Submitted:
23 June 2025
Posted:
24 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
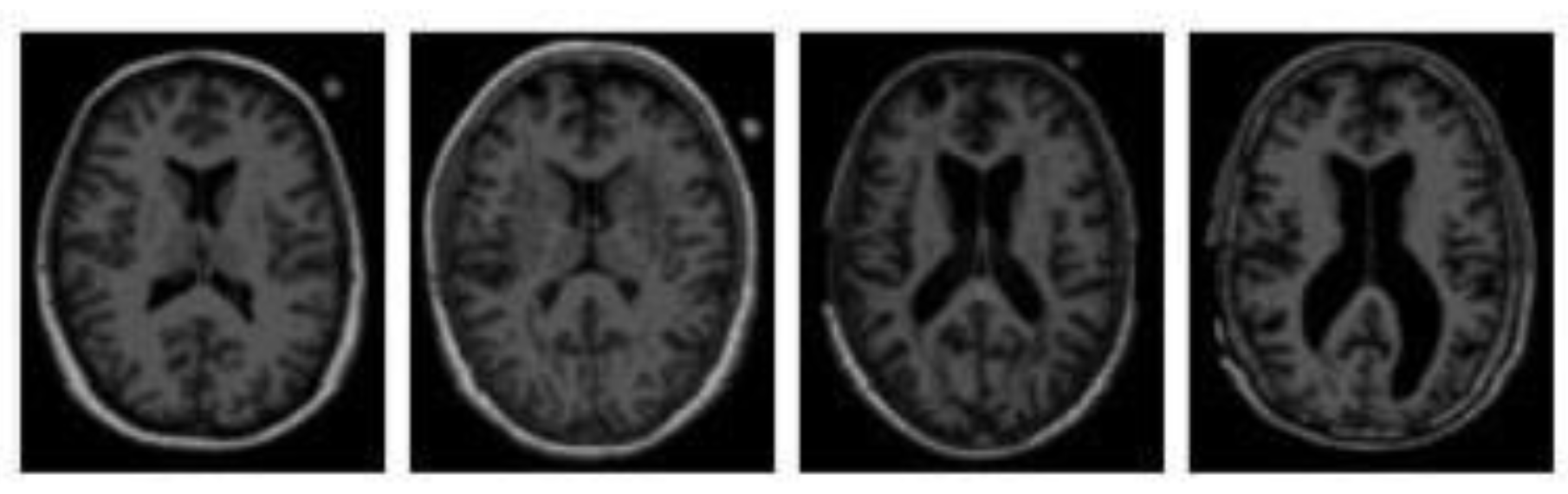
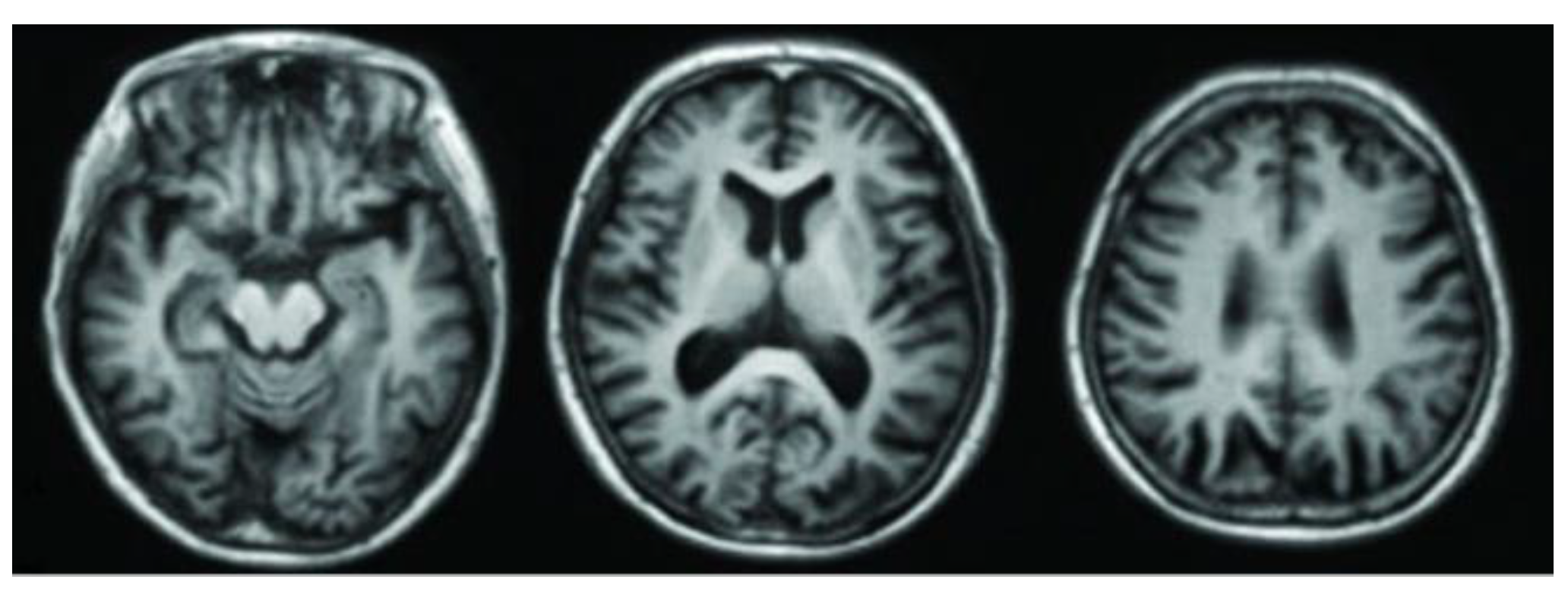
2. Alzheimer's Disease Stages
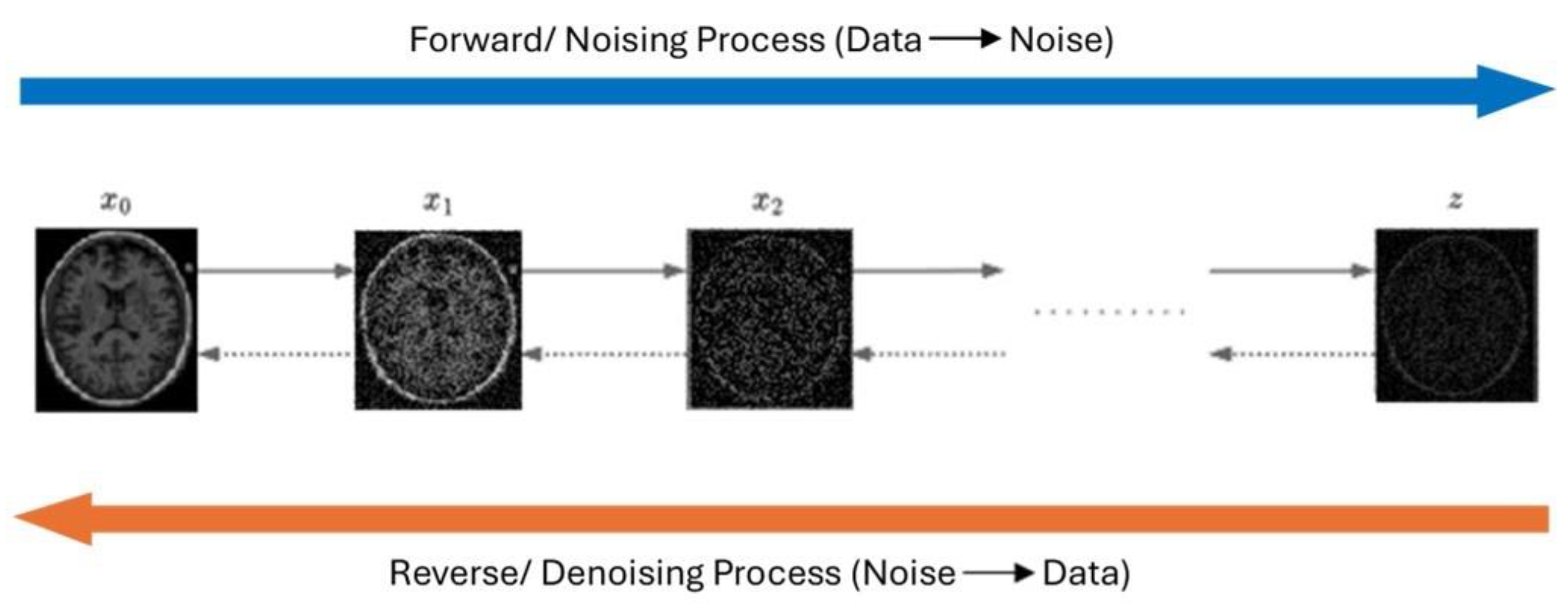
3. Neuroimaging Modalities
3.1. Magnetic Resonance Imaging (MRI)
3.2. Functional Magnetic Resonance Imaging (fMRI)
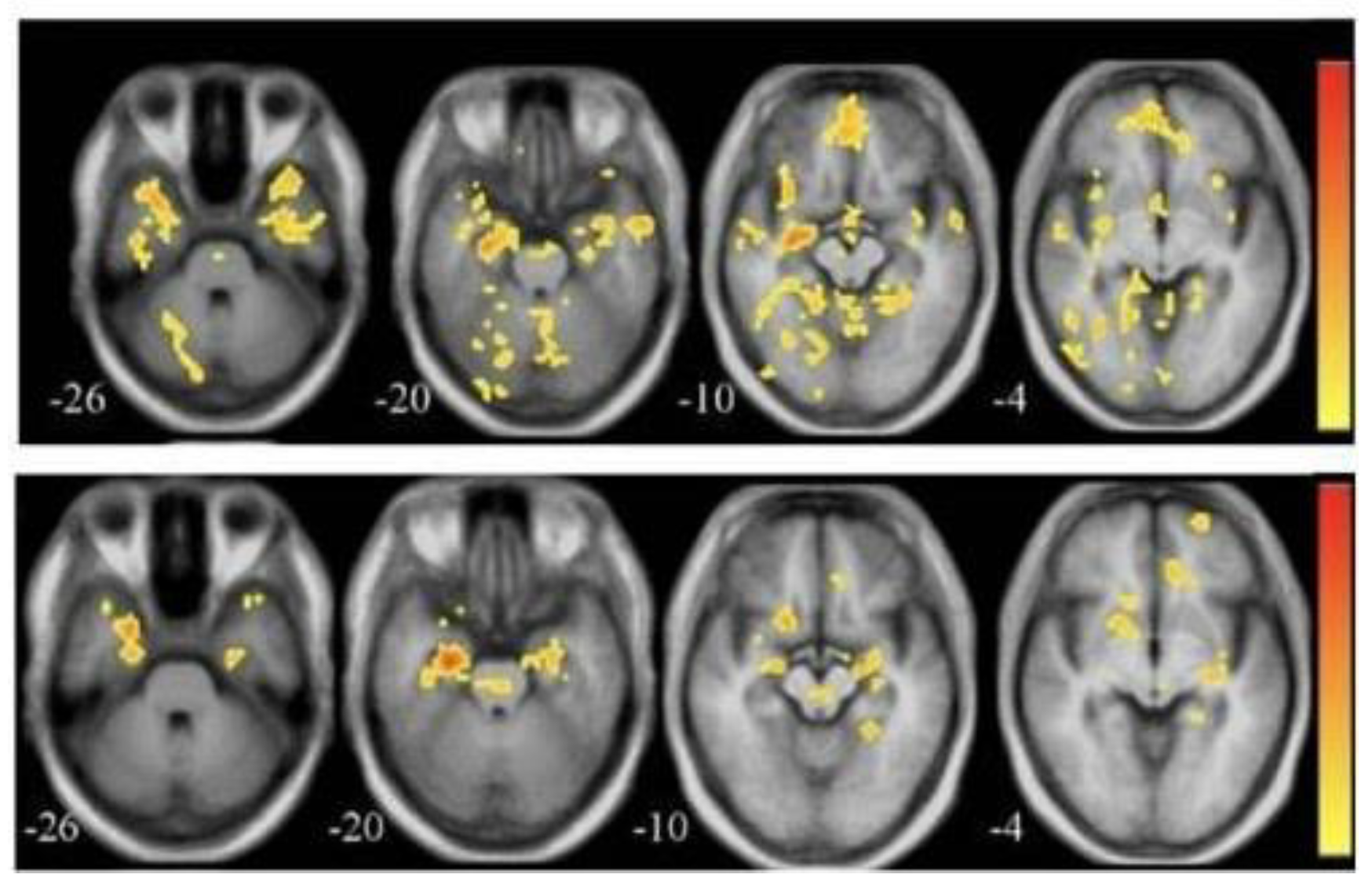
3.3. Positron Emission Tomography (PET)
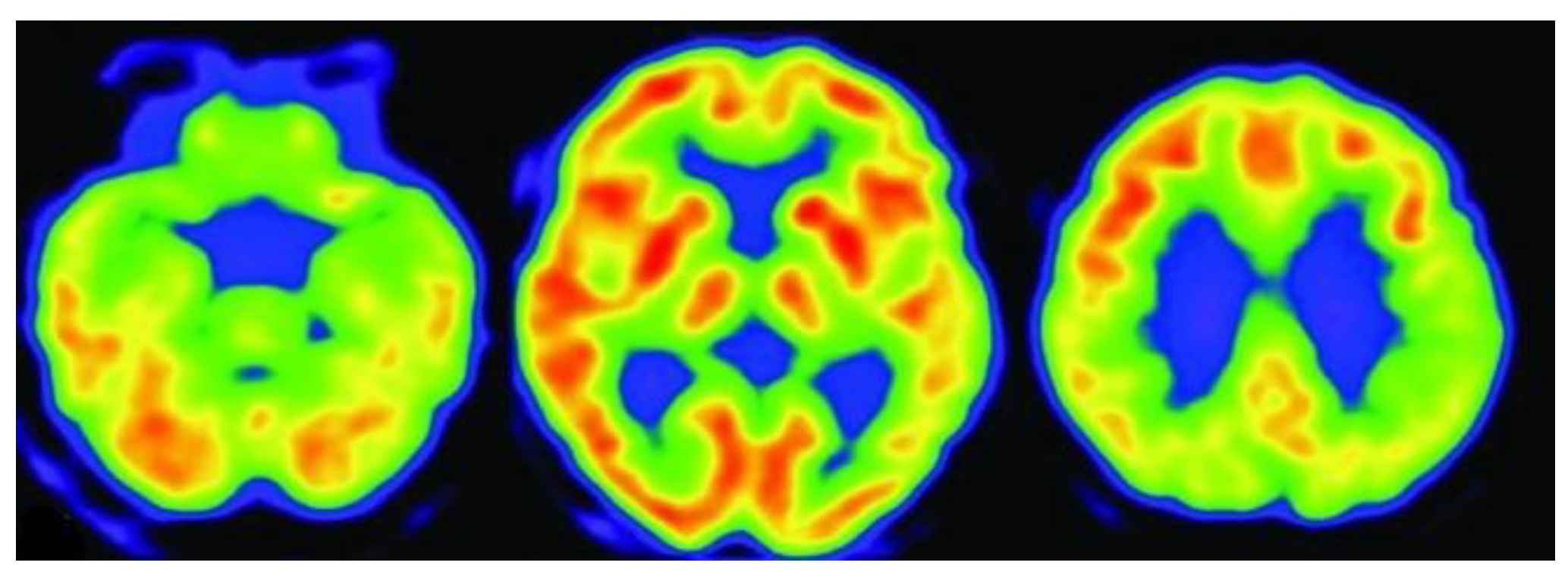
3.4. Fluorodeoxyglucose Positron Emission Tomography (FDG-PET)
3.5. Computed Tomography (CT)
3.6. Diffusion Tensor Imaging (DTI)
| Modality | Type | What It Detects | Strengths | Limitations |
|---|---|---|---|---|
| MRI | Structural | Brain atrophy, hippocampal and cortical shrinkage | Non-invasive, high-resolution, and widely available. | Limited functional insight, relatively expensive |
| fMRI | Functional | Brain activity and connectivity between regions. | Real-time brain function | Sensitive to motion and requires complex analysis |
| PET | Molecular / Functional |
Amyloid plaques, tau proteins, glucose metabolism | Identifies biochemical changes early, aids in staging | High cost, uses radioactive tracers |
| FDG-PET | Metabolic Imaging | Glucose metabolism, hypometabolic regions in AD | Detects early metabolic dysfunction in AD-affected areas | Radiation exposure, lower spatial resolution than MRI |
| CT | Structural | Structural abnormalities, bleeding or lesions | Fast, accessible in emergency settings | Lower soft tissue contrast than MRI, less specific for AD |
| DTI | Microstructural | White matter integrity, neural pathway disruptions | Subtle white matter changes, supports early diagnosis | Requires complex processing, susceptible to motion and noise. |
4. Background Study
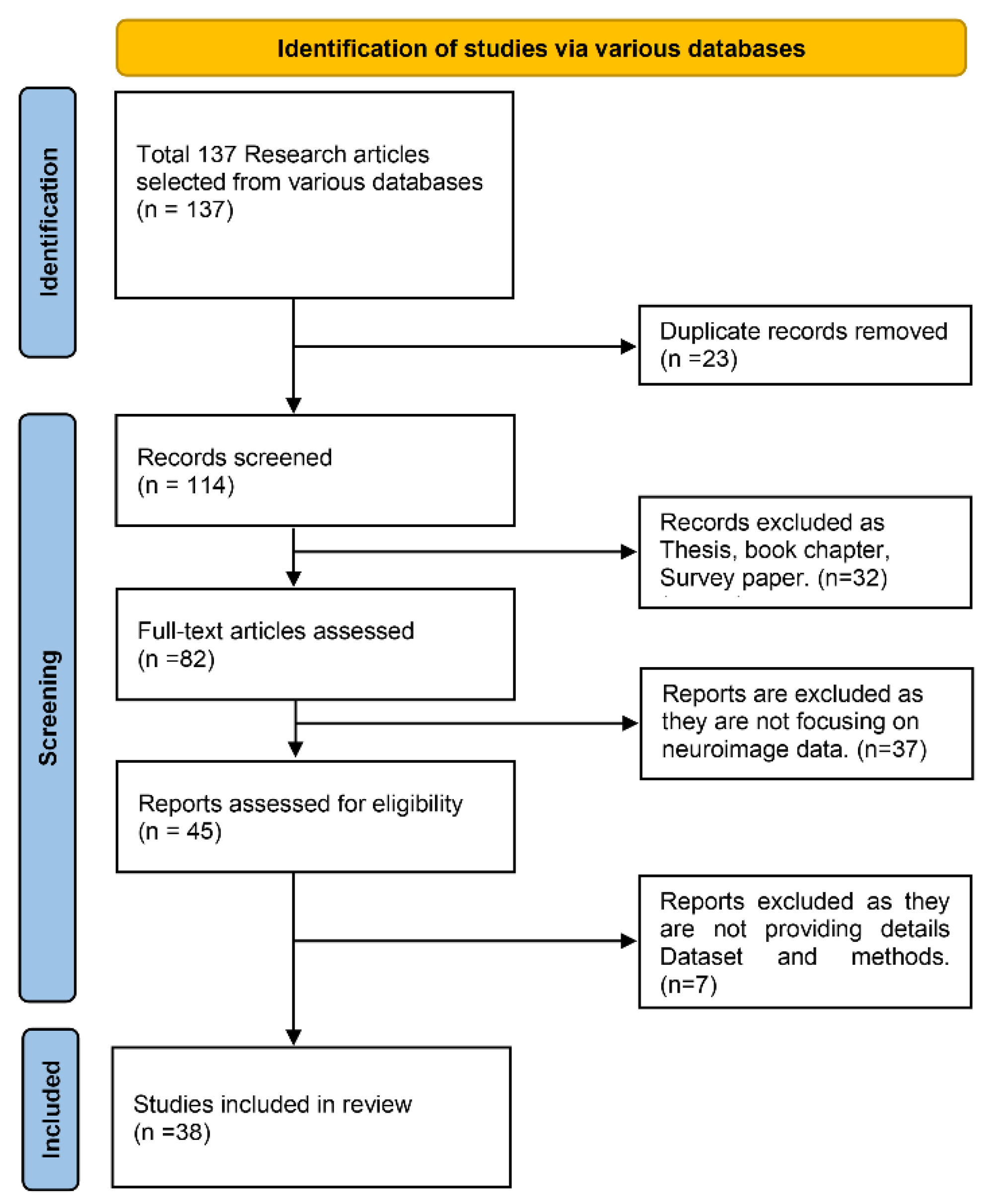
4.1. Paper Selection Strategy
4.2. Literature Review
| Reference | Dataset | Modality | Total number of participants |
|---|---|---|---|
| [21] | ADNI-like MRI data (T1w), Diffusion MRI of healthy subjects | Structural MRI | 18 AD, 18 bvFTD, 19 control: 14 healthy for connectome |
| [23] | ADNI-1 & ADNI-2 | MRI, PET | ADNI-1: 821; ADNI-2: 636 |
| [24] | OASIS-3 | MRI | Training: 408 subjects; Test: 113 healthy, 99 (CDR 0.5), 61 (CDR 1), 4 (CDR 2) |
| [25] | ADNI | MRI, PET | 1,033 (722 train, 104 val, 207 test) |
| [26] | ADNI | PET | 411 PET scans (98 AD, 105 NC, 208 MCI) |
| [27] | ADNI | MRI, PET | 680 subjects |
| [28] | ADNI-1, ADNI-2 | MRI, PET | ADNI-1 (PET AD only), ADNI-2 (100 NC, 20 AD, 80 AD MRI-only) |
| [29] | ADNI-GO, ADNI-2, OASIS | 3D MRI | 210 (mi-GAN), 603 (classifier), 48 (validation) |
| [30] | OASIS-3, Internal dataset | T1, T1c MRI | 408 (T1), 135 (T1c healthy) |
| [31] | ADNI, AIBL, NACC | MRI (1.5-T and 3-T) | ADNI: 151 (training), AIBL: 107, NACC: 565 |
| [32] | ADNI-1, ADNI-2 | MRI + PET | ADNI-1: 821; ADNI-2: 534 |
| [33] | ADNI-1 | T1 MRI | 833 (221 AD, 297 MCI, 315 NC) |
| [34] | ADNI (268 subjects) | rs-fMRI + DTI | 268 |
| [35] | ADNI (13,500 3D MRI images after augmentation) | 3D Structural MRI | 138 (original), 13,500 (augmented scans) |
| [36] | ADNI (1732 scan-pairs, 873 subjects) | MRI → Synthesized PET | 873 |
| [37] | ADNI | T1-weighted MRI | 632 participants |
| [38] | ADNI2 | T1-weighted MRI | 169 participants, 27,600 image pairs |
| [39] | Custom MRI dataset (Kaggle) | T1-weighted Brain MRI | 6400 images (approx.) |
| [40] | ADNI2, NIFD (in-domain), NACC (external) | T1-weighted MRI | 3,319 MRI scans |
| [41] | ADNI | MRI and PET (multimodal) | ~2,400 (14,800 imaging sessions) |
| [42] | ADNI (Discovery), SMC (Practice) | T1-weighted MRI, Demographics, Cognitive scores | 538 (ADNI) + 343 (SMC) |
| [43] | ADNI | T1-weighted MRI | 362 (CN: 87, MCI: 211, AD: 64) |
| [44] | ADNI1, ADNI3, AIBL | 1.5T & 3T MRI | ~168 for SR cohort, ~1,517 for classification |
| [45] | ADNI | T1-weighted MRI | 6,400 images across 4 stages (Non-Demented, Very Mild, Mild, Moderate Demented) |
| [46] | ADNI | Cognitive Features | 819 participants (5013 records) |
| [47] | ADNI, OASIS-3, Centiloid | Low-res PET + MRI → High-res PET | ADNI: 334; OASIS-3: 113; Centiloid: 46 |
| [48] | ADNI | MRI T1WI, FDG PET (Synth.) | 332 subjects, 1035 paired scans |
| [49] | ADNI, OASIS, UK Biobank | 3D T1-weighted MRI | ADNI: 1,188, OASIS: 600, UKB: 38,703 |
| [50] | Alzheimer MRI (6,400 images) | T1-weighted MRI | Alzheimer: 6,400 images |
| [51] | OASIS-3 | T1-weighted MRI | 300 (100 AD, 100 MCI, 100 NC) |
| [52] | ADNI-3, In-house | Siemens ASL MRI (T1, M0, CBF) | ADNI Siemens: 122; GE: 52; In-house: 58 |
| [53] | ADNI | MRI + Biospecimen (Aβ, t-tau, p-tau) | 50 subjects |
| [54] | OASIS | MRI | 300 subjects (100 AD, 100 MCI, 100 NC) |
| [55] | ADNI | MRI | 311 (AD: 65, MCI: 67, NC: 102, cMCI: 77) |
| [56] | ADNI | Structural MRI → Aβ-PET, Tau-PET (synthetic) | 1,274 |
| [57] | Kaggle | MRI | 6,400 images (4 AD classes) |
| [58] | ADNI | sMRI, DTI, fMRI (multimodal) | 5 AD stages (NC, SMC, EMCI, LMCI, AD) |
| Reference | Technique/Method | Model | Results |
|---|---|---|---|
| [21] | Network Diffusion Model | Network eigenmode diffusion model | Strong correlation between predicted and actual atrophy maps; eigenmodes accurately classified AD/bvFTD; ROC AUC higher than PCA |
| [23] | 3D CycleGAN + LM3IL | Two-stage: PET synthesis (3D-cGAN) + classification (LM3IL) | AD vs HC – Accuracy: 92.5%, Sensitivity: 89.94%, Specificity: 94.53%; PSNR: 24.49 ± 3.46 |
| [24] | WGAN-GP + L1 loss (MRI slice reconstruction) | WGAN-GP-based unsupervised reconstruction + anomaly detection using L2 loss | AUC: 0.780 (CDR 0.5), 0.833 (CDR 1), 0.917 (CDR 2) |
| [25] | GANDALF: GAN with discriminator-adaptive loss for MRI-to-PET synthesis and AD classification | GAN + Classifier | Binary (AD/CN): 85.2% Acc3-class: 78.7% Acc, F2: 0.69, Prec: 0.83, Rec: 0.664-class: 37.0% Acc |
| [26] | DCGAN to generate PET images for NC, MCI, and AD | DCGAN | PSNR: 32.83, SSIM: 77.48, CNN classification accuracy improved to 71.45% with synthetic data |
| [27] | Bidirectional GAN with ResU-Net generator, ResNet-34 encoder, PatchGAN discriminator | Bidirectional GAN | PSNR: 27.36, SSIM: 0.88; AD vs. CN classification accuracy: 87.82% with synthetic PET |
| [28] | DCGAN for PET synthesis from noise; DenseNet classifier for AD vs. NC | DCGAN + DenseNet | Accuracy improved from 67% to 74%; MMD: 1.78, SSIM: 0.53 |
| [29] | 3D patch-based mi-GAN with baseline MRI + metadata; 3D DenseNet with focal loss for classification | mi-GAN + DenseNet | SSIM: 0.943, Multi-class Accuracy: 76.67%, pMCI vs. sMCI Accuracy: 78.45% |
| [30] | MADGAN: GAN with multiple adjacent slice reconstruction using WGAN-GP + ℓ1 loss and self-attention | 7-SA MADGAN | AUC for AD: 0.727 (MCI), 0.894 (late AD); AUC for brain metastases: 0.921 |
| [31] | Generative Adversarial Network (GAN), Fully Convolutional Network (FCN) | GAN + FCN | Improved AD classification with accuracy increases up to 5.5%. SNR, BRISQUE, and NIQE metrics showed significant image quality improvements. |
| [32] | TPA-GAN for PET imputation, PT-DCN for classification | TPA-GAN + PT-DCN | AD vs CN: ACC 90.7%, SEN 91.2%, SPE 90.3%, F1 90.9%, AUC 0.95; pMCI vs sMCI: ACC 85.2%, AUC 0.89 |
| [33] | THS-GAN: Tensor-train semi-supervised GAN with high-order pooling and 3D-DenseNet | THS-GAN | AD vs NC: AUC 95.92%, Acc 95.92%; MCI vs NC: AUC 88.72%, Acc 89.29%; AD vs MCI: AUC 85.35%, Acc 85.71% |
| [34] | CT-GAN with Cross-Modal Transformer and Bi-Attention | GAN + Transformer with Bi-Attention | AD vs NC: Acc=94.44%, Sen=93.33%, Spe=95.24%LMCI vs NC: Acc=93.55%, Sen=90.0%, Spe=95.24%EMCI vs NC: Acc=92.68%, Sen=90.48%, Spe=95.0% |
| [35] | WGANGP-DTL (Wasserstein GAN with Gradient Penalty + Deep Transfer Learning using Inception v3 and DBN) | WGANGP + Inception v3 + DBN | Accuracy: 99.70%Sensitivity: 99.09%Specificity: 99.82%F1-score: >99% |
| [36] | BPGAN (3D BicycleGAN with Multiple Convolution U-Net, Hybrid Loss) | 3D BicycleGAN (BPGAN) with MCU Generator | Dataset-A: MAE=0.0318, PSNR=26.92, SSIM=0.7294Dataset-B: MAE=0.0396, PSNR=25.08, SSIM=0.6646Diagnosis Acc=85.03% (multi-class, MRI + Synth. PET) |
| [37] | ReMiND (Diffusion-based MRI Imputation) | Denoising Diffusion Probabilistic Model (DDPM) with modified U-Net | SSIM: 0.895, PSNR: 28.96; no classification metrics reported |
| [38] | Wavelet-guided Denoising Diffusion Probabilistic Model (Wavelet Diffusion) | Wavelet Diffusion with Wavelet U-Net | SSIM: 0.8201, PSNR: 27.15, FID: 13.15 (×4 scale); Recall ~90% (AD vs NC); improved classification performance overall |
| [39] | CNN + GAN (DCGAN to augment data; CNN for classification) | CNN + DCGAN (data augmentation) | Accuracy: 96% (with GAN), 69% (without GAN); classification across 4 AD stages |
| [40] | Deep Grading + Multi-layer Perceptron + SVM Ensemble (Structure Grading + Atrophy) | 125 3D U-Nets + Ensemble (MLP + SVM) | In-domain (3-class): Accuracy: 86.0%, BACC: 84.7%, AUC: 93.8%, Sensitivity (CN/AD/FTD): 89.6/83.2/81.3; Out-of-domain: Accuracy: 87.1%, BACC: 81.6%, AUC: 91.6%, Sensitivity (CN/AD/FTD): 89.6/76.9/78.4 |
| [41] | GAN for synthetic MRI generation + Ensemble deep learning classifiers | GAN + CNN, LSTM, Ensemble Networks | GAN results: Precision: 0.84, Recall: 0.76, F1-score: 0.80, AUC-ROC: 0.91, Proposed Ensemble: Precision: 0.85, Recall: 0.79, F1-score: 0.82, AUC-ROC: 0.93 |
| [42] | Modified HexaGAN (Deep Generative Framework) | Modified HexaGAN (GAN + Semi-supervised + Imputation) | ADNI: AUROC 0.8609, Accuracy 0.8244, F1-score 0.7596, Sensitivity 0.8415, Specificity 0.8178; SMC: AUROC 0.9143, Accuracy 0.8528, Sensitivity 0.9667, Specificity 0.8286. |
| [43] | Conditional Diffusion Model for Data Augmentation | Conditional DDPM + U-Net | Best result (Combine 900): Accuracy: 74.73%, Precision: 77.28%, Recall (Sensitivity): 66.52%, F1-score: 0.6968, AUC: 0.8590; Specificity: not reported |
| [44] | Latent Diffusion Model (d3T*) for MRI super-resolution + DenseNet Siamese Network for AD/MCI/NC classification | Latent Diffusion-based SR + Siamese DenseNet | AD classification: Accuracy 92.3%, AUROC 93.1%, F1-score 91.9%; Significant improvement over 1.5T and c3T*; Comparable to real 3T MRI |
| [45] | GAN-based data augmentation + hybrid CNN-InceptionV3 model for multiclass AD classification | GAN + Transfer Learning (CNN + InceptionV3) | Accuracy: 90.91%; metrics like precision, recall, and F1-score also reported high performance |
| [46] | DeepCGAN (GAN + BiGRU with Wasserstein Loss) | Encoder-Decoder GAN with BiGRU layers | Accuracy: 97.32%, Recall (Sensitivity): 95.43%, Precision: 95.31%, F1-Score: 95.61%, AUC: 99.51% |
| [47] | Latent Diffusion Model for Resolution Recovery (LDM-RR) | Latent Diffusion Model (LDM-RR) | Recovery coefficient: 0.96; Longitudinal p-value: 1.3×10⁻¹⁰; Cross-tracer correlation: r = 0.9411; Harmonization p = 0.0421 |
| [48] | Diffusion-based multi-view learning (one-way & two-way synthesis) | U-NET-based Diffusion Model with MLP Classifier | Accuracy: 82.19%, SSIM: 0.9380, PSNR: 26.47, Sensitivity: 95.19%, Specificity: 92.98%, Recall: 82.19% |
| [49] | Conditional DDPM and LDM with counterfactual generation and DenseNet121 classifier | LDM + 3D DenseNet121 CNN | AUC: 0.870, F1-score: 0.760, Sensitivity: 0.889, Specificity: 0.837 (ADNI test set after fine-tuning) |
| [50] | GANs, VAEs, Diffusion (DDIM) models for MRI generation + DenseNet/ResNet classifiers | DDIM (Diffusion Model) + DenseNet | Accuracy: 80.84%, Precision: 86.06%, Recall: 78.14%, F1-Score: 80.98% (Alzheimer’s, DenseNet + DDIM) |
| [51] | GAN-based data generation + EfficientNet for multistage classification | GAN for data augmentation + EfficientNet CNN | Accuracy: 88.67% (1:0), 87.17% (9:1), 82.50% (8:2), 80.17% (7:3); Recall/Sensitivity/Specificity not separately reported |
| [52] | Conditional Latent Diffusion Model (LDM) for M0 image synthesis from Siemens PASL | Conditional LDM + ML classifier | SSIM: 0.924, PSNR: 33.35, CBF error: 1.07 ± 2.12 ml/100g/min; AUC: 0.75 (Siemens), 0.90 (GE) in AD vs CN classification |
| [53] | Multi-modal conditional diffusion model for image-to-image translation (prognosis prediction) | Conditional Diffusion Model + U-Net | PSNR: 31.99 dB, SSIM: 0.75, FID: 11.43 |
| [54] | GAN for synthetic MRI image generation + EfficientNet for multi-stage classification | GAN + EfficientNet | Validation accuracy improved from 78.48% to 85.11%, training accuracy from 90.16% to 98.68% with GAN data |
| [55] | Dual GAN + Pyramid Attention + CNN | Dual GAN with Pyramid Attention and CNN | Accuracy: 98.87%, Recall/Sensitivity: 95.67%, Specificity: 98.78%, Precision: 99.78%, F1-score: 99.67% |
| [56] | Prior-information-guided residual diffusion model with CLIP module and intra-domain difference loss | Residual Diffusion Model with CLIP guidance | SSIM: 92.49% (Aβ), 91.44% (Tau); PSNR: 26.38 dB (Aβ), 27.78 dB (Tau); AUC: 90.74%, F1: 82.74% (Aβ); AUC: 90.02%, F1: 76.67% (Tau) |
| [57] | Hybrid of Deep Super-Resolution GAN (DSR-GAN) for image enhancement + CNN for classification | DSR-GAN + CNN | Accuracy: 99.22%, Precision: 99.01%, Recall: 99.01%, F1-score: 99.01%, AUC: 100%, PSNR: 29.30 dB, SSIM: 0.847, MS-SSIM: 96.39% |
| [58] | Bidirectional Graph GAN (BG-GAN) + Inner Graph Convolution Network + Balancer for stable multimodal connectivity generation | BG-GAN + InnerGCN | Accuracy >96%, Precision/Recall/F1 ≈ 0.98–1.00, synthetic data outperformed real in classification |
| Reference | Dataset | Model | Challenges and Limitations |
|---|---|---|---|
| [21] | ADNI-like MRI data (T1w), Diffusion MRI of healthy subjects | Network eigenmode diffusion model | Small sample size; no conventional ML metrics (accuracy, F1); assumes static connectivity; limited resolution in tractography; noise in MRI volumetrics. |
| [23] | ADNI-1 & ADNI-2 | Two-stage: PET synthesis (3D-cGAN) + classification (LM3IL) | Requires accurate MRI–PET alignment; patch-based learning may limit generalization. |
| [24] | OASIS-3 | WGAN-GP-based unsupervised reconstruction + anomaly detection using L2 loss | Region-limited detection (hippocampus/amygdala); may miss anomalies outside selected areas. |
| [25] | ADNI | GAN + Classifier | Binary classification was not better than CNN-only models; requires more tuning and architectural exploration. |
| [26] | ADNI | DCGAN | Trained separate GANs per class; used 2D slices only; lacks unified 3D modeling approach. |
| [27] | ADNI | Bidirectional GAN | Limited fine detail in some outputs; latent vector injection mechanism could be improved for better synthesis. |
| [28] | ADNI-1, ADNI-2 | DCGAN + DenseNet | Used 2D image generation; manual filtering of outputs; lacks 3D modeling and automation. |
| [29] | ADNI-GO, ADNI-2, OASIS | mi-GAN + DenseNet | Lower performance on gray matter prediction; limited short-term progression prediction; improvement possible with better feature modeling. |
| [30] | OASIS-3, Internal dataset | 7-SA MADGAN | Reconstruction instability on T1c scans; limited generalization; fewer healthy T1c scans; needs optimized attention modules. |
| [31] | ADNI, AIBL, NACC | GAN + FCN | Small sample size for GAN training (151 participants). Limited to AD vs. normal cognition (no MCI). |
| [32] | ADNI-1, ADNI-2 | TPA-GAN + PT-DCN | Requires paired modalities; model trained/tested on ADNI-1/2 independently; limited generalization. |
| [33] | ADNI-1 | THS-GAN | Requires careful TT-rank tuning; performance varies with GSP block position; validation limited to ADNI dataset. |
| [34] | ADNI (268 subjects) | GAN + Transformer with Bi-Attention | Limited dataset size, dependency on predefined ROIs, potential overfitting; lacks validation on other neurodegenerative disorders. |
| [35] | ADNI (13,500 3D MRI images after augmentation) | WGANGP + Inception v3 + DBN | Heavy reliance on data augmentation, complex pipeline requiring multiple preprocessing and tuning steps. |
| [36] | ADNI (1732 scan-pairs, 873 subjects) | 3D BicycleGAN (BPGAN) with MCU Generator | High preprocessing complexity, marginal diagnostic gains, requires broader validation and adaptive ROI exploration. |
| [37] | ADNI | Denoising Diffusion Probabilistic Model (DDPM) with modified U-Net | Uses only adjacent timepoints; assumes fixed intervals; no classification; no sensitivity/specificity; computationally intensive. |
| [38] | ADNI2 | Wavelet Diffusion with Wavelet U-Net | High computational cost; limited to T1 MRI; does not incorporate multi-modal data or longitudinal timepoints. |
| [39] | Custom MRI dataset (Kaggle) | CNN + DCGAN (data augmentation) | Risk of overfitting due to small original dataset; no reporting of sensitivity/specificity; limited to image data. |
| [40] | ADNI2, NIFD (in-domain), NACC (external) | 125 3D U-Nets + Ensemble (MLP + SVM) | High computational cost (393M parameters, 25.9 TFLOPs); inference time ~1.6s; only baseline MRI used; limited by class imbalance and absence of multimodal or longitudinal data. |
| [41] | ADNI | GAN + CNN, LSTM, Ensemble Networks | Limited real Alzheimer’s samples; reliance on synthetic augmentation; needs more external validation and data diversity. |
| [42] | ADNI (Discovery), SMC (Practice) | Modified HexaGAN (GAN + Semi-supervised + Imputation) | High model complexity; requires fine-tuning across datasets; limited to MRI and tabular inputs. |
| [43] | ADNI | Conditional DDPM + U-Net | Small dataset, imbalanced classes; specificity not reported; limited to static MRI slices; no multi-modal or longitudinal data. |
| [44] | DNI1, ADNI3, AIBL | Latent Diffusion-based SR + Siamese DenseNet | High computational cost; needs advanced infrastructure for training; diffusion SR takes longer than CNN-based methods. |
| [45] | ADNI | GAN + Transfer Learning (CNN + InceptionV3) | Class imbalance still impacts performance slightly; more detailed metrics (sensitivity/specificity) not reported. |
| [46] | ADNI | Encoder-Decoder GAN with BiGRU layers | Computational complexity, GAN training instability, underutilization of multimodal data (e.g., neuroimaging). |
| [47] | ADNI, OASIS-3, Centiloid | Latent Diffusion Model (LDM-RR) | High computational cost; trained on synthetic data; limited interpretability; real-time deployment needs optimization. |
| [48] | ADNI | U-NET-based Diffusion Model with MLP Classifier | One-way synthesis introduces variability; computational intensity; requires improvement in generalization and speed. |
| [49] | ADNI, OASIS, UK Biobank | LDM + 3D DenseNet121 CNN | High computational cost, requires careful fine-tuning, limited by resolution/memory constraints. |
| [50] | Alzheimer MRI (6,400 images) | DDIM (Diffusion Model) + DenseNet | Diffusion models are computationally intensive; VAE had low image quality; fine-tuning reduced accuracy in some cases; computational cost vs. performance tradeoff. |
| [51] | OASIS-3 | GAN for data augmentation + EfficientNet CNN | GAN training instability; performance drop when synthetic data exceeds real data; no separate sensitivity/specificity metrics reported. |
| [52] | ADNI-3, In-house | Conditional LDM + ML classifier | No ground truth M0 for Siemens data; SNR difference between PASL/pCASL; class imbalance; vendor variability. |
| [53] | ADNI | Conditional Diffusion Model + U-Net | Small sample size; no classification metrics reported; requires broader validation with more diverse data. |
| [54] | OASIS | GAN + EfficientNet | GAN training instability; overfitting in CNN; limited dataset size; scope to explore alternate GAN models for robustness. |
| [55] | ADNI | Dual GAN with Pyramid Attention and CNN | Dependent on ADNI dataset quality; generalization affected by population diversity; limited interpretability; reliance on image features for AD detection. |
| [56] | ADNI | Residual Diffusion Model with CLIP guidance | Dependent on accurate prior info (e.g., age, gender); high computational cost; needs optimization for broader demographic generalization. |
| [57] | Kaggle | DSR-GAN + CNN | High computational complexity; SR trained on only 1,700 images; generalizability and real-time scalability remain open challenges. |
| [58] | ADNI | BG-GAN + InnerGCN | Difficulty in precise structure-function mapping due to fMRI variability; biological coordination model can be improved. |
5. Dataset
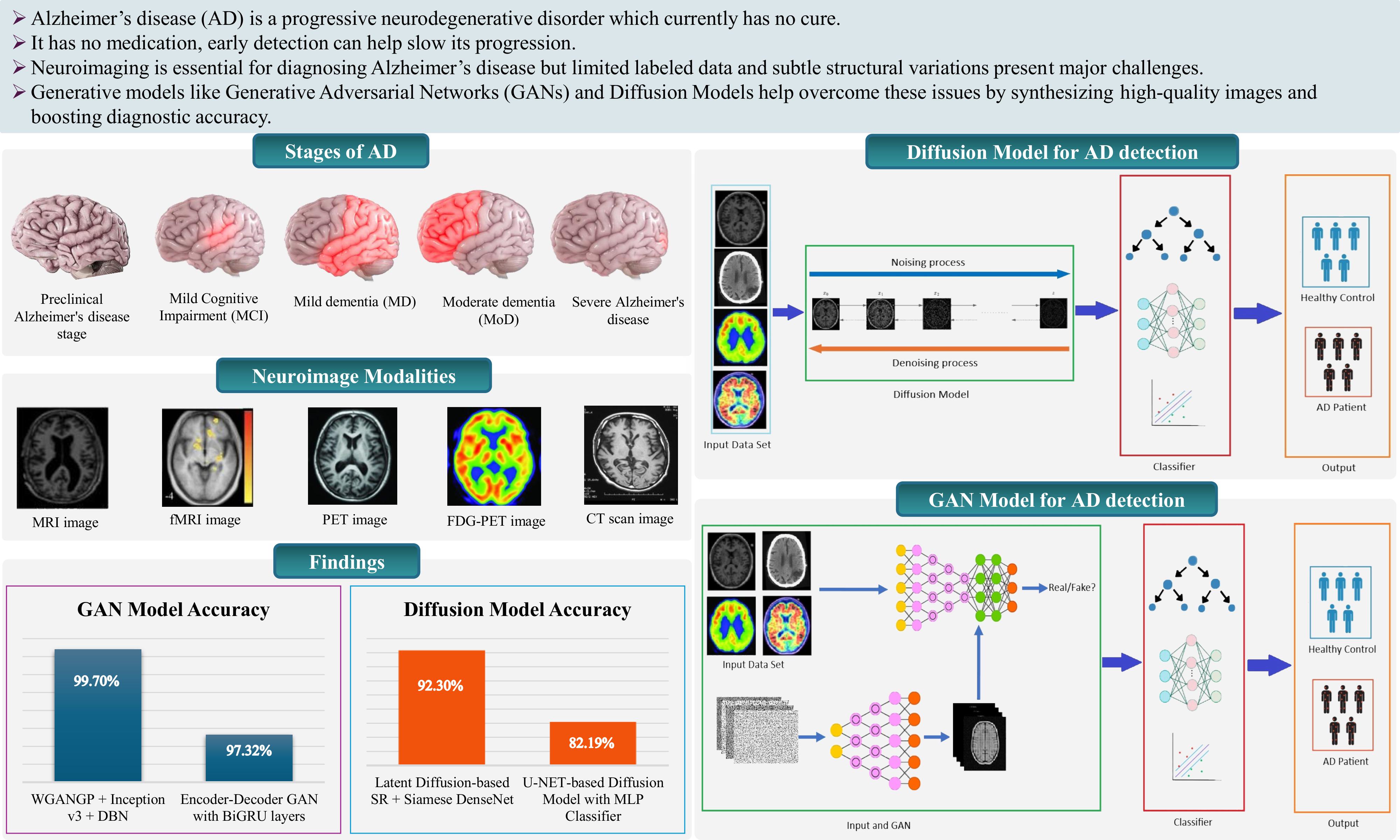
6. Generative Adversarial Networks (GANs)
6.1. Deep Convolutional Gan (DCGAN)
6.2. Convolutional Gan (CGAN)
6.3. CycleGAN
6.4. StyleGAN
6.5. Wasserstein GAN (WGAN)
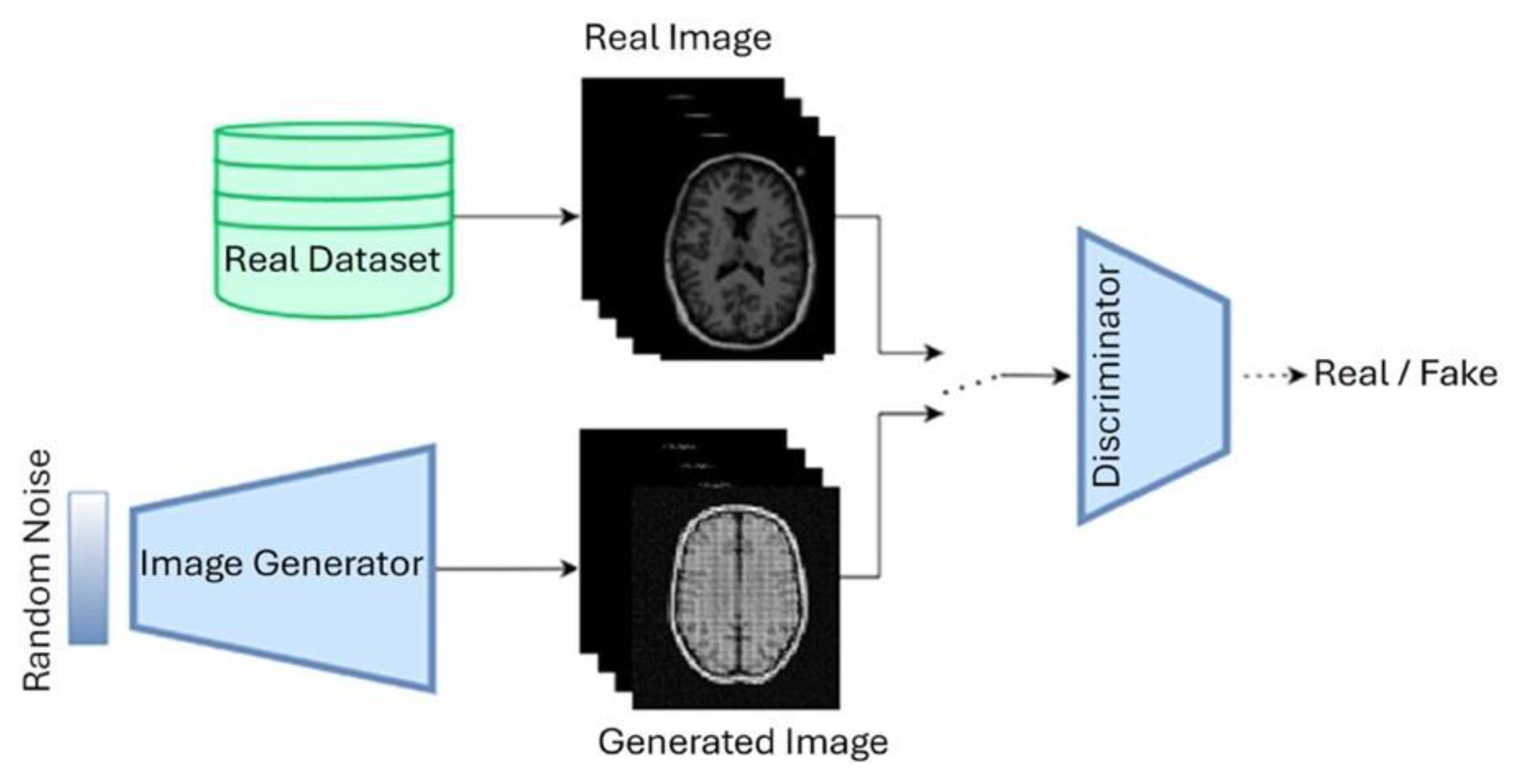
7. Diffusion Models
7.1. Denoising Diffusion Probabilistic Models (DDPM)
7.2. Convolutional Diffusion Models (CDM)
7.3. Latentl Diffusion Models (LDM)
7.4. Score-Based Generative Models (SGMS)
8. Discussion and Conclusion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| GAN | Generative Adversarial Network |
| MRI | Magnetic Resonance Imaging |
| MCI | Mild Cognitive Impairment |
| AD | Alzheimer’s Disease |
References
- World Health Organization, “Ageing and health,” WHO (World Health Organization).
- M. R. Bronzuoli, A. Iacomino, L. Steardo, and C. Scuderi, “Targeting neuroinflammation in Alzheimer’s disease,” J Inflamm Res, vol. Volume 9, pp. 199–208, Nov. 2016. [CrossRef]
- Alzheimer’s Disease International, “Alzheimer’s Disease International,” Alzheimer’s Disease International.
- Alzheimer and Association, “American Perspectives on Early Detection of Alzheimer’s Disease in the Era of Treatment.
- J. Uddin, S. Sultana Shumi, F. Khandoker, and T. Fariha, “Factors Contributing to Alzheimer’s disease in Older Adult Populations: A Narrative Review,” International Journal of Scientific Research in Multidisciplinary Studies, vol. 10, no. 7, pp. 76–82, 2024.
- H. Braak and E. Braak, “Neuropathological stageing of Alzheimer-related changes,” Acta Neuropathol, vol. 82, no. 4, pp. 239–259, Sep. 1991. [CrossRef]
- M. Ávila-Villanueva, A. Marcos Dolado, J. Gómez-Ramírez, and M. Fernández-Blázquez, “Brain Structural and Functional Changes in Cognitive Impairment Due to Alzheimer’s Disease,” Front Psychol, vol. 13, Jun. 2022. [CrossRef]
- C. R. Jack et al., “NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease,” Alzheimer’s & Dementia, vol. 14, no. 4, pp. 535–562, Apr. 2018. [CrossRef]
- J. Goodfellow et al., “Generative Adversarial Networks,” Jun. 2014, [Online]. Available online: http://arxiv.org/abs/1406.2661.
- C. Bowles et al., “GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks,” Oct. 2018, [Online]. Available online: http://arxiv.org/abs/1810.10863.
- Chartsias, T. Joyce, M. V. Giuffrida, and S. A. Tsaftaris, “Multimodal MR Synthesis via Modality-Invariant Latent Representation,” IEEE Trans Med Imaging, vol. 37, no. 3, pp. 803–814, Mar. 2018. [CrossRef]
- You et al., “CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE),” IEEE Trans Med Imaging, vol. 39, no. 1, pp. 188–203, Jan. 2020. [CrossRef]
- J. Ho, A. Jain, and P. Abbeel, “Denoising Diffusion Probabilistic Models,” Jun. 2020, [Online]. Available online: http://arxiv.org/abs/2006.11239.
- F. Khandoker, J. Uddin, T. Fariha, and S. Sultana Shumi, “Importance of Psychological Well-being after disasters in Bangladesh: A Narrative Review,” International Journal of Scientific Research in Multidisciplinary Studies, vol. 10, no. 10, 2024. [CrossRef]
- “Alzheimer’s stages: How the disease progresses,” Mayo Clinic. [Online]. Available online: https://www.mayoclinic.org/diseases-conditions/alzheimers-disease/in-depth/alzheimers-stages/art-20048448.
- M. R. Ahmed, Y. Zhang, Z. Feng, B. Lo, O. T. Inan, and H. Liao, “Neuroimaging and Machine Learning for Dementia Diagnosis: Recent Advancements and Future Prospects,” IEEE Rev Biomed Eng, vol. 12, pp. 19–33, 2019. [CrossRef]
- P. Scheltens, N. Fox, F. Barkhof, and C. De Carli, “Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion,” Lancet Neurol, vol. 1, no. 1, pp. 13–21, May 2002. [CrossRef]
- J. Kim, M. Jeong, W. R. Stiles, and H. S. Choi, “Neuroimaging Modalities in Alzheimer’s Disease: Diagnosis and Clinical Features,” Int J Mol Sci, vol. 23, no. 11, p. 6079, May 2022. [CrossRef]
- B. Yu, Y. Shan, and J. Ding, “A literature review of MRI techniques used to detect amyloid-beta plaques in Alzheimer’s disease patients,” Ann Palliat Med, vol. 10, no. 9, pp. 10062–10074, Sep. 2021. [CrossRef]
- G. S. Alves et al., “Integrating Retrogenesis Theory to Alzheimer’s Disease Pathology: Insight from DTI-TBSS Investigation of the White Matter Microstructural Integrity,” Biomed Res Int, vol. 2015, pp. 1–11, 2015. [CrossRef]
- Raj, A. Kuceyeski, and M. Weiner, “A Network Diffusion Model of Disease Progression in Dementia,” Neuron, vol. 73, no. 6, pp. 1204–1215, Mar. 2012. [CrossRef]
- W. Lee, B. Park, and K. Han, “Classification of diffusion tensor images for the early detection of Alzheimer’s disease,” Comput Biol Med, vol. 43, no. 10, pp. 1313–1320, Oct. 2013. [CrossRef]
- Y. Pan, M. Liu, C. Lian, T. Zhou, Y. Xia, and D. Shen, “Synthesizing missing PET from MRI with cycle-consistent generative adversarial networks for Alzheimer’s disease diagnosis,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Springer Verlag, 2018, pp. 455–463. [CrossRef]
- C. Han et al., “GAN-based Multiple Adjacent Brain MRI Slice Reconstruction for Unsupervised Alzheimer’s Disease Diagnosis,” Jun. 2019, [Online]. Available online: http://arxiv.org/abs/1906.06114.
- H. C. Shin et al., “GANDALF: Generative Adversarial Networks with Discriminator-Adaptive Loss Fine-Tuning for Alzheimer’s Disease Diagnosis from MRI,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Springer Science and Business Media Deutschland GmbH, 2020, pp. 688–697. [CrossRef]
- J. Islam and Y. Zhang, “GAN-based synthetic brain PET image generation,” Brain Inform, vol. 7, no. 1, Dec. 2020. [CrossRef]
- S. Hu, Y. Shen, S. Wang, and B. Lei, “Brain MR to PET Synthesis via Bidirectional Generative Adversarial Network,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Springer Science and Business Media Deutschland GmbH, 2020, pp. 698–707. [CrossRef]
- S. Hu, W. Yu, Z. Chen, and S. Wang, “Medical Image Reconstruction Using Generative Adversarial Network for Alzheimer Disease Assessment with Class-Imbalance Problem,” in 2020 IEEE 6th International Conference on Computer and Communications, ICCC 2020, Institute of Electrical and Electronics Engineers Inc., Dec. 2020, pp. 1323–1327. [CrossRef]
- Y. Zhao, B. Ma, P. Jiang, D. Zeng, X. Wang, and S. Li, “Prediction of Alzheimer’s Disease Progression with Multi-Information Generative Adversarial Network,” IEEE J Biomed Health Inform, vol. 25, no. 3, pp. 711–719, Mar. 2021. [CrossRef]
- C. Han et al., “MADGAN: unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction,” BMC Bioinformatics, vol. 22, Apr. 2021. [CrossRef]
- X. Zhou et al., “Enhancing magnetic resonance imaging-driven Alzheimer’s disease classification performance using generative adversarial learning,” Alzheimers Res Ther, vol. 13, no. 1, Dec. 2021. [CrossRef]
- X. Gao, F. Shi, D. Shen, M. Liu, and T. Alzheimer’s Disease, “Task-Induced Pyramid and Attention GAN for Multimodal Brain Image Imputation and Classification in Alzheimer’s Disease,” IEEE J Biomed Health Inform, vol. 26, no. 1, 2022. [CrossRef]
- W. Yu, B. Lei, M. K. Ng, A. C. Cheung, Y. Shen, and S. Wang, “Tensorizing GAN with High-Order Pooling for Alzheimer’s Disease Assessment,” Aug. 2020, [Online]. Available online: http://arxiv.org/abs/2008.00748.
- J. Pan and S. Wang, “Cross-Modal Transformer GAN: A Brain Structure-Function Deep Fusing Framework for Alzheimer’s Disease,” Jun. 2022, [Online]. Available online: http://arxiv.org/abs/2206.13393.
- N. Rao Thota and D. Vasumathi, “WASSERSTEIN GAN-GRADIENT PENALTY WITH DEEP TRANSFER LEARNING BASED ALZHEIMER DISEASE CLASSIFICATION ON 3D MRI SCANS By.
- J. Zhang, X. He, L. Qing, F. Gao, and B. Wang, “BPGAN: Brain PET synthesis from MRI using generative adversarial network for multi-modal Alzheimer’s disease diagnosis,” Comput Methods Programs Biomed, vol. 217, Apr. 2022. [CrossRef]
- C. Yuan, J. Duan, N. J. Tustison, K. Xu, R. A. Hubbard, and K. A. Linn, “ReMiND: Recovery of Missing Neuroimaging using Diffusion Models with Application to Alzheimer’s Disease,” Aug. 21, 2023. [CrossRef]
- G. Huang, X. Chen, Y. Shen, and S. Wang, “MR Image Super-Resolution Using Wavelet Diffusion for Predicting Alzheimer’s Disease,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Springer Science and Business Media Deutschland GmbH, 2023, pp. 146–157. [CrossRef]
- N. Boyapati et al., “Alzheimer’s Disease Prediction using Convolutional Neural Network (CNN) with Generative Adversarial Network (GAN),” in 2023 International Conference on Data Science, Agents and Artificial Intelligence, ICDSAAI 2023, Institute of Electrical and Electronics Engineers Inc., 2023. [CrossRef]
- H. D. Nguyen, M. Clément, V. Planche, B. Mansencal, and P. Coupé, “Deep grading for MRI-based differential diagnosis of Alzheimer’s disease and Frontotemporal dementia,” Artif Intell Med, vol. 144, Oct. 2023. [CrossRef]
- U. S. Sekhar, N. Vyas, V. Dutt, and A. Kumar, “Multimodal Neuroimaging Data in Early Detection of Alzheimer’s Disease: Exploring the Role of Ensemble Models and GAN Algorithm,” in Proceedings of the International Conference on Circuit Power and Computing Technologies, ICCPCT 2023, Institute of Electrical and Electronics Engineers Inc., 2023, pp. 1664–1669. [CrossRef]
- U. Hwang et al., “Real-world prediction of preclinical Alzheimer’s disease with a deep generative model,” Artif Intell Med, vol. 144, Oct. 2023. [CrossRef]
- W. Yao, Y. Shen, F. Nicolls, and S. Q. Wang, “Conditional Diffusion Model-Based Data Augmentation for Alzheimer’s Prediction,” in Communications in Computer and Information Science, Springer Science and Business Media Deutschland GmbH, 2023, pp. 33–46. [CrossRef]
- D. Yoon et al., “Latent diffusion model-based MRI superresolution enhances mild cognitive impairment prognostication and Alzheimer’s disease classification,” Neuroimage, vol. 296, Aug. 2024. [CrossRef]
- H. Tufail, A. Ahad, I. Puspitasari, I. Shayea, P. J. Coelho, and I. M. Pires, “Deep Learning in Smart Healthcare: A GAN-based Approach for Imbalanced Alzheimer’s Disease Classification,” Procedia Comput Sci, vol. 241, pp. 146–153, 2024. [CrossRef]
- Saleem, M. Alhussein, B. Zohra, K. Aurangzeb, and Q. M. ul Haq, “DeepCGAN: early Alzheimer’s detection with deep convolutional generative adversarial networks,” Front Med (Lausanne), vol. 11, 2024. [CrossRef]
- J. Shah et al., “Enhancing Amyloid PET Quantification: MRI-Guided Super-Resolution Using Latent Diffusion Models,” Life, vol. 14, no. 12, Dec. 2024. [CrossRef]
- K. Chen et al., “A multi-view learning approach with diffusion model to synthesize FDG PET from MRI T1WI for diagnosis of Alzheimer’s disease,” Alzheimer’s and Dementia, Feb. 2024. [CrossRef]
- N. J. Dhinagar, S. I. Thomopoulos, E. Laltoo, and P. M. Thompson, “Counterfactual MRI Generation with Denoising Diffusion Models for Interpretable Alzheimer’s Disease Effect Detection,” in 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, Jul. 2024, pp. 1–6. [CrossRef]
- P. Gajjar et al., “An Empirical Analysis of Diffusion, Autoencoders, and Adversarial Deep Learning Models for Predicting Dementia Using High-Fidelity MRI,” IEEE Access, 2024. [CrossRef]
- P. C. Wong, S. S. Abdullah, and M. I. Shapiai, “Exceptional performance with minimal data using a generative adversarial network for alzheimer’s disease classification,” Sci Rep, vol. 14, no. 1, Dec. 2024. [CrossRef]
- Q. Shou et al., “Diffusion model enables quantitative CBF analysis of Alzheimer’s Disease,” Jul. 03, 2024. [CrossRef]
- S. Hwang and J. Shin, “Prognosis Prediction of Alzheimer’s Disease Based on Multi-Modal Diffusion Model,” in Proceedings of the 2024 18th International Conference on Ubiquitous Information Management and Communication, IMCOM 2024, Institute of Electrical and Electronics Engineers Inc., 2024. [CrossRef]
- W. P. Ching, S. S. Abdullah, M. I. Shapiai, and A. K. M. Muzahidul Islam, “Performance Enhancement of Alzheimer’s Disease Diagnosis Using Generative Adversarial Network,” Journal of Advanced Research in Applied Sciences and Engineering Technology, vol. 45, no. 2, pp. 191–201, Mar. 2025. [CrossRef]
- Y. Zhang and L. Wang, “Early diagnosis of Alzheimer’s disease using dual GAN model with pyramid attention networks,” 2024, Taylor and Francis Ltd. [CrossRef]
- Z. Ou, C. Jiang, Y. Pan, Y. Zhang, Z. Cui, and D. Shen, “A Prior-information-guided Residual Diffusion Model for Multi-modal PET Synthesis from MRI,” 2024.
- S. Oraby, A. Emran, B. El-Saghir, and S. Mohsen, “Hybrid of DSR-GAN and CNN for Alzheimer disease detection based on MRI images,” Sci Rep, vol. 15, no. 1, Dec. 2025. [CrossRef]
- T. Zhou et al., “BG-GAN: Generative AI Enable Representing Brain Structure-Function Connections for Alzheimer’s Disease,” IEEE Transactions on Consumer Electronics, 2025. [CrossRef]
- G. Gavidia-Bovadilla, S. Kanaan-Izquierdo, M. Mataroa-Serrat, and A. Perera-Lluna, “Early prediction of Alzheimer’s disease using null longitudinal model-based classifiers,” PLoS One, vol. 12, no. 1, Jan. 2017. [CrossRef]
- M. Liu, J. Zhang, E. Adeli, and D. Shen, “Landmark-based deep multi-instance learning for brain disease diagnosis,” Med Image Anal, vol. 43, pp. 157–168, Jan. 2018. [CrossRef]
- Radford, L. Metz, and S. Chintala, “Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks,” Nov. 2015, [Online]. Available online: http://arxiv.org/abs/1511.06434.
- M. Mirza and S. Osindero, “Conditional Generative Adversarial Nets,” Nov. 2014, [Online]. Available online: http://arxiv.org/abs/1411.1784.
- S. Kazeminia et al., “GANs for medical image analysis,” Artif Intell Med, vol. 109, p. 101938, Sep. 2020. [CrossRef]
- T. Karras, S. Laine, and T. Aila, “A Style-Based Generator Architecture for Generative Adversarial Networks,” in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, Jun. 2019, pp. 4396–4405. [CrossRef]
- M. Arjovsky, S. Chintala, and L. Bottou, “Wasserstein Generative Adversarial Networks,” 2017.
- Y. Song and S. Ermon, “Generative Modeling by Estimating Gradients of the Data Distribution,” Jul. 2019, [Online]. Available online: http://arxiv.org/abs/1907.05600.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





