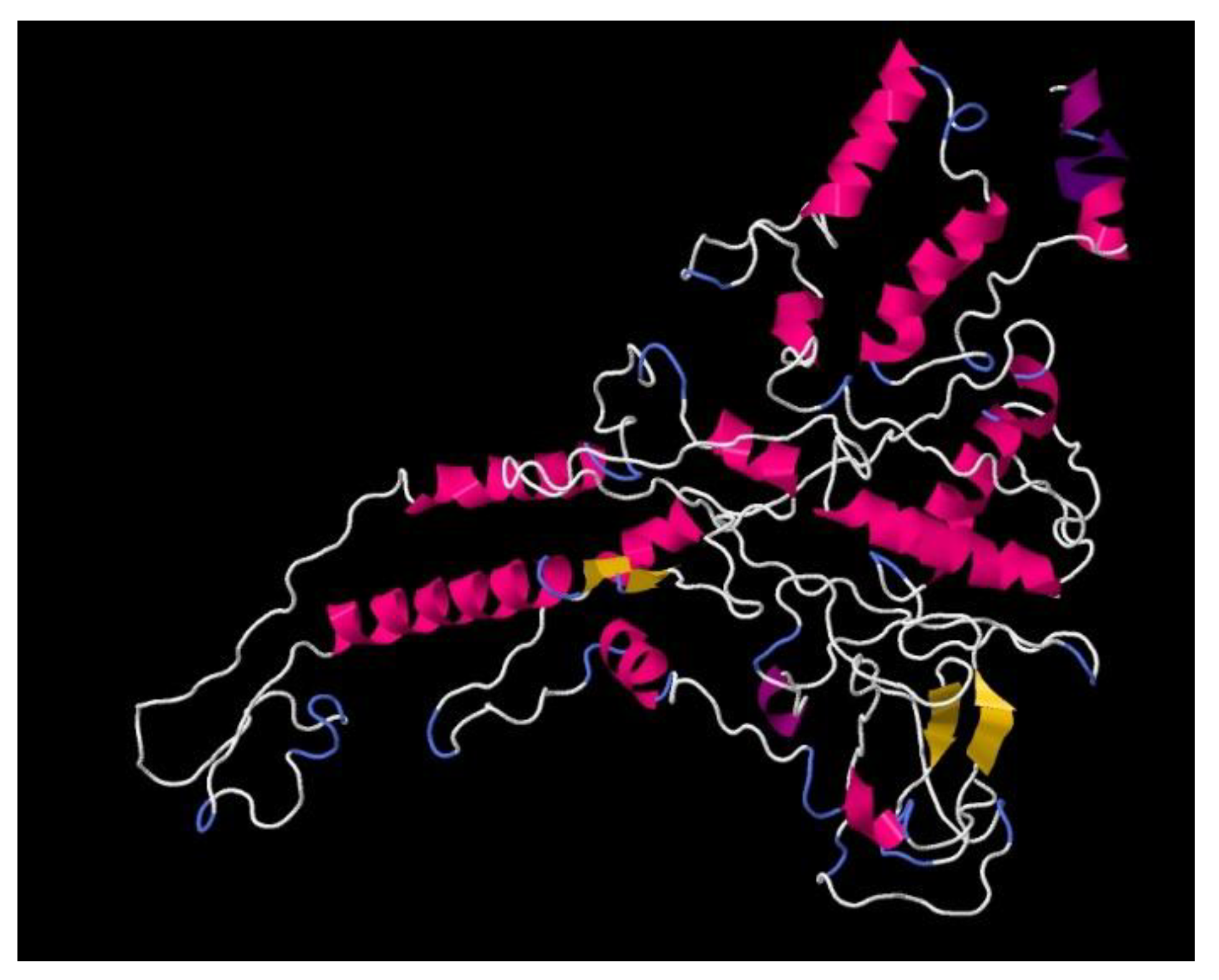
2.1. Development of the Recombinant Antigen Preparation CoronaDerm-PS
2.2. Preclinical Study Phase in Animals
2.3. Integrated Analysis of CoronaDerm-PS Safety and Specific Activity in Volunteer Groups
3. Results
3.1. Finalized Recombinant Antigen
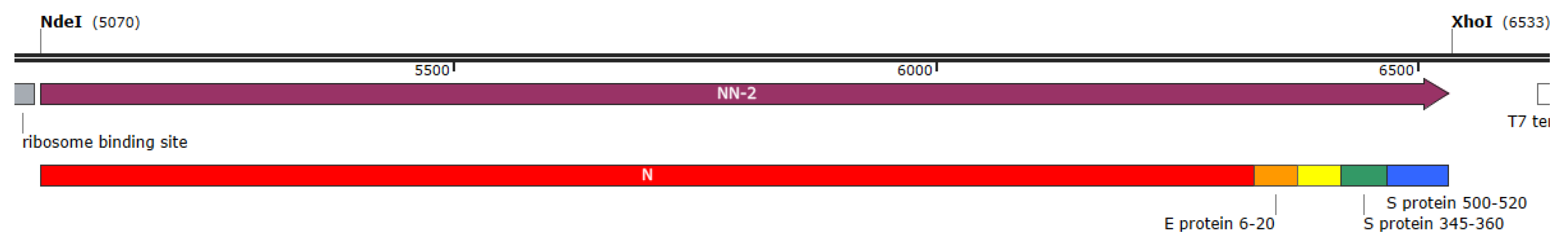
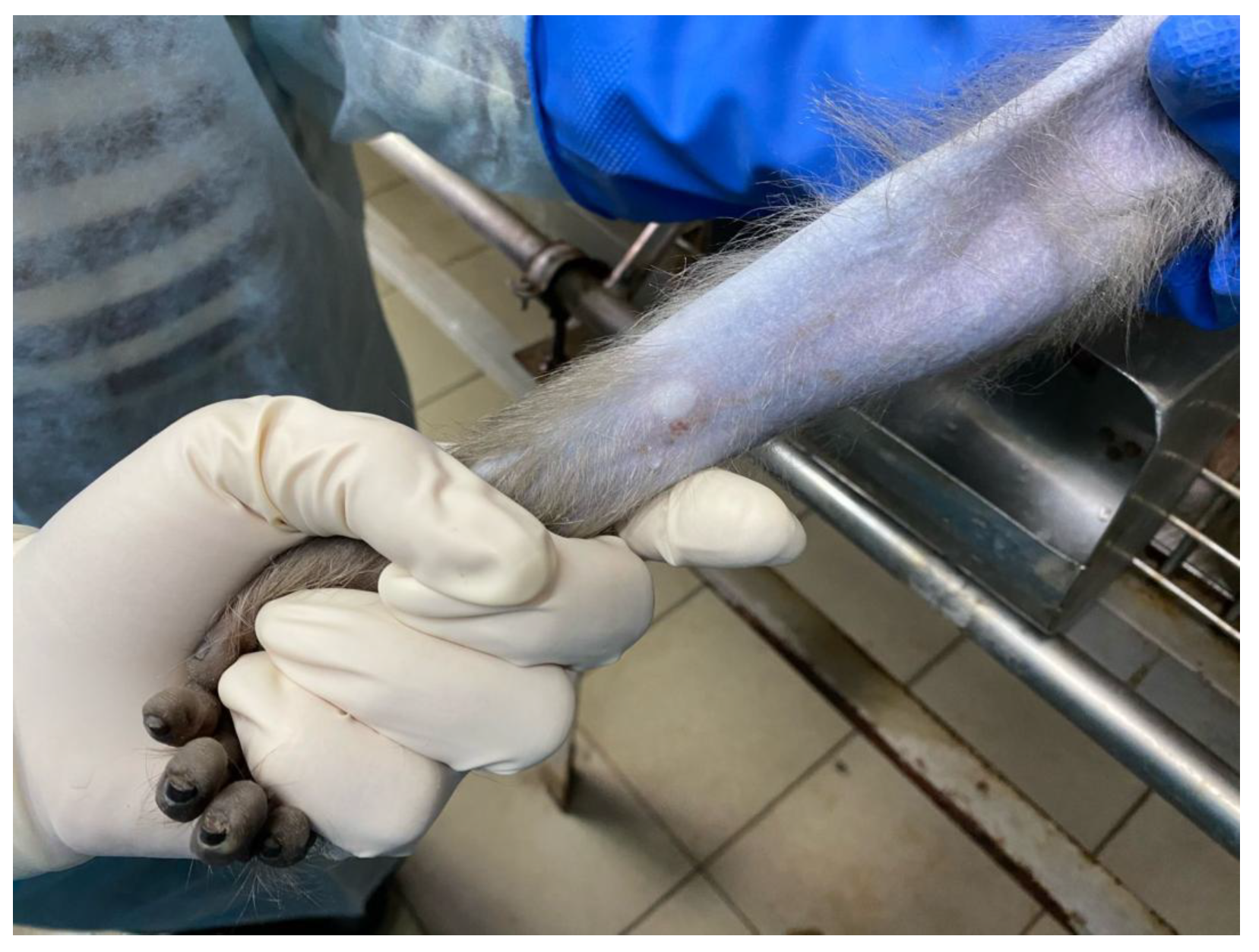
3.2. Preclinical Study Results with CoronaDerm-PS in Animals
3.3. Integrated Analysis of CoronaDerm-PS Safety and Specific Activity in Volunteer Groups

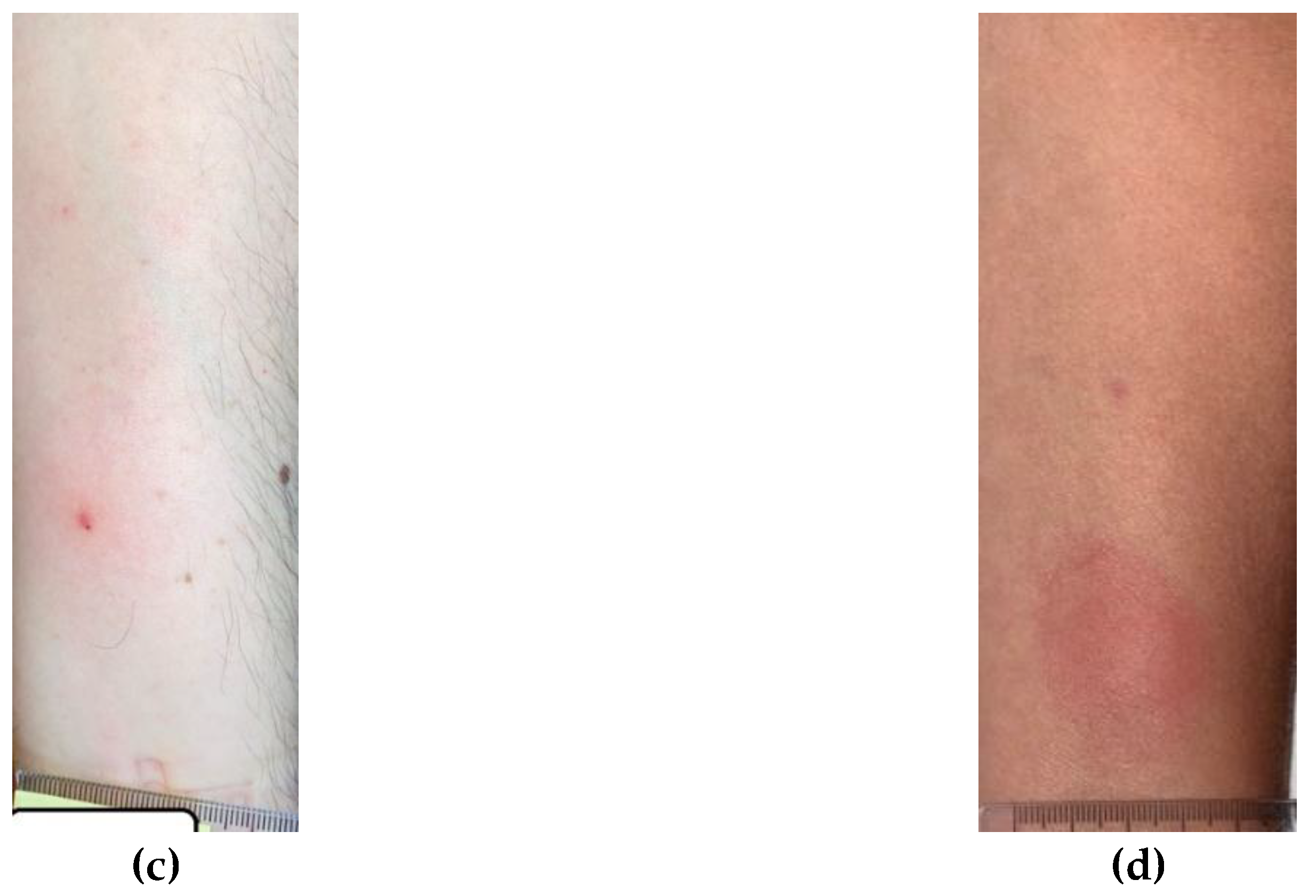
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; Bi, Y.; Ma, X.; Zhan, F.; Wang, L.; Hu, T.; Zhou, H.; Hu, Z.; Zhou, W.; Zhao, L.; Chen, J.; Meng, Y.; Wang, J.; Lin, Y.; Yuan, J.; Xie, Z.; Ma, J.; Liu, W.J.; Wang, D.; Xu, W.; Holmes, E.C.; Gao, G.F.; Wu, G.; Chen, W.; Shi, W.; Tan, W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduction and Targeted Therapy 2020, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; Müller, M.A.; Drosten, C.; Pöhlmann, S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews Molecular Cell Biology 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Zhou, R.; To, K.K.; Wong, Y.C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.Y.; Lau, T.T.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e5. [Google Scholar] [CrossRef]
- Domingo, P.; Mur, I.; Pomar, V.; Corominas, H.; Casademont, J.; de Benito, N. The four horsemen of a viral Apocalypse: The pathogenesis of SARS-CoV-2 infection (COVID-19). EBioMedicine 2020, 58, 102887. [Google Scholar] [CrossRef]
- Marsh, S.G.E. The HLA FactsBook. Academic Press: Amsterdam, 2000; 564. [Google Scholar]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nature Immunology 2020, 21, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Reports 2021, 34, 108728. [Google Scholar] [CrossRef]
- Lucas, C.; Klein, J.; Sundaram, M.E.; Liu, F.; Wong, P.; Silva, J.; Mao, T.; Oh, J.E.; Mohanty, S.; Huang, J.; et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nature Medicine 2021, 27, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Tavakolpour, S.; Rakhshandehroo, T.; Wei, E.X.; Rashidian, M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunology Letters 2020, 225, 31–32. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Frontiers in Immunology 2020, 11, 827. [Google Scholar] [CrossRef]
- Shouman, S.; El-Kholy, N.; Hussien, A.E.; El-Derby, A.M.; Magdy, S.; Abou-Shanab, A.M.; Elmehrath, A.O.; Abdelwaly, A.; Helal, M.; El-Badri, N. SARS-CoV-2-associated lymphopenia: possible mechanisms and the role of CD147. Cell Communication and Signaling 2024, 22, 349. [Google Scholar] [CrossRef]
- MacLeod, M.K.; Clambey, E.T.; Kappler, J.W.; Marrack, P. CD4 memory T cells: what are they and what can they do? Seminars in Immunology 2009, 21, 53–61. [Google Scholar] [CrossRef]
- Iyer, S.S.; Latner, D.R.; Zilliox, M.J.; McCausland, M.; Akondy, R.S.; Penaloza-Macmaster, P.; Hale, J.S.; Ye, L.; Mohammed, A.U.; Yamaguchi, T.; et al. Identification of novel markers for mouse CD4(+) T follicular helper cells. European Journal of Immunology 2013, 43, 3219–3232. [Google Scholar] [CrossRef]
- Hale, J.S.; Ahmed, R. Memory T follicular helper CD4 T cells. Frontiers in Immunology 2015, 6, 16. [Google Scholar] [CrossRef]
- Baumjohann, D.; Okada, T.; Ansel, K.M. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. Journal of Immunology 2011, 187, 2089–2092. [Google Scholar] [CrossRef] [PubMed]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Sagar; Daul, F. ; Salvat Lago, M.; Decker, A.; et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nature Medicine 2021, 27, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Dowell, A.C.; Pearce, H.; Verma, K.; Long, H.M.; Begum, J.; Aiano, F.; Amin-Chowdhury, Z.; Hoschler, K.; Brooks, T.; et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nature Immunology 2021, 22, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nature Immunology 2021, 22, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Faliti, C.E.; Ramirez, S.I.; Frazier, A.; Yu, E.D.; Grifoni, A.; Rawlings, S.A.; et al. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Norddahl, G.L.; Melsted, P.; Gunnarsdottir, K.; Holm, H.; Eythorsson, E.; Arnthorsson, A.O.; Helgason, D.; Bjarnadottir, K.; Ingvarsson, R.F.; et al. Humoral Immune Response to SARS-CoV-2 in Iceland. New England Journal of Medicine 2020, 383, 1724–1734. [Google Scholar] [CrossRef]
- Lipsitch, M.; Grad, Y.H.; Sette, A.; Crotty, S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nature Reviews Immunology 2020, 20, 709–713. [Google Scholar] [CrossRef]
- Villemonteix, J.; Cohen, L.; Guihot, A.; Guérin, V.; Moulin, C.; Caseris, M.; Carol, A.; Bonacorsi, S.; Carcelain, G. Comparison between enzyme-linked immunospot assay and intracellular cytokine flow cytometry assays for the evaluation of T cell response to SARS-CoV-2 after symptomatic COVID-19. Immunity, Inflammation and Disease 2022, 10, e617. [Google Scholar] [CrossRef]
- Aleksova, M.; Todorova, Y.; Emilova, R.; Baymakova, M.; Yancheva, N.; Andonova, R.; Zasheva, A.; Grifoni, A.; Weiskopf, D.; Sette, A.; et al. Virus-Specific Stem Cell Memory CD8+ T Cells May Indicate a Long-Term Protection against Evolving SARS-CoV-2. Diagnostics 2023, 13, 1280. [Google Scholar] [CrossRef]
- Tormo, N.; Giménez, E.; Martínez-Navarro, M.; Albert, E.; Navalpotro, D.; Torres, I.; Gimeno, C.; Navarro, D. Performance comparison of a flow cytometry immunoassay for intracellular cytokine staining and the QuantiFERON SARS-CoV-2 test for detection and quantification of SARS-CoV-2-Spike-reactive-IFN-γ-producing T cells after COVID-19 vaccination. European Journal of Clinical Microbiology and Infectious Diseases 2022, 41, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Ning, B.; Chandra, S.; Rosen, J.; Multala, E.; Argrave, M.; Pierson, L.; Trinh, I.; Simone, B.; Escarra, M.D.; Drury, S.; et al. Evaluation of SARS-CoV-2-Specific T-Cell Activation with a Rapid On-Chip IGRA. ACS Nano 2023, 17, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Black, C.A. Delayed type hypersensitivity: current theories with an historic perspective. Dermatology Online Journal 1999, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Kalish, R.S.; Askenase, P.W. Molecular mechanisms of CD8+ T cell-mediated delayed hypersensitivity: implications for allergies, asthma, and autoimmunity. Journal of Allergy and Clinical Immunology 1999, 103, 192–199. [Google Scholar] [CrossRef]
- Huebner, R.E.; Schein, M.F.; Bass, J.B. The tuberculin skin test. Clinical Infectious Diseases 1993, 17, 968–975. [Google Scholar] [CrossRef]
- Barrios, Y.; Franco, A.; Sánchez-Machín, I.; Poza-Guedes, P.; González-Pérez, R.; Matheu, V. The Beauty of Simplicity: Delayed-Type Hypersensitivity Reaction to Measure Cellular Immune Responses in RNA-SARS-Cov-2 Vaccinated Individuals. Vaccines 2021, 9, 575. [Google Scholar] [CrossRef]
- Barrios, Y.; Alava-Cruz, C.; Franco, A.; Matheu, V. Long Term Cell Immune Response to COVID-19 Vaccines Assessment Using a Delayed-Type Hypersensitivity (DTH) Cutaneous Test. Diagnostics 2022, 12, 1421. [Google Scholar] [CrossRef]
- Barrios, Y.; Alava-Cruz, C.; Marrero-Miranda, D.; Matheu, V. Early riser specific immune cell response by delayed-type hypersensitivity in a kidney transplant patient vaccinated against COVID-19. BMJ Case Reports 2022, 15, e250509. [Google Scholar] [CrossRef]
- Barrios, Y.; Franco, A.; Sanchez-Machin, I.; Poza-Guedes, P.; Gonzalez-Perez, R.; Matheu, V. A novel application of delayed-type hypersensitivity reaction to measure cellular immune response in SARS-CoV-2 exposed individuals. Clinical Immunology 2021, 226, 108730. [Google Scholar] [CrossRef]
- Barrios, Y.; Franco, A.; Alava-Cruz, C.; Cuesta-Martin, R.; Camara, C.; Matheu, V. Easy approach to detect cell immunity to COVID vaccines in common variable immunodeficiency patients. Allergologia et Immunopathologia 2022, 50, 101–105. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Paul, S.; Sidney, J.; Sette, A.; Peters, B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Current Protocols in Immunology 2016, 114, 18.19.1–18.19.24. [Google Scholar] [CrossRef]
- Reynisson, B.; Barra, C.; Kaabinejadian, S.; Hildebrand, W.H.; Peters, B.; Nielsen, M. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. Journal of Proteome Research 2020, 19, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Sidney, J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 1999, 50, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Kopat, V.V.; Riabchenkova, A.A.; Chirak, E.L.; Chirak, E.R.; Saenko, A.I.; Kolmakov, N.N.; Simbirtsev, A.S.; Dukhovlinov, I.V.; Totolian, A.A. Designing structure and E. coli strain-producer bearing SARS-CoV-2 N, S, M, E protein-related sequence antigen. Russian Journal of Infection and Immunity 2023, 13, 653–662. [Google Scholar] [CrossRef]
- Springer, I.; Besser, H.; Tickotsky-Moskovitz, N.; Dvorkin, S.; Louzoun, Y. Prediction of Specific TCR-Peptide Binding From Large Dictionaries of TCR-Peptide Pairs. Frontiers in Immunology 2020, 11, 1803. [Google Scholar] [CrossRef]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Walker, J.M. (Ed.) The Proteomics Protocols Handbook. Humana Press: New York, 2005. [Google Scholar]
- Kozlowski, L.P. IPC - Isoelectric Point Calculator. Biology Direct 2016, 11, 55. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics 2019, 20, 405. [Google Scholar] [CrossRef]
- Kopat, V.V.; Riabchenkova, A.A.; Chirak, E.L.; Chirak, E.R.; Saenko, A.I.; Kudryavtsev, I.V.; Trulioff, A.S.; Savin, T.V.; Zueva, E.V.; Simbirtsev, A.S.; et al. Purification technology design, biochemical and immunological characteristics of the recombinant chimeric antigen for evaluation of T cell immunity against coronavirus infection. Medical Immunology 2024, 26, 591–606. [Google Scholar] [CrossRef]
- Chang, C.K.; Hou, M.H.; Chang, C.F.; Hsiao, C.D.; Huang, T.H. The SARS coronavirus nucleocapsid protein—forms and functions. Antiviral Research 2014, 103, 39–50. [Google Scholar] [CrossRef]
- Savin, T.V.; Kopat, V.V.; Riabchenkova, A.A.; Chirak, E.L.; Chirak, E.R.; Saenko, A.I.; Dukhovlinov, I.V.; Sysoeva, G.M.; Gamaley, S.G.; Shimina, G.G.; et al. Experimentally investigated “CoronaDerm-PS”-driven SARS-CoV-2-specific cellular immunity and safety. Russian Journal of Infection and Immunity 2024, 14, 238–250. [Google Scholar] [CrossRef]
- Savin, T.V.; Milichkina, A.M.; Krasnov, A.A.; Kuznetsova, R.N.; Shchederkina, E.E.; Svarval, A.V.; Sharova, A.А.; Reingardt, D.E.; Ostankova, Y.V.; Gubanova, A.V.; et al. Safety and specific activity of the recombinant SARS-CoV-2 allergen (“CoronaDerm-PS”) based on phase I–II clinical trial results. Russian Journal of Infection and Immunity 2024, 14, 900–916. [Google Scholar] [CrossRef]
- Ding, X.; Du, W.; Liu, Q.; Tao, L.; Shao, Y.; Lu, P.; Yang, H.; Teng, X.; Chen, C.; Li, Z.; et al. Accuracy of ESAT6-CFP10 skin test compared with tuberculin skin test in a healthy population: a randomized, blind, parallel controlled phase III clinical study. BMC Infectious Diseases 2024, 24, 1479. [Google Scholar] [CrossRef]
- Dukhovlinov, I.V. , Simbirtsev A.S., Totolian Areg A. Method for assessing cellular immune response against coronavirus infection. Eurasian Patent No. 047119, 03 June 2024. [Google Scholar]
| Analyses | Group 1 | Group 2 | Group 3 | Group 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Group 2a | Group 2b | Group 2c | Group 3a | Group 3b | Group 3c | |||
| receipt of written informed consent | Х | Х | Х | Х | Х | Х | Х | Х |
| collection and registration of medical history | Х | Х | Х | Х | Х | Х | Х | Х |
| physical examination | Х | Х | Х | Х | Х | Х | Х | Х |
| vital sign assessment (BP, HR, RR, temperature) | Х | Х | Х | Х | Х | Х | Х | Х |
| blood analysis (biochemical, clinical), coagulogram, total IgE | Х | Х | Х | Х | Х | Х | ||
| serological analysis (HIV, hepatitis B/C, syphilis) |
Х | Х | Х | Х | Х | Х | ||
| general urine analysis | Х | Х | Х | Х | Х | Х | ||
| anti-SARS-CoV-2 IgG determination (ELISA) | Х | Х | Х | Х | Х | Х | Х | Х |
| analysis of nasopharyngeal, oropharyngeal swabs for SARS-CoV-2 RNA (PCR) | Х | Х | Х | Х | Х | Х | ||
| evaluation of T-cell immunity by flow cytometry (ex vivo) | Х | Х | Х | Х | Х | Х | ||
| lung fluorography | Х | Х | Х | Х | Х | Х | ||
| pregnancy test | Х | Х | Х | Х | Х | Х | Х | Х |
| ECG | Х | Х | Х | Х | Х | Х | ||
| CoronaDerm-PS injection | Х | Х | Х | Х | Х | Х | Х | Х |
| adverse event assessment | Х | Х | Х | Х | Х | Х | Х | Х |
| Group | Positive (volunteers) |
Inconclusive (volunteers) |
Negative (volunteers) |
|---|---|---|---|
| Group 1 (no history of illness or vaccination), phase I |
0 | 0† | 20 |
| Group 2а (EpiVacCorona) |
61 | 0† | 18 |
| Group 2b (Gam-COVID-Vac) |
67 | 0† | 15 |
| Group 2c (CoviVac) |
21 | 0† | 4 |
| Group 3а (Wuhan strain and Alpha variant) |
27 | 5 | 1 |
| Group 3b (Delta variant) |
63 | 0† | 17 |
| Group 3c (Omicron subvariants) |
40 | 7 | 5 |
| Group 4 (no history of illness or vaccination), phases I, II |
3 | 0† | 20 |
| Group | AUC (95% CI) |
SE | p | Sensitivity (95% CI) |
Specificity (95% CI) |
|---|---|---|---|---|---|
| Group 2а (EpiVacCorona) |
0.782 (0.678–0.887) |
0.053 | <0.001 | 76.60% (95% CI: 67.36%–83.85%) |
80.00% (95% CI: 71.07%–86.69%) |
| Group 2b (Gam-COVID-Vac) |
0.843 (0.751–0.935) |
0.047 | <0.001 | 81.70% (95%CI: 73.09%–88.01%) |
87.00% (95% CI: 79.11%–92.20%) |
| Group 2c (CoviVac) |
0.87 (0.764–0.975) |
0.054 | <0.001 | 84.00% (95% CI: 70.70%–91.95%) |
87.00% (95% CI: 74.22%–93.96%) |
| Group 3а (Wuhan strain and Alpha genetic variant) |
0.844 (0.733–0.955) |
0.057 | <0.001 | 81.80% (95% CI: 69.39%–89.91%) |
86.95% (95% CI: 75.35%–93.56%) |
| Group 3b (Delta variant) |
0.844 (0.760–0.928) |
0.043 | <0.001 | 79.70% (95% CI: 70.79%–86.42%) |
87.50% (95% CI: 79.60%–92.62%) |
| Group 3c (Omicron subvariants) |
0.799 (0.689–0.909) |
0.056 | <0.001 | 76.47% (95% CI: 65.49%–84.77%) |
86.95% (95% CI: 77.27%–92.89%) |
| DeLong test, Group 2 | 2a vs 2b | Z=0.861 | p=0.39 | ||
| 2a vs 2c | Z=1.162 | p=0.246 | |||
| 2b vs 2c | Z=0.377 | p=0.706 | |||
| DeLong test, Group 3 | 3a vs 3b | Z=0 | p=1 | ||
| 3a vs 3c | Z=0.563 | p=0.574 | |||
| 3b vs 3c | Z=0.637 | p=0.524 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





