Submitted:
30 June 2025
Posted:
02 July 2025
You are already at the latest version
Abstract
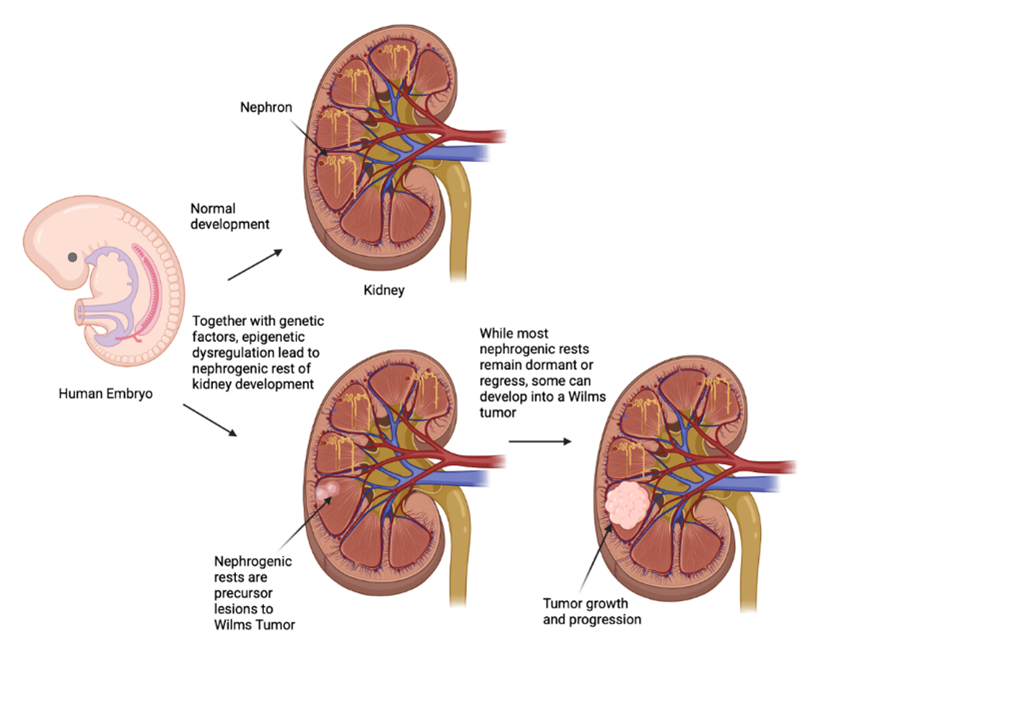
Keywords:
Introduction
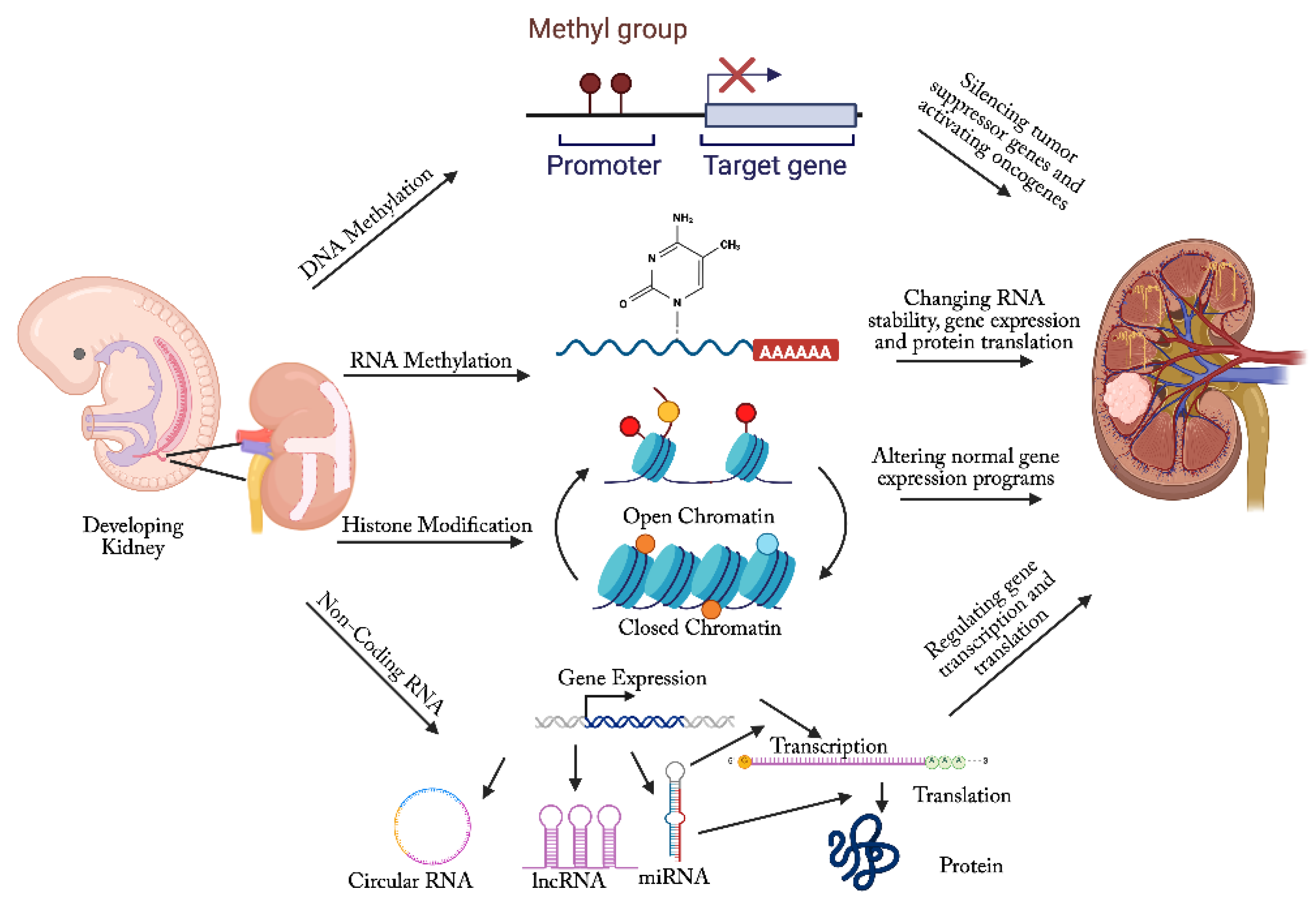
Epigenetic Alterations in Wilms Tumor
DNA methylation in Wilms Tumor
RNA Methylation in Wilms Tumor
Histone Modifications in Wilms Tumor
Non-Coding RNA
Therapeutic Implications of Epigenetic Regulation in WT
Conclusions and Perspective
Acknowledgments
References
- Neagu, M.C. , et al., Wilms' Tumor: A Review of Clinical Characteristics, Treatment Advances, and Research Opportunities. Medicina (Kaunas) 2025, 61. [Google Scholar]
- Hohenstein, P., K. Pritchard-Jones, and J. Charlton, The yin and yang of kidney development and Wilms' tumors. Genes Dev 2015, 29, 467–82. [Google Scholar] [CrossRef] [PubMed]
- Wilms Tumor and Other Childhood Kidney Tumors Treatment (PDQ(R)): Health Professional Version, in PDQ Cancer Information Summaries. 2002: Bethesda (MD).
- Jia, C. , et al., Identification of the expression patterns and potential prognostic role of m6A-RNA methylation regulators in Wilms Tumor. BMC Med Genomics 2023, 16, 222. [Google Scholar] [CrossRef] [PubMed]
- Faria, P. , et al., Focal versus diffuse anaplasia in Wilms tumor--new definitions with prognostic significance: a report from the National Wilms Tumor Study Group. Am J Surg Pathol 1996, 20, 909–20. [Google Scholar] [CrossRef] [PubMed]
- Aiden, A.P. , et al., Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell 2010, 6, 591–602. [Google Scholar] [CrossRef]
- Treger, T.D. , et al., The genetic changes of Wilms tumour. Nat Rev Nephrol 2019, 15, 240–251. [Google Scholar] [CrossRef]
- Young, M.D. , et al., Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 2018, 361, 594–599. [Google Scholar] [CrossRef]
- Huff, V. , Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 2011, 11, 111–21. [Google Scholar] [CrossRef]
- Ruteshouser, E.C., S. M. Robinson, and V. Huff, Wilms tumor genetics: mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 2008, 47, 461–70. [Google Scholar] [CrossRef]
- Scott, R.H. , et al., Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 2012, 3, 327–35. [Google Scholar] [CrossRef]
- Fiala, E.M. , et al., 11p15.5 epimutations in children with Wilms tumor and hepatoblastoma detected in peripheral blood. Cancer 2020, 126, 3114–3121. [Google Scholar] [CrossRef]
- Fiala, E.M. , et al., 11p15.5 epimutations in children with Wilms tumor and hepatoblastoma detected in peripheral blood. Cancer 2020, 126, 3114–3121. [Google Scholar] [CrossRef]
- Filbin, M. and M. Monje, Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat Med 2019, 25, 367–376. [Google Scholar] [CrossRef]
- Murphy, A.J. , et al., Genetic and epigenetic features of bilateral Wilms tumor predisposition in patients from the Children's Oncology Group AREN18B5-Q. Nat Commun 2023, 14, 8006. [Google Scholar] [CrossRef] [PubMed]
- Wegert, J. , et al., Distinct pathways for genetic and epigenetic predisposition in familial and bilateral Wilms tumor. Genome Med 2025, 17, 49. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C. and D. Bourc'his, The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Sonar, S. , et al., Role of DNA methylation in cancer development and its clinical applications. Clinical and Translational Discovery 2024, 4. [Google Scholar] [CrossRef]
- Guerra, J.V.D. , et al., Genes Controlled by DNA Methylation Are Involved in Wilms Tumor Progression. Cells 2019, 8. [Google Scholar]
- Anvar, Z. , et al., Origins of DNA methylation defects in Wilms tumors. Cancer Letters 2019, 457, 119–128. [Google Scholar] [CrossRef]
- Tang, F. , et al., DNA Methylation Data-Based Classification and Identification of Prognostic Signature of Children With Wilms Tumor. Front Cell Dev Biol 2021, 9, 683242. [Google Scholar] [CrossRef]
- Albasha, S.A. , et al., In Silico Analysis of DNA Methylation Status of T-Cell Inflamed Signature Reveals a Therapeutic Strategy to Improve Immune Checkpoint Inhibitors Efficacy in Wilms Tumor. Journal of Pharmacology and Experimental Therapeutics 2023, 385. [Google Scholar] [CrossRef]
- Guerra, J. , et al., Genes Controlled by DNA Methylation Are Involved in Wilms Tumor Progression. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.W. , et al., Insights into the mA demethylases FTO and ALKBH5: structural, biological function, and inhibitor development. Cell and Bioscience 2024, 14. [Google Scholar] [CrossRef]
- Wang, Z.T. , et al. , RNA m<SUP>6</SUP>A methylation in cancer. Molecular Oncology 2023, 17, 195–229. [Google Scholar]
- Sun, T., R. Y. Wu, and L. Ming, The role of m6A RNA methylation in cancer. Biomedicine & Pharmacotherapy 2019, 112. [Google Scholar]
- Zhuo, Y. , et al., Identification of m6A-associated genes as prognostic and immune-associated biomarkers in Wilms tumor. Discov Oncol 2023, 14, 201. [Google Scholar] [CrossRef]
- Huang, Q. , et al., The RNA m(6)A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis 2022, 13, 852. [Google Scholar] [CrossRef]
- Du, Y.M. , et al., WTAP-mediated abnormal m6A modification promotes cancer progression by remodeling the tumor microenvironment: bibliometric and database analyses. Translational Cancer Research 2024, 13. [Google Scholar] [CrossRef]
- You, L.L. , et al., The role of N6-methyladenosine (m>A) in kidney diseases. Frontiers in Medicine. 2023, 10. [Google Scholar]
- Chen, X.H. , et al., Regulations of m(6)A and other RNA modifications and their roles in cancer. Front Med 2024, 18, 622–648. [Google Scholar] [CrossRef]
- Vujanic, G.M. and W. Mifsud, Anaplasia in Wilms tumor: A critical review. Pediatr Blood Cancer 2024, 71, e31000. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y., M. Zhang, and Y. Wang, The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J Natl Cancer Cent 2022, 2, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F. and Q. Yan, The roles of epigenetics in cancer progression and metastasis. Biochem J 2021, 478, 3373–3393. [Google Scholar] [CrossRef] [PubMed]
- Liu, R. , et al., Molecular Mechanisms of MYCN Dysregulation in Cancers. Front Oncol 2020, 10, 625332. [Google Scholar] [CrossRef]
- Jimenez Martin, O. , et al., MYCN and MAX alterations in Wilms tumor and identification of novel N-MYC interaction partners as biomarker candidates. Cancer Cell Int 2021, 21, 555. [Google Scholar] [CrossRef]
- Ruiz-Perez, M.V. B. Henley, and M. Arsenian-Henriksson, The MYCN Protein in Health and Disease. Genes (Basel) 2017, 8. [Google Scholar] [CrossRef]
- Chen, S. , et al., Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J Biol Chem 2011, 286, 32775–89. [Google Scholar] [CrossRef]
- Liu, H. , The roles of histone deacetylases in kidney development and disease. Clin Exp Nephrol 2021, 25, 215–223. [Google Scholar] [CrossRef]
- Liu, H. , et al., Histone deacetylases 1 and 2 regulate the transcriptional programs of nephron progenitors and renal vesicles. Development 2018, 145. [Google Scholar] [CrossRef]
- Liu, H. , et al., Histone deacetylases 1 and 2 target gene regulatory networks of nephron progenitors to control nephrogenesis. Biochem Pharmacol 2022, 206, 115341. [Google Scholar] [CrossRef]
- Cao, X. , et al., Histone deacetylase 5 promotes Wilms' tumor cell proliferation through the upregulation of c-Met. Mol Med Rep 2016, 13, 2745–50. [Google Scholar] [CrossRef]
- Chen, C.H. , et al., Overactivation of histone deacetylases and EZH2 in Wilms tumorigenesis. Genes Dis 2023, 10, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Ma, G. , et al., Modeling high-risk Wilms tumors enables the discovery of therapeutic vulnerability. Cell Rep Med 2024, 101770. [Google Scholar] [CrossRef]
- Su, X. , et al., Delineating the interplay between oncogenic pathways and immunity in anaplastic Wilms tumors. Nat Commun 2023, 14, 7884. [Google Scholar] [CrossRef] [PubMed]
- Makki, M.S., T. Heinzel, and C. Englert, TSA downregulates Wilms tumor gene 1 (Wt1) expression at multiple levels. Nucleic Acids Res 2008, 36, 4067–78. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y. , et al., Histone acetyltransferase p300 promotes the activation of human WT1 promoter and intronic enhancer. Arch Biochem Biophys 2005, 436, 62–8. [Google Scholar] [CrossRef]
- Wang, W. , et al., A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J Biol Chem 2001, 276, 16810–6. [Google Scholar] [CrossRef]
- Arroyo-Parejo Drayer, P. , et al., Spectrum of Clinical Manifestations in Children With WT1 Mutation: Case Series and Literature Review. Front Pediatr 2022, 10, 847295. [Google Scholar] [CrossRef]
- Lee, S.C. , et al., Essential role of insulin-like growth factor 2 in resistance to histone deacetylase inhibitors. Oncogene 2016, 35, 5515–5526. [Google Scholar] [CrossRef]
- Liu, H. , et al., The polycomb proteins EZH1 and EZH2 co-regulate chromatin accessibility and nephron progenitor cell lifespan in mice. J Biol Chem 2020, 295, 11542–11558. [Google Scholar] [CrossRef]
- Straining, R. and W. Eighmy, Tazemetostat: EZH2 Inhibitor. J Adv Pract Oncol 2022, 13, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Cai, L. , et al., Bioinformatical analysis of the key differentially expressed genes for screening potential biomarkers in Wilms tumor. Sci Rep 2023, 13, 15404. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J. , et al., Comparative methylome analysis identifies new tumour subtypes and biomarkers for transformation of nephrogenic rests into Wilms tumour. Genome Med 2015, 7, 11. [Google Scholar] [CrossRef]
- Epp, S., S. M. Chuah, and M. Halasz, Epigenetic Dysregulation in -Amplified Neuroblastoma. International Journal of Molecular Sciences 2023, 24. [Google Scholar] [CrossRef]
- Maschietto, M. , et al., The IGF signalling pathway in Wilms tumours - A report from the ENCCA Renal Tumours Biology-driven drug development workshop. Oncotarget 2014, 5, 8014–8026. [Google Scholar] [CrossRef]
- Kazanets, A. , et al., Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochimica Et Biophysica Acta-Reviews on Cancer 2016, 1865, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Waehle, V. , et al., The tumor suppressor WT1 drives progenitor cell progression and epithelialization to prevent Wilms tumorigenesis in human kidney organoids. Stem Cell Reports 2021, 16, 2107–2117. [Google Scholar] [CrossRef]
- Chan, K. and X. Li, Current Epigenetic Insights in Kidney Development. Genes (Basel) 2021, 12. [Google Scholar] [CrossRef]
- Hilliard, S.A. and S.S. El-Dahr, Epigenetics mechanisms in renal development. Pediatr Nephrol 2016, 31, 1055–60. [Google Scholar] [CrossRef]
- Slack, F.J. and A.M. Chinnaiyan, The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Alfaifi, J. , miRNAs Role in Wilms tumor pathogenesis: Signaling pathways interplay. Pathol Res Pract 2024, 256, 155254. [Google Scholar] [CrossRef] [PubMed]
- Bekes, M., D. R. Langley, and C.M. Crews, PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Perez-Linares, F.J. , et al., MicroRNA Profiling in Wilms Tumor: Identification of Potential Biomarkers. Front Pediatr 2020, 8, 337. [Google Scholar] [CrossRef] [PubMed]
- Yu, X. , et al., The roles of microRNAs in Wilms' tumors. Tumour Biol 2016, 37, 1445–50. [Google Scholar] [CrossRef]
- Orgen Calli, A. , et al., The association of miR-204 and mir-483 5p expression with clinicopathological features of Wilms tumor: Could this provide foresight? Jpn J Clin Oncol 2023, 53, 1170–1176. [Google Scholar] [CrossRef]
- Urbach, A. , et al., Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 2014, 28, 971–82. [Google Scholar] [CrossRef] [PubMed]
- Benlhachemi, S. , et al., Circulating microRNA profiles in Wilms tumour (WT): A systematic review and meta-analysis of diagnostic test accuracy. Noncoding RNA Res 2023, 8, 413–425. [Google Scholar] [CrossRef]
- Mohamed, F.S. , et al., Profiling of the serum MiRNAome in pediatric egyptian patients with wilms tumor. Front Mol Biosci 2024, 11, 1453562. [Google Scholar] [CrossRef]
- Ludwig, N. , et al., Circulating serum miRNAs as potential biomarkers for nephroblastoma. Pediatr Blood Cancer 2015, 62, 1360–7. [Google Scholar] [CrossRef]
- Andersen, G.B. and J. Tost, Circulating miRNAs as Biomarker in Cancer. Recent Results Cancer Res 2020, 215, 277–298. [Google Scholar]
- Liu, Q. , The Emerging Landscape of Long Non-Coding RNAs in Wilms Tumor. Frontiers in Oncology 2022, 11. [Google Scholar] [CrossRef]
- Liu, F. , et al., Roles of lncRNAs in childhood cancer: Current landscape and future perspectives. Front Oncol 2023, 13, 1060107. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.B. , et al., Investigating the dysfunctional pathogenesis of Wilms' tumor through a multidimensional integration strategy. Annals of Translational Medicine 2019, 7. [Google Scholar]
- Zhou, H., L. N. Sun, and F.S. Wan, Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncology Letters 2019, 18, 4393–4402. [Google Scholar] [PubMed]
- Sharma, D. , et al., Exosomal long non-coding RNA MALAT1: a candidate of liquid biopsy in monitoring of Wilms' tumor. Pediatr Surg Int 2024, 40, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R. , et al., CircCDYL Acts as a Tumor Suppressor in Wilms' Tumor by Targeting miR-145-5p. Front Cell Dev Biol 2021, 9, 668947. [Google Scholar]
- Cao, J. , et al., circ0093740 Promotes Tumor Growth and Metastasis by Sponging miR-136/145 and Upregulating DNMT3A in Wilms Tumor. Front Oncol 2021, 11, 647352. [Google Scholar] [CrossRef]
- Tian, X.M. , et al., The Regulatory Network and Role of the circRNA-miRNA-mRNA ceRNA Network in the Progression and the Immune Response of Wilms Tumor Based on RNA-Seq. Frontiers in Genetics 2022, 13. [Google Scholar] [CrossRef]
- Bhutani, N., P. Kajal, and U. Sharma, Many faces of Wilms Tumor: Recent advances and future directions. Ann Med Surg (Lond) 2021, 64, 102202. [Google Scholar]
- Perlman, E.J. , Pediatric renal tumors: practical updates for the pathologist. Pediatr Dev Pathol 2005, 8, 320–38. [Google Scholar] [CrossRef]
- Gnyszka, A., Z. Jastrzebski, and S. Flis, DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res 2013, 33, 2989–96. [Google Scholar] [PubMed]
- Hu, C. , et al., DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin Epigenetics 2021, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Brok, J. , et al., Unmet needs for relapsed or refractory Wilms tumour: Mapping the molecular features, exploring organoids and designing early phase trials - A collaborative SIOPRTSG, COG and ITCC session at the first SIOPE meeting. European Journal of Cancer 2021, 144, 113–122. [Google Scholar] [CrossRef] [PubMed]
| Role | Details | Reference(s) |
| Diagnostic Biomarker | EZH2 overexpression is common in WT compared to normal kidney tissue. It can help distinguish tumor tissue from normal or benign kidney lesions in some cases. | [43] |
| Prognostic Biomarker | High levels of EZH2 correlate with poor prognosis in many cancers. In WT, higher EZH2 expression has been associated with worse differentiation and potentially more aggressive tumor behavior, but this link needs more clinical validation. | [53] |
| Predictive Biomarker | EZH2 expression could predict response to EZH2 inhibitors in the future, though this is still at an experimental stage for WT. | [1,53] |
| Therapeutic Target | EZH2 is a marker and an active target: Inhibiting it might reverse the block in differentiation that drives WT growth. | [43] |
| Name |
Target(s) | Mechanism | Potential Relevance in Wilms Tumor | Development Status |
| Panobinostat | HDAC (Class I, II, IV) | Broad-spectrum HDAC inhibitor; induces apoptosis, cell cycle arrest | May target aberrant epigenetics in Wilms tumor; preclinical studies needed | FDA-approved (myeloma) |
| Vorinostat (SAHA) | HDAC (Class I, II) | Promotes histone acetylation, reactivates tumor suppressor genes | Potential for differentiation therapy in Wilms tumor | FDA-approved (myeloma) |
| Romidepsin | HDAC (Class I) | Selective HDAC1/2 inhibition; disrupts oncogenic pathways | Possible synergy with chemotherapy in pediatric tumors | FDA-approved (myeloma) |
| Tazemetostat (EPZ-6438) | EZH2 (H3K27me3 methyltransferase) | Blocks PRC2-mediated silencing of tumor suppressors | EZH2 overexpression linked to poor prognosis; may inhibit Wilms tumor progression | FDA-approved (epithelioid sarcoma) |
| GSK126 | EZH2 inhibitor | Reduces H3K27me3 marks, reactivates silenced genes | Preclinical potential for Wilms tumors with EZH2 dysregulation | Investigational |
| JQ1 | BET (BRD4) inhibitor | Targets bromodomains; suppresses MYC and other oncogenes | MYC is implicated in Wilms tumor; may disrupt oncogenic transcription | Preclinical |
| CPI-203 | BET inhibitor | Downregulates pro-proliferative genes | Potential for high-risk Wilms tumor subtypes | Investigational |
| Valproic Acid | HDAC (Class I, IIa) | Weak HDAC inhibitor; induces differentiation | Possible adjunct therapy due to low toxicity in pediatrics | FDA-approved (epilepsy) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





