Submitted:
17 July 2025
Posted:
17 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. WGS Data of African Patient-Derived Tumours
2.1. Cohort Information of African Patients
2.2. Cancer Discoveries from African Genomic Studies
2.3. Challenges of Analysing WGS Data of African Patients
3. Rapid and Scalable HPC Workflow for African Genomic Studies
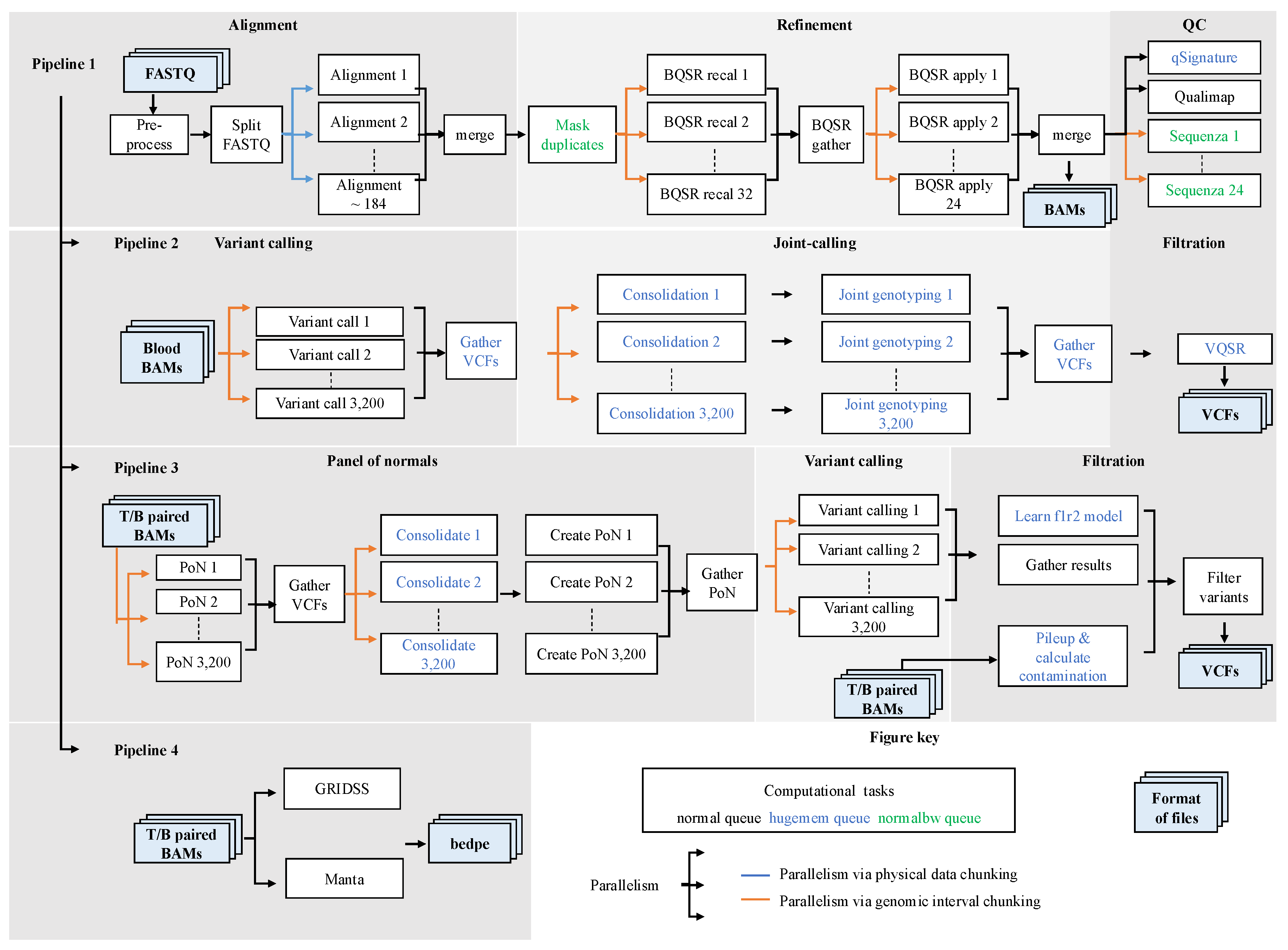
3.1. SAPCS Workflow Overview
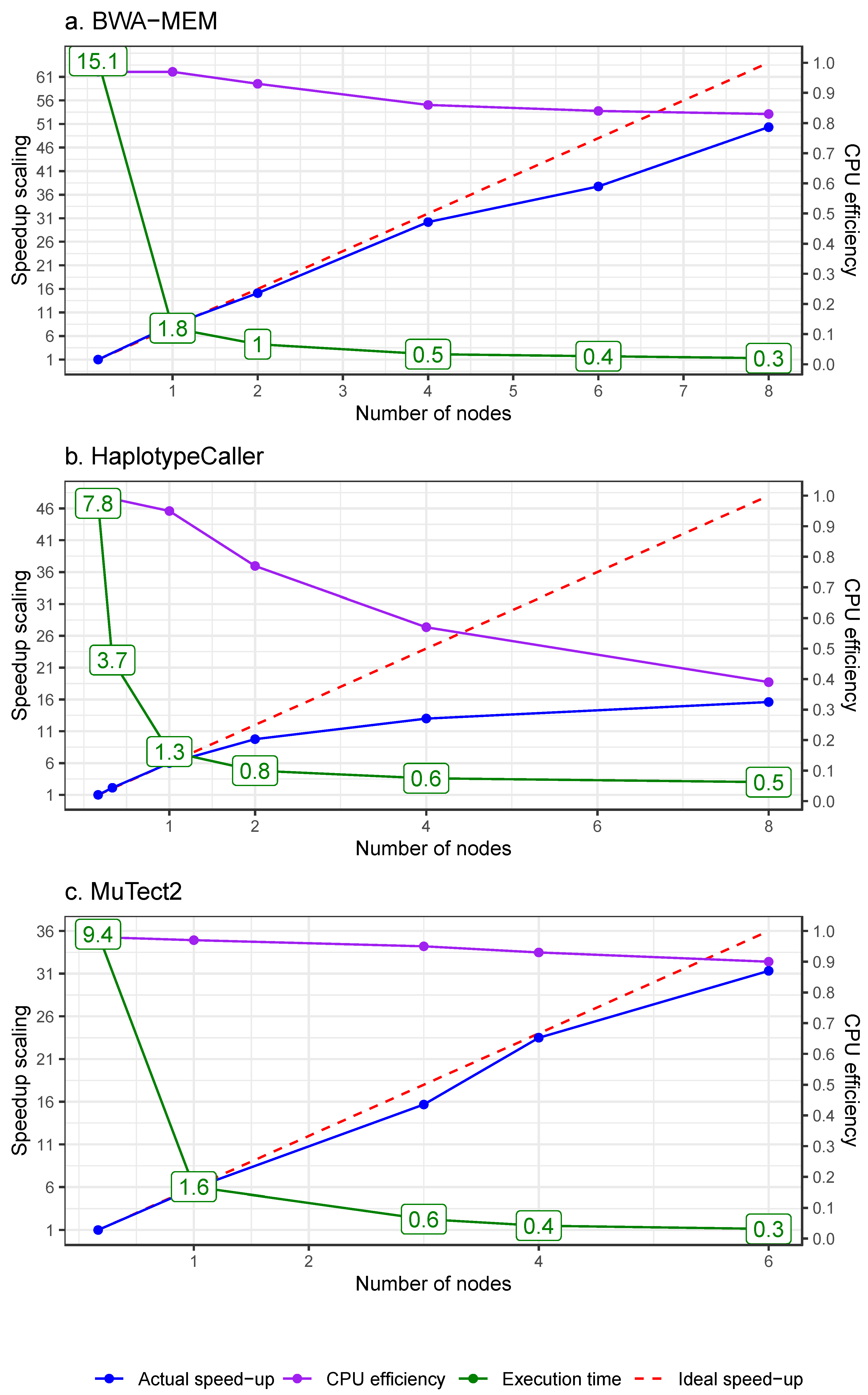
3.2. High-Level Parallelism
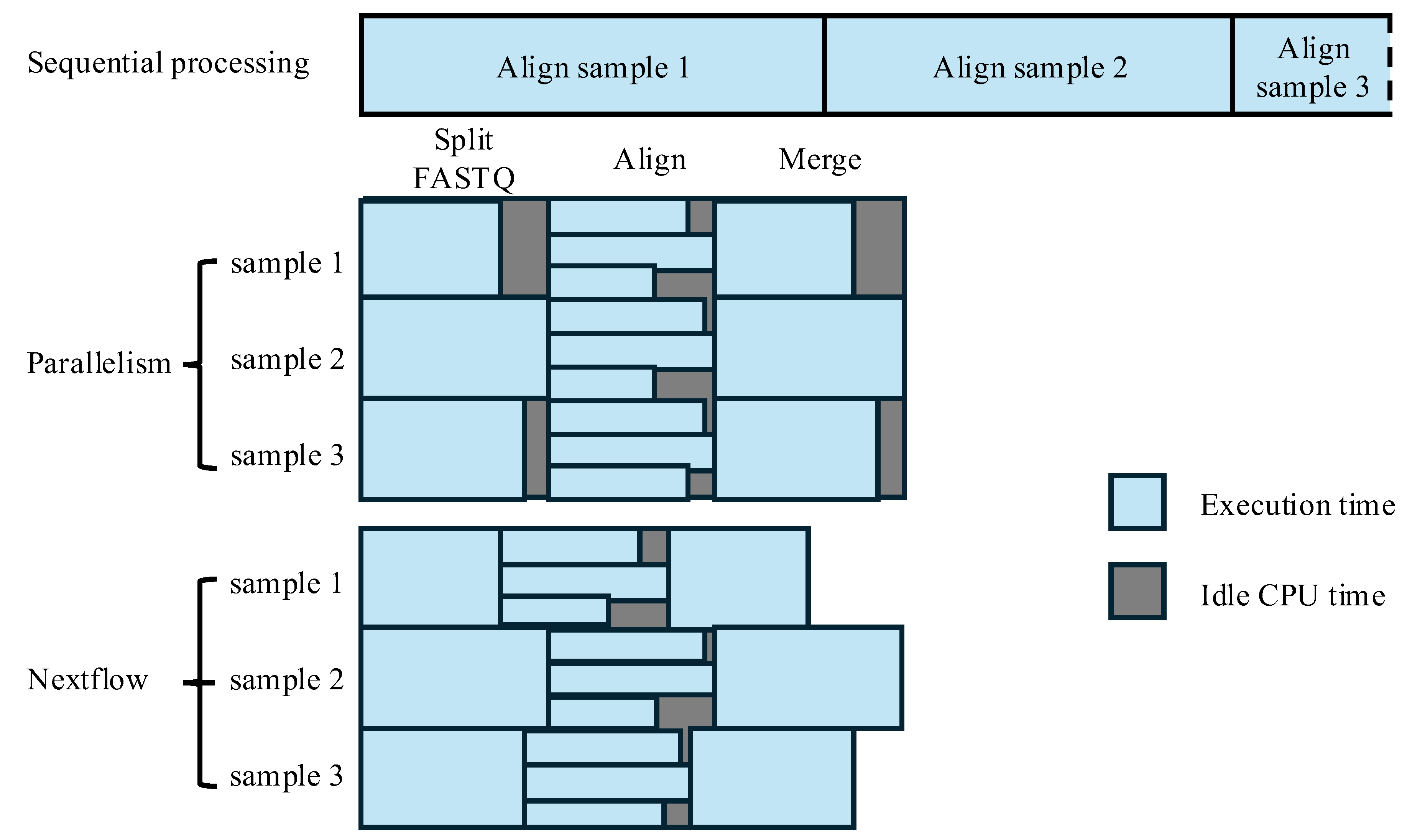
3.2.1. Parallelism via Physical Data Chunking for Alignment
3.2.2. Parallelism via Genomic Interval Chunking
3.3. Integration with Workflow Management Tools
4. Emerging Technologies and Resources to be Integrated to African Genomic Studies
5. Conclusions and Challenges
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R. L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024, 74 (3), 229-263. [CrossRef]
- Hayes, V. M.; Gong, T.; Mutambirwa, S. B.; Jaratlerdsiri, W.; Bornman, M. R. African inclusion in prostate cancer genomic studies provides the first glimpses into addressing health disparities through tailored clinical care. Clinical and Translational Medicine 2023, 13 (1), e1142. [CrossRef]
- Rubagumya, F.; Carson, L.; Mushonga, M.; Manirakiza, A.; Murenzi, G.; Abdihamid, O.; Athman, A.; Mungo, C.; Booth, C.; Hammad, N. An analysis of the African cancer research ecosystem: tackling disparities. BMJ Global Health 2023, 8 (2), e011338. [CrossRef]
- Drake, T. M.; Knight, S. R.; Harrison, E. M.; Søreide, K. Global inequities in precision medicine and molecular cancer research. Frontiers in Oncology 2018, 8, 346. [CrossRef]
- Pereira, L.; Mutesa, L.; Tindana, P.; Ramsay, M. African genetic diversity and adaptation inform a precision medicine agenda. Nature Reviews Genetics 2021, 22 (5), 284-306. [CrossRef]
- Omotoso, O.; Teibo, J. O.; Atiba, F. A.; Oladimeji, T.; Paimo, O. K.; Ataya, F. S.; Batiha, G. E.-S.; Alexiou, A. Addressing cancer care inequities in sub-Saharan Africa: current challenges and proposed solutions. International journal for equity in health 2023, 22 (1), 189. [CrossRef]
- Lawson, D. J.; Van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nature communications 2018, 9 (1), 3258. [CrossRef]
- Liu, W.; Zheng, S. L.; Na, R.; Wei, L.; Sun, J.; Gallagher, J.; Wei, J.; Resurreccion, W. K.; Ernst, S.; Sfanos, K. S. Distinct genomic alterations in prostate tumors derived from African American men. Molecular Cancer Research 2020, 18 (12), 1815-1824. [CrossRef]
- Kittles, R. A.; Baffoe-Bonnie, A. B.; Moses, T. Y.; Robbins, C. M.; Ahaghotu, C.; Huusko, P.; Pettaway, C.; Vijayakumar, S.; Bennett, J.; Hoke, G. A common nonsense mutation in EphB2 is associated with prostate cancer risk in African American men with a positive family history. Journal of medical genetics 2006, 43 (6), 507-511. [CrossRef]
- Khani, F.; Mosquera, J. M.; Park, K.; Blattner, M.; O'Reilly, C.; MacDonald, T. Y.; Chen, Z.; Srivastava, A.; Tewari, A. K.; Barbieri, C. E. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clinical Cancer Research 2014, 20 (18), 4925-4934. [CrossRef]
- Huang, F. W.; Mosquera, J. M.; Garofalo, A.; Oh, C.; Baco, M.; Amin-Mansour, A.; Rabasha, B.; Bahl, S.; Mullane, S. A.; Robinson, B. D. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer discovery 2017, 7 (9), 973-983. [CrossRef]
- Blattner, M.; Lee, D. J.; O'Reilly, C.; Park, K.; MacDonald, T. Y.; Khani, F.; Turner, K. R.; Chiu, Y.-L.; Wild, P. J.; Dolgalev, I. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 2014, 16 (1), 14-W10.
- Yuan, J.; Kensler, K. H.; Hu, Z.; Zhang, Y.; Zhang, T.; Jiang, J.; Xu, M.; Pan, Y.; Long, M.; Montone, K. T. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS genetics 2020, 16 (2), e1008641. [CrossRef]
- Lindquist, K. J.; Paris, P. L.; Hoffmann, T. J.; Cardin, N. J.; Kazma, R.; Mefford, J. A.; Simko, J. P.; Ngo, V.; Chen, Y.; Levin, A. M. Mutational landscape of aggressive prostate tumors in African American men. Cancer research 2016, 76 (7), 1860-1868. [CrossRef]
- Xiao, Q.; Sun, Y.; Dobi, A.; Srivastava, S.; Wang, W.; Srivastava, S.; Ji, Y.; Hou, J.; Zhao, G.-P.; Li, Y. Systematic analysis reveals molecular characteristics of ERG-negative prostate cancer. Scientific reports 2018, 8 (1), 12868. [CrossRef]
- Petrovics, G.; Li, H.; Stümpel, T.; Tan, S.-H.; Young, D.; Katta, S.; Li, Q.; Ying, K.; Klocke, B.; Ravindranath, L. A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. EBioMedicine 2015, 2 (12), 1957-1964. [CrossRef]
- Consortium, I. C. G. International network of cancer genome projects. Nature 2010, 464 (7291), 993.
- Weinstein, J. N.; Collisson, E. A.; Mills, G. B.; Shaw, K. R.; Ozenberger, B. A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J. M. The cancer genome atlas pan-cancer analysis project. Nature genetics 2013, 45 (10), 1113-1120. [CrossRef]
- Aaltonen, L. A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D. J.; Agrawal, N.; Ahn, K. S.; Ahn, S.-M.; Aikata, H.; Akbani, R.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578 (7793), 82-93. [CrossRef]
- Jiagge, E.; Jin, D. X.; Newberg, J. Y.; Perea-Chamblee, T.; Pekala, K. R.; Fong, C.; Waters, M.; Ma, D.; Dei-Adomakoh, Y.; Erb, G. Tumor sequencing of African ancestry reveals differences in clinically relevant alterations across common cancers. Cancer cell 2023, 41 (11), 1963-1971. e1963. [CrossRef]
- Brown, L. M.; Hagenson, R. A.; Koklič, T.; Urbančič, I.; Qiao, L.; Strancar, J.; Sheltzer, J. M. An elevated rate of whole-genome duplications in cancers from Black patients. Nature Communications 2024, 15 (1), 8218. [CrossRef]
- Johnson, J. A.; Moore, B. J.; Syrnioti, G.; Eden, C. M.; Wright, D.; Newman, L. A. Landmark series: the cancer genome atlas and the study of breast cancer disparities. Annals of Surgical Oncology 2023, 30 (11), 6427-6440. [CrossRef]
- Van der Auwera, G. A.; Carneiro, M. O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Current protocols in bioinformatics 2013, 43 (1), 11.10. 11-11.10. 33. [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research 2010, 20 (9), 1297-1303. [CrossRef]
- Huang, Z.; Rustagi, N.; Veeraraghavan, N.; Carroll, A.; Gibbs, R.; Boerwinkle, E.; Venkata, M. G.; Yu, F. A hybrid computational strategy to address WGS variant analysis in> 5000 samples. BMC bioinformatics 2016, 17, 1-12. ttps://doi.org/10.1186/s12859-016-1211-6.
- Meggendorfer, M.; Jobanputra, V.; Wrzeszczynski, K. O.; Roepman, P.; de Bruijn, E.; Cuppen, E.; Buttner, R.; Caldas, C.; Grimmond, S.; Mullighan, C. G. Analytical demands to use whole-genome sequencing in precision oncology. In Seminars in cancer biology, 2022; Elsevier: Vol. 84, pp 16-22. [CrossRef]
- Jaratlerdsiri, W.; Jiang, J.; Gong, T.; Patrick, S. M.; Willet, C.; Chew, T.; Lyons, R. J.; Haynes, A.-M.; Pasqualim, G.; Louw, M.; et al. African-specific molecular taxonomy of prostate cancer. Nature 2022, 609 (7927), 552-559. [CrossRef]
- Jaratlerdsiri, W.; Chan, E. K.; Gong, T.; Petersen, D. C.; Kalsbeek, A. M.; Venter, P. A.; Stricker, P. D.; Bornman, M. R.; Hayes, V. M. Whole-genome sequencing reveals elevated tumor mutational burden and initiating driver mutations in African men with treatment-naïve, high-risk prostate cancer. Cancer research 2018, 78 (24), 6736-6746.
- Moody, S.; Senkin, S.; Islam, S. M. A.; Wang, J.; Nasrollahzadeh, D.; Cortez Cardoso Penha, R.; Fitzgerald, S.; Bergstrom, E. N.; Atkins, J.; He, Y.; et al. Mutational signatures in esophageal squamous cell carcinoma from eight countries with varying incidence. Nature Genetics 2021, 53 (11), 1553-1563. [CrossRef]
- Van Loon, K.; Mmbaga, E. J.; Mushi, B. P.; Selekwa, M.; Mwanga, A.; Akoko, L. O.; Mwaiselage, J.; Mosha, I.; Ng, D. L.; Wu, W. A Genomic Analysis of Esophageal Squamous Cell Carcinoma in Eastern Africa. Cancer Epidemiology, Biomarkers & Prevention 2023, 32 (10), 1411-1420. [CrossRef]
- Grande, B. M.; Gerhard, D. S.; Jiang, A.; Griner, N. B.; Abramson, J. S.; Alexander, T. B.; Allen, H.; Ayers, L. W.; Bethony, J. M.; Bhatia, K. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood, The Journal of the American Society of Hematology 2019, 133 (12), 1313-1324. [CrossRef]
- Thomas, N.; Dreval, K.; Gerhard, D. S.; Hilton, L. K.; Abramson, J. S.; Ambinder, R. F.; Barta, S.; Bartlett, N. L.; Bethony, J.; Bhatia, K. Genetic subgroups inform on pathobiology in adult and pediatric Burkitt lymphoma. Blood 2023, 141 (8), 904-916. [CrossRef]
- Ansari-Pour, N.; Zheng, Y.; Yoshimatsu, T. F.; Sanni, A.; Ajani, M.; Reynier, J.-B.; Tapinos, A.; Pitt, J. J.; Dentro, S.; Woodard, A. Whole-genome analysis of Nigerian patients with breast cancer reveals ethnic-driven somatic evolution and distinct genomic subtypes. Nature communications 2021, 12 (1), 6946. [CrossRef]
- Tindall, E. A.; Monare, L. R.; Petersen, D. C.; Van Zyl, S.; Hardie, R. A.; Segone, A. M.; Venter, P. A.; Bornman, M. R.; Hayes, V. M. Clinical presentation of prostate cancer in black South Africans. The Prostate 2014, 74 (8), 880-891. [CrossRef]
- Hayes, V. M.; Patrick, S. M.; Shirinde, J.; Jaratlerdsiri, W.; Nenzhelele, M.; Radzuma, M. B.; Gheybi, K.; Mokua, W.; Oyaro, M. O.; Moreira, D. M. Health equity research outcomes and improvement Consortium Prostate Cancer Health Precision Africa1K: closing the health equity gap through rural community inclusion. Journal of Urologic Oncology 2024, 22 (2), 144-149. [CrossRef]
- Zhang, R.; Li, C.; Wan, Z.; Qin, J.; Li, Y.; Wang, Z.; Zheng, Q.; Kang, X.; Chen, X.; Li, Y. Comparative genomic analysis of esophageal squamous cell carcinoma among different geographic regions. Frontiers in Oncology 2023, 12, 999424. [CrossRef]
- Li, M.; Zhang, Z.; Wang, Q.; Yi, Y.; Li, B. Integrated cohort of esophageal squamous cell cancer reveals genomic features underlying clinical characteristics. Nature Communications 2022, 13 (1), 5268. [CrossRef]
- Cui, Y.; Chen, H.; Xi, R.; Cui, H.; Zhao, Y.; Xu, E.; Yan, T.; Lu, X.; Huang, F.; Kong, P. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell research 2020, 30 (10), 902-913. [CrossRef]
- Gong, T.; Jaratlerdsiri, W.; Jiang, J.; Willet, C.; Chew, T.; Patrick, S. M.; Lyons, R. J.; Haynes, A.-M.; Pasqualim, G.; Brum, I. S. Genome-wide interrogation of structural variation reveals novel African-specific prostate cancer oncogenic drivers. Genome medicine 2022, 14 (1), 100. [CrossRef]
- Huang, R.; Bornman, M. R.; Stricker, P. D.; Simoni Brum, I.; Mutambirwa, S. B.; Jaratlerdsiri, W.; Hayes, V. M. The impact of telomere length on prostate cancer aggressiveness, genomic instability and health disparities. Scientific Reports 2024, 14 (1), 7706. [CrossRef]
- Soh, P. X.; Adams, A.; Bornman, M. R.; Jiang, J.; Stricker, P. D.; Mutambirwa, S. B.; Jaratlerdsiri, W.; Hayes, V. M. Y chromosome variation and prostate cancer ancestral disparities. iScience 2025, 28 (5). [CrossRef]
- Hayes, V.; Jiang, J.; Tapinos, A.; Huang, R.; Bornman, R.; Stricker, P.; Mutambirwa, S.; Wedge, D.; Jaratlerdsiri, W. Kataegis associated mutational processes linked to adverse prostate cancer presentation in African men. Research Square 2024, rs. 3. rs-4597464.
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 2013.
- Chen, Z.; Yuan, Y.; Chen, X.; Chen, J.; Lin, S.; Li, X.; Du, H. Systematic comparison of somatic variant calling performance among different sequencing depth and mutation frequency. Scientific Reports 2020, 10 (1), 3501. [CrossRef]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M. A.; Fennell, T. J.; Carneiro, M. O.; Van der Auwera, G. A.; Kling, D. E.; Gauthier, L. D.; Levy-Moonshine, A.; Roazen, D. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv 2017, 201178.
- Cibulskis, K.; Lawrence, M. S.; Carter, S. L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E. S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 2013, 31 (3), 213-219. [CrossRef]
- Cameron, D. L.; Baber, J.; Shale, C.; Valle-Inclan, J. E.; Besselink, N.; van Hoeck, A.; Janssen, R.; Cuppen, E.; Priestley, P.; Papenfuss, A. T. GRIDSS2: comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biology 2021, 22, 1-25. [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A. J.; Kruglyak, S.; Saunders, C. T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2015, 32 (8), 1220-1222. [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A. L.; Bekritsky, M. A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P. Strelka2: fast and accurate calling of germline and somatic variants. Nature methods 2018, 15 (8), 591-594. [CrossRef]
- Jones, D.; Raine, K. M.; Davies, H.; Tarpey, P. S.; Butler, A. P.; Teague, J. W.; Nik-Zainal, S.; Campbell, P. J. cgpCaVEManWrapper: simple execution of CaVEMan in order to detect somatic single nucleotide variants in NGS data. Current protocols in bioinformatics 2016, 56 (1), 15.10. 11-15.10. 18. [CrossRef]
- Raine, K. M.; Hinton, J.; Butler, A. P.; Teague, J. W.; Davies, H.; Tarpey, P.; Nik-Zainal, S.; Campbell, P. J. cgpPindel: identifying somatically acquired insertion and deletion events from paired end sequencing. Current protocols in bioinformatics 2015, 52 (1), 15.17. 11-15.17. 12. [CrossRef]
- Radenbaugh, A. J.; Ma, S.; Ewing, A.; Stuart, J. M.; Collisson, E. A.; Zhu, J.; Haussler, D. RADIA: RNA and DNA integrated analysis for somatic mutation detection. PloS one 2014, 9 (11), e111516. [CrossRef]
- Wilm, A.; Aw, P. P. K.; Bertrand, D.; Yeo, G. H. T.; Ong, S. H.; Wong, C. H.; Khor, C. C.; Petric, R.; Hibberd, M. L.; Nagarajan, N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic acids research 2012, 40 (22), 11189-11201. [CrossRef]
- Rimmer, A.; Phan, H.; Mathieson, I.; Iqbal, Z.; Twigg, S. R. F.; Wilkie, A. O. M.; McVean, G.; Lunter, G.; Consortium, W. G. S. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nature Genetics 2014, 46 (8), 912-918. [CrossRef]
- Saunders, C. T.; Wong, W. S.; Swamy, S.; Becq, J.; Murray, L. J.; Cheetham, R. K. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012, 28 (14), 1811-1817. [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A. M.; Benes, V.; Korbel, J. O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28 (18), i333-i339. [CrossRef]
- Layer, R. M.; Chiang, C.; Quinlan, A. R.; Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biology 2014, 15 (6), R84. [CrossRef]
- Sadsad, R., Samaha, G., & Chew, T. . Fastq-to-bam @ NCI-Gadi. WorkflowHub 2021. [CrossRef]
- Sadsad, R., Samaha, G., & Chew, T. . Germline-ShortV @ NCI-Gadi. WorkflowHub 2021. [CrossRef]
- Sadsad, R., & Chew, T. Somatic-ShortV @ NCI-Gadi. WorkflowHub 2021. [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34 (17), i884-i890. [CrossRef]
- Tarasov, A.; Vilella, A. J.; Cuppen, E.; Nijman, I. J.; Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015, 31 (12), 2032-2034. [CrossRef]
- Faust, G. G.; Hall, I. M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014, 30 (17), 2503-2505. [CrossRef]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L. M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T. F.; Conesa, A. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28 (20), 2678-2679. [CrossRef]
- Favero, F.; Joshi, T.; Marquard, A. M.; Birkbak, N. J.; Krzystanek, M.; Li, Q.; Szallasi, Z.; Eklund, A. C. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Annals of Oncology 2015, 26 (1), 64-70. [CrossRef]
- Gong, T.; Hayes, V. M.; Chan, E. K. Detection of somatic structural variants from short-read next-generation sequencing data. Briefings in bioinformatics 2021, 22 (3), bbaa056. [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E. W.; Barja, P. P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nature biotechnology 2017, 35 (4), 316-319. [CrossRef]
- Chan, E. K.; Cameron, D. L.; Petersen, D. C.; Lyons, R. J.; Baldi, B. F.; Papenfuss, A. T.; Thomas, D. M.; Hayes, V. M. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome research 2018, 28 (5), 726-738. [CrossRef]
- Sakamoto, Y.; Sereewattanawoot, S.; Suzuki, A. A new era of long-read sequencing for cancer genomics. Journal of human genetics 2020, 65 (1), 3-10. [CrossRef]
- Rodriguez, I.; Rossi, N. M.; Keskus, A. G.; Xie, Y.; Ahmad, T.; Bryant, A.; Lou, H.; Paredes, J. G.; Milano, R.; Rao, N.; et al. Insights into the mechanisms and structure of breakage-fusion-bridge cycles in cervical cancer using long-read sequencing. Am J Hum Genet 2024, 111 (3), 544-561. [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A. V.; Mikheenko, A.; Vollger, M. R.; Altemose, N.; Uralsky, L.; Gershman, A. The complete sequence of a human genome. Science 2022, 376 (6588), 44-53.
- Liao, W.-W.; Asri, M.; Ebler, J.; Doerr, D.; Haukness, M.; Hickey, G.; Lu, S.; Lucas, J. K.; Monlong, J.; Abel, H. J.; et al. A draft human pangenome reference. Nature 2023, 617 (7960), 312-324. [CrossRef]
- Rhie, A.; McCarthy, S. A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J. Towards complete and error-free genome assemblies of all vertebrate species. Nature 2021, 592 (7856), 737-746. [CrossRef]
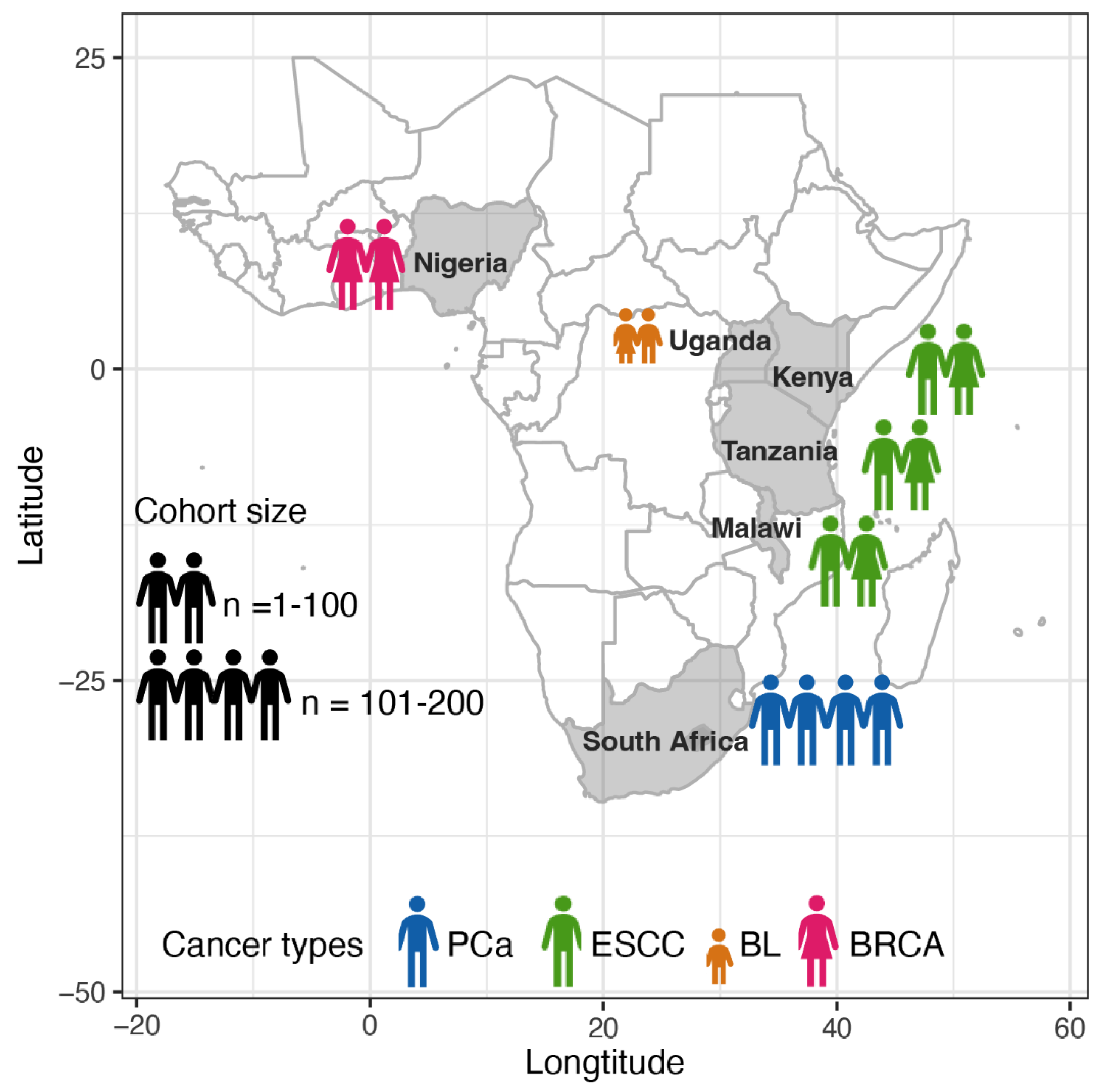

| Consortium or project | Cancer type | Country | Cohort size a | Tissue fixation b | Coverage of tumour, normal (median/mean) |
Recruitment time | Recruitment hospitals |
| SAPCS [27,28] | PCa | South Africa | 123 | FF | 88.69X, 44.3X (median) | 2013 – 2018 | Polokwane Urology Clinic, Limpopo; Tshilidzini Hospital, Limpopo; Pretoria's Steve Biko Academic Hospitals, Gauteng; Dr. George Mukhari Academic Hospitals, Gauteng; and Kalafong Academic Hospital, Gauteng |
| ESCCAPE [29] | ESCC | Kenya | 68 | FF | 49X, 26X (mean c) | 2014 - 2020 | Moi Teaching and Referral Hospital, Eldoret; |
| Malawi | 59 | Queen Elizabeth Central Hospital, Blantyre; | |||||
| Tanzania | 35 | Kilimanjaro Clinical Research Institute, Moshi | |||||
| AfrECC [30] | ESCC | Tanzania | 61 | FFPE | 60X, 30X (targeted coverage, de facto values unavailable) | 2016 - 2018 | Muhimbili National Hospital, Dar es Salaam, |
| BLGSP [31,32] | BL | Uganda | 87 | 83 FF, 4 FFPE | 82X, 41X (mean c); 72.6X (mean across sample types c) |
Unavailable | Uganda Cancer Institute, Kampala; St. Mary's Hospital, Gulu |
| NBCS [33] | BRCA | Nigeria | 97 | FPAX | 103.2X, 35.1X (mean) | 2013 - 2015 | Lagos State University Teaching Hospital, Lagos |
| Cancer type | Measurement | Values or odds ratios | P-value | Comparison b |
| Short variants (nucleotide variants, insertion and deletion variants less than 50bp) | ||||
| PCa | Tumour mutational burden (TMB, mutations per Mb) | 1.197 versus 1.061 | 0.013 | EUR |
| PCa | Predicted damaging mutations (count) | 14 versus 11 | 0.022 | EUR |
| BRCA | Insertions and deletions (indels) | N/A | 6.510−5, 210−4 | EUR, AA |
| Driver genes | ||||
| BRCA | GATA3 | 6.3-fold | FDR=0.038 | EUR, AA |
| BRCA | Non-coding region, upstream of ZNF217 (frequency) | 42.3% versus 4.3% | FDR=0.037 | EUR, AA |
| BRCA | Non-coding region, spanning SYPL1 (frequency | 28.9% versus 0% | FDR=0.097 | EUR, AA |
| ESCC | TP53 (frequency) | 72% versus 74.8% - 87% [36,37,38] | EUR, AA | |
| BL | SIN3A (frequency) | 18.4% versus 9.1% | patients from the USA | |
| BL | HIST1H1E (frequency) | 9.2% versus 4.5% | ||
| BL | CHD8 (frequency) | 9.2% versus 4.5% | ||
| Somatic copy number alteration (SCNA) | ||||
| PCa | Percentage of genome alteration (PGA) | 7.26% versus 2.82% | 0.021 | EUR |
| BRCA | Whole-genome duplications (WGD) | 3-fold | FDR=0.02 | EUR, AA |
| Structural variants (SV) | ||||
| PCa | Duplication (relative frequency, count) [39] | 1.6-fold, 2.5-fold | EUR | |
| PCa | A single type hyper-SV frequency [39] a | 2-fold | EUR | |
| PCa | PCAT1 | 9.09-fold | 0.012 | EUR |
| PCa | TMPRSS2-ERG | 0.26-fold | 0.0004 | EUR |
| Several types of variants combined | ||||
| BRCA | intra-tumoral heterogeneity (ITH, increase %) | 3.4%, 5.7 % | 0.005, 0.00017 | EUR, AA |
| PCa | NCOA2 | 5.81-fold | 3.1410−6 | EUR |
| PCa | DDX11L1 | 4.17-fold | 0.0001 | EUR |
| PCa | STK19 | 4.65-fold | 0.004 | EUR |
| PCa | SETBP1 | 2.80-fold | 0.012 | EUR |
| Consortium or project | Genome | Variant callers | ||
| Short variants | Structural variants | |||
| Germline | Somatic | |||
| SAPCS | GRCh38 | GATK HaplotypeCaller [45] | GATK MuTect2 [46] | GRIDSS [47], Manta [48] |
| ESCCAPE | GRCh37 | Strelka2 [49] | Strelka2, and cgpCaVEMan [50] for SNVs; cgpPindel [51] for INDELs | BRASSa |
| AfrECC | GRCh37 | - | RADIA [52] | - |
| BLGSP | GRCh38 | - | Strelka2, GATK Mutect2, Lofreq [53], and SAGEb | GRIDSS, Manta |
| NBCS | GRCh37 |
Platypus [54] | GATK MuTect and Strelka [55] | Manta, DELLY [56], and Lumpy [57] |
| Steps | Sample type a | CPU/task | Total tasks | Batches | CPUs/batch | Execution time (hr) | Main algorithm with version |
| Pipeline 1 Data pre-processing for variant discovery | 14.4 | ||||||
| Split FASTQ | Bood | 4 | 20 | 1 | 96 | 0.9 | fastp [61] v0.20.0 |
| Tumour | 4 | 20 | 1 | 96 | 1.8 | ||
| Alignment | Both | 6 | 11,040 | 3 | 3,840 | 0.5 | BWA-MEM v0.7.15 |
| Merge | Bood | 24 | 20 | 1 | 480 | 0.4 | SAMBAMBA [62] v0.7.1 |
| Tumour | 24 | 20 | 1 | 480 | 0.8 | ||
| Mask duplicate | Bood | 14 | 20 | 1 | 280 | 1.3 | SAMBLASTER [63] v0.1.24 |
| Tumour | 14 | 20 | 1 | 280 | 2.6 | ||
| BQSR recal | Bood | 1 | 640 | 1 | 640 | 0.2 | GATK v4.4.0.0 b BaseRecalibrator |
| Tumour | 1 | 640 | 1 | 640 | 0.3 | ||
| BQSR apply | Bood | 2 | 480 | 1 | 960 | 0.3 | GATK ApplyBQSR |
| Tumour | 2 | 480 | 1 | 960 | 0.6 | ||
| qSignature | Bood | 24 | 20 | 1 | 480 | 0.7 | QSignature c v0.1pre (75) |
| Tumour | 24 | 20 | 1 | 480 | 1.4 | ||
| Qualimap | Bood | 6 | 20 | 2 | 144 | 1.4 | Qualimap [64] v.2.2.1 |
| Tumour | 6 | 20 | 2 | 144 | 2.8 | ||
| Sequenza | Pair | 2 | 480 | 1 | 504 | 3.6 | Sequenza [65] v3.0.0 |
| Pipeline 2 Germline short variant discovery | 8.1 | ||||||
| Variant call | Bood | 1 | 64,000 | 1 | 480 | 1.8 | GATK HaplotypeCaller |
| Consolidation | Bood | 1 | 3,200 | 11 | 144 | 1.3 | GATK GenomicsDBImport |
| Joint genotyping | Bood | 1 | 3,200 | 1 | 144 | 2 | GATK GenotypeGVCFs |
| VQSR | Blood | 16 | 1 | 1 | 16 | 3 | GATK VariantFiltration, MakeSitesOnlyVcf, VariantRecalibrator, CollectVariantCallingMetrics, ApplyVQSR, CollectVariantCallingMetrics |
| Pipeline 3 Somatic short variant discovery | 3.3 | ||||||
| PoN | Bood | 1 | 64,000 | 1 | 2,880 | 0.6 | GATK Mutect2 |
| Consolidate | Blood | 2 | 3,200 | 1 | 96 | 0.3 | GATK GenomicsDBImport |
| Create PoN | Blood | 1 | 3,200 | 1 | 960 | 1.6 | GATK CreateSomaticPON |
| Variant call | Pair | 1 | 64,000 | 1 | 2,880 | 0.8 | GATK Mutect2 |
| Pipeline 4 Structural variant discovery | 23 | ||||||
| GRIDSS | Pair | 8 | 20 | 20 | 8 | Range, 10 - 20 | GRIDSS v2.8.3 |
| Manta | Pair | 24 | 20 | 2 | 48 | 3.0 | Manta v1.6.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





