Submitted:
13 May 2025
Posted:
14 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Background and Importance of Disease Detection in Horticulture
1.2. Overview of Machine Learning (ML) and Deep Learning (DL) Applications
1.3. Challenges and Research Gaps
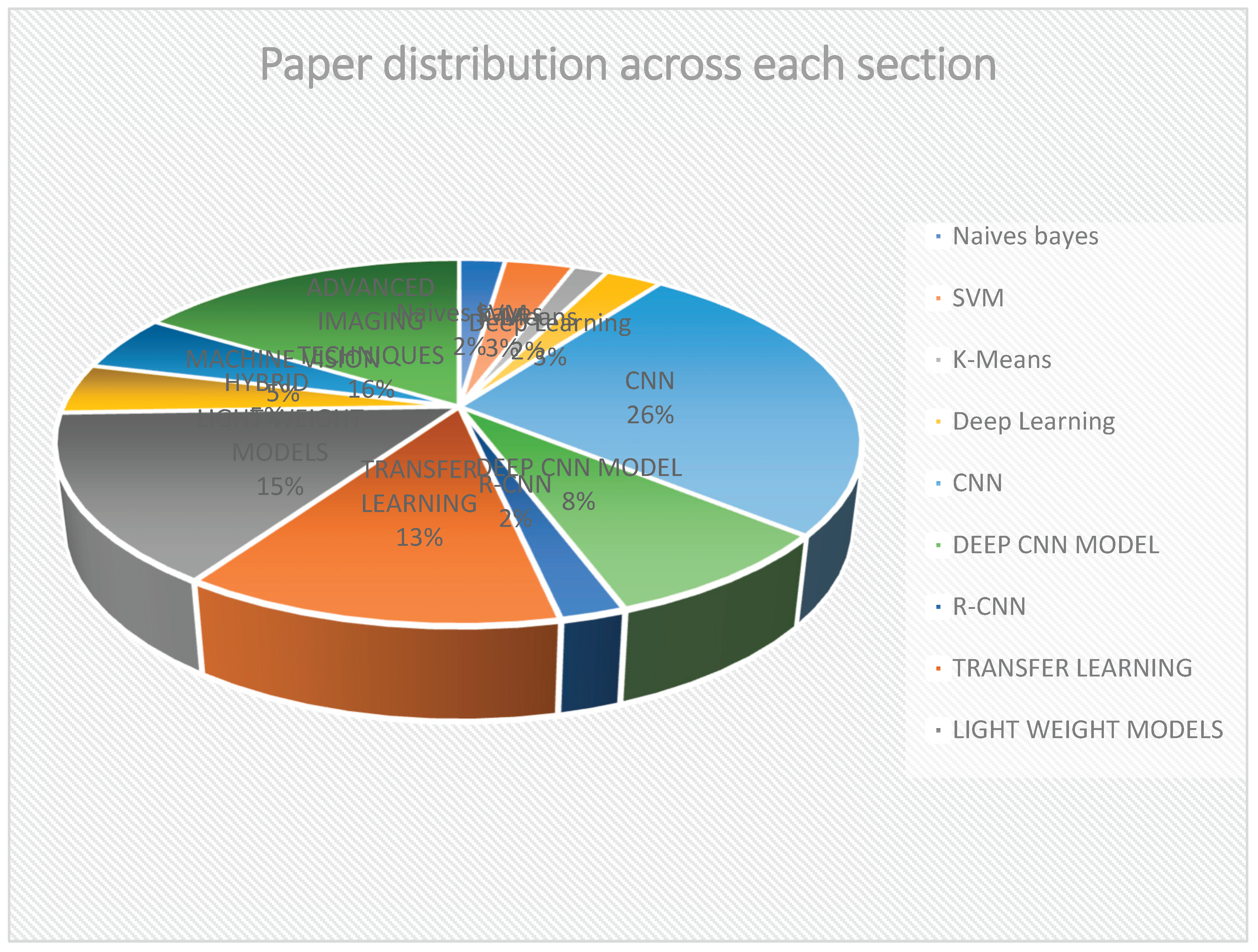
2. Machine Learning and Deep Learning Techniques for Disease Detection
2.1. Traditional Machine Learning Techniques:
2.1.1. Naïve Bayes:
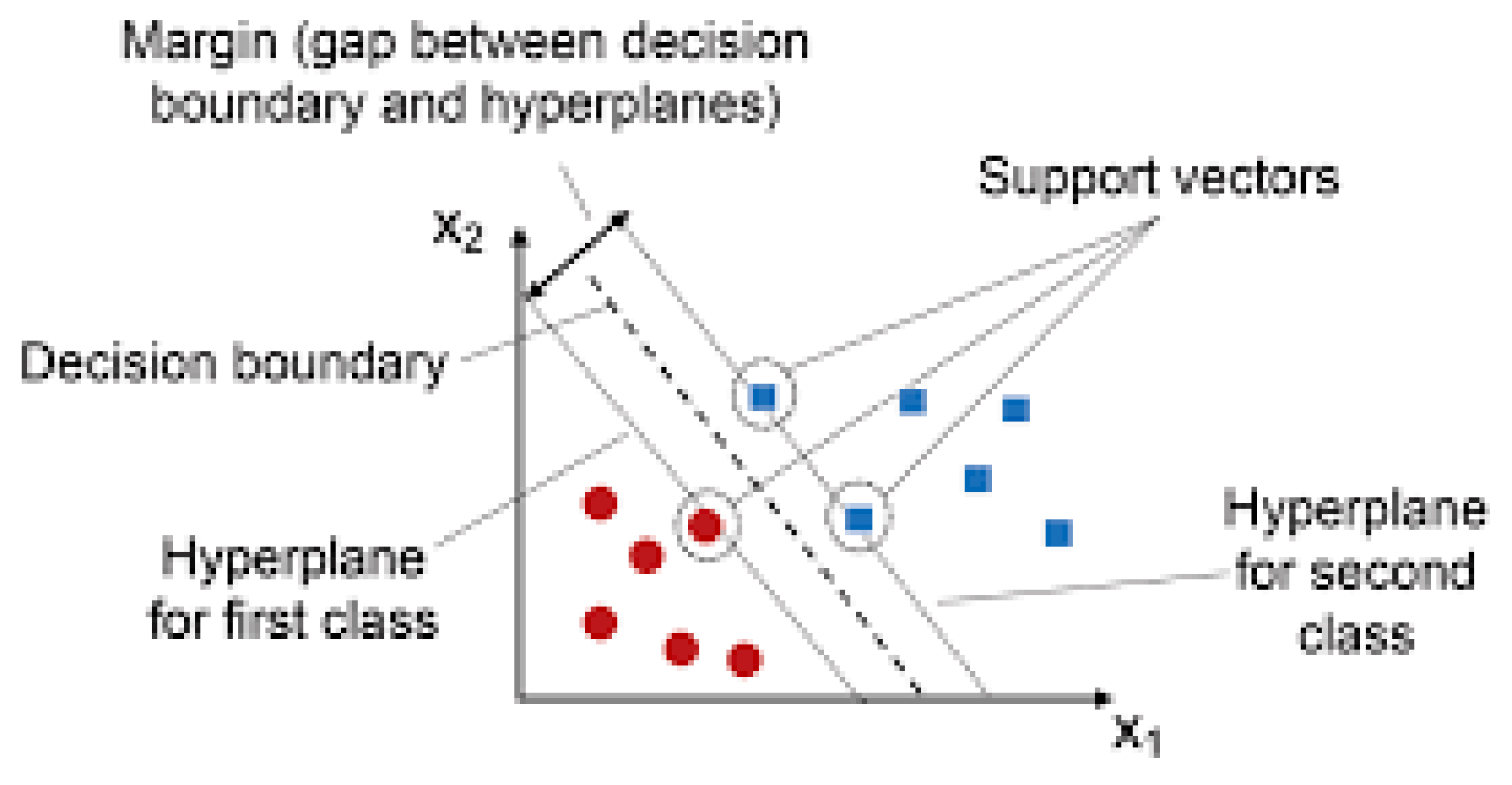
2.1.2. Support Vector Machine (SVM):
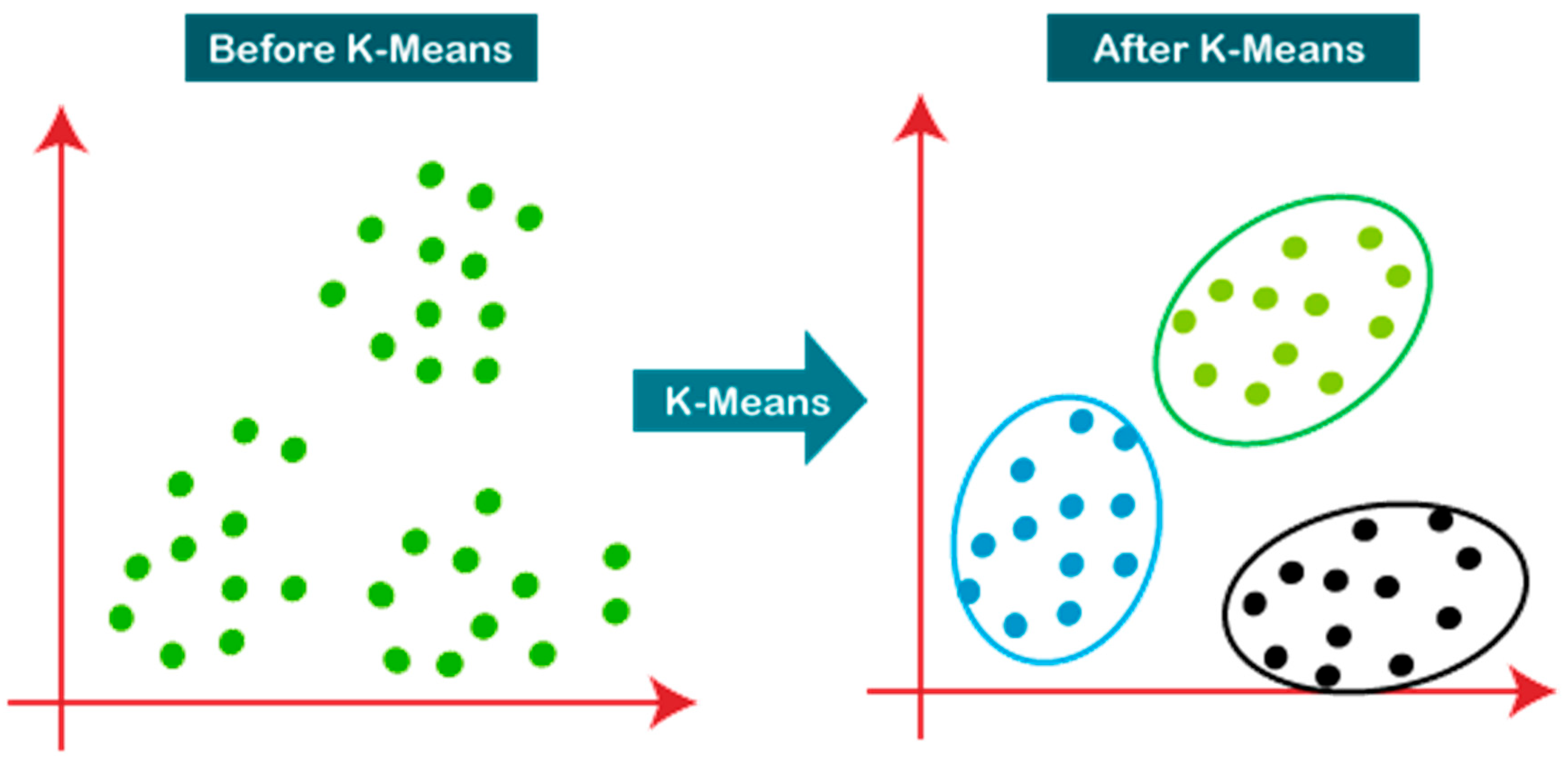
2.1.3 K-Means Clustering:
2.2. Deep Learning:
2.2.1. Convolution Neural Network (CNN):
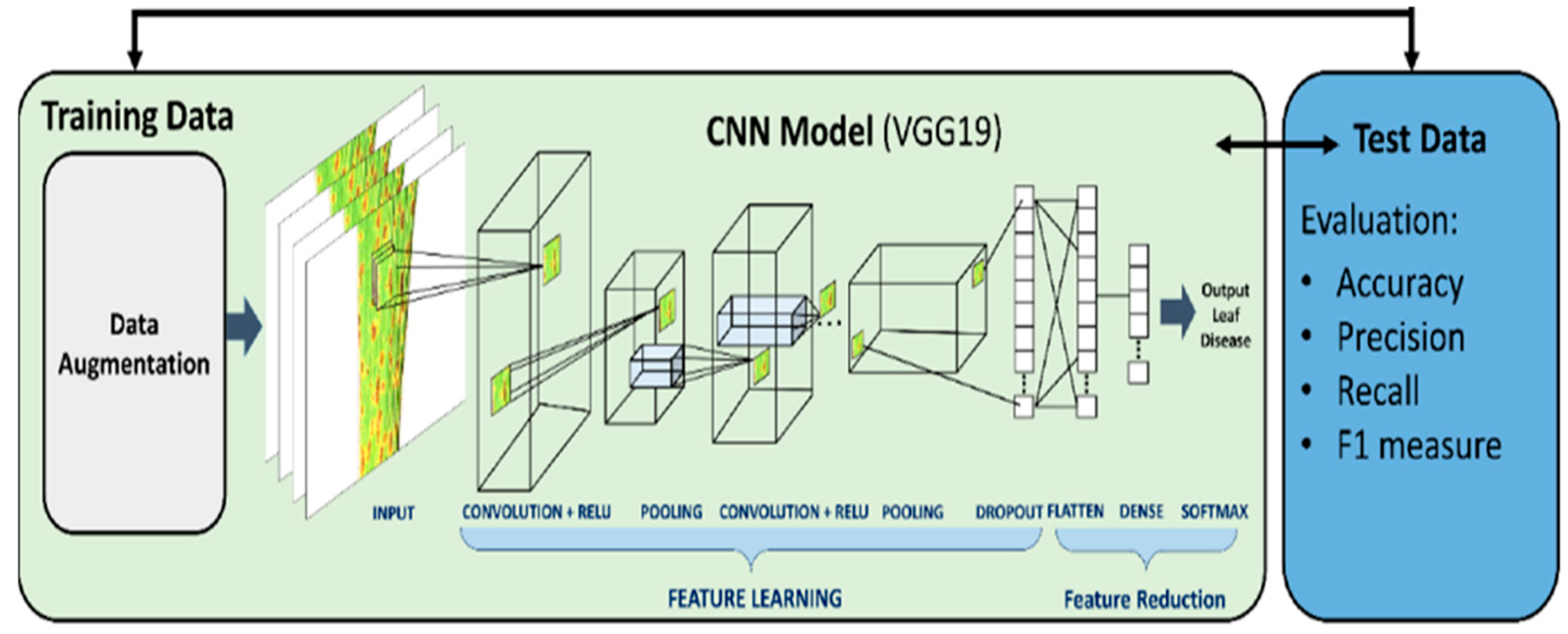
2.2.2. Deep CNN Models:
2.2.3. Region-Based Convolutional Neural Network (R-CNN):
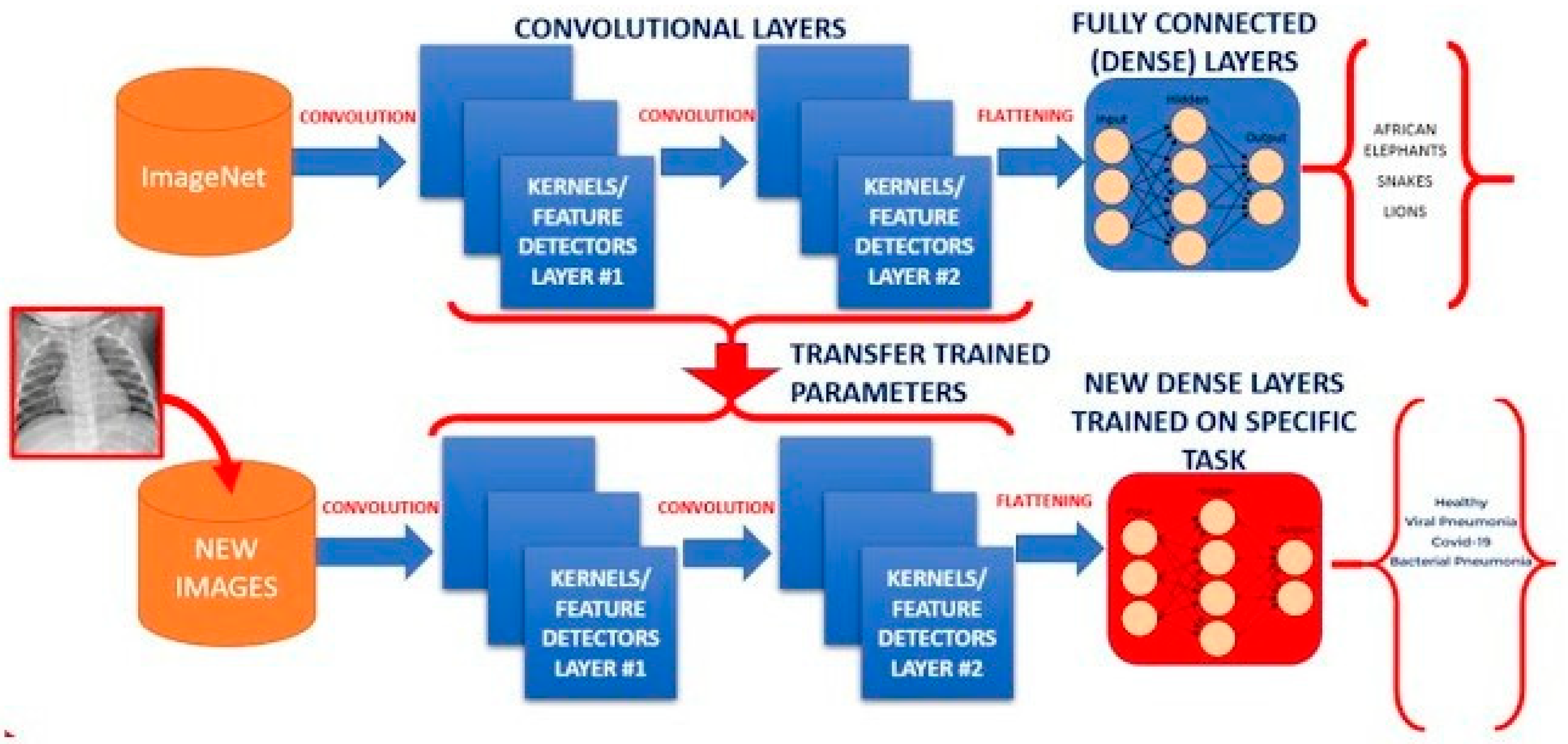
2.2.4. Transfer-Learning:
2.2.5. Lightweight Models:
2.2.6. Hybrid Models
| Approach | Model / Paper | Accuracy (% | F1-score (% | Precision |
| CNN | CNN baseline(various) | 91–97 | 88–96 | 90-95 |
| YOLOv5 | MEAN-SSD, YOLOv5 variations | 97.9-99.6 | 97.7-99.5 | 97-99.7 |
| Transfer Learning | EfficientNet, MobileNet | 98.6-99.7 | 97.7-99.6 | 97.1-99.8 |
| Hybrid Lightweight | Efficient-ECANet, PDICNet | 99.71 | 97.77 | 99.41 |
| SVM | Traditional ML + Features | 85-93 | 80-91 | 83-90 |
| Naive Bayes | Traditional ML | 80-88 | 75-86 | 78-87 |
| Machine Vision | Color/texture/shape-based | 75-90 | 70-88 | 72-89 |
3. Machine Vision and Image Processing:
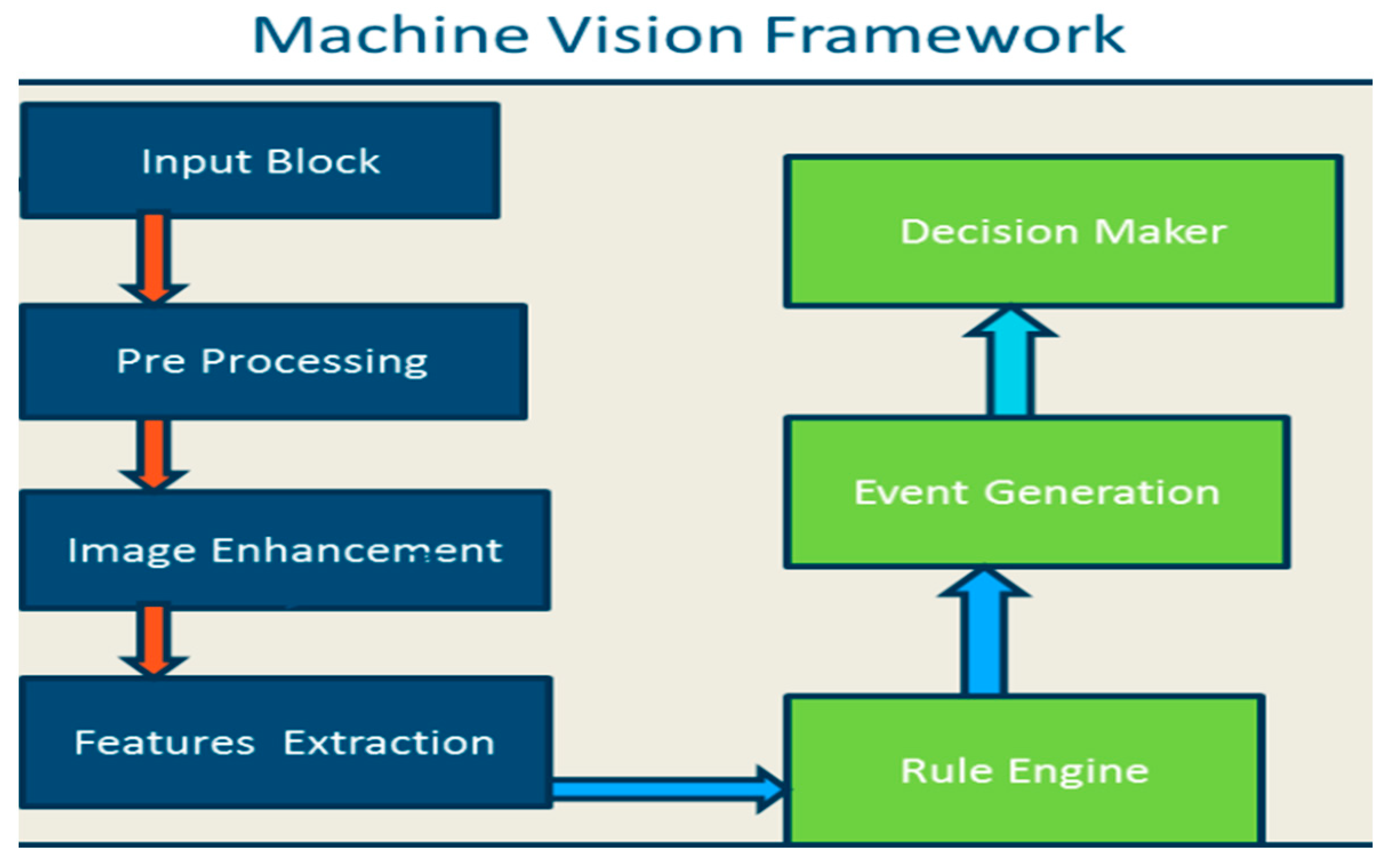
4. Applications of Advanced Imaging Techniques:
5. Future Trends and Research Directions:
5.1. The Convergence of IoT with Edge Computing:
5.2. Resource-Constrained Environments’ Lightweight Models:
5.3. Explainable AI for Better Decision-Making:
6. Conclusion:
References
- Dharm, Padaliya & Pandya, Parth & Bhavik, Patel & Vatsal, Salkiya & Yuvraj, Solanki & Darji, Mittal. (2019). A Review of Apple Diseases Detection and Classification. International Journal of Engineering and Technical Research. 8. 382-387.
- Zhang, Daping & Yang, Hongyu & Cao, Jiayu. (2021). Identify Apple Leaf Diseases Using a Deep Learning Algorithm. arXiv:10.48550/arXiv.2107.12598.
- Al-Wesabi, Fahd & Albraikan, Amani & Hilal, Anwer & Eltahir, Majdy & Hamza, Manar & Zamani, Abu. (2022). Artificial Intelligence Enabled Apple Leaf Disease Classification for Precision Agriculture. Computers, Materials and Continua. 70. 6223-6238. [CrossRef]
- S. Kumar, R. Kumar and M. Gupta, "Analysis of Apple Plant Leaf Diseases Detection and Classification: A Review," 2022 Seventh International Conference on Parallel, Distributed and Grid Computing (PDGC), Solan, Himachal Pradesh, India, 2022, pp. 361-365. [CrossRef]
- Singh, Swati & Gupta, Sheifali. (2018). Apple Scab and Marsonina Coronaria Diseases Detection in Apple Leaves Using Machine Learning. International Journal of Pure and Applied Mathematics. 1151-1166.
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN COMPUT. SCI. 2, 160 (2021). [CrossRef]
- Meshram, V., Patil, K., Meshram, V., Hanchate, D., & Ramkteke, S. D. (2021). Machine learning in agriculture domain: A state-of-art survey. Artificial Intelligence in the Life Sciences, 1, 100010. [CrossRef]
- Sandhu, K. K. Sandhu, K. K. (2021). Apple leaves disease detection using machine learning approach. International Journal of Computer Science and Information Technology Research, 9(1), 127-135. Retrieved from www.researchpublish.com.
- Sugiarti, Y., Supriyatna, A., Carolina, I., Amin, R., & Yani, A. (2021, September). Model naïve Bayes classifiers for detection apple diseases. In 2021 9th International Conference on Cyber and IT Service Management (CITSM) (pp. 1-4). IEEE.
- Miriti, E. (2016). Classification of selected apple fruit varieties using Naive Bayes (Doctoral dissertation, University of Nairobi).
- Sumanto, Sumanto & Sugiarti, Yuni & Supriyatna, Adi & Carolina, Irmawati & Amin, Ruhul & Yani, Ahmad. (2021). Model Naïve Bayes Classifiers For Detection Apple Diseases. 1-4.
- Misigo, Ronald & Kirimi, Evans. (2016). CLASSIFICATION OF SELECTED APPLE FRUIT VARIETIES USING NAIVE BAYES. Indian Journal of Computer Science and Engineering (IJCSE). 7. 13.
- Aravind, K.R.N.V.V.D. & Shyry, S.Prayla & Felix, A Yovan. (2019). Classification of Healthy and Rot Leaves of Apple Using Gradient Boosting and Support Vector Classifier. International Journal of Innovative Technology and Exploring Engineering. 8. 2868-2872.
- S. Chakraborty, S. Paul and M. Rahat-uz-Zaman, "Prediction of Apple Leaf Diseases Using Multiclass Support Vector Machine," 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), DHAKA, Bangladesh, 2021, pp. 147-151. [CrossRef]
- Omrani, E., Khoshnevisan, B., Shamshirband, S., Saboohi, H., Anuar, N. B., & Nasir, M. H. N. M. (2014). Potential of radial basis function-based support vector regression for apple disease detection. Measurement, 55, 512-519.
- Sivakamasundari, G., & Seenivasagam, V. (2018). Classification of leaf diseases in apple using support vector machine. International Journal of Advanced Research in Computer Science, 9(1), 261-265.
- Alagu, S., & BhoopathyBagan, K. Apple Fruit disease detection and classification using Multiclass SVM classifier and IP Webcam APP. International Journal of Management, Technology, and Engineering, Volume Number (Issue Number), Page Range. ISSN, 2249-7455.
- Pujari, D., Yakkundimath, R., & Byadgi, A. S. (2016). SVM and ANN based classification of plant diseases using feature reduction technique. IJIMAI, 3(7), 6-14.
- Anam, S. (2020, June). Segmentation of leaf spots disease in apple plants using particle swarm optimization and K-means algorithm. In Journal of Physics: Conference Series (Vol. 1562, No. 1, p. 012011). IOP Publishing.
- Tiwari, R., & Chahande, M. (2021). Apple fruit disease detection and classification using k-means clustering method. In Advances in Intelligent Computing and Communication: Proceedings of ICAC 2020 (pp. 71-84). Springer Singapore.
- Al Bashish, D., Braik, M., & Bani-Ahmad, S. (2011). Detection and classification of leaf diseases using K-means-based segmentation and. Information technology journal, 10(2), 267-275.
- Zhang, Daping & Yang, Hongyu & Cao, Jiayu. (2021). Identify Apple Leaf Diseases Using Deep Learning Algorithm. arXiv:10.48550/arXiv.2107.12598.
- Doutoum AS, Tugrul B. 2025. A systematic review of deep learning techniques for apple leaf diseases classification and detection. PeerJ Computer Science 11:e2655. [CrossRef]
- Banjar, A., Javed, A., Nawaz, M. et al. E-AppleNet: An Enhanced Deep Learning Approach for Apple Fruit Leaf Disease Classification. Applied Fruit Science 67, 18 (2025). [CrossRef]
- Alsayed, Ashwaq & Alsabei, Amani & Arif, Muhammad. (2021). Classification of Apple Tree Leaves Diseases using Deep Learning Methods. International Journal of Computer Network and Information Security. 21. 324. [CrossRef]
- Y. Luo, J. Sun, J. Shen, X. Wu, L. Wang and W. Zhu, "Apple Leaf Disease Recognition and Sub-Class Categorization Based on Improved Multi-Scale Feature Fusion Network," in IEEE Access, vol. 9, pp. 95517-95527, 2021. [CrossRef]
- Yang, Q., Duan, S., & Wang, L. (2022). Efficient Identification of Apple Leaf Diseases in the Wild Using Convolutional Neural Networks. Agronomy, 12(11), 2784. [CrossRef]
- Pradhan, Priyanka & Kumar, Brajesh & Mohan, Shashank. (2022). Comparison of various deep convolutional neural network models to discriminate apple leaf diseases using transfer learning. Journal of Plant Diseases and Protection. 129. [CrossRef]
- Bansal, P., Kumar, R., & Kumar, S. (2021). Disease Detection in Apple Leaves Using Deep Convolutional Neural Network. Agriculture, 11(7), 617. [CrossRef]
- Ni, J. (2024). Smart agriculture: An intelligent approach for apple leaf disease identification based on convolutional neural network. Journal of Phytopathology, 172(4). [CrossRef]
- Srivastav, Somya & Guleria, Kalpna & Sharma, Shagun. (2024). Apple Leaf Disease Detection using Deep Learning-based Convolutional Neural Network. 1-5. [CrossRef]
- Kaur, Arshleen & Chadha, Raman. (2023). An Optimized Ant Gardient Convolutional Neural Network for Disease Detection in Apple Leaves. 1-8. [CrossRef]
- B. Biswas and R. K. Yadav, "Multilayer Convolutional Neural Network Based Approach to Detect Apple Foliar Disease," 2023 2nd International Conference for Innovation in Technology (INOCON), Bangalore, India, 2023, pp. 1-5. [CrossRef]
- V. K. Vishnoi, K. Kumar, B. Kumar, S. Mohan and A. A. Khan, "Detection of Apple Plant Diseases Using Leaf Images Through Convolutional Neural Network," in IEEE Access, vol. 11, pp. 6594-6609, 2023. [CrossRef]
- Firdous, Saba & Akbar, Shahzad & Hassan, Syed Ale & Khalid, Aima & Gull, Sahar. (2023). Deep Convolutional Neural Network-based Framework for Apple Leaves Disease Detection. 1-6. [CrossRef]
- C. Thakur, N. Kapoor and R. Saini, "A Novel Framework of Apple Leaf Disease Detection using Convolutional Neural Network," 2023 International Conference on Inventive Computation Technologies (ICICT), Lalitpur, Nepal, 2023, pp. 491-496. [CrossRef]
- Tanwar, V.K., Sharma, B., & Anand, V. (2023). A Sophisticated Deep Convolutional Neural Network for Multiple Classification of Apple Leaf Diseases. 2023 International Conference on Research Methodologies in Knowledge Management, Artificial Intelligence and Telecommunication Engineering (RMKMATE), 1-6.
- Chen, Y., Pan, J. & Wu, Q. Apple leaf disease identification via improved CycleGAN and convolutional neural network. Soft Comput 27, 9773–9786 (2023). [CrossRef]
- Mahato, D.K., Pundir, A. & Saxena, G.J. An Improved Deep Convolutional Neural Network for Image-Based Apple Plant Leaf Disease Detection and Identification. J. Inst. Eng. India Ser. A 103, 975–987 (2022). [CrossRef]
- Sharma, V., Verma, A., Goel, N. (2022). A Modified Feature Optimization Approach with Convolutional Neural Network for Apple Leaf Disease Detection. In: Abraham, A., et al. Innovations in Bio-Inspired Computing and Applications. IBICA 2021. Lecture Notes in Networks and Systems, vol 419. Springer, Cham. [CrossRef]
- Fu L, Li S, Sun Y, Mu Y, Hu T, Gong H. Lightweight-Convolutional Neural Network for Apple Leaf Disease Identification. Front Plant Sci. 2022 May 24;13:831219. [CrossRef] [PubMed] [PubMed Central]
- Tugrul, B., Elfatimi, E., & Eryigit, R. (2022). Convolutional Neural Networks in Detection of Plant Leaf Diseases: A Review. Agriculture, 12(8), 1192. [CrossRef]
- Liu, B., Zhang, Y., He, D., & Li, Y. (2017). Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry, 10(1), 11.
- Khan, A. I., Quadri, S. M. K., Banday, S., & Shah, J. L. (2022). Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. computers and Electronics in Agriculture, 198, 107093.
- V. K. Vishnoi, K. Kumar, B. Kumar, S. Mohan and A. A. Khan, "Detection of Apple Plant Diseases Using Leaf Images Through Convolutional Neural Network," in IEEE Access, vol. 11, pp. 6594-6609, 2023. [CrossRef]
- Bansal, P., Kumar, R., & Kumar, S. (2021). Disease detection in apple leaves using deep convolutional neural network. Agriculture, 11(7), 617.
- Gong, X., & Zhang, S. (2023). A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN. Agriculture, 13(2), 240. [CrossRef]
- Baranwal, S., Khandelwal, S., & Arora, A. (2019, February). Deep learning convolutional neural network for apple leaves disease detection. In Proceedings of international conference on sustainable computing in science, technology and management (SUSCOM), Amity University Rajasthan, Jaipur-India.
- Yan, Q., Yang, B., Wang, W., Wang, B., Chen, P., & Zhang, J. (2020). Apple leaf diseases recognition based on an improved convolutional neural network. Sensors, 20(12), 3535.
- Parashar, N., & Johri, P. (2024). Enhancing apple leaf disease detection: A CNN-based model integrated with image segmentation techniques for precision agriculture. International Journal of Mathematical, Engineering and Management Sciences, 9(4), 943.
- Agarwal, M., Kaliyar, R. K., & Gupta, S. K. (2022, July). Differential Evolution based compression of CNN for Apple fruit disease classification. In 2022 International Conference on Inventive Computation Technologies (ICICT) (pp. 76-82). IEEE.
- Çetiner, İ. (2025). AppleCNN: A new CNN-based deep learning model for classification of apple leaf diseases. Gümüşhane Üniversitesi Fen Bilimleri Dergisi, 15(1), 51-63.
- Kaur, A., Kukreja, V., Aggarwal, P., Thapliyal, S., & Sharma, R. (2024). Amplifying apple mosaic illness detection: Combining CNN and random forest models. In 2024 IEEE International Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation (IATMSI), Gwalior, India (pp. 1-5). [CrossRef]
- S. Mehta, V. Kukreja and R. Gupta, "Empowering Precision Agriculture: Detecting Apple Leaf Diseases and Severity Levels with Federated Learning CNN," 2023 3rd International Conference on Intelligent Technologies (CONIT), Hubli, India, 2023, pp. 1-6. [CrossRef]
- Fu, Longsheng & Majeed, Yaqoob & Zhang, Xin & Karkee, Manoj & Zhang, Qin. (2020). Faster R–CNN–based apple detection in dense-foliage fruiting-wall trees using RGB and depth features for robotic harvesting. Biosystems Engineering. 197. 245-256.
- Kodors, Sergejs & Lacis, Gunars & Sokolova, Olga & Zhukov, Vitaliy & Apeinans, Ilmars & Bartulsons, Toms. (2021). Apple scab detection using CNN and Transfer Learning. Agronomy Research. 19. 507-519. [CrossRef]
- K. Sujatha, K. Gayatri, M. S. Yadav, N. C. Sekhara Rao and B. S. Rao, "Customized Deep CNN for Foliar Disease Prediction Based on Features Extracted from Apple Tree Leaves Images," 2022 International Interdisciplinary Humanitarian Conference for Sustainability (IIHC), Bengaluru, India, 2022, pp. 193-197. [CrossRef]
- Ziyi Yang and Minchen Yang "Apple leaf scab recognition using CNN and transfer learning", Proc. SPIE 13486, Fourth International Conference on Computer Vision, Application, and Algorithm (CVAA 2024), 134860D (9 January 2025). [CrossRef]
- Si, H., Li, M., Li, W., Zhang, G., Wang, M., Li, F., & Li, Y. (2024). A Dual-Branch Model Integrating CNN and Swin Transformer for Efficient Apple Leaf Disease Classification. Agriculture, 14(1), 142. [CrossRef]
- Agarwal, M., Kaliyar, R.K., Singal, G., & Gupta, S.K. (2019). FCNN-LDA: A Faster Convolution Neural Network model for Leaf Disease identification on Apple's leaf dataset. 2019 12th International Conference on Information & Communication Technology and System (ICTS), 246-251.
- Yu, Hee-Jin & Son, Chang-Hwan & Lee, Dong. (2020). Apple Leaf Disease Identification Through Region-of-Interest-Aware Deep Convolutional Neural Network. Journal of Imaging Science and Technology. 64. [CrossRef]
- Baranwal, Saraansh & Khandelwal, Siddhant & Arora, Anuja. (2019). Deep Learning Convolutional Neural Network for Apple Leaves Disease Detection. SSRN Electronic Journal. [CrossRef]
- Di, Jie & Li, Qing. (2022). A method of detecting apple leaf diseases based on improved convolutional neural network. PLOS ONE. 17. [CrossRef]
- Garcia Nachtigall, Lucas & Araujo, Ricardo & Nachtigall, Gilmar. (2016). Classification of Apple Tree Disorders Using Convolutional Neural Networks. 472-476. [CrossRef]
- Liu, B., Zhang, Y., He, D., & Li, Y. (2018). Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry, 10(1), 11. [CrossRef]
- türkoğlu, Muammer & Hanbay, Davut & Sengur, Abdulkadir. (2022). Multi-model LSTM-based convolutional neural networks for detection of apple diseases and pests. Journal of Ambient Intelligence and Humanized Computing. 13. [CrossRef]
- P. Jiang, Y. Chen, B. Liu, D. He and C. Liang, "Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks," in IEEE Access, vol. 7, pp. 59069-59080, 2019. [CrossRef]
- Yan, Q., Yang, B., Wang, W., Wang, B., Chen, P., & Zhang, J. (2020). Apple leaf diseases recognition based on an improved convolutional neural network. Sensors, 20(12), 3535. [CrossRef]
- Yadav, D., Akanksha, Yadav, A.K. (2020). A novel convolutional neural network based model for recognition and classification of apple leaf diseases. Traitement du Signal, Vol. 37, No. 6, pp. 1093-1101. [CrossRef]
- Di, J., & Li, Q. (2022). A method of detecting apple leaf diseases based on improved convolutional neural network. PLOS ONE, 17(2), e0262629. [CrossRef]
- Albogamy, F. R. (2021). A Deep Convolutional Neural Network with Batch Normalization Approach for Plant Disease Detection. International Journal of Computer Science and Network Security, 21(9), 51–62. [CrossRef]
- Srinidhi, V & Sahay, Apoorva & Deeba, K.. (2021). Plant Pathology Disease Detection in Apple Leaves Using Deep Convolutional Neural Networks: Apple Leaves Disease Detection using EfficientNet and DenseNet. 1119-1127. [CrossRef]
- Sun, H., Xu, H., Liu, B., He, D., He, J., Zhang, H., & Geng, N. (2021). MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Computers and Electronics in Agriculture, 189, 106379. [CrossRef]
- Yatoo, A. (2024). Empowering Precision Agriculture: A Novel ResNet50 based PDICNet for Automated Apple Leaf Disease Detection. Journal of Electrical Systems, 20(7s), 2211–2220. [CrossRef]
- Yongjun Ding, Wentao Yang, Jingjing Zhang, An improved DeepLabV3+ based approach for disease spot segmentation on apple leaves, Computers and Electronics in Agriculture, Volume 231, 2025, 110041, ISSN 0168-1699. [CrossRef]
- Čirjak, D., Aleksi, I., Lemic, D., & Pajač Živković, I. (2023). EfficientDet-4 Deep Neural Network-Based Remote Monitoring of Codling Moth Population for Early Damage Detection in Apple Orchard. Agriculture, 13(5), 961. [CrossRef]
- X. Li and L. Rai, "Apple Leaf Disease Identification and Classification using ResNet Models," 2020 IEEE 3rd International Conference on Electronic Information and Communication Technology (ICEICT), Shenzhen, China, 2020, pp. 738-742. [CrossRef]
- Lin, Renyi. (2024). Apple leaf diseases recognition based on ResNet-101 and CBAM. Applied and Computational Engineering. 51. 256-266. [CrossRef]
- Banarase, S., & Shirbahadurkar, S. (2024). The Orchard Guard: Deep Learning powered apple leaf disease detection with MobileNetV2 model. Journal of Integrated Science and Technology, 12(4), 799. [CrossRef]
- Bin Liu, Xulei Huang, Leiming Sun, Xing Wei, Zeyu Ji, Haixi Zhang, MCDCNet: Multi-scale constrained deformable convolution network for apple leaf disease detection, Computers and Electronics in Agriculture, Volume 222, 2024, 109028, ISSN 0168-1699. [CrossRef]
- Nain, S., Mittal, N., Jain, A. (2024). Recognition of Apple Leaves Infection Using DenseNet121 with Additional Layers. In: Sharma, D.K., Peng, SL., Sharma, R., Jeon, G. (eds) Micro-Electronics and Telecommunication Engineering. ICMETE 2023. Lecture Notes in Networks and Systems, vol 894. Springer, Singapore. [CrossRef]
- Gao, Y., Cao, Z., Cai, W., Gong, G., Zhou, G., & Li, L. (2023). Apple Leaf Disease Identification in Complex Background Based on BAM-Net. Agronomy, 13(5), 1240. [CrossRef]
- Gao, X., Tang, Z., Deng, Y., Hu, S., Zhao, H., & Zhou, G. (2023). HSSNet: A End-to-End Network for Detecting Tiny Targets of Apple Leaf Diseases in Complex Backgrounds. Plants, 12(15), 2806. [CrossRef]
- Bhat, I.R., & Wani, M.A. (2023). Modified Grouped Convolution-Based EfficientNet Deep Learning Architecture for Apple Disease Detection. 2023 International Conference on Machine Learning and Applications (ICMLA), 1465-1472.
- Bi, C., Wang, J., Duan, Y. et al. MobileNet Based Apple Leaf Diseases Identification. Mobile Netw Appl 27, 172–180 (2022). [CrossRef]
- Yukai Zhang, Guoxiong Zhou, Aibin Chen, Mingfang He, Johnny Li, Yahui Hu, A precise apple leaf diseases detection using BCTNet under unconstrained environments, Computers and Electronics in Agriculture, Volume 212, 2023, 108132, ISSN 0168-1699. [CrossRef]
- Liu, S., Qiao, Y., Li, J., Zhang, H., Zhang, M., & Wang, M. (2022). An improved lightweight network for real-time detection of apple leaf diseases in natural scenes. Agronomy, 12(10), 2363. [CrossRef]
- Upadhyay, Nidhi & Gupta, Neeraj. (2024). Diagnosis of fungi affected apple crop disease using improved ResNeXt deep learning model. Multimedia Tools and Applications. 83. 1-20.
- Gong, X., & Zhang, S. (2023). A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN. Agriculture, 13(2), 240. [CrossRef]
- Zia Ur Rehman, Muhammad & Khan, M. & Ahmed, Fawad & Damaševičius, Robertas & Naqvi, Syed & Nisar, Muhammad & Javed, Kashif. (2021). Recognizing apple leaf diseases using a novel parallel real-time processing framework based on MASK RCNN and transfer learning: An application for smart agriculture. IET Image Processing. 15. [CrossRef]
- Gao, F., Fu, L., Zhang, X., Majeed, Y., Li, R., Karkee, M., & Zhang, Q. (2020). Multi-class fruit-on-plant detection for apple in SNAP system using Faster R-CNN. Computers and Electronics in Agriculture, 176, 105634. [CrossRef]
- Alwaseela Abdalla et al., "Fine-tuning convolutional neural network with transfer learning for semantic segmentation of ground-level oilseed rape images in a field with high weed pressure," Computers and Electronics in Agriculture, vol. 167, 2019, 105091. [CrossRef]
- Assad, A., Bhat, M. R., Bhat, Z. A., Ahanger, A. N., Kundroo, M., Dar, R. A., ... & Dar, B. N. (2023). Apple diseases: detection and classification using transfer learning. Quality Assurance and Safety of Crops & Foods, 15(SP1), 27-37.
- Sulaiman, A., Anand, V., Gupta, S., Alshahrani, H., Reshan, M. S. A., Rajab, A., ... & Azar, A. T. (2023). Sustainable apple disease management using an intelligent fine-tuned transfer learning-based model. Sustainability, 15(17), 13228.
- Fan, X., Luo, P., Mu, Y., Zhou, R., Tjahjadi, T., & Ren, Y. (2022). Leaf image-based plant disease identification using transfer learning and feature fusion. Computers and Electronics in agriculture, 196, 106892.
- Rawat, P., & Singh, S. K. (2024, February). Apple leaf disease detection using transfer learning. In 2024 International Conference on Integrated Circuits and Communication Systems (ICICACS) (pp. 1-6). IEEE.
- Özden, C. (2021). Apple leaf disease detection and classification based on transfer learning. Turkish Journal of Agriculture and Forestry, 45(6), 775-783.
- Bhat, M. R., Assad, A., Dar, B. N., Ahanger, A. N., Kundroo, M., Dar, R. A., ... & Bhat, Z. A. (2023). Apple diseases: detection and classification using transfer learning. QUALITY ASSURANCE AND SAFETY OF CROPS & FOODS, 15, 27-37.
- Kodors, S., Lacis, G., Sokolova, O., Zhukovs, V., Apeinans, I., & Bartulsons, T. (2021). Apple scab detection using CNN and Transfer Learning.
- Rehman, Z. U., Khan, M. A., Ahmed, F., Damaševičius, R., Naqvi, S. R., Nisar, W., & Javed, K. (2021). Recognizing apple leaf diseases using a novel parallel real-time processing framework based on MASK RCNN and transfer learning: An application for smart agriculture. IET Image Processing, 15(10), 2157-2168.
- Kumar, A., Nelson, L., & Gomathi, S. (2024, January). Transfer learning of vgg19 for the classification of apple leaf diseases. In 2024 2nd International Conference on Intelligent Data Communication Technologies and Internet of Things (IDCIoT) (pp. 1643-1648). IEEE.
- Nagaraju, Y., Swetha, S., & Stalin, S. (2020, December). Apple and grape leaf diseases classification using transfer learning via fine-tuned classifier. In 2020 IEEE International Conference on Machine Learning and Applied Network Technologies (ICMLANT) (pp. 1-6). IEEE.
- Wani, O. A., Zahoor, U., Shah, S. Z. A., & Khan, R. (2024). Apple leaf disease detection using transfer learning. Annals of Data Science, 1-10.
- Si, H., Wang, Y., Zhao, W., Wang, M., Song, J., Wan, L., ... & Sun, C. (2023). Apple surface defect detection method based on weight comparison transfer learning with MobileNetV3. Agriculture, 13(4), 824.
- Chao, X., Sun, G., Zhao, H., Li, M., & He, D. (2020). Identification of apple tree leaf diseases based on deep learning models. Symmetry, 12(7), 1065.
- Su, J., Zhang, M., & Yu, W. (2022, April). An identification method of apple leaf disease based on transfer learning. In 2022 7th International Conference on Cloud Computing and Big Data Analytics (ICCCBDA) (pp. 478-482). IEEE.
- Mahmud, M. S., He, L., Zahid, A., Heinemann, P., Choi, D., Krawczyk, G., & Zhu, H. (2023). Detection and infected area segmentation of apple fire blight using image processing and deep transfer learning for site-specific management. Computers and Electronics in Agriculture, 209, 107862.
- Jesupriya, J., Mageswari, P. U., & Alli, A. (2025, January). Deep Learning-Based Transfer Learning with MobileNetV2 for Crop Disease Detection. In 2025 International Conference on Intelligent and Innovative Technologies in Computing, Electrical and Electronics (IITCEE) (pp. 1-9). IEEE.
- Singh, R., Sharma, N., & Gupta, R. (2023, November). Apple leaf disease detection using densenet121 transfer learning model. In 2023 International Conference on Research Methodologies in Knowledge Management, Artificial Intelligence and Telecommunication Engineering (RMKMATE) (pp. 1-5). IEEE.
- Hassan, S. M., Maji, A. K., Jasiński, M., Leonowicz, Z., & Jasińska, E. (2021). Identification of plant-leaf diseases using CNN and transfer-learning approach. Electronics, 10(12), 1388.
- Polder, G., Blok, P. M., van Daalen, T., Peller, J., & Mylonas, N. (2025). A smart camera with integrated deep learning processing for disease detection in open field crops of grape, apple, and carrot. Journal of Field Robotics. [CrossRef]
- Reddy, T. Reddy, T. & Rekha, K. (2021). Deep Leaf Disease Prediction Framework (DLDPF) with Transfer Learning for Automatic Leaf Disease Detection. 1408-1415. [CrossRef]
- Kumar, A., Nelson, L., & Gomathi, S. (2024). Transfer learning of VGG19 for the classification of apple leaf diseases. In 2024 2nd International Conference on Intelligent Data Communication Technologies and Internet of Things (IDCIoT), Bengaluru, India (pp. 1643-1648). [CrossRef]
- Ozden, Cevher. (2021). Apple leaf disease detection and classification based on transfer learning. TURKISH JOURNAL OF AGRICULTURE AND FORESTRY. 45. 775-783. [CrossRef]
- Santoso, C., Singadji, M., Purnama, D., Abdel, S., & Kharismawardani, A. (2024). Enhancing Apple Leaf Disease Detection with Deep Learning: From Model Training to Android App Integration. Journal of Applied Data Sciences, 6(1), 377-390. [CrossRef]
- Wang, Yunlu & Sun, Fenggang & Wang, Zhijun & Zhou, Zhongchang & Lan, Peng. (2022). Apple Leaf Disease Identification Method Based on Improved YoloV5. [CrossRef]
- Rishitha, T., Krishna Mohan, G. (2023). Apple Leaf Disease Prediction Using Deep Learning Technique. In: Jacob, I.J., Kolandapalayam Shanmugam, S., Izonin, I. (eds) Data Intelligence and Cognitive Informatics. Algorithms for Intelligent Systems. Springer, Singapore. [CrossRef]
- Zhu, S., Ma, W., Wang, J., Yang, M., Wang, Y., & Wang, C. (2023). EADD-YOLO: An efficient and accurate disease detector for apple leaf using improved lightweight YOLOv5. Frontiers in Plant Science, 14, 1120724. [CrossRef]
- Vivek Sharma, Ashish Kumar Tripathi, Himanshu Mittal, "DLMC-Net: Deeper lightweight multi-class classification model for plant leaf disease detection," Ecological Informatics, Vol. 75, 2023, 102025, ISSN 1574-9541. [CrossRef]
- Xu W and Wang R (2023) ALAD-YOLO:an lightweight and accurate detector for apple leaf diseases. Front. Plant Sci. 14:1204569. [CrossRef]
- Fu, L., Li, S., Sun, Y., Mu, Y., Hu, T., & Gong, H. (2022). Lightweight-convolutional neural network for apple leaf disease identification. Frontiers in Plant Science, 13, 831219.
- Li, L., Zhang, S., & Wang, B. (2021). Apple leaf disease identification with a small and imbalanced dataset based on lightweight convolutional networks. Sensors, 22(1), 173.
- Sun, H., Xu, H., Liu, B., He, D., He, J., Zhang, H., & Geng, N. (2021). MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Computers and Electronics in Agriculture, 189, 106379.
- Gao, L., Zhao, X., Yue, X., Yue, Y., Wang, X., Wu, H., & Zhang, X. (2024). A Lightweight YOLOv8 Model for Apple Leaf Disease Detection. Applied Sciences, 14(15), 6710.
- Wang, Y., Wang, Y., & Zhao, J. (2022). MGA-YOLO: A lightweight one-stage network for apple leaf disease detection. Frontiers in plant science, 13, 927424.
- Zhu, X., Li, J., Jia, R., Liu, B., Yao, Z., Yuan, A., ... & Zhang, H. (2022). Lad-net: A novel light weight model for early apple leaf pests and diseases classification. IEEE/ACM transactions on computational biology and bioinformatics, 20(2), 1156-1169.
- Zheng, J., Li, K., Wu, W., & Ruan, H. (2023). RepDI: A light-weight CPU network for apple leaf disease identification. Computers and Electronics in Agriculture, 212, 108122.
- Wang, B., Yang, H., Zhang, S., & Li, L. (2024). Identification of Multiple Diseases in Apple Leaf Based on Optimized Lightweight Convolutional Neural Network. Plants, 13(11), 1535.
- Xu, W., & Wang, R. (2023). ALAD-YOLO: An lightweight and accurate detector for apple leaf diseases. Frontiers in Plant Science, 14, 1204569.
- Wang, G., Sang, W., Xu, F., Gao, Y., Han, Y., & Liu, Q. (2025). An enhanced lightweight model for apple leaf disease detection in complex orchard environments. Frontiers in Plant Science, 16, 1545875.
- Zeng, W., Pang, J., Ni, K., Peng, P., & Hu, R. (2024). Apple leaf disease detection based on lightweight YOLOv8-GSSW. Applied Engineering in Agriculture, 40(5), 589-598.
- Sun, Z., Feng, Z., & Chen, Z. (2024). Highly Accurate and Lightweight Detection Model of Apple Leaf Diseases Based on YOLO. Agronomy, 14(6), 1331.
- Zhu, R., Zou, H., Li, Z., & Ni, R. (2022). Apple-Net: A model based on improved YOLOv5 to detect the apple leaf diseases. Plants, 12(1), 169.
- Liu, Z., Li, X. An improved YOLOv5-based apple leaf disease detection method. Sci Rep 14, 17508 (2024). [CrossRef]
- Li, Fengmei & Zheng, Yuhui & Liu, Song & Sun, Fengbo & Bai, Haoran. (2024). A Multi-objective Apple Leaf Disease Detection Algorithm Based on Improved TPH-YOLOV5. Applied Fruit Science. 66. 1-17. [CrossRef]
- Lv, M., & Su, W.-H. (2024). YOLOV5-CBAM-C3TR: an optimized model based on transformer module and attention mechanism for apple leaf disease detection. Frontiers in Plant Science, 14. [CrossRef]
- Li, H., Shi, L., Fang, S., & Yin, F. (2023). Real-Time Detection of Apple Leaf Diseases in Natural Scenes Based on YOLOv5. Agriculture, 13(4), 878. [CrossRef]
- Praveen Kumar S, Naveen Kumar K, Drone-based apple detection: Finding the depth of apples using YOLOv7 architecture with multi-head attention mechanism, Smart Agricultural Technology, Volume 5, 2023, 100311, ISSN 2772-3755. [CrossRef]
- Yan, Chunman & Yang, Kangyi. (2024). FSM-YOLO: Apple leaf disease detection network based on adaptive feature capture and spatial context awareness. Digital Signal Processing. 155. 104770. [CrossRef]
- Mathew, Midhun & Mahesh, Therese Yamuna. (2021). Determining The Region of Apple Leaf Affected by Disease Using YOLO V3. 1-4. [CrossRef]
- Yan, Bin & Pan, Fan & Lei, Xiaoyan & Liu, Zhijie & Yang, Fuzeng. (2021). A Real-Time Apple Targets Detection Method for Picking Robot Based on Improved YOLOv5. Remote Sensing. 13. 1619. [CrossRef]
- Chunman Yan, Kangyi Yang, FSM-YOLO: Apple leaf disease detection network based on adaptive feature capture and spatial context awareness, Digital Signal Processing, Volume 155, 2024, 104770, ISSN 1051-2004. [CrossRef]
- Li, F., Zheng, Y., Liu, S. et al. A Multi-objective Apple Leaf Disease Detection Algorithm Based on Improved TPH-YOLOV5. Applied Fruit Science 66, 399–415 (2024). [CrossRef]
- A. Haruna, I. A. Badi, L. J. Muhammad, A. Abuobieda and A. Altamimi, "CNN-LSTM Learning Approach for Classification of Foliar Disease of Apple," 2023 1st International Conference on Advanced Innovations in Smart Cities (ICAISC), Jeddah, Saudi Arabia, 2023, pp. 1-6. [CrossRef]
- Abeyrathna, R.M.Rasika D. & Nakaguchi, Victor & Minn, Arkar & Ahamed, Tofael. (2023). Apple Position Estimation for Robotic Harvesting Using YOLO and Deep-SORT Algorithms.
- Sun, Henan & Xu, Haowei & Bin, Liu & He, Dongjian & He, Jinrong & Zhang, Haixi & Geng, Nan. (2021). MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Computers and Electronics in Agriculture. 189. 106379. [CrossRef]
- Tian, Yunong & Yang, Guodong & Wang, Zhe & Li, En & Liang, Zize. (2019). Detection of Apple Lesions in Orchards Based on Deep Learning Methods of CycleGAN and YOLOV3-Dense. Journal of Sensors. 2019. [CrossRef]
- Mahmoud, Yasmin & Sakr, Nehal & Elmogy, Mohammed. (2023). Plant Disease Detection and Classification Using Machine Learning and Deep Learning Techniques: Current Trends and Challenges. 197-217. [CrossRef]
- Gou, C., Zafar, S., Hasnain, Z., Aslam, N., Iqbal, N., Abbas, S., Li, H., Li, J., Chen, B., Ragauskas, A. J., & Abbas, M. (2024). Machine and Deep Learning: Artificial Intelligence Application in Biotic and Abiotic Stress Management in Plants. Frontiers in bioscience (Landmark edition), 29(1), 20. [CrossRef]
- Hasan, S., Mahbub, R., & Islam, M. (2022). Disease detection of apple leaf with combination of color segmentation and modified DWT. Journal of King Saud University - Computer and Information Sciences, 34(9), 7212–7224. [CrossRef]
- A. Bracino, R. S. Concepcion, R. A. R. Bedruz, E. P. Dadios and R. R. P. Vicerra, "Development of a Hybrid Machine Learning Model for Apple (Malus domestica) Health Detection and Disease Classification," 2020 IEEE 12th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Manila, Philippines, 2020, pp. 1-6. [CrossRef]
- Sharma, M., & Jindal, V. (2023). Approximation techniques for apple disease detection and prediction using computer enabled technologies: A review. Remote Sensing Applications: Society and Environment, 32, 101038.
- Logashov, D., Shadrin, D., Somov, A., Pukalchik, M., Uryasheva, A., Gupta, H. P., & Rodichenko, N. (2021, June). Apple trees diseases detection through computer vision in embedded systems. In 2021 IEEE 30th International Symposium on Industrial Electronics (ISIE) (pp. 1-6). IEEE.
- Dubey, Shiv Ram & Jalal, Anand. (2016). Apple disease classification using color, texture and shape features from images. Signal Image and Video Processing. 10. 819-826. [CrossRef]
- A. Gargade and S. A. Khandekar, "A Review: Custard Apple Leaf Parameter Analysis and Leaf Disease Detection using Digital Image Processing," 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 2019, pp. 267-271. [CrossRef]
- Li, J., Huang, W., & Guo, Z. (2013). Detection of defects on apple using B-spline lighting correction method. In PIAGENG 2013: Image Processing and Photonics for Agricultural Engineering (Vol. 8761, p. 87610L). SPIE. [CrossRef]
- Hossein Azgomi, Fatemeh Roshannia Haredasht, Mohammad Reza Safari Motlagh, "Diagnosis of some apple fruit diseases by using image processing and artificial neural network," Food Control, Vol. 145, 2023, 109484, ISSN 0956-7135. [CrossRef]
- Singh, Swati & Gupta, Isha & Gupta, Sheifali & Koundal, Deepika & Aljahdali, Sultan & Mahajan, Shubham & Pandit, Amit. (2021). Deep Learning Based Automated Detection of Diseases from Apple Leaf Images. Computers, Materials & Continua. 71. 1849-1866. [CrossRef]
- Kang, H., & Chen, C. (2020). Fruit detection, segmentation and 3D visualisation of environments in apple orchards. Computers and Electronics in Agriculture, 171, 105302. [CrossRef]
- Manish Sharma, Vikas Jindal, "Approximation techniques for apple disease detection and prediction using computer enabled technologies: A review," Remote Sensing Applications: Society and Environment, Vol. 32, 2023, 101038, ISSN 2352-9385. [CrossRef]
- Qiu, Z., Xu, Y., Chen, C., Zhou, W., & Yu, G. (2024). Enhanced Disease Detection for Apple Leaves with Rotating Feature Extraction. Agronomy, 14(11), 2602. [CrossRef]
- Delalieux, Stephanie & van Aardt, Jan & Keulemans, Wannes & Coppin, Pol. (2005). Detection of biotic stress (Venturia inaequalis) in apple trees using hyperspectral analysis.
- Knauer, U., Warnemünde, S., Menz, P., Thielert, B., Klein, L., Holstein, K., Runne, M., & Jarausch, W. (2024). Detection of Apple Proliferation Disease Using Hyperspectral Imaging and Machine Learning Techniques. Sensors (Basel, Switzerland), 24(23), 7774. [CrossRef]
- Kim, Yunseop & Glenn, D.M. & Park, Johnny & Ngugi, Henry & Lehman, Brian. (2011). Hyperspectral image analysis for water stress detection of apple trees. Computers and Electronics in Agriculture - COMPUT ELECTRON AGRIC. 77. 155-160. [CrossRef]
- Shuaibu, Mubarakat & Lee, W. S. & Schueller, John & Gader, Paul & Hong, Young & Kim, Sangcheol. (2018). Unsupervised hyperspectral band selection for apple Marssonina blotch detection. Computers and Electronics in Agriculture. 148. 45-53. [CrossRef]
- N. Gorretta, M. Nouri, A. Herrero, A. Gowen and J. -M. Roger, "Early detection of the fungal disease "apple scab" using SWIR hyperspectral imaging," 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, Netherlands, 2019, pp. 1-4. [CrossRef]
- Ye, X., Abe, S. & Zhang, S. Estimation and mapping of nitrogen content in apple trees at leaf and canopy levels using hyperspectral imaging. Precision Agric 21, 198–225 (2020). [CrossRef]
- Chandel, Abhilash & Khot, Lav & Sallato, Bernardita. (2021). Apple powdery mildew infestation detection and mapping using high-resolution visible and multispectral aerial imaging technique. Scientia Horticulturae. 287. 110228. [CrossRef]
- Karpyshev, Pavel & Ilin, Valery & Kalinov, Ivan & Petrovsky, Alexander & Tsetserukou, Dzmitry. (2021). Autonomous Mobile Robot for Apple Plant Disease Detection based on CNN and Multi-Spectral Vision System. 157-162. [CrossRef]
- Blok, P. M., Polder, G., Peller, J., & van Daalen, T. (2022). OPTIMA - RGB colour images and multispectral images (including LabelImg annotations) (Version 1) [Data set]. Zenodo. [CrossRef]
- Alexander J. Bleasdale, J. Duncan Whyatt, Classifying early apple scab infections in multispectral imagery using convolutional neural networks, Artificial Intelligence in Agriculture, Volume 15, Issue 1, 2025, Pages 39-51, ISSN 2589-7217. [CrossRef]
- Barthel D, Cullinan C, Mejia-Aguilar A, Chuprikova E, McLeod BA, Kerschbamer C, Trenti M, Monsorno R, Prechsl UE, Janik K. Identification of spectral ranges that contribute to phytoplasma detection in apple trees - A step towards an on-site method. Spectrochim Acta A Mol Biomol Spectrosc. 2023 Dec 15;303:123246. Epub 2023 Aug 8. [CrossRef] [PubMed]
- Jiang, D., Chang, Q., Zhang, Z., Liu, Y., Zhang, Y., & Zheng, Z. (2023). Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images. Remote Sensing, 15(10), 2504. [CrossRef]
- Jang, S., Han, J., Cho, J., Jung, J., Lee, S., Lee, D., & Kim, J. (2024). Estimation of Apple Leaf Nitrogen Concentration Using Hyperspectral Imaging-Based Wavelength Selection and Machine Learning. Horticulturae, 10(1), 35. [CrossRef]
- Barthel, D., Dordevic, N., Fischnaller, S., Kerschbamer, C., Messner, M., Eisenstecken, D., Robatscher, P., & Janik, K. (2021). Detection of apple proliferation disease in Malus × domestica by near infrared reflectance analysis of leaves. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 263, 120178. [CrossRef]
- Prechsl, U.E., Mejia-Aguilar, A. & Cullinan, C.B. In vivo spectroscopy and machine learning for the early detection and classification of different stresses in apple trees. Sci Rep 13, 15857 (2023). [CrossRef]
- Neware, Rahul. (2019). Comparative Analysis of Land Cover Classification Using ML and SVM Classifier for LISS-iv Data. [CrossRef]
- Nguyen, T. T., Vandevoorde, K., Wouters, N., Kayacan, E., De Baerdemaeker, J. G., & Saeys, W. (2016). Detection of red and bicoloured apples on tree with an RGB-D camera. Biosystems Engineering, 146, 33-44.
- Yin, Z., Zhao, C., Zhang, W., Guo, P., Ma, Y., Wu, H., ... & Lu, Q. (2025). Nondestructive detection of apple watercore disease content based on 3D watercore model. Industrial Crops and Products, 228, 120888.
- Lin, G., Tang, Y., Zou, X., Xiong, J., & Fang, Y. (2020). Color-, depth-, and shape-based 3D fruit detection. Precision Agriculture, 21, 1-17.
- Schatzki, T. F., Haff, R. P., Young, R., Can, I., Le, L. C., & Toyofuku, N. (1997). Defect detection in apples by means of X-ray imaging. Transactions of the ASAE, 40(5), 1407-1415.
- Tempelaere, A., Van Doorselaer, L., He, J., Verboven, P., Tuytelaars, T., & Nicolai, B. (2023). Deep Learning for Apple Fruit Quality Inspection using X-Ray Imaging. In Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 552-560).
- Liangliang, Y. A. N. G., & Fuzeng, Y. A. N. G. (2011). Apple internal quality classification using X-ray and SVM. IFAC Proceedings Volumes, 44(1), 14145-14150.
- He, J., Van Doorselaer, L., Tempelaere, A., Vignero, J., Saeys, W., Bosmans, H., ... & Nicolai, B. (2024). Nondestructive internal disorders detection of ‘Braeburn’apple fruit by X-ray dark-field imaging and machine learning. Postharvest Biology and Technology, 214, 112981.
- Delalieux, S., Auwerkerken, A., Verstraeten, W. W., Somers, B., Valcke, R., Lhermitte, S., ... & Coppin, P. (2009). Hyperspectral reflectance and fluorescence imaging to detect scab induced stress in apple leaves. Remote sensing, 1(4), 858-874.
- Ariana, D., Guyer, D. E., & Shrestha, B. (2006). Integrating multispectral reflectance and fluorescence imaging for defect detection on apples. Computers and electronics in agriculture, 50(2), 148-161.
- Heyens, K., & Valcke, R. (2004, July). Fluorescence imaging of the infection pattern of apple leaves with Erwinia amylovora. In X International Workshop on Fire Blight 704 (pp. 69-74).
- Liu, Y., Zhang, Y., Jiang, D., Zhang, Z., & Chang, Q. (2023). Quantitative assessment of apple mosaic disease severity based on hyperspectral images and chlorophyll content. Remote Sensing, 15(8), 2202.
- Baranowski, P., Mazurek, W., Wozniak, J., & Majewska, U. (2012). Detection of early bruises in apples using hyperspectral data and thermal imaging. Journal of Food Engineering, 110(3), 345-355.
- M. M. U. Saleheen, M. S. Islam, R. Fahad, M. J. B. Belal and R. Khan, "IoT-Based Smart Agriculture Monitoring System," 2022 IEEE International Conference on Artificial Intelligence in Engineering and Technology (IICAIET), Kota Kinabalu, Malaysia, 2022, pp. 1–6. [CrossRef]
- Neware, Rahul & Khan, Amreen. (2018). SOFTWARE DEVELOPMENT FOR SATELLITE DATA ANALYTICS IN AGRICULTURE AREAS.
- Jiang, He & Li, Xiaoru & Safara, Fatemeh. (2021). IoT-based Agriculture: Deep Learning in Detecting Apple Fruit Diseases. Microprocessors and Microsystems. 104321. [CrossRef]
- R. Neware and A. Khan, "Survey on Classification Techniques Used in Remote Sensing for Satellite Images," 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 2018, pp. 1860–1863. [CrossRef]
- Ryo, M. (2022). Explainable artificial intelligence and interpretable machine learning for agricultural data analysis. Artificial Intelligence in Agriculture, 6, 257–265. [CrossRef]
- Coussement, K., Abedin, M. Z., Kraus, M., Maldonado, S., & Topuz, K. (2024). Explainable AI for enhanced decision-making. Decision Support Systems, 184, 114276. [CrossRef]
- Ali, S., Abuhmed, T., El-Sappagh, S., Muhammad, K., Alonso-Moral, J. M., Confalonieri, R., Guidotti, R., Del Ser, J., Díaz-Rodríguez, N., & Herrera, F. (2023). Explainable Artificial Intelligence (XAI): What we know and what is left to attain Trustworthy Artificial Intelligence. Information Fusion, 99, 101805. [CrossRef]
- El Sakka, M., Ivanovici, M., Chaari, L., & Mothe, J. (2025). A Review of CNN Applications in Smart Agriculture Using Multimodal Data. Sensors, 25(2), 472. [CrossRef]
- Kondaveeti, Hari & Vatsavayi, Valli Kumari & Mangapathi, Srileakhana & Yasaswini, Reddy. (2023). Lightweight Deep Learning: Introduction, Advancements, and Applications. [CrossRef]
- Wiley, Victor & Lucas, Thomas. (2018). Computer Vision and Image Processing: A Paper Review. International Journal of Artificial Intelligence Research. 2. 22. [CrossRef]
- Isaac Christopher, L. (2023). The Internet of Things: Connecting a Smarter World.
| Algorithm used | Feature type used | Accuracy | Advantages | Limitations |
| Naive Bayes | Texture (GLCM) | 91 - 96.43 | Simple, fast, effective on small data | Assumes feature independence |
| Support Vector Machine (SVM) | Texture, color, shape | 87 - 96 | Robust, good generalization | Computationally expensive for large datasets |
| k-Nearest Neighbors (k-NN) | Texture, color | 90 | Simple, interpretable | Sensitive to noise, slow on large data |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





