Submitted:
31 May 2025
Posted:
02 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Selection and Structural Profiling of Antiviral Peptides (AVPs)
2.2. Molecular Docking of Antiviral Peptides (AVPs) to the HR2 Target Site
2.3. Structural Dynamics and MM/PBSA-Based Evaluation of Antiviral Peptide (AVP)–HR2 Complexes
2.4. Predicted Hemolytic Activity Profiles of Antiviral Peptides (AVPs)
2.5. Predicted Physicochemical Properties of Antiviral Peptides (AVPs)
3. Discussion
4. Limitations, Clinical Implications, and Future Works
5. Materials and Methods
5.1. Selection and Preparation of Antiviral Peptides (AVPs)
5.2. Peptide-Protein Docking Simulation
5.3. Molecular Dynamics (MD) Simulation
5.4. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
5.5. Haemolytic Activity Prediction of Antiviral Peptides (AVPs)
5.6. Physicochemical Characterization of Antiviral Peptides (AVPs)
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 6-HB | Six-helix bundle |
| APD3 | Antimicrobial peptide database 3 |
| CASTpFold | Computed atlas of surface topography of the universe of protein folds |
| DPP4 | Dipeptidyl peptidase 4 |
| GRAVY | Grand average of hydropathicity |
| HADDOCK | High ambiguity driven protein-protein docking |
| HD5 | Human defensin 5 |
| HR1 | Heptad repeat 1 |
| HR2 | Heptad repeat 2 |
| ICs | Intermolecular contacts |
| MD | Molecular dynamics |
| MERCI | Motif-emerging and with classes-identification |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| MM/PBSA | Molecular mechanics/Poisson-Boltzmann surface area |
| ML | Machine learning |
| NIS | Non-interacting surface areas |
| NPT | Number of particles, pressure, and temperature |
| NVT | Number of particles, volume, and temperature |
| OPLS-AA/L | Optimized potentials for liquid simulations |
| pI | Isoelectric point |
| pLDDT | Predicted local distance difference test |
| PME | Particle mesh Ewald |
| PRODIGY | PROtein binDIng enerGY prediction |
| RBD | Receptor-binding domain |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| RoG | Radius of gyration |
| SPCE | Single point charge extended |
References
- Fehr, A.R., R. Channappanavar, and S. Perlman, Middle East Respiratory Syndrome: Emergence of a Pathogenic Human Coronavirus. Annu Rev Med, 2017. 68: p. 387-399.
- Al-Raddadi, R.M. , et al., Burden of Middle East respiratory syndrome coronavirus infection in Saudi Arabia. Journal of Infection and Public Health, 2020. 13(5): p. 692-696.
- Wang, Q. , et al., MERS-CoV spike protein: Targets for vaccines and therapeutics. Antiviral Res, 2016. 133: p. 165-77.
- Goyal, R. , et al., Comparative highlights on MERS-CoV, SARS-CoV-1, SARS-CoV-2, and NEO-CoV. Excli j, 2022. 21: p. 1245-1272.
- Li, Y.H. , et al., Molecular Characteristics, Functions, and Related Pathogenicity of MERS-CoV Proteins. Engineering (Beijing), 2019. 5(5): p. 940-947.
- Abdelrahman, Z., M. Li, and X. Wang, Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses. Front Immunol, 2020. 11: p. 552909.
- Bassendine, M.F. , et al., COVID-19 and comorbidities: A role for dipeptidyl peptidase 4 (DPP4) in disease severity? Journal of Diabetes, 2020. 12(9): p. 649-658.
- Yuan, H.-W. and H.-L. Wen, Research progress on coronavirus S proteins and their receptors. Archives of Virology, 2021. 166(7): p. 1811-1817.
- Forni, D. , et al., The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Scientific Reports, 2015. 5(1): p. 14480.
- Xu, Y. , et al., Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem, 2004. 279(47): p. 49414-9.
- Wang, C. , et al., Discovery of Hydrocarbon-Stapled Short α-Helical Peptides as Promising Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Fusion Inhibitors. Journal of Medicinal Chemistry, 2018. 61(5): p. 2018-2026.
- Lu, L. , et al., Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun, 2014. 5: p. 3067.
- Xia, S. , et al., Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res, 2014. 194: p. 200-10.
- Xia, S. , et al., Potent MERS-CoV Fusion Inhibitory Peptides Identified from HR2 Domain in Spike Protein of Bat Coronavirus HKU4. Viruses, 2019. 11(1).
- Du, L. , et al., MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets, 2017. 21(2): p. 131-143.
- Safiriyu, A.A. , et al., The fusion peptide of the spike protein S2 domain may be a mimetic analog of β-coronaviruses and serve as a novel virus-host membrane fusion inhibitor. Antiviral Research, 2025. 237: p. 106144.
- Alotaiq, N., D. Dermawan, and N.E. Elwali, Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations. International Journal of Molecular Sciences, 2024. 25(19): p. 10818.
- Dermawan, D. and N. Alotaiq, Computational analysis of antimicrobial peptides targeting key receptors in infection-related cardiovascular diseases: molecular docking and dynamics insights. Scientific Reports, 2025. 15(1): p. 8896.
- Alotaiq, N. and D. Dermawan Evaluation of Structure Prediction and Molecular Docking Tools for Therapeutic Peptides in Clinical Use and Trials Targeting Coronary Artery Disease. International Journal of Molecular Sciences, 2025. 26. [CrossRef]
- Doni Dermawan, F.A. , Nasr Eldin Elwali, Nasser Alotaiq, Therapeutic potential of earthworm-derived proteins: Targeting NEDD4 for cardiovascular disease intervention. Vol. Volume: 15. 2024: Issue: 1. 216-232.
- Rosson, E. , et al., Focus on therapeutic peptides and their delivery. International Journal of Pharmaceutics, 2025. 675: p. 125555.
- Xiao, W. , et al., Advance in peptide-based drug development: delivery platforms, therapeutics and vaccines. Signal Transduction and Targeted Therapy, 2025. 10(1): p. 74.
- Ricardo, M.G. , et al., Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions. Angew Chem Int Ed Engl, 2020. 59(13): p. 5235-5241.
- Yang, S. and P. Xu, HemoDL: Hemolytic peptides prediction by double ensemble engines from Rich sequence-derived and transformer-enhanced information. Analytical Biochemistry, 2024. 690: p. 115523.
- Xia, S. , et al., A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Science Advances, 2019. 5: p. eaav4580.
- Xia, S. , et al., A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Science Advances. 5(4): p. eaav4580.
- Santopolo, S., A. Riccio, and M.G. Santoro, The biogenesis of SARS-CoV-2 spike glycoprotein: multiple targets for host-directed antiviral therapy. Biochemical and Biophysical Research Communications, 2021. 538: p. 80-87.
- Zhu, Y. , et al., Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike. 2021.
- Channappanavar, R. , et al., Protective Effect of Intranasal Regimens Containing Peptidic Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor Against MERS-CoV Infection. J Infect Dis, 2015. 212(12): p. 1894-903.
- Lee, C. , Griffithsin, a Highly Potent Broad-Spectrum Antiviral Lectin from Red Algae: From Discovery to Clinical Application. Mar Drugs, 2019. 17(10).
- Bains, A., et al. The Antiviral Activity of the Lectin Griffithsin against SARS-CoV-2 Is Enhanced by the Presence of Structural Proteins. Viruses, 2023. 15. [CrossRef]
- Decker, J., R. Menacho Melgar, and M. Lynch, Low-Cost, Large-Scale Production of the Anti-viral Lectin Griffithsin. Frontiers in Bioengineering and Biotechnology, 2020. 8: p. 1020.
- Ahan, R.E. , et al., A Highly Potent SARS-CoV-2 Blocking Lectin Protein. ACS Infectious Diseases, 2022. 8(7): p. 1253-1264.
- Xiong, W. , et al., Brevinin-2GHk, a Peptide Derived from the Skin of Fejervarya limnocharis, Inhibits Zika Virus Infection by Disrupting Viral Integrity. Viruses, 2021. 13(12).
- Loffredo, M.R. , et al., Antimicrobial peptides for novel antiviral strategies in the current post-COVID-19 pandemic. J Pept Sci, 2024. 30(1): p. e3534.
- Kallal, L.E. , et al., CCL20/CCR6 blockade enhances immunity to RSV by impairing recruitment of DC. Eur J Immunol, 2010. 40(4): p. 1042-52.
- Bayat, M. , et al., An overview of some potential immunotherapeutic options against COVID-19. Int Immunopharmacol, 2021. 95: p. 107516.
- Wang, G., X. Li, and Z. Wang, APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res, 2016. 44(D1): p. D1087-93.
- Šali, A. and T.L. Blundell, Comparative Protein Modelling by Satisfaction of Spatial Restraints. Journal of Molecular Biology, 1993. 234(3): p. 779-815.
- Jumper, J. , et al., Highly accurate protein structure prediction with AlphaFold. Nature, 2021. 596(7873): p. 583-589.
- Science-Gateway. Protein Molecular Weight Calculator. 2025 [cited 2025 10th February]; Available from: https://www.sciencegateway.org/tools/proteinmw.htm.
- Ye, B. , et al., CASTpFold: Computed Atlas of Surface Topography of the universe of protein Folds. Nucleic Acids Research, 2024. 52(W1): p. W194-W199.
- Kandeel, M. , et al., Discovery of New Potent anti-MERS CoV Fusion Inhibitors. Front Pharmacol, 2021. 12: p. 685161.
- Laskowski, R.A. , et al., PDBsum: Structural summaries of PDB entries. Protein Sci, 2018. 27(1): p. 129-134.
- Johansson, M.U. , et al., Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics, 2012. 13(173): p. 1-10.
- Dominguez, C., R. Boelens, and A.M.J.J. Bonvin, HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. Journal of the American Chemical Society, 2003. 125(7): p. 1731-1737.
- van Zundert, G.C.P. , et al., The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol, 2016. 428(4): p. 720-725.
- Vangone, A. and A. Bonvin, PRODIGY: A Contact-based Predictor of Binding Affinity in Protein-protein Complexes. BIO-PROTOCOL, 2017. 7.
- Pronk, S. , et al., GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 2013. 29(7): p. 845-854.
- Robertson, M.J., J. Tirado-Rives, and W.L. Jorgensen, Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J Chem Theory Comput, 2015. 11(7): p. 3499-509.
- Yuet, P. and D. Blankschtein, Molecular Dynamics Simulation Study of Water Surfaces: Comparison of Flexible Water Models. The journal of physical chemistry. B, 2010. 114: p. 13786-13795.
- Schrödinger, The PyMOL Molecular Graphics System. 2020.
- Pettersen, E.F. , et al., UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 2004. 25(13): p. 1605-1612.
- Tian, S. , et al., Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model, 2014. 54(10): p. 2664-2679.
- Valdés-Tresanco, M.S. , et al., gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. Journal of Chemical Theory and Computation, 2021. 17(10): p. 6281-6291.
- Miller, B.R. , 3rd, et al., MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. Journal of chemical theory and computation, 2012. 8(9): p. 3314-3321.
- Panday, S.K. and E. Alexov, Protein-Protein Binding Free Energy Predictions with the MM/PBSA Approach Complemented with the Gaussian-Based Method for Entropy Estimation. ACS Omega, 2022. 7(13): p. 11057-11067.
- Rathore, A.S. , et al., Prediction of hemolytic peptides and their hemolytic concentration. Communications Biology, 2025. 8(1): p. 176.
- Gasteiger, E. , et al., Protein Identification and Analysis Tools on the ExPASy Server, in The Proteomics Protocols Handbook, J.M. Walker, Editor. 2005, Humana Press: Totowa, NJ. p. 571-607.
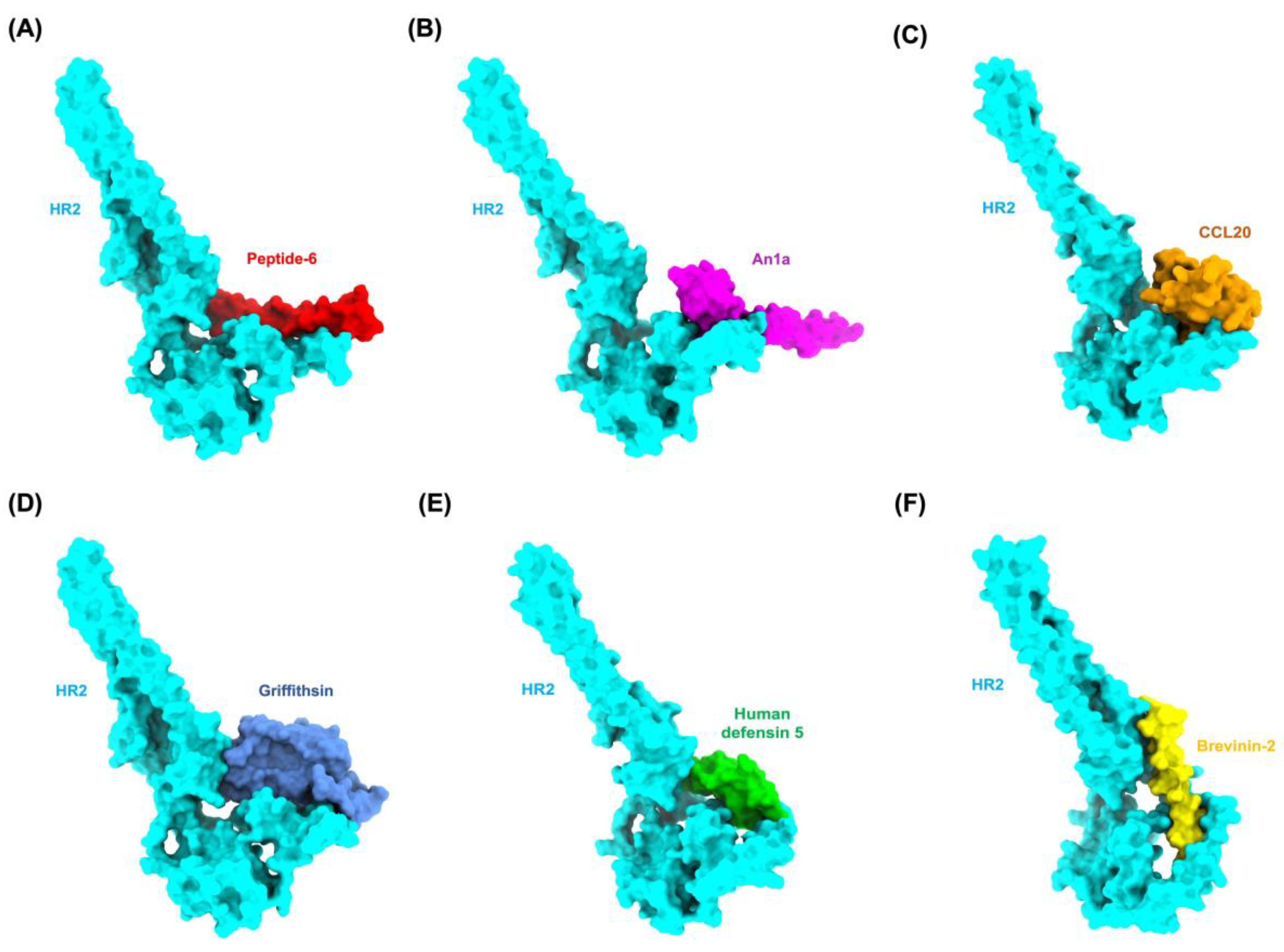
| Anviral Peptide | APD ID | SwissProt ID/ PDB ID | Size (kDa) | Average pLDDT | Peptide Binding Sites (Position of Residues) |
|---|---|---|---|---|---|
| An1a | AP03266 | A0A5Q1NCA8 | 6.93 | 83.62 | 20, 21, 23, 24, 25, 27, 28, 30, 55 |
| Brevinin-2 | AP00599 | P32424 | 2.32 | 81.49 | 1, 2, 12, 13, 21 ,22 |
| CCL20 | AP02075 | 1M8A | 7.96 | N/A | 5, 9, 11, 14, 15, 16, 17, 20, 25, 29, 37, 40, 46, 48, 51, 55, 63 |
| Griffithsin | AP02133 | 2GTY | 12.69 | N/A | 4, 6, 12, 16, 17, 18, 26, 27, 28, 35, 56, 57, 58 |
| Human defensin 5 | AP00180 | 1ZMP | 3.59 | N/A | 3, 6, 7, 8, 9, 15, 31 |
| Lactoferricin B | AP00026 | 1LFC | 3.13 | N/A | 11, 12, 13, 15 |
| Neutrophil cationic peptide 1 type B | AP00174 | Q64365 | 3.84 | 63.00 | 1, 2, 3, 6, 7, 13, 14, 17, 18 |
| Piscidin 2 | AP01649 | Q8UUG2 | 9.11 | 74.76 | 14, 20, 23 |
| Shepherin II | AP00512 | Q9FR52 | 3.26 | 81.04 | 7, 9 |
| Varv peptide E | AP01030 | P83835 | 2.92 | 90.55 | 20, 27, 28, 29 |
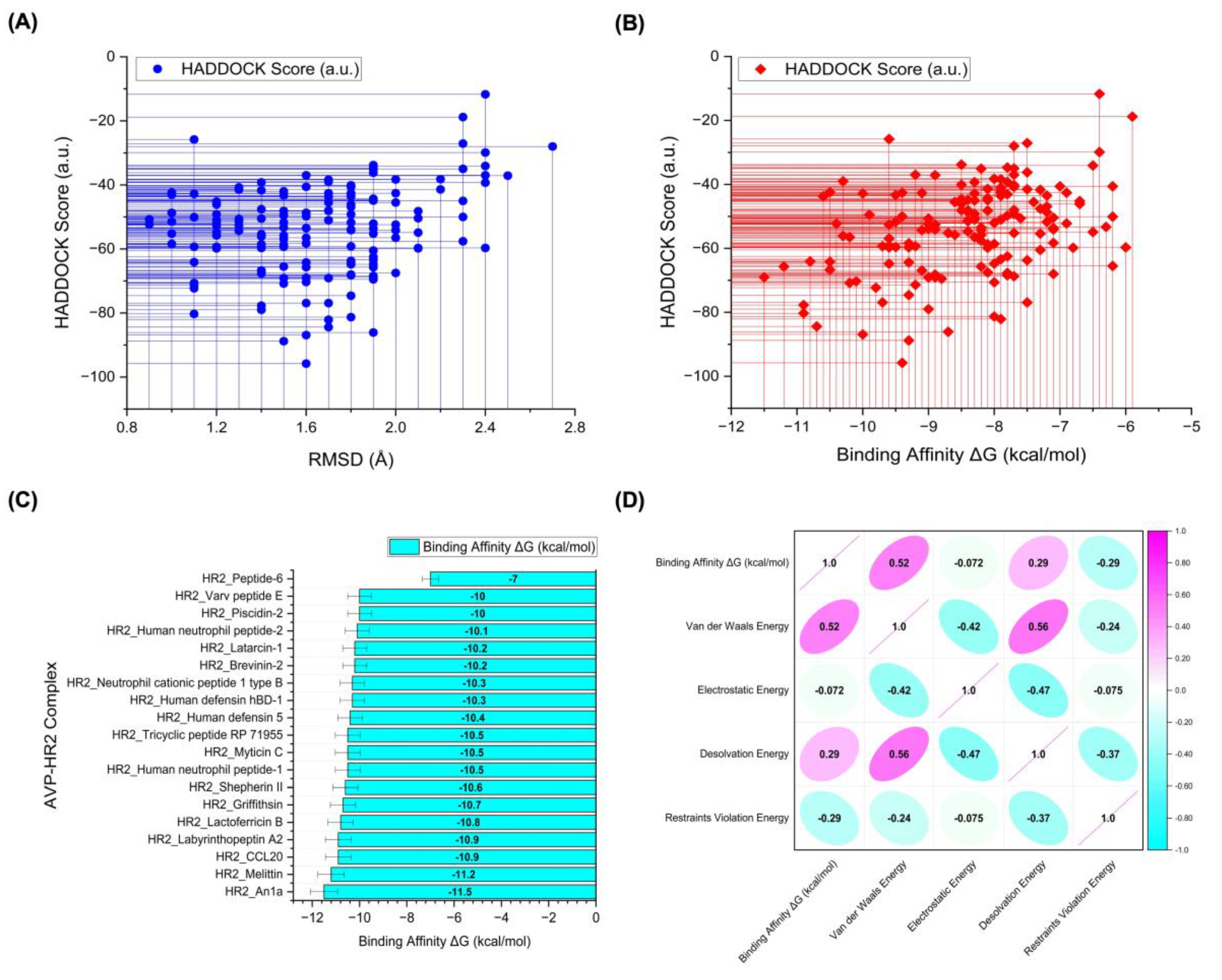
| AVP-HR2 Complex | HADDOCK Score (a.u.) |
Binding Affinity ΔG (kcal/mol) | Kd (M) |
Cluster size | RMSD (Å) |
|---|---|---|---|---|---|
| HR2_Peptide-6 (standard inhibitor) | -45.5 ± 4.6 | -7.0 | 1.10E-05 | 6 | 1.8 ± 0.3 |
| HR2_An1a | -69.0 ± 10.8 | -11.5 | 8.00E-09 | 19 | 1.6 ± 0.3 |
| HR2_Melittin | -65.7 ± 11.0 | -11.2 | 1.20E-08 | 17 | 1.9 ± 1.2 |
| HR2_CCL20 | -77.7 ± 12.2 | -10.9 | 1.90E-08 | 10 | 1.4 ± 0.2 |
| HR2_Labyrinthopeptin A2 | -80.3 ± 16.7 | -10.9 | 2.20E-08 | 9 | 1.1 ± 0.1 |
| HR2_Lactoferricin B | -64.1 ± 11.3 | -10.8 | 2.40E-08 | 8 | 1.1 ± 0.1 |
| HR2_Griffithsin | -84.4 ± 12.2 | -10.7 | 2.80E-08 | 16 | 1.7 ± 0.9 |
| HR2_Shepherin II | -43.7 ± 3.7 | -10.6 | 3.50E-08 | 9 | 1.7 ± 0.3 |
| HR2_Human neutrophil peptide-1 | -42.4 ± 10.6 | -10.5 | 3.70E-08 | 8 | 1.0 ± 0.7 |
| HR2_Myticin C | -64.2 ± 5.1 | -10.5 | 4.00E-08 | 27 | 1.6 ± 0.1 |
| HR2_Tricyclic peptide RP 71955 | -66.7 ± 5.6 | -10.5 | 3.70E-08 | 6 | 1.4 ± 0.1 |
| HR2_Human defensin 5 | -52.3 ± 18.5 | -10.4 | 4.60E-08 | 5 | 0.9 ± 0.6 |
| HR2_Human defensin hBD-1 | -56.1 ± 34.7 | -10.3 | 5.40E-08 | 5 | 1.4 ± 0.9 |
| HR2_Neutrophil cationic peptide 1 type B | -39.0 ± 16.4 | -10.3 | 5.80E-08 | 5 | 1.7 ± 0.3 |
| HR2_Brevinin-2 | -70.8 ± 8.9 | -10.2 | 6.20E-08 | 5 | 1.5 ± 0.5 |
| HR2_Latarcin 1 | -56.5 ± 16.3 | -10.2 | 6.50E-08 | 8 | 2.0 ± 0.1 |
| HR2_Human neutrophil peptide-2 | -70.3 ± 9.4 | -10.1 | 7.70E-08 | 6 | 1.6 ± 0.2 |
| HR2_Piscidin 2 | -86.9 ± 15.8 | -10.0 | 9.40E-08 | 5 | 1.6 ± 1.6 |
| HR2_Varv peptide E | -42.8 ± 10.1 | -10.0 | 8.60E-08 | 7 | 1.1 ± 0.6 |
| AVP-HR2 Complex | ICs charged-charged | ICs charged-polar | ICs charged-apolar | ICs polar-polar | ICs polar-apolar | ICs apolar-apolar | NIS charged | NIS apolar |
|---|---|---|---|---|---|---|---|---|
| HR2_Peptide-6 (standard inhibitor) | 2 | 5 | 3 | 2 | 7 | 3 | 16.13 | 44.84 |
| HR2_An1a | 3 | 3 | 17 | 1 | 20 | 21 | 15.20 | 46.50 |
| HR2_Melittin | 3 | 4 | 12 | 4 | 24 | 21 | 16.11 | 46.31 |
| HR2_CCL20 | 4 | 5 | 19 | 3 | 18 | 20 | 17.48 | 44.79 |
| HR2_Labyrinthopeptin A2 | 2 | 9 | 19 | 8 | 22 | 12 | 16.39 | 44.59 |
| HR2_Lactoferricin B | 3 | 8 | 13 | 0 | 18 | 16 | 17.06 | 45.15 |
| HR2_Griffithsin | 3 | 11 | 25 | 7 | 17 | 23 | 15.90 | 44.47 |
| HR2_Shepherin II | 1 | 6 | 13 | 1 | 19 | 21 | 13.78 | 48.08 |
| HR2_Human neutrophil peptide-1 | 0 | 6 | 11 | 2 | 20 | 20 | 16.05 | 45.15 |
| HR2_Myticin C | 1 | 5 | 6 | 7 | 27 | 17 | 15.45 | 46.88 |
| HR2_Tricyclic peptide RP 71955 | 0 | 0 | 12 | 3 | 21 | 26 | 14.78 | 46.74 |
| HR2_Human defensin 5 | 3 | 4 | 11 | 2 | 18 | 16 | 16.78 | 44.41 |
| HR2_Human defensin hBD-1 | 3 | 10 | 10 | 4 | 20 | 21 | 16.07 | 45.25 |
| HR2_Neutrophil cationic peptide 1 type B | 1 | 4 | 8 | 3 | 22 | 20 | 17.76 | 45.63 |
| HR2_Brevinin-2 | 0 | 2 | 12 | 1 | 18 | 31 | 15.74 | 46.23 |
| HR2_Latarcin 1 | 5 | 9 | 25 | 2 | 11 | 12 | 18.46 | 44.30 |
| HR2_Human neutrophil peptide-2 | 1 | 5 | 10 | 1 | 17 | 28 | 15.72 | 45.15 |
| HR2_Piscidin 2 | 2 | 6 | 17 | 0 | 13 | 22 | 17.00 | 45.33 |
| HR2_Varv peptide E | 1 | 2 | 2 | 6 | 25 | 19 | 15.15 | 46.13 |
| AVP-HR2 Complex | Residue (Receptor) | Protein Atom (Receptor) |
Residue (Interacting Protein/Peptide) |
Protein Atom (Interacting Protein/Peptide) |
Interaction Distance (Å) |
|---|---|---|---|---|---|
| HR2_Peptide-6 (standard inhibitor) | Thr1258 | N | Glu15 | OE2 | 2.98 |
| Thr1258 | OG1 | Glu15 | OE2 | 2.74 | |
| Leu1260 | N | Tyr14 | OH | 2.96 | |
| Asp1261 | O | Ser18 | OG | 2.93 | |
| Glu1265 | OE2 | Ser18 | OG | 2.87 | |
| Ser1268 | OG | Lys24 | NZ | 2.79 | |
| Gln1271 | NE2 | Glu28 | OE1 | 2.72 | |
| HR2_An1a | Asp1261 | N | Leu21 | O | 3.21 |
| Glu1265 | OE2 | Phe26 | N | 3.11 | |
| Glu1265 | OE2 | Gly27 | N | 2.67 | |
| Ser1268 | OG | His38 | NE2 | 2.99 | |
| HR2_Griffithsin | Thr1253 | OG1 | Gly36 | O | 3.26 |
| Leu1260 | N | Ser19 | OG | 2.85 | |
| Tyr1264 | OH | His4 | ND1 | 2.85 | |
| Tyr1264 | OH | Glu119 | OE2 | 3.21 | |
| Glu1265 | OE2 | Gln102 | NE2 | 2.68 | |
| Ser1268 | OG | Arg5 | N | 2.90 | |
| HR2_Brevinin-2 | Thr1258 | O | Lys31 | NZ | 2.67 |
| Glu1265 | SD | Lys28 | NZ | 3.12 | |
| Ser1268 | O | Ser5 | OG | 2.70 | |
| Ser1268 | OG1 | Leu2 | N | 3.17 |
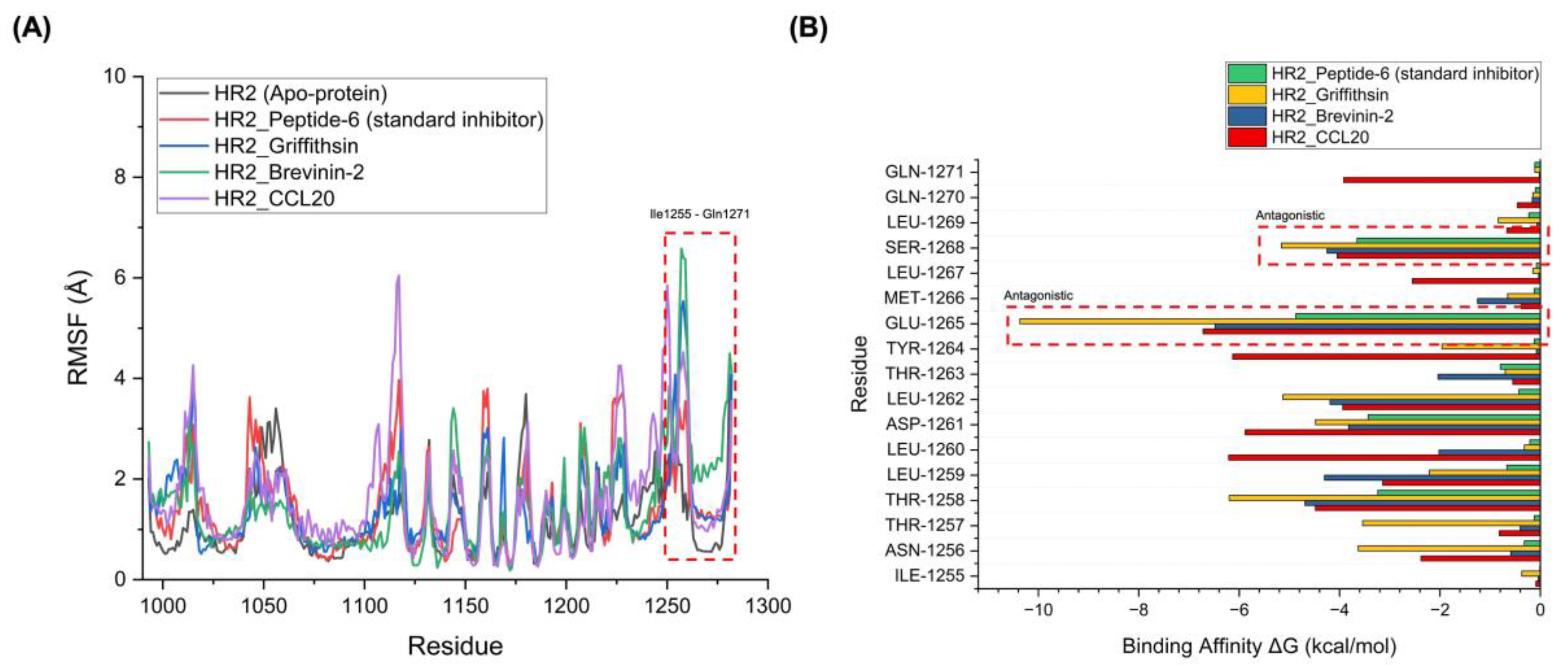
| AVP-HR2 Complex | Average RMSD (Å) | Average RMSF (Å) | Average RoG (Å) |
Number of Hydrogen Bonds Between the Two Proteins |
|---|---|---|---|---|
| HR2_Peptide-6 (standard inhibitor) | 2.572 | 0.976 | 2.282 | 24 |
| HR2_An1a | 2.347 | 0.942 | 2.176 | 35 |
| HR2_Melittin | 2.341 | 0.936 | 2.174 | 37 |
| HR2_CCL20 | 2.330 | 0.924 | 2.171 | 40 |
| HR2_Labyrinthopeptin A2 | 2.433 | 0.926 | 2.172 | 39 |
| HR2_Lactoferricin B | 2.455 | 0.952 | 2.181 | 33 |
| HR2_Griffithsin | 2.301 | 0.888 | 2.161 | 43 |
| HR2_Shepherin II | 2.378 | 0.971 | 2.194 | 29 |
| HR2_Human neutrophil peptide-1 | 2.359 | 0.956 | 2.184 | 32 |
| HR2_Myticin C | 2.411 | 1.115 | 2.298 | 33 |
| HR2_Tricyclic peptide RP 71955 | 2.365 | 1.021 | 2.188 | 30 |
| HR2_Human defensin 5 | 2.244 | 0.938 | 2.176 | 36 |
| HR2_Human defensin hBD-1 | 2.325 | 0.919 | 2.170 | 41 |
| HR2_Neutrophil cationic peptide 1 type B | 2.543 | 1.022 | 2.187 | 33 |
| HR2_Brevinin-2 | 2.322 | 0.914 | 2.168 | 42 |
| HR2_Latarcin 1 | 2.351 | 0.948 | 2.179 | 34 |
| HR2_Human neutrophil peptide-2 | 2.362 | 0.959 | 2.186 | 31 |
| HR2_Piscidin 2 | 2.469 | 1.124 | 2.298 | 24 |
| HR2_Varv peptide E | 2.495 | 0.989 | 2.201 | 26 |
| AVP-HR2 Complex | MM/PBSA Calculation Results ΔGbinding (kcal/mol) | |
|---|---|---|
| HR2_Peptide-6 (standard inhibitor) | −49.73 ± 4.08 | |
| HR2_An1a | −138.97 ± 2.13 | |
| HR2_Melittin | −145.99 ± 3.56 | |
| HR2_CCL20 | −165.17 ± 2.91 | |
| HR2_Labyrinthopeptin A2 | −165.13 ± 1.87 | |
| HR2_Lactoferricin B | −130.55 ± 3.42 | |
| HR2_Griffithsin | −213.69 ± 4.73 | |
| HR2_Shepherin II | −106.17 ± 2.59 | |
| HR2_Human neutrophil peptide-1 | −127.17 ± 1.96 | |
| HR2_Myticin C | −154.63 ± 2.77 | |
| HR2_Tricyclic peptide RP 71955 | −121.32 ± 1.85 | |
| HR2_Human defensin 5 | −142.05 ± 3.21 | |
| HR2_Human defensin hBD-1 | −166.63 ± 4.15 | |
| HR2_Neutrophil cationic peptide 1 type B | −153.91 ± 3.94 | |
| HR2_Brevinin-2 | −168.83 ± 2.66 | |
| HR2_Latarcin 1 | −133.42 ± 1.78 | |
| HR2_Human neutrophil peptide-2 | −124.60 ± 2.35 | |
| HR2_Piscidin 2 | −147.04 ± 1.42 | |
| HR2_Varv peptide E | −98.99 ± 4.92 |
| Antiviral Peptide | Sequence | ESM Score | MERCI Score | Hybrid Scores | Prediction |
|---|---|---|---|---|---|
| Peptide-6 (standard inhibitor) |
SLTQINWTLLDLTYEMESLQQVVKALNEYYIDLKHL | 0.675 | −1.0 | 0.0 | Non-Haemolytic |
| An1a | METAHVFLLSFLLLCVFAVDLIEAGFGCPLDQMQCHNHCQSVRYRGGYCTNFLKMTCKCYG | 0.679 | −1.0 | 0.0 | Non-Haemolytic |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | 0.764 | −1.0 | 0.0 | Non-Haemolytic |
| CCL20 | SNFDCCLGYTDRILHPKFIVGFTRQLANEGCDINAIIFHTKKKLSVCANPKQTWVKYIVRLLSKKVKNM | 0.674 | −0.5 | 0.174 | Non-Haemolytic |
| Labyrinthopeptin A2 | MASILELQNLDVEHARGENRSDWSLWECCSTGSLFACC | 0.724 | −1.0 | 0.0 | Non-Haemolytic |
| Lactoferricin B | FKCRRWQWRMKKLGAPSITCVRRAF | 0.479 | −1.0 | 0.0 | Non-Haemolytic |
| Griffithsin | SLTHRKFGGSGGSPFSGLSSIAVRSGSYLDAIIIDGVHHGGSGGNLSPTFTFGSGEYISNMTIRSGDYIDNISFETNMGRRFGPYGGSGGSANTLSNVKVIQINGSAGDYLDSLDIYYEQY | 0.636 | −1.0 | 0.0 | Non-Haemolytic |
| Shepherin II | GYHGGHGGHGGGYNGGGGHGGHGGGYNGGGHHGGGGHG | 0.706 | −0.5 | 0.206 | Non-Haemolytic |
| Human neutrophil peptide-1 | ACYCRIPACIAGERRYGTCIYQGRLWAFCC | 0.742 | −1.0 | 0.0 | Non-Haemolytic |
| Myticin C | MKATILLAVVVAVIVGVQEAQSVACTSYYCSKFCGSAGCSLYGCYLLHPGKICYCLHCSRAESPLALSGSARNVNDKNNEMENSPLMNEVVNLDQEMNMF | 0.666 | −0.5 | 0.166 | Non-Haemolytic |
| Tricyclic peptide RP 71955 | CLGIGSCNDFAGCGYAVVCFW | 0.755 | −1.0 | 0.0 | Non-Haemolytic |
| Human defensin 5 | ATCYCRTGRCATRESLSGVCEISGRLYRLCCR | 0.709 | −1.0 | 0.0 | Non-Haemolytic |
| Human defensin hBD-1 | DHYNCVSSGGQCLYSACPIFTKIQGTCYRGKAKCCK | 0.684 | −1.0 | 0.0 | Non-Haemolytic |
| Neutrophil cationic peptide 1 type B | RRCICTTRTCRFPYRRLGTCIFQNRVYTFCC | 0.729 | −1.0 | 0.0 | Non-Haemolytic |
| Brevinin-2 | GIWDTIKSMGKVFAGKILQNL | 0.722 | −1.0 | 0.0 | Non-Haemolytic |
| Latarcin 1 | SMWSGMWRRKLKKLRNALKKKLKGE | 0.221 | −1.0 | 0.0 | Non-Haemolytic |
| Human neutrophil peptide-2 | CYCRIPACIAGERRYGTCIYQGRLWAFCC | 0.741 | −1.0 | 0.0 | Non-Haemolytic |
| Piscidin 2 | MKCATLSLVLSMVVLMAEPGDAFFHHIFRGIVHVGKTIHKLVTGGKAEQDQQDQQYQQDQQDQQAQQYQRFNRERAAFD | 0.679 | −1.0 | 0.0 | Non-Haemolytic |
| Varv peptide E | GLPICGETCVGGTCNTPGCSCSWPVCTRN | 0.742 | 0.0 | 0.742 | Hemolytic |
| Antiviral Peptide | Theoretical pI | Extinction Coefficients* | Estimated Half-life** | Instability index | Aliphatic index | GRAVY*** |
|---|---|---|---|---|---|---|
| Peptide-6 (standard inhibitor) | 4.50 | 9970 | 1.9 hours | 16.41 | 127.22 | −0.083 |
| An1a | 6.78 | 4845 | 30 hours | 25.56 | 81.48 | 0.413 |
| Melittin | 12.02 | 5500 | 30 hours | 44.73 | 135.00 | 0.273 |
| CCL20 | 9.70 | 8730 | 1.9 hours | 8.19 | 93.19 | −0.106 |
| Labyrinthopeptin A2 | 4.35 | 11250 | 30 hours | 59.12 | 77.11 | −0.084 |
| Lactoferricin B | 11.84 | 11125 | 1.1 hours | 77.92 | 50.80 | −0.576 |
| Griffithsin | 5.39 | 11920 | 1.9 hours | 39.86 | 70.91 | −0.240 |
| Shepherin II | 7.28 | 4470 | 30 hours | 27.98 | 0.00 | −1.224 |
| Human neutrophil peptide-1 | 8.68 | 10345 | 4.4 hours | 55.71 | 65.33 | 0.300 |
| Myticin C | 5.52 | 7950 | 30 hours | 39.38 | 88.70 | 0.242 |
| Tricyclic peptide RP 71955 | 3.80 | 7240 | 1.2 hours | 33.82 | 74.29 | 1.157 |
| Human defensin 5 | 8.96 | 3355 | 4.4 hours | 13.79 | 64.06 | −0.113 |
| Human defensin hBD-1 | 8.87 | 4845 | 1.1 hours | 34.49 | 46.11 | −0.272 |
| Neutrophil cationic peptide 1 type B | 9.80 | 3355 | 1 hour | 53.98 | 47.10 | −0.200 |
| Brevinin-2 | 9.70 | 5500 | 30 hours | 2.80 | 111.43 | 0.286 |
| Latarcin 1 | 11.77 | 11000 | 1.9 hours | 18.36 | 66.40 | −1.248 |
| Human neutrophil peptide-2 | 8.67 | 10345 | 1.2 hours | 42.13 | 64.14 | 0.248 |
| Piscidin 2 | 6.38 | 2980 | 30 hours | 42.36 | 70.38 | −0.605 |
| Varv peptide E | 5.96 | 5875 | 30 hours | 39.95 | 46.90 | 0.159 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





