Submitted:
16 July 2025
Posted:
17 July 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Vaccination Coverage Data
Geospatial Covariates, Gridded Population and Boundary Data
Model Development
Bayesian Inference, Model Fitting and Prediction
Model Choice and Validation
Results
Model Choice
Parameter Estimates
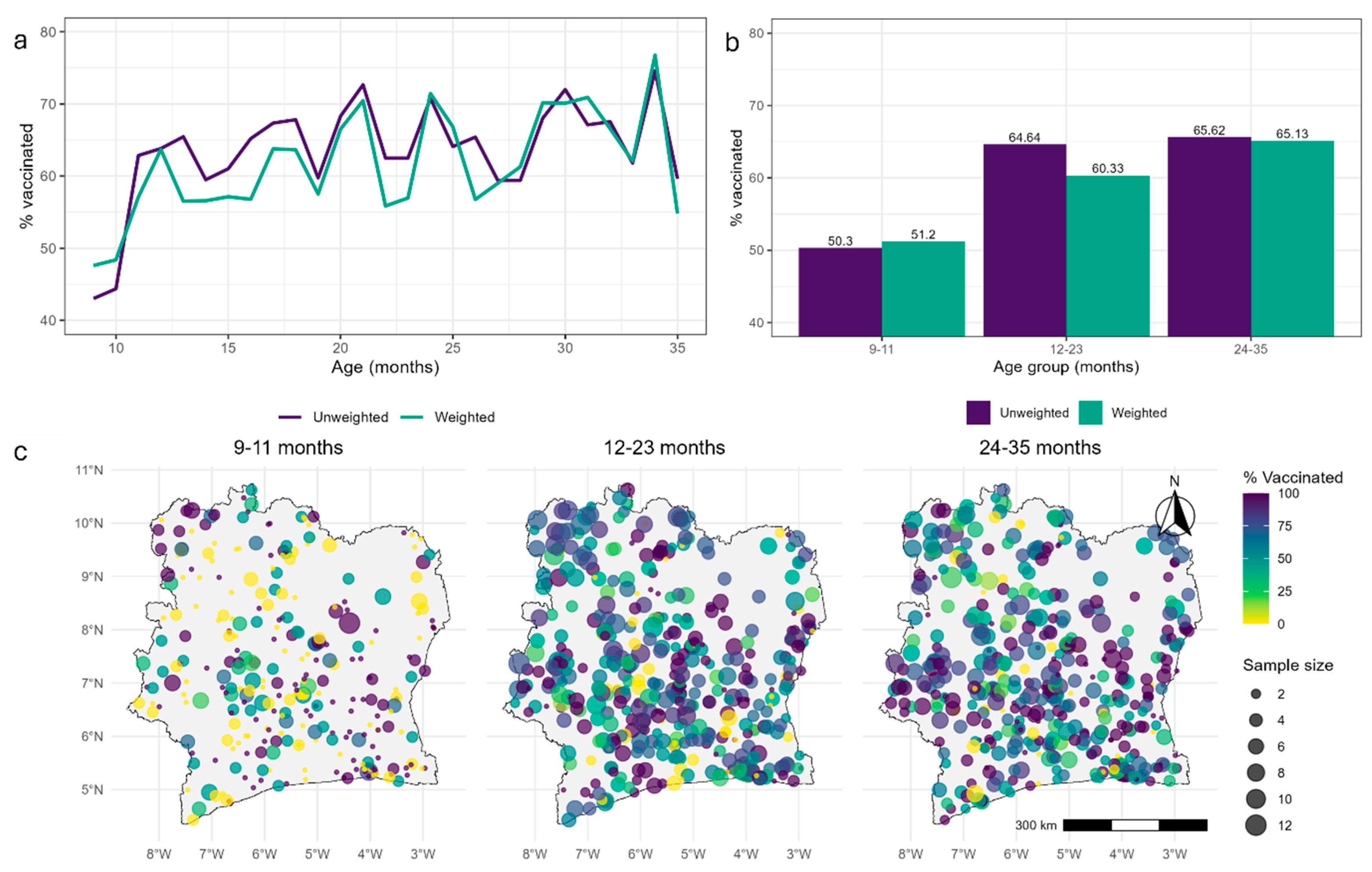
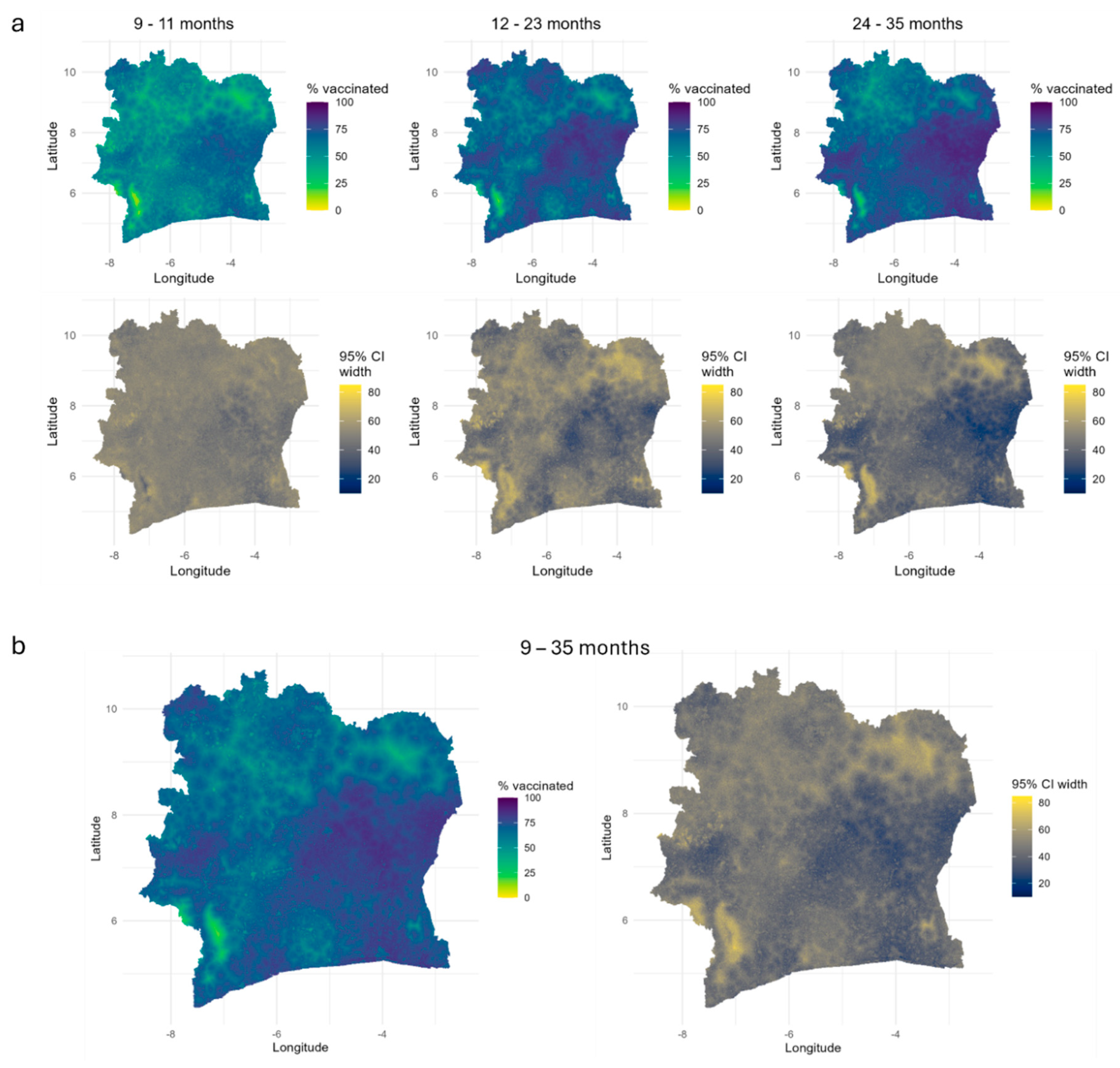
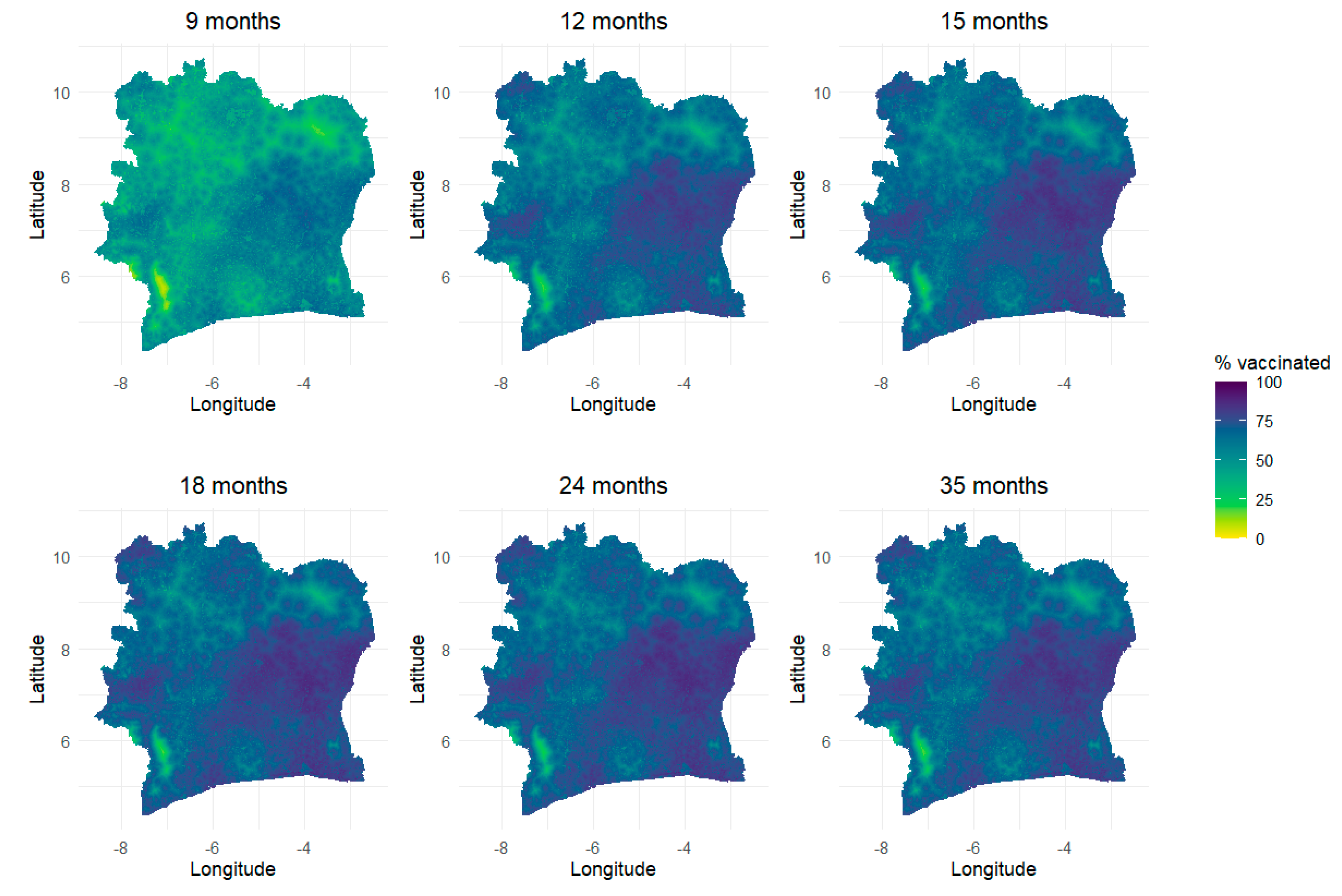
Predicted 1x1 km Maps of Vaccination Coverage for Age Groups and Single age Points
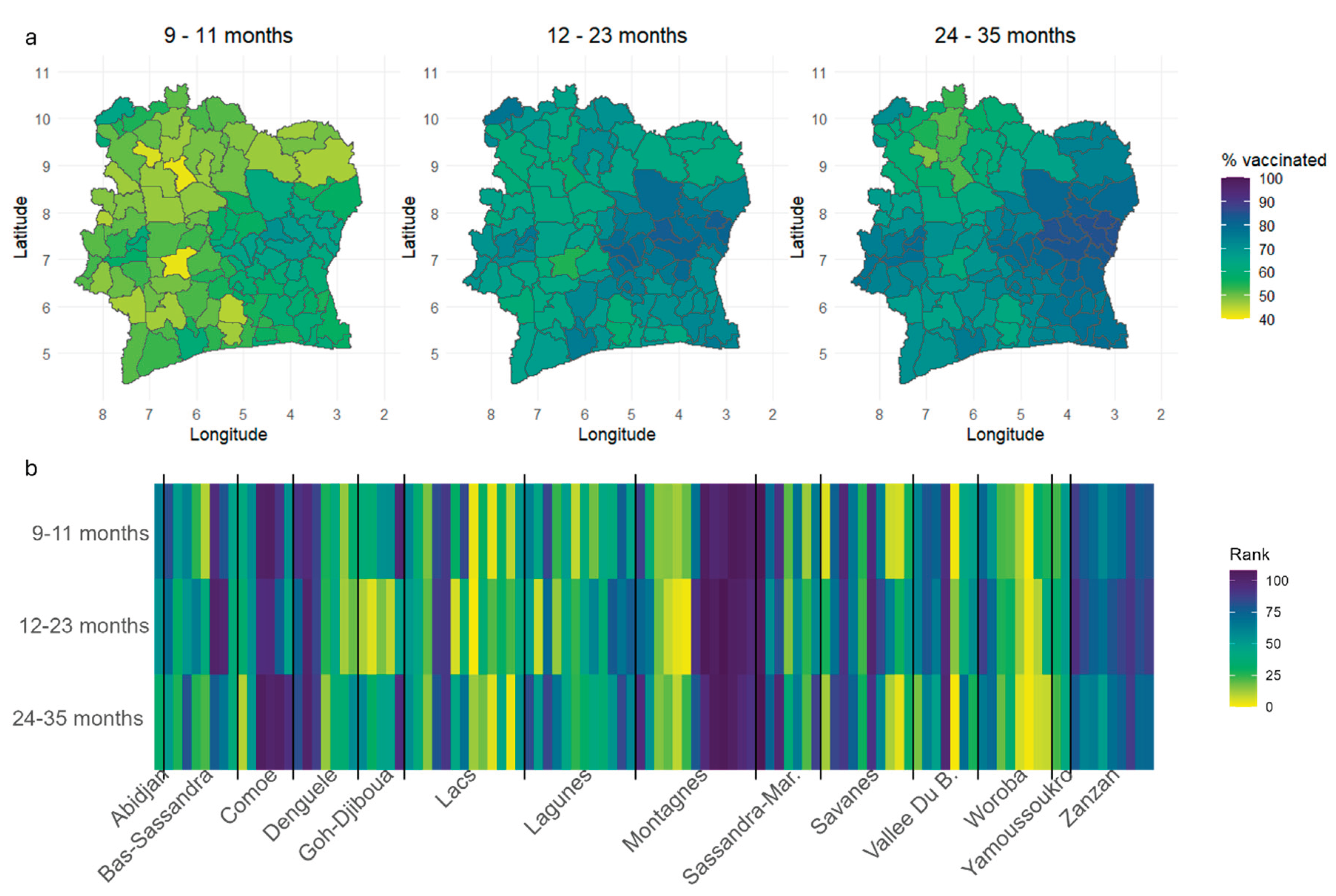
Estimation of Vaccination Coverage at the Administrative Level
Discussion
Supplementary Materials
Data and code availability
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- World Health Organization. Immunization Agenda 2030: A global strategy to leave no one behind; 2020. Available online: https://www.who.int/immunization/immunization_agenda_2030/en/ (accessed on 25 June 2020).
- Cutts, F.T.; Claquin, P.; Danovaro-Holliday, M.C.; Rhoda, D.A. Monitoring vaccination coverage: Defining the role of surveys. Vaccine 2016, 34, 4103–9. [Google Scholar] [CrossRef] [PubMed]
- Utazi, C.E.; Thorley, J.; Alegana, V.A.; Ferrari, M.J.; Takahashi, S.; Metcalf, C.J.E.; et al. Mapping vaccination coverage to explore the effects of delivery mechanisms and inform vaccination strategies. Nature Communications 2019, 10, 1633. [Google Scholar] [CrossRef] [PubMed]
- Auzenbergs, M.; Fu, H.; Abbas, K.; Procter, S.R.; Cutts, F.T.; Jit, M. Health effects of routine measles vaccination and supplementary immunisation activities in 14 high-burden countries: a Dynamic Measles Immunization Calculation Engine (DynaMICE) modelling study. The Lancet Global Health 2023, 11, e1194–e204. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.N.; Wallace, A.S.; Sodha, S.V.; Daniels, D.; Dietz, V. Assessing strategies for increasing urban routine immunization coverage of childhood vaccines in low and middle-income countries: A systematic review of peer-reviewed literature. Vaccine 2016, 34, 5495–503. [Google Scholar] [CrossRef] [PubMed]
- Gavi The Vaccine Alliance. Gavi Strategy 5.0, 2021-2025; 2020. Available online: https://www.gavi.org/our-alliance/strategy/phase-5-2021-2025 (accessed on 25 June 2021).
- Haeuser, E.; Byrne, S.; Nguyen, J.; Raggi, C.; McLaughlin, S.A.; Bisignano, C.; et al. Global, regional, and national trends in routine childhood vaccination coverage from 1980 to 2023 with forecasts to 2030: a systematic analysis for the Global Burden of Disease Study 2023. The Lancet 2025. [Google Scholar] [CrossRef] [PubMed]
- Cutts, F.T.; Ferrari, M.J.; Krause, L.K.; Tatem, A.J.; Mosser, J.F. Vaccination strategies for measles control and elimination: time to strengthen local initiatives. BMC Medicine 2021, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Wariri, O.; Utazi, C.E.; Okomo, U.; Metcalf, C.J.E.; Sogur, M.; Fofana, S.; et al. Mapping the timeliness of routine childhood vaccination in The Gambia: A spatial modelling study. Vaccine 2023, 41, 5696–705. [Google Scholar] [CrossRef] [PubMed]
- Manaksha, M.; Danya Arif, S.; Vijay Kumar, D.; Mubarak Taighoon, S.; Sundus, I.; Hamidreza, S.; et al. Coverage, timeliness of measles immunisation and its predictors in Pakistan: an analysis of 6.2 million children enrolled in the Provincial Electronic Immunisation Registry. BMJ Global Health 2025, 10, e016717. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization, United Nations Children’s Fund (UNICEF), Gavi the Vaccine Alliance. The Big Catch-Up: An Essential Immunization Recovery Plan for 2023 and Beyond; 2023. Available online: https://www.who.int/publications/i/item/9789240075511 (accessed on 12 July 2025).
- O'Brien, K.L.; Lemango, E. The big catch-up in immunisation coverage after the COVID-19 pandemic: progress and challenges to achieving equitable recovery. The Lancet 2023, 402, 510–2. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, D.; Hosseinpoor, A.R.; Bergen, N.; Danovaro-Holliday, M.C.; Wallace, A.; Johnson, H.L. Inequality in Immunization: Holding on to Equity as We ‘Catch Up’. Vaccines 2023, 11, 913. [Google Scholar] [CrossRef] [PubMed]
- Sbarra, A.N.; Rolfe, S.; Nguyen, J.Q.; Earl, L.; Galles, N.C.; Marks, A.; et al. Mapping routine measles vaccination in low- and middle-income countries. Nature 2021, 589, 415–9. [Google Scholar]
- Takahashi, S.; Metcalf, C.J.E.; Ferrari, M.J.; Tatem, A.J.; Lessler, J. The geography of measles vaccination in the African Great Lakes region. Nature Communications 2017, 8, 15585. [Google Scholar] [CrossRef] [PubMed]
- Utazi, C.E.; Nilsen, K.; Pannell, O.; Dotse-Gborgbortsi, W.; Tatem, A.J. District-level estimation of vaccination coverage: Discrete vs continuous spatial models. Stat Med. 2021, 40, 2197–211. [Google Scholar] [CrossRef]
- Utazi, C.E.; Thorley, J.; Alegana, V.A.; Ferrari, M.J.; Takahashi, S.; Metcalf, C.J.E.; et al. High resolution age-structured mapping of childhood vaccination coverage in low and middle income countries. Vaccine 2018, 36, 1583–91. [Google Scholar] [CrossRef] [PubMed]
- Utazi, C.E.; Thorley, J.; Alegana, V.A.; Ferrari, M.J.; Nilsen, K.; Takahashi, S.; et al. A spatial regression model for the disaggregation of areal unit based data to high-resolution grids with application to vaccination coverage mapping. Statistical Methods in Medical Research 2018, 28, 3226–41. [Google Scholar] [CrossRef] [PubMed]
- Utazi, C.E.; Aheto, J.M.K.; Chan, H.M.T.; Tatem, A.J.; Sahu, S.K. Conditional probability and ratio-based approaches for mapping the coverage of multi-dose vaccines. Statistics in Medicine 2022, 41, 5662–78. [Google Scholar] [CrossRef] [PubMed]
- Utazi, C.; Yankey, O.; Chaudhuri, S.; Olowe, I.; Danovaro-Holliday, C.; Lazar, A.; et al. Geostatistical and Machine Learning Approaches for High-Resolution Mapping of Vaccination Coverage. Preprints 2024. [Google Scholar]
- Gelfand, A.E.; Kim, H.-J.; Sirmans, C.F.; Banerjee, S. Spatial Modeling With Spatially Varying Coefficient Processes. Journal of the American Statistical Association 2003, 98, 387–96. [Google Scholar] [CrossRef] [PubMed]
- Finley, A.O. Comparing spatially-varying coefficients models for analysis of ecological data with non-stationary and anisotropic residual dependence. Methods in Ecology and Evolution 2011, 2, 143–54. [Google Scholar] [CrossRef]
- Meehan, T.D.; Michel, N.L.; Rue, H. Spatial modeling of Audubon Christmas Bird Counts reveals fine-scale patterns and drivers of relative abundance trends. Ecosphere 2019, 10, e02707. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Brunsdon, C.; Charlton, M. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships. Chichester: Wiley; 2003.
- Utazi, C.E.; Aheto, J.M.K.; Wigley, A.; Tejedor-Garavito, N.; Bonnie, A.; Nnanatu, C.C.; et al. Mapping the distribution of zero-dose children to assess the performance of vaccine delivery strategies and their relationships with measles incidence in Nigeria. Vaccine 2023, 41, 170–81. [Google Scholar] [CrossRef] [PubMed]
- Institut National de la Statistique, I.C.F. Côte d’Ivoire Enquête Démographique et de Santé 2021 Rapport final. Rockville, Maryland, USA et la Côte d’Ivoire: INS et ICF; 2023.
- UNICEF Reaching the unreached with life-saving vaccines through data science and geospatial technologies; 2025. Available online: https://data.unicef.org/resources/reaching-the-unreached-with-life-saving-vaccines-through-data-science-and-geospatial-technologies/ (accessed on 12 July 2025).
- World Health Organisation. Vaccination schedule for Measles (Cote d'Ivoire); 2025. 2025. Available online: https://immunizationdata.who.int/global/wiise-detail-page/vaccination-schedule-for-measles?ISO_3_CODE=CIV&TARGETPOP_GENERAL=. (accessed on 12 July 2025).
- Cressie, N. Statistics for Spatial Data: Wiley; 2015.
- Tatem, A.J. WorldPop, open data for spatial demography. Scientific Data 2017, 4, 170004. [Google Scholar] [CrossRef] [PubMed]
- Runfola, D.; Anderson, A.; Baier, H.; Crittenden, M.; Dowker, E.; Fuhrig, S.; et al. geoBoundaries: A global database of political administrative boundaries. PLoS One 2020, 15, e0231866. [Google Scholar] [CrossRef] [PubMed]
- Matérn, B. Spatial Variation. 2nd ed. Berlin, Germany: Springer-Verlag; 1960.
- Lindgren, F.; Rue, H.; Lindström, J. An explicit link between Gaussian fields and Gaussian Markov random fields: the stochastic partial differential equation approach. J Roy Stat Soc Series B (Stat Methodol) 2011, 73, 423–98. [Google Scholar] [CrossRef]
- Wang, X.; Yue, Y.R.; Faraway, J.J. Bayesian regression modeling with INLA: Chapman and Hall/CRC; 2018.
- Browne, W.J.; Subramanian, S.V.; Jones, K.; Goldstein, H. Variance partitioning in multilevel logistic models that exhibit overdispersion. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2005, 168, 599–613. [Google Scholar] [CrossRef]
- Simpson, D.; Rue, H.; Riebler, A.; Martins, T.G.; Sørbye, S.H. Penalising model component complexity: A principled, practical approach to constructing priors. Statistical Science 2017, 32, 1–28. [Google Scholar] [CrossRef]
- Fuglstad, G.-A.; Simpson, D.; Lindgren, F.; Rue, H. Constructing priors that penalize the complexity of Gaussian random fields. Journal of the American Statistical Association 2019, 114, 445–52. [Google Scholar] [CrossRef]
- Rue, H.; Martino, S.; Chopin, N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2009, 71, 319–92. [Google Scholar] [CrossRef]
- Lindgren, F.; Bachl, F.; Illian, J.; Suen, M.H.; Rue, H.; Seaton, A.E. inlabru: software for fitting latent Gaussian models with non-linear predictors. arXiv preprint, 2024; arXiv:240700791. [Google Scholar]
- Watanabe, S. Asymptotic equivalence of Bayes cross validation and widely applicable information criterion in singular learning theory. Journal of machine learning research 2010, 11, 3571–94. [Google Scholar]
- Gelman, A.; Hwang, J.; Vehtari, A. Understanding predictive information criteria for Bayesian models. Statistics and Computing 2014, 24, 997–1016. [Google Scholar] [CrossRef]
- Gneiting, T.; Raftery, A.E. Strictly proper scoring rules, prediction, and estimation. Journal of the American Statistical Association 2007, 102, 359–78. [Google Scholar] [CrossRef]
- Brier, G.W. Verification of forecasts expressed in terms of probability. Monthly Weather Review 1950, 78, 1–3. [Google Scholar] [CrossRef]
- Ogundele, O.A.; Ogunwemimo, H.S.; Fehintola, F.O.; Ogundele, T.; Olorunsola, A.; Bello, O.E.; et al. Predictors of incomplete childhood vaccination in four West African countries: a population based cross-sectional study. Sci Rep. 2025, 15, 17119. [Google Scholar] [CrossRef] [PubMed]
- Douba, A.; Aka Lepri Bernadin, N.; Attoh-Toure, H.; Bangaman Akani, C.; Yao, G.H.A.; Konan Ng et, a.l. An analysis of risk factors for incomplete immunization for children in Côte d’Ivoire: Examination of 1998-1999 and 2011-2012 Demographic and Health Survey. HEALTH SCIENCES AND DISEASE 2016, 17. [Google Scholar]
- Ahoussou, E.M.K.; Ekra, K.D.; Dananche, C.; Aka, L.B.N.; Rabilloud, M.; Vanhems, P. Factors associated with detection of measles-specific immunoglobulin M (IgM-positive) in Côte d'Ivoire from 2015 to 2019: A nested case-control study from a measles national surveillance programme. Journal of Public Health and Epidemiology 2023, 15, 97–105. [Google Scholar]
- World Health Organization. WHO/UNICEF Estimates of National Immunization Coverage (WUENIC) 2024 revision; 2024. Available online: https://www.who.int/teams/immunization-vaccines-and-biologicals/immunization-analysis-and-insights/global-monitoring/immunization-coverage/who-unicef-estimates-of-national-immunization-coverage (accessed on 5 July 2025).
- World Health Organisation Côte d'Ivoire Reported cases of vaccine-preventable diseases, (.V.P.D.s.).; 2025 Available online: http://www.w3.org/1999/xlink" xlink:href="h.t.t.p.s.:././.i.m.m.u.n.i.z.a.t.i.o.n.d.a.t.a.w.h.o.i.n.t./.d.a.s.h.b.o.a.r.d./.r.e.g.i.o.n.s./.a.f.r.i.c.a.n.-r.e.g.i.o.n./.C.I.V (accessed on 12 July 2025).
- Carcelen, A.C.; Winter, A.K.; Moss, W.J.; Chilumba, I.; Mutale, I.; Chongwe, G.; et al. Leveraging a national biorepository in Zambia to assess measles and rubella immunity gaps across age and space. Scientific Reports 2022, 12, 10217. [Google Scholar] [CrossRef] [PubMed]
- Lee, D. A comparison of conditional autoregressive models used in Bayesian disease mapping. Spat Spatiotemporal Epidemiol. 2011, 2, 79–89. [Google Scholar] [CrossRef] [PubMed]
| Model | WAIC | |
|---|---|---|
| MODsvc1 | 210 | 5191 |
| MODsvc2 | 219 | 5197 |
| MODnosvc | 186 | 5215 |
| MODsmooth | 187 | 5204 |
| MODall | 184 | 5262 |
| Model | Age group | RMSE | MAE | AVG_BIAS | CRPS |
|---|---|---|---|---|---|
| RANDOM | |||||
| MODsvc1 | 9 -11 | 0.439 | 0.403 | 0.006 | 0.332 |
| MODsvc2 | 9 -11 | 0.440 | 0.404 | 0.006 | 0.335 |
| MODnosvc | 9 -11 | 0.440 | 0.403 | 0.008 | 0.334 |
| MODsvc1 | 12 - 23 | 0.315 | 0.263 | 0.008 | 0.206 |
| MODsvc2 | 12 - 23 | 0.315 | 0.262 | 0.009 | 0.203 |
| MODnosvc | 12 - 23 | 0.314 | 0.262 | 0.012 | 0.208 |
| MODsvc1 | 24 - 35 | 0.298 | 0.249 | -0.015 | 0.197 |
| MODsvc2 | 24 - 35 | 0.299 | 0.249 | -0.015 | 0.196 |
| MODnosvc | 24 - 35 | 0.300 | 0.251 | -0.016 | 0.198 |
| STRATIFIED | |||||
| MODsvc1 | 9 - 11 | 0.444 | 0.408 | 0.004 | 0.337 |
| MODsvc2 | 9 - 11 | 0.445 | 0.408 | 0.004 | 0.341 |
| MODnosvc | 9 - 11 | 0.445 | 0.409 | 0.004 | 0.338 |
| MODsvc1 | 12 - 23 | 0.318 | 0.266 | 0.003 | 0.206 |
| MODsvc2 | 12 - 23 | 0.320 | 0.267 | 0.002 | 0.207 |
| MODnosvc | 12 - 23 | 0.319 | 0.265 | 0.009 | 0.209 |
| MODsvc1 | 24 - 35 | 0.304 | 0.252 | -0.017 | 0.196 |
| MODsvc2 | 24 - 35 | 0.302 | 0.253 | -0.017 | 0.198 |
| MODnosvc | 24 - 35 | 0.304 | 0.255 | -0.018 | 0.200 |
| Parameter | Mean | Odds ratio | Std. Dev. | 2.5% | 97.5% |
|---|---|---|---|---|---|
| -3.835 | 0.022 | 5.557 | -14.727 | 7.057 | |
| Urban | -0.611 | 0.543 | 0.143 | -0.892 | -0.33 |
| Veg_index | -3.384 | 0.034 | 2.143 | -7.585 | 0.816 |
| Wetdays | -0.102 | 0.903 | 0.102 | -0.303 | 0.099 |
| Dist_conf | 0.214 | 1.239 | 0.091 | 0.035 | 0.393 |
| Elevation | 1.000 | ||||
| Urban_access | 1.000 | -0.001 | 0.001 | ||
| Walking_tt | -0.002 | 0.998 | 0.001 | -0.004 | -0.001 |
| Mal_prev | -0.558 | 0.572 | 0.834 | -2.192 | 1.077 |
| Max_temp | 0.117 | 1.124 | 0.144 | -0.166 | 0.4 |
| Mat_educ | 1.035 | 2.815 | 0.236 | 0.572 | 1.498 |
| Health_card | 1.824 | 6.197 | 0.302 | 1.231 | 2.417 |
| Media | 0.552 | 1.737 | 0.272 | 0.020 | 1.085 |
| Wealth | -0.135 | 0.874 | 0.247 | -0.619 | 0.349 |
| 0.716 | 2.046 | 0.124 | 0.473 | 0.959 | |
| 0.827 | 2.286 | 0.305 | 0.228 | 1.425 | |
| 0.844 | - | 0.456 | 0.293 | 2.028 | |
| 0.367 | - | 0.094 | 0.216 | 0.583 | |
| 0.702 | - | 0.344 | 0.283 | 1.595 | |
| 0.399 | - | 0.112 | 0.214 | 0.649 | |
| 6.697 | - | 5.659 | 1.561 | 21.783 | |
| 0.400 | - | 0.17 | 0.171 | 0.828 | |
| 4.056 | - | 1.228 | 2.151 | 6.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





