1. Introduction
The use of imaging for diagnosis in healthcare is substantial, amounting to about 100 billion dollars globally per year [
1]. Mounting pressures on the healthcare facilities and the market for imaging diagnosis have led to increasing demands for diagnostic excellence in the clinical setting due to rising number of clinical images, image complexities and faster results, as demanded by clinicians. As a result, the need for new technologies is centred around providing solutions that will increase the effectiveness of the clinical process, improving the healthcare systems and provide accurate diagnosis for the patients, while improving care quality. Therefore, there have been high demands for technologies that can aid the automation of workflows associated with the use of medical imaging for diagnosis, leading to advances in the use of artificial intelligence (AI) methods such as deep learning to assist radiologists in analysing complex image datasets [
2].
1.1. Deep Learning Overview
Deep learning is a subfield of machine learning which leverages the artificial neural networks (ANN) architecture to acquire knowledge from large datasets and perform intricate operations. One of the main advantages of deep learning techniques is their ability to mimic the information processing complexity of the human brain [
2,
3]. The field has been a studied since the 1980s but gained prominence in recent years. This is because of access to large datasets for model training, improved algorithm development and increased processing power of microprocessors. The structure of an ANN is an interconnection of nodes (sometimes referred to as processing elements or neurons) which can span up to multiple layers, depending on the intricacy of the tasks and capabilities of hardware resources. Each node gathers information from the previous layer and then transmits to the subsequent layer based on the configured characteristics and set parameters. The values of the parameters used are normally initially randomised but then iteratively updated during training based on a set learning rate [
4]. A deep learning network extracts deeper and intricate information as its number of architectural layers increases, resulting in optimized performance with large datasets and training iterations. This in turn ensures precisive recognition of patterns in the data [
3].
1.2. Deep Learning in Medical Imaging, Classification and Segmentation
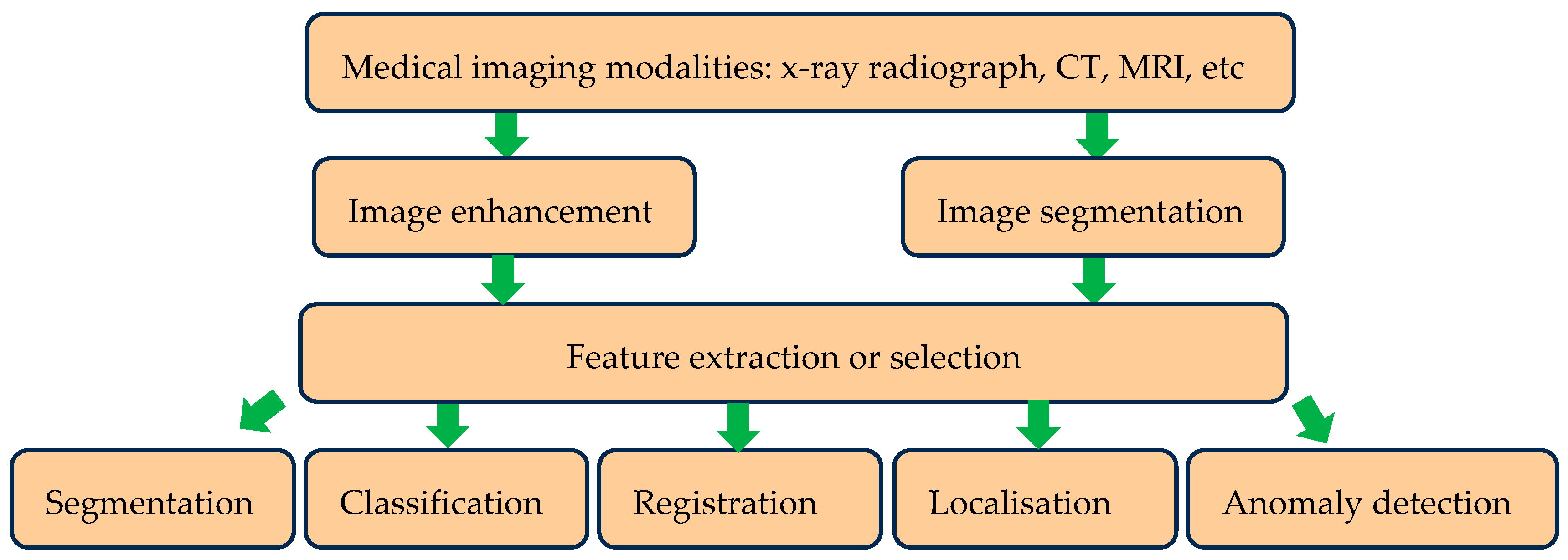
Deep learning has seen extensive applications in medical image analysis. It has been used for different medical image modalities including X-ray radiographs, CT and MRI scans to provide predictive diagnosis and treatment. Subtle and intricate patterns presented by these medical images are effectively identified by the adapted deep learning approach, thereby providing a means to automate the feature extraction process. Image feature extraction or selection is a process that can be performed manually by a qualified specialist, but this can be time consuming and subjective. When deep learning models are properly trained, they have an ability to accurately identify lesion or tumours, examine membranes and tissues for differences and do diverse medical related tasks thereby providing accelerated diagnostic outcomes. Therefore, deep learning is emerging into the realm of medical images analysis assisting with diagnosis. A summary of deep learning applications is shown in
Figure 1.
1.3. Challenges in Utilizing Deep learning in the Medical Field
Despite the significant advancements and capabilities of deep learning techniques in medical image analysis, there are certain limitations and challenges in their implementation and acceptance. For instance, most deep learning algorithms often lack explainability, i.e., they typically operate as black boxes [
5]
. In the medical field where decision making processes are required for diagnosis and provision of treatment, the deep learning method of convolutional neural network (CNN), for instance, does not provide an insight into the manner it came up with a decision. Other challenges that applications of deep learning techniques can face include:
Overfitting: This causes poor accuracy by the deep learning artificial neural networks in recognising images not included in the training set (i.e., the unseen images) even though the images in the training set were accurately recognised. Overfitting has several causes that include insufficient training imaging examples and excessive model parameters for the architecture. However, there are techniques that could be valuable to deal with overfitting occurs in constrained training data set scenarios. These include the drop out technique whereby some nodes in the architecture are temporarily left out during training [
6]. Another approach is to artificially extend the training data set through a process known as data augmentation [
7].
Image annotation: Many deep learning algorithms are supervised, i.e. they require labelled images indicating their categories during their training phase. The labelling requires annotation of images by qualified medical practitioners. Because deep learning requires large data for training, this process can be time consuming.
Noisy images: Medical images can be noisy. This distorts the quality of images being used to train the deep learning networks.
Image variations: Different medical imaging equipment can cause variations in the quality of the image they produce that can cause inconsistencies during training.
Privacy and ethics: There are concerns around ethics and privacy if the medical images are not properly anonymised.
Trust: There is an ongoing issue around relying on critical medical diagnostic results generated by when the manner of their generation is not sufficiently transparent [
4].
Computational requirement and environmental issues: training deep learning algorithms typically require high computational ability and long durations. Many general-purpose computers do not have the means of delivering the required computational resources and there is also the issue of the environmental aspects of using so much electrical energy to perform the required deep learning training.
This article explores deep learning techniques for medical image analysis, highlighting their roles in assisting diagnostic tasks. It examines CNNs for classification and segmentation, recurrent neural networks (RNNs) for sequential data, autoencoders for feature extraction, and generative adversarial networks (GANs) for data augmentation. It also discusses U-Net models for segmentation, vision transformers (ViTs) for long-range dependencies, and hybrid models that integrate multiple architectures. Different algorithms and their architectures are discussed. The associated mathematical models to demonstrate the manner different image modalities operate are presented. The study emphasizes the transformative impact of deep learning in medical diagnostics and suggests future improvements in efficiency and interpretability. In the following sections the materials and methods are explained, and the results are discussed.
2. Materials and Methods
A systematic review was undertaken to explore the latest developments in deep learning ANNs for analysing medical images. As this field is growing with many related articles published daily, a rapid review approach was undertaken [
6]. The necessary constraints and criteria for inclusion were used when searching for literature that provided information about recent deep learning techniques used for medical image analysis. The following keywords were combined as part of the literature search “deep learning” and “medical image analysis”. We also included image modalities and used “OR” and “AND” operators as illustrated below:
“Deep learning” AND “Medical image analysis”
“Deep learning” AND “Medical image analysis” OR “CT”
“Deep learning” AND “Medical image analysis” OR “MRI”
“Deep learning” AND “Medical image analysis” OR X-ray”
“Deep learning” AND “Medical image analysis” OR “Infrared Thermal Image”
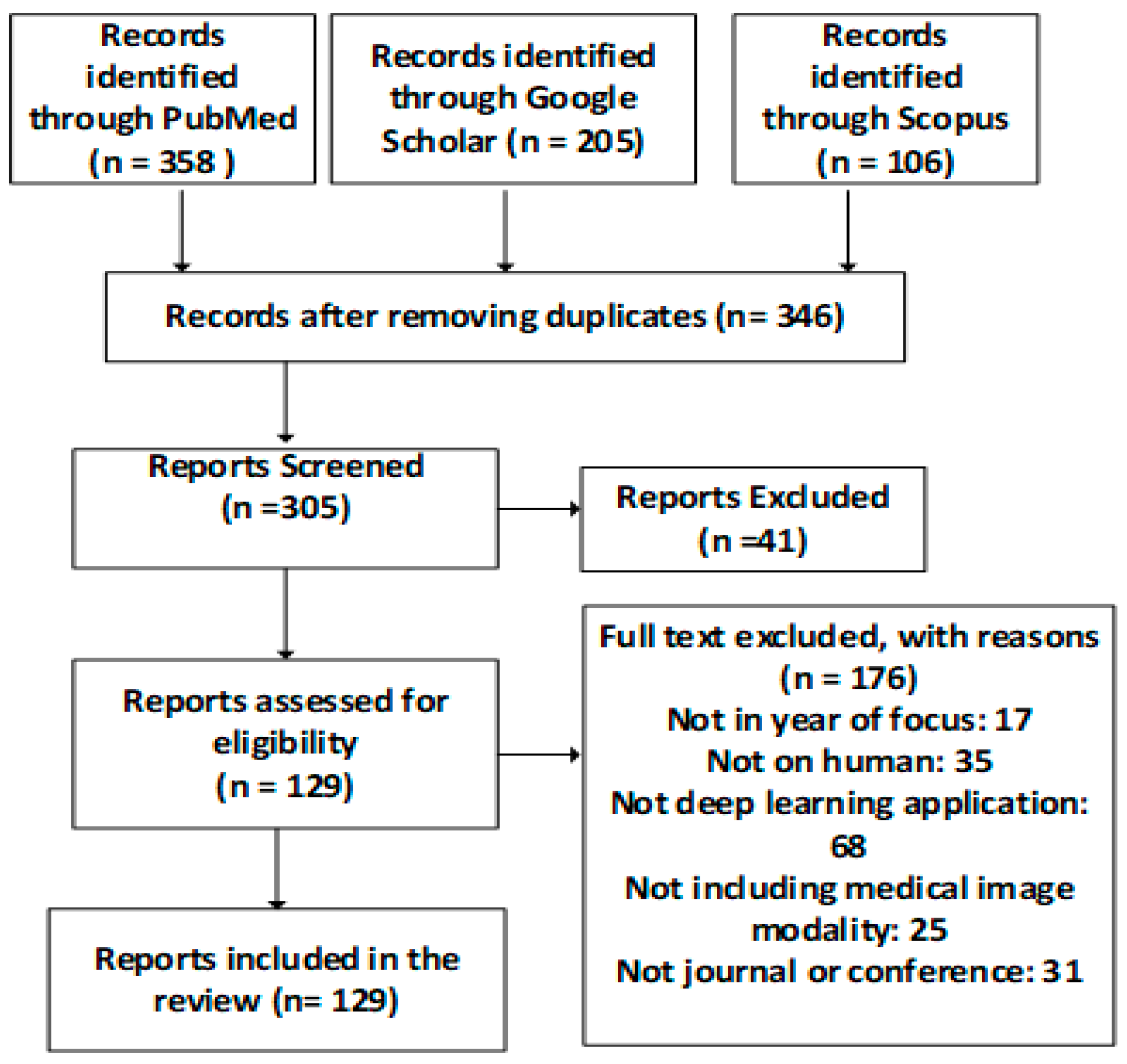
The search was performed on different scientific databases including Scopus, PubMed and Google Scholar and the inclusion and exclusion criteria as shown in
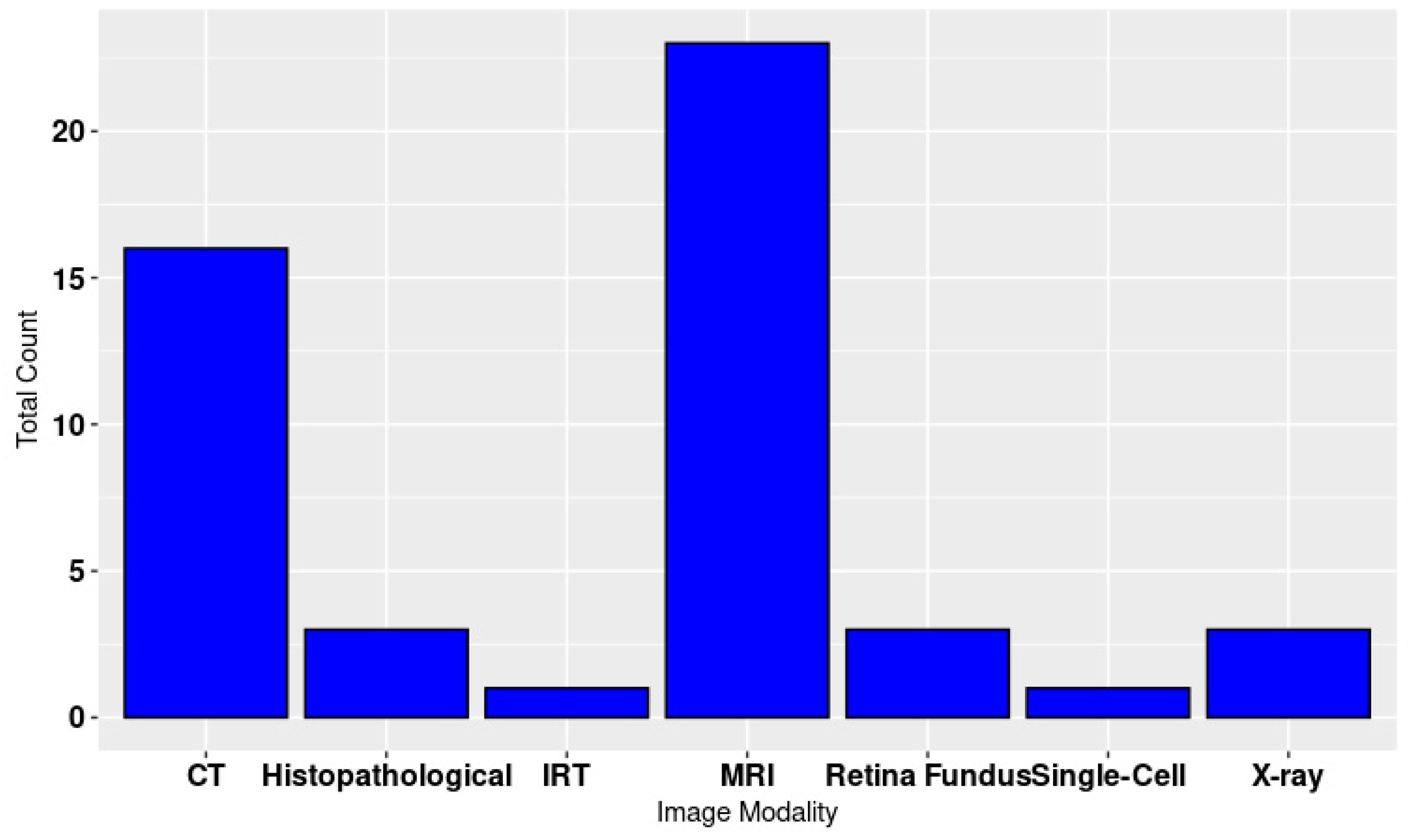
Figure 2. The image modalities are shown in
Figure 3.
We examined each article mainly focussing on the deep learning techniques deployed and the image modalities adopted. The articles that did not report either a deep learning algorithm or a medical imaging modalities were excluded. The remaining articles were then filtered to include the deep learning techniques in whole or in part as many works used a combination of deep learning techniques. The categories included were: CNN, RNN, autoencoders, GANs, transfer learning (TL), ViTs and hybrid models. We also considered studies that combined multi-deep learning algorithms, i.e. hybrid systems further refined our search to include the terms “Hybrid”. For completeness, a summary description of each of method is included as part of the review. As CNN architectures were widely deployed, mostly with other deep learning techniques, we further narrowed the search terms to include only publications that specifically used CNN by updating the search terms to include: (“Convolutional neural network” AND “Medical image analysis”) OR (“CNN” AND “Medical image analysis”).
Over 500 different studies were first identified across the databases during the initial criteria with some duplicated across the databases, especially in PubMed and Google Scholar. After applying all the constraints, this was refined to 129 articles that was eventually reviewed in this study. The 129 articles were distributed over all the identified categories, and these were based on the applied constraints. We have included a broad imaging modality. The focus was to identify the deep learning techniques most suitable for medical imaging tasks, the processes employed to improve the monitoring and diagnosis performance of the deep learning techniques, and the optimisation performed.
3. Results
This section contains an overview of deep learning neural network concepts.
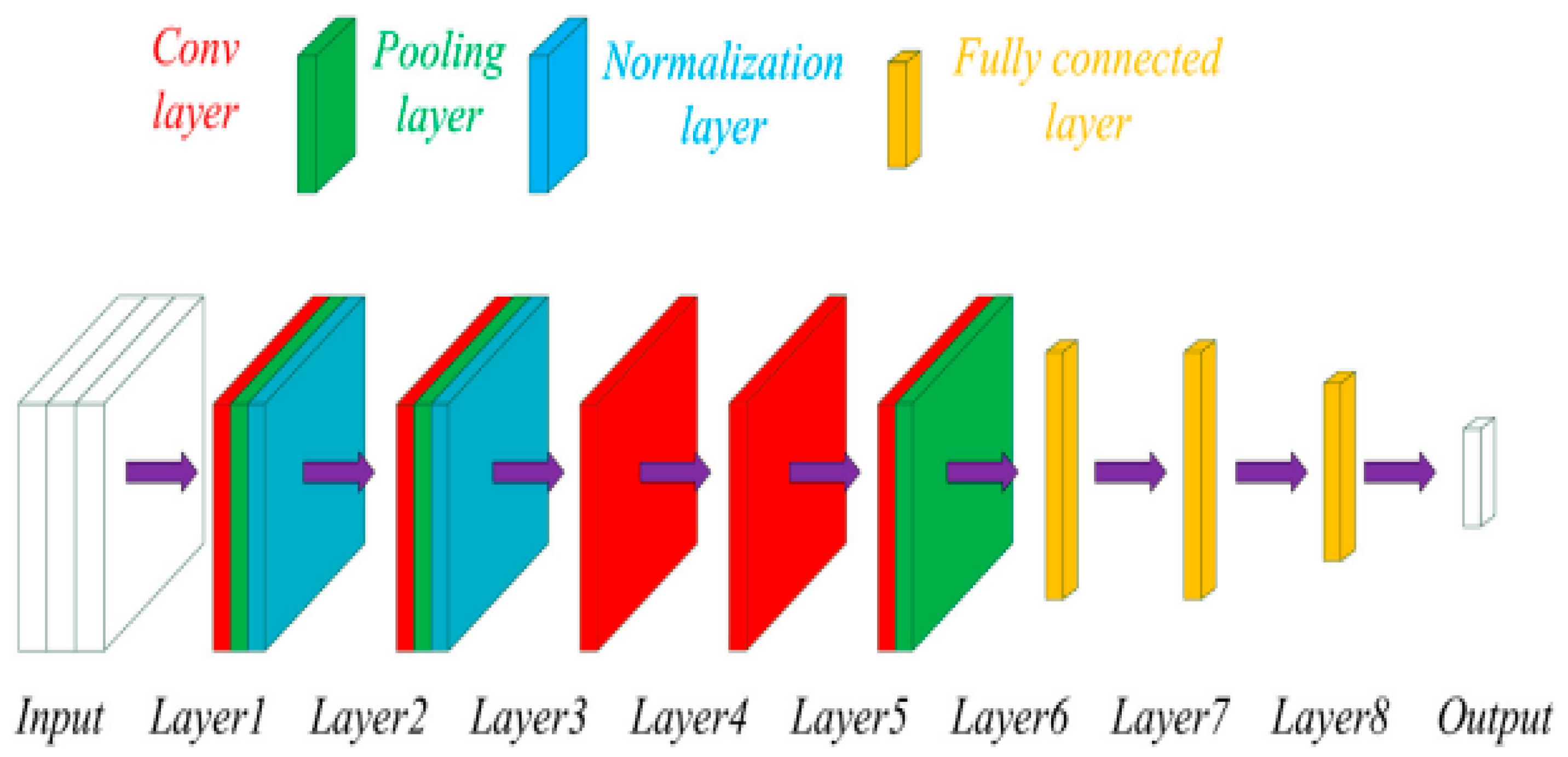
3.1. Convolutioal Neural Network
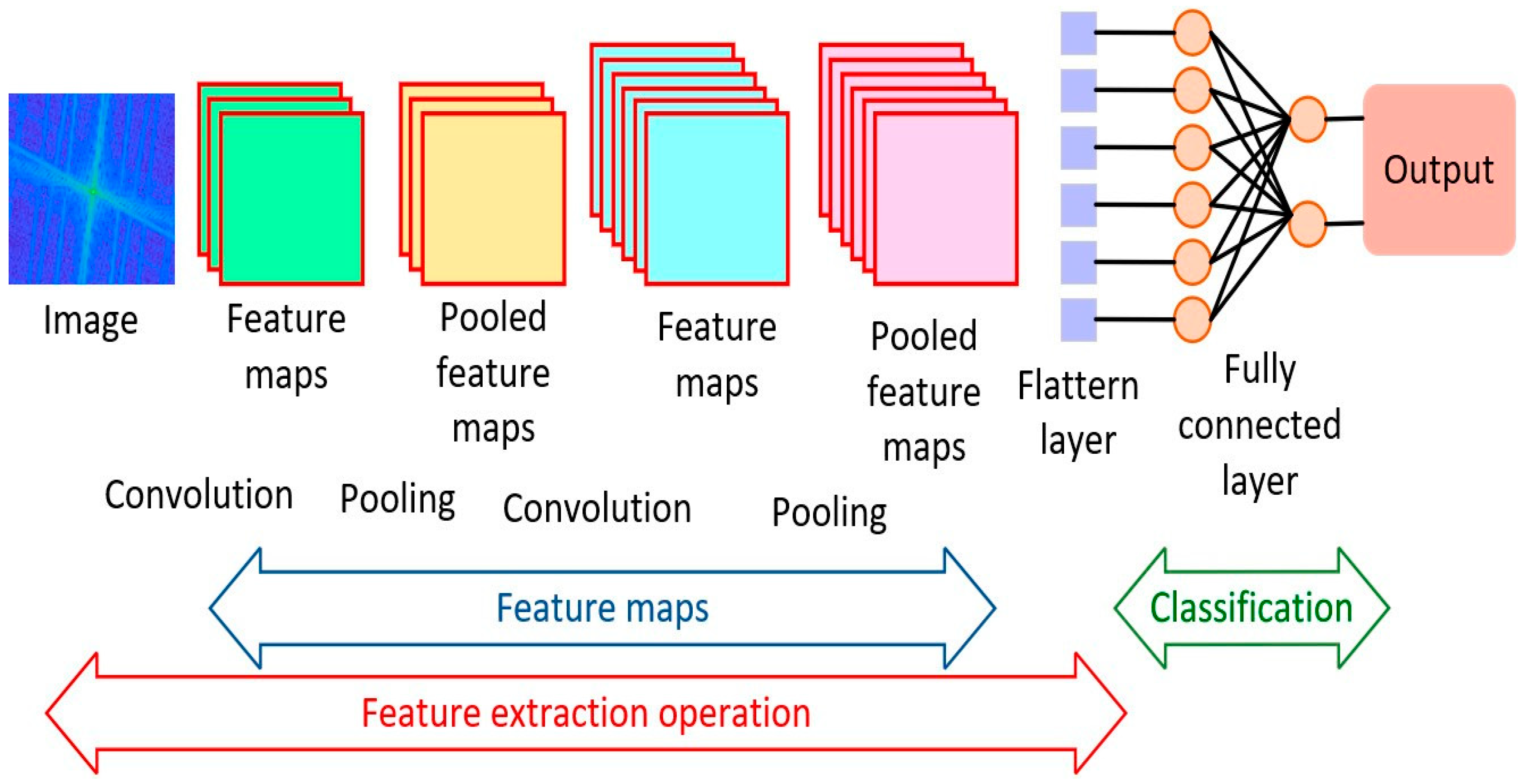
The CNN were the dominant technique for analysing medical images for diagnostic purposes [
7]. It has gained immense popularity since 2012 when high performance computing (HPC) became more accessible. This led to the ImageNet competition for different combination of the deep CNN networks to achieve better diagnostics results when compared to the human experts. CNN has been effective for several tasks such as image segmentation, detection, registration, localisation, and classification [
5]. They consist of numerous layers of convolutional filters with nonlinear activation functions, combined with pooling layers, dropout layers and fully connected layers. Their ability to extract complex spatial relationships and patterns in images has seen them used in various medical imaging modalities such as magnetic resonance imaging (MRI), computed tomography (CT), X-ray radiographs, ultrasounds (US), histopathology and more recently in infrared thermal imaging (IRT) [
8]. The images associated with multiple diseases have been segmented, classified, registered and interpreted, ranging from bone fractures, cancer, liver diseases, pneumonia, Covid 19, etc. The CNN architecture applied to the discrete Fourier transformed infrared thermal images is shown in
Figure 4 [
10]. Although the study was a pilot, the model was effective in screening wrist fracture in children.
The concept of convolution is simply a mathematical operation where a filter, also known as kernel, is applied on an input image for feature extraction. The convolution process is described mathematically next. Let the input image to the CNN be represented by a 3D matrix such that
where
is the height of the image,
is the width of the image and
is the depth of the image (it represents the number of channels which is usually 3 for an RGB image). When applied to a set of convolutional filters for each input, the output of the convolutional filter can be represented by:
where
is the output feature map for the filter
at position
,
is the convolutional filter (kernel)
are the height and width of the kernel which is usually a sample of the size of the input. This process helps to identify local patterns such as the edges, shapes and textures from the input image.
When the CNN algorithm is used for classification purposes, it is combined with other layers such as the activation layers, pooling layer and the fully connected layer. The output from the convolutional filter is passed through an activation function
. If the rectified linear unit (ReLU) activation function is applied, the output from the convolution layer becomes:
This operation helps to add nonlinearity by ensuring only positive values are retained thereby helping to learn complex patterns from the image.
The pooling layer is then applied to the output of the activation layers. The effect of the pooling operation is to reduce spatial dimensions from the convolution operation thus help the network to capture small translations in the image. This process also helps to reduce the computational complexity of the network. Assuming the
max (maximum) pooling function is applied to the activation layer, with a
window size, the output from the pooling layer is represented by:
The fully connected layer is connected to the output of a flattened
max pooling layer output. This layer takes a vector value and is a standard neural network with each input connected to all the neurons in the next layer. The output of the fully connected (FC) layer is given by:
where
is the input into the FC layer and
and
represent the weights and biases of the FC layer. The final or output layer which is also a vector is mostly passed through a SoftMax function for a classification task. This function helps to convert the output classes into probabilities with the following expression:
where
represents the probability of predicting the class
and
represents the total number of classes being differentiated. The full CNN network undergoes training to correctly learn the features of the image input. This training uses a loss function, usually a cross-entropy loss when considering a classification task. The cross-entropy loss is given by:
where
and
is the predicted probability for class
and the target label respectively.
The gradient of the loss function is calculated with respect to the weights through back propagation formula,
The weights are updated using the gradient descent, i.e.,
where
(
) represents the learning rate controlling convergence rate.
3.1.1. Literature Review Findings for CNN
Most studies have used CNN individually while there were studies that applied it in combination with another algorithm such as UNET and GANs for medical image diagnostics. However, in this section the focus is on CNN application on its own. A lightweight CNN algorithm was used to detect Covid-19 from chest X-ray radiograph images [
9]. Their proposed CNN network was inspired by the ResNet model [
10] in which all layers were not connected in sequence, creating a skip connection whereby neurons in a particular layer can be connected to another neuron further ahead. This arrangement helped to create a lightweight model that was effective in edge detection applications. Their study was compared with other CNN models such as CVDNet and Deep GRU-CNN and showed a similar performance with the models compared with a reduced computational complexity. A deeper CNN in their work was proposed that used CT scans and X-ray radiographs for the detection of several pulmonary diseases including Covid-19 and viral pneumonia [
11]. They used varied datasets, consisting of different image modalities for their work which provided another dimension of the efficacy of CNN in diagnostic imaging. They proposed a 26-layer deep CNN network which was inspired by a wide residual network (WRN) [
12]. It provided a faster training time when considering the deep nature of the architecture. This was achievable based on the sophistication of their hardware. Their model was effective in terms of the accuracy in detecting the different pulmonary diseases when compared to the traditional methods. A CNN model was used to classify ultrasonic images of fatty liver [
15]. A pertinent problem in their research was the similarity in the pathological ultrasonic images used for training the CNN algorithm. This sort of challenge might pose a problem for the CNN architecture as it may struggle to extract distinct features for the different pathological images. Therefore, there will be a need for deep convolution layers leading to computational complexities. They however used pixel-level feature extraction as a preprocessing step and then proposed a CNN architecture comprising of two convolutional layers, a pooling layer and a fully connected layer. They also experimented the proposed method with a skip connection and improved the accuracy when compared to other algorithms such as VGGNet.
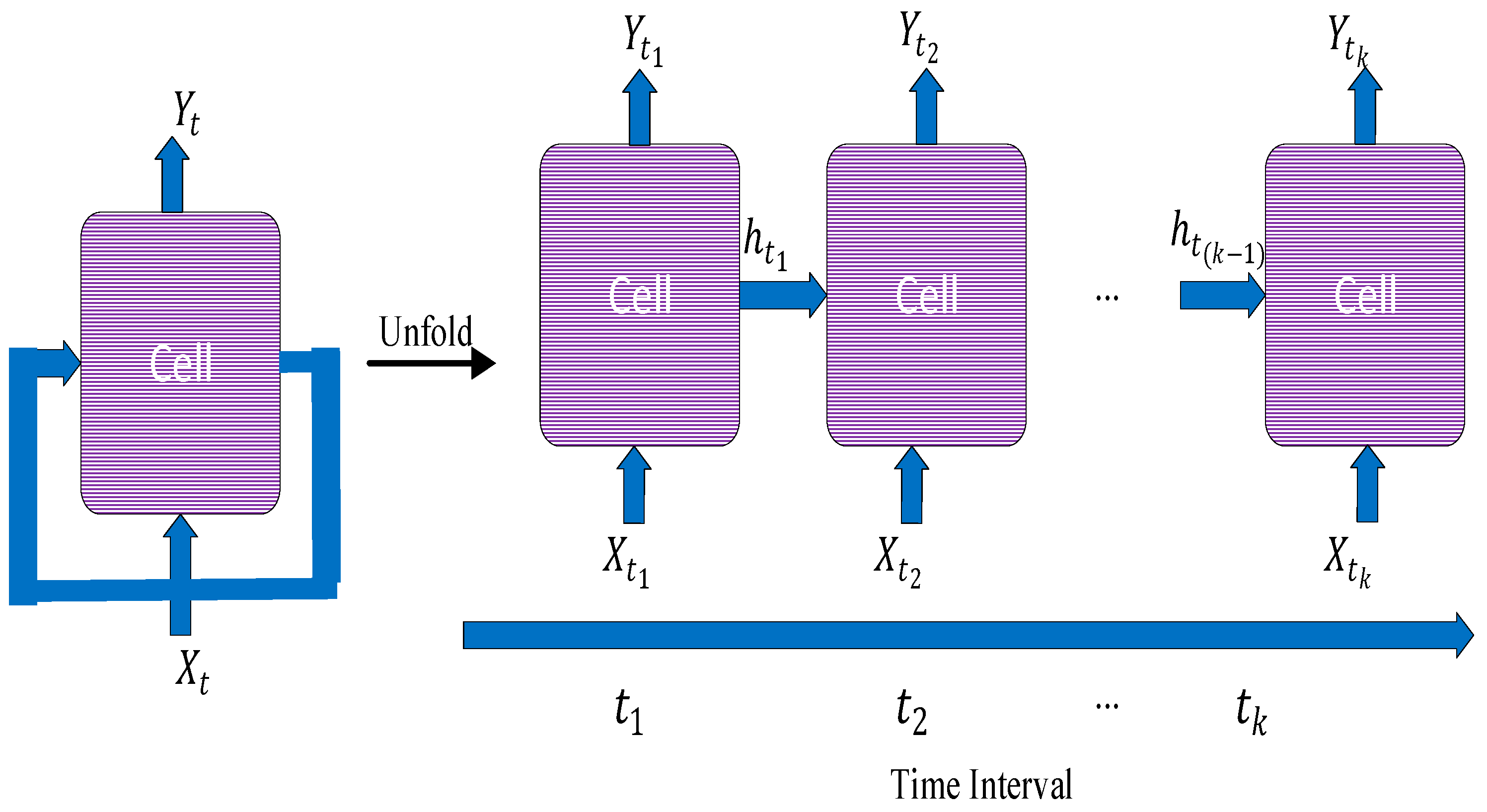
3.2. Recurrent Neural Network
Recurrent neural networks (RNNs) are another class of neural networks that have gained significant research consideration for modelling of sequential data. Their variability of the length of their input and output also makes them suitable for natural language processing tasks [
1]. A RNN has an internal memory unlike feedforward networks which helps to keep memory of the hidden states across different time stamps. In RNN, there is a feedback loop between the outputs of the hidden layers [
13]. This arrangement allows the RNNs to learn sequential patterns, making them suitable for tasks such as time series prediction and video analysis [
14]. RNN architecture normally suffers from the issue of vanishing gradients [
15]. Their use transcends sequential and textual data. They can also be applied to image modalities with time-series characteristic information such dynamic imaging in functional MRIs to monitor progress of disease in a patient and to check how they respond to treatments. This functionality has been made popular by a variant of the network known as Long Short-Term Memory Network (LSTM). The LSTM works by introducing self-loops to allow for the flow of gradients for long durations [
16]. The recurrent structure of the neural network is always enforced by the LSTM by introducing gating functions on the neurons in the hidden layer. The architecture of the RNN is shown in
Figure 5.
The mathematical representation of RNN is described next. Let the sequential input to the RNN at time step
be
where
is a vector. Hence the input sequence becomes
where
represents numbers of time steps in the sequence. For every time step
, a hidden state
is maintained. Each hidden state is updated as
where
represents the hidden states or recurrent weights,
is the weight of the connection between the input layer and the hidden layer and
is the bias for the hidden state. The function
represents the activation function, typically the ReLU or the hyperbolic tangent. To obtain the output of the RNN at each time step, the output from each hidden state is multiplied by its associated weights and added with a chosen bias vector. This is represented as
where
represents the weight matrix between the hidden state and the output, and
represents the output bias vector. The function
is the activation function for the output, which could be task dependent. SoftMax activation function is mostly employed for classification tasks.
3.2.1. Literature Review Findings for RNN
RNN is widely used for machine learning or deep learning tasks that are time related. For image diagnosing, it can be used to monitor the disease progression and patient response to treatments, based on different time stamp that the images are being taken. Most works that have based their classification task on RNN, typically combined it with CNN for the feature extraction stage as shown in
Table 1.
MRI images were used for classification of Alzheimer’s disease by analysing the longitudinal sequence of the MRI images taken at different time steps to measure the disease progression [
17]. They combined CNN for feature extraction and RNN for classification. For the RNN architecture, they cascaded three bidirectional gated recurrent units (BGRU) with the inputs from the CNN network at multiple points, providing a longitudinal analysis. RNN architectures can suffer from vanishing gradients when the sequence of the images becomes too long as they do not contain memory units to store sequences [
23] , hence the reason for developing its variants such as LSTM and GRU. RNN can also be used on their own without the addition of gated memory units. An RNN was used, with slight modification, for the classification of breast cancer with microscopic histopathological image [
17]. The RNN architecture was gain modulated using the honey badger algorithm (HBA) by updating the weights of the RNN during training. The capacity in which RNN was used in their study is for classification of different stages of breast cancer and not disease progression hence why the gated memory approaches were not used. RNN can also be used for image segmentation tasks, especially when the images of the part of the body are a moving part, requiring sequencing the time frame when image is taken for segmenting anomalies. An RNN-based LSTM model was proposed together with U-NET to label aortic MRI images [
19]. This sort of tasks use the capacity of the RNN architecture being able to segment temporal images as most annotated images normally used for classification are static. The U-NET algorithm was combined with CNN for feature extraction with the sequencing part achieved with the RNN algorithm. The ability of the neural network to visualise the different thousands of images compared to the human makes the RNNs suitable for segmenting image labels for disease identification. RNNs can also been used to denoise diagnostic images. Images used for diagnosis can be susceptible to noise such as white, salt and pepper noise. A LSTM model was combined with particle swarm optimisation (PSO) algorithms to optimise the batch normalisation process of the RNN training to remove noise from CT images of the lungs [
18]. The technique provided improved peak signal to noise ratio (PSNR) when compared to the traditional noise removal techniques such filtering-based and diffusion-based techniques.
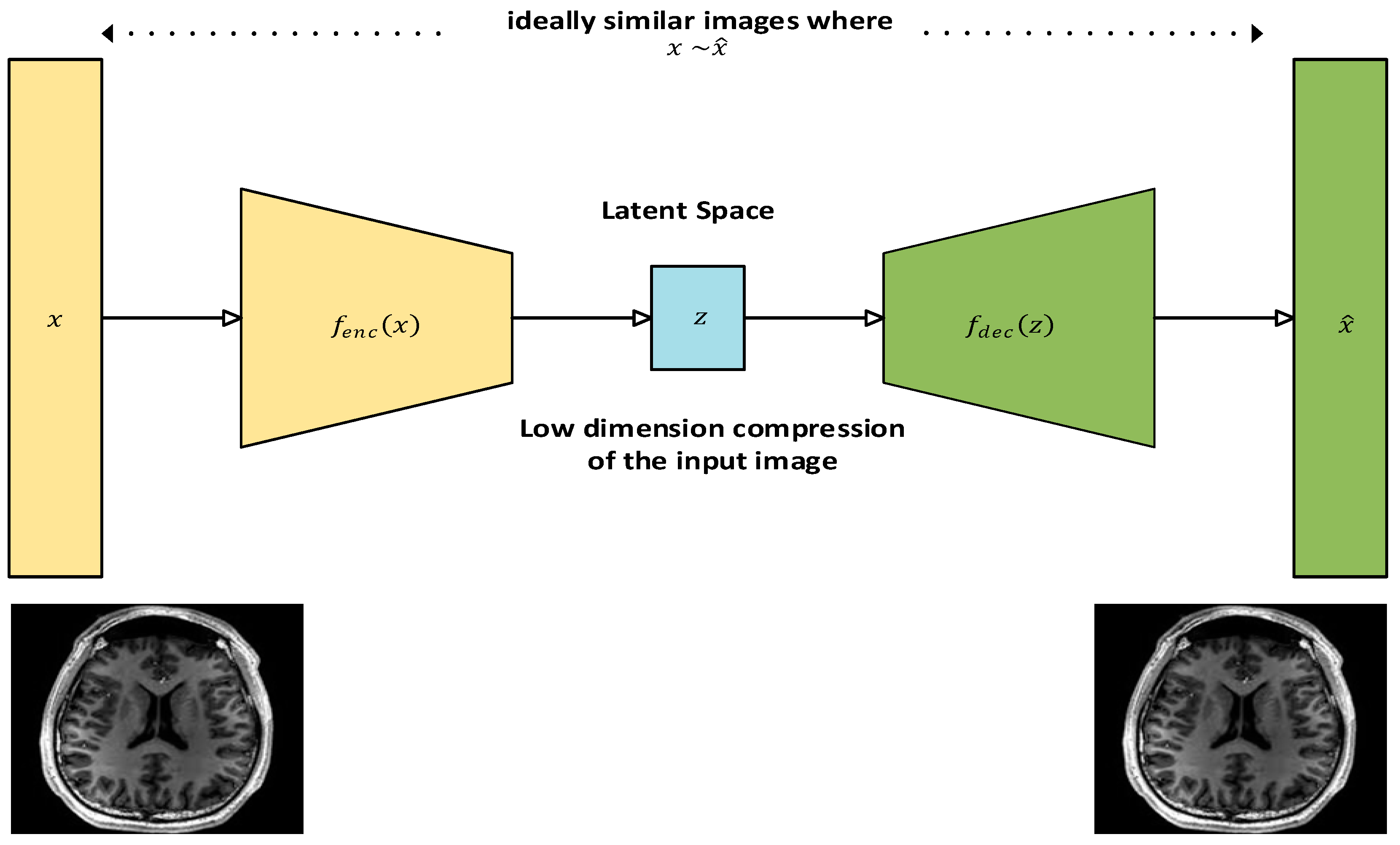
3.3. Autoencoders
Autoencoders are unsupervised learning models used for dimensionality reduction and feature extraction with minimal distortion when their input is compared to their output [
24]. They play an important role in the deep learning paradigm for medical image analysis [
25]. They can help denoise or compress medical images and are useful in anomaly detection, where unusual patterns in images indicate potential medical issues. They can also be used as a semi-supervised deep learning model to produce annotated data in situations where there is a lack of substantial amount of annotated dataset available for training any deep learning network for tasks such as classification or segmentation [
26]. Their architecture consists of an encoder and decoder structure with a latent space to store the value of the compressed data. Both the encoder and the decoder comprise of a fully connected feedforward neural network. The encoder converts the input image into a low dimension compressed version, which is referred to as latent space or the encoder. The latent space contains only essential features of the input from the encoder and is kept as shallow as possible in terms of the number of neurons used to retain the compressed version of the input and computational efficiency. The encoder in turn transforms the latent space to a reconstruction of the input. A loss function is generally used during training to compare the input with its reconstruction [
27]. The architecture of an autoencoder with an MRI image as its input is shown in
Figure 6.
The mathematical representation for the dimensionality reduction function of the autoencoder’s encoder and decoder is described next. Let the input image to the encoder be represented by
The encoder in turn transforms the input
into a lower-dimensional latent representation
where
.
The transformation function that produced the latent information is given as:
where
is the encoder weight matrix,
represented the encoder’s bias vector and
represents the selected activation function of the encoder which is commonly ReLU or sigmoid function. The latent representation
of the input
provides compressed information by only capturing the essential features of the input image, a feature referred to as bottleneck in which the dimension of the latent feature is smaller than the input vector. This facilitates the autoencoder to learn a better way to efficiently represent the input vectors. The decoder network tries to transform the latent space
back to the reconstructed input
to match the input vectors
. The decoder function can be represented as thus:
The loss function is used to minimize the difference between the input and the reconstructed input . This difference is normally quantified by binary cross-entropy error (BCE) when the dataset is binary or Mean Squared Error (MSE) for multi-class data.
The MSE loss computed with the following function
And the BCE loss is computed as follows
Training of the autoencoder requires the finding of the parameters for the weights and biases of the encoder and decoder such that the reconstruction loss is minimised over the training data. Gradient based optimization techniques such as stochastic gradient descent (SGD) or Adam can be used for this purpose.
Table 2.
Autoencoder based techniques.
Table 2.
Autoencoder based techniques.
| Article |
Image
modality |
Task |
Disease/
body part |
| [26] |
MRI |
Augmentation/
segmentation |
Brain |
| [28] |
MRI |
Denoising |
Prostate |
| [29] |
CT + others |
Classification |
Face |
| [30] |
CT |
Augmentation |
Various |
| [31] |
MRI & CT |
Classification |
Intracerebral
hemorrhage |
| [32] |
X-ray/digital Histopathology |
Anomaly detection |
Various |
| [33] |
Single-cell images |
Classification |
Myeloid Leukemia |
| [34] |
None |
Anomaly detection |
None |
| [35] |
CT |
Classification |
Covid-19 |
| [36] |
MRI |
Denoising/classification |
Autism/Brain |
3.3.1. Literature Review Findings for Autoencoder
Autoencoders are associated with both unsupervised and semi-supervised deep learning tasks. These tasks are normally preferred in the absence of ample annotated datasets required for deep learning activities especially in imaging analytics. Autoencoders are used to augment small number of annotated datasets often before a segmentation task. For example, a method known as GenSeg was proposed which combines the generative aspects of the autoencoders to generate the latent representation of the tumour cells from a labelled health image and using U-NET architecture to obtain the unique information of tumours present in the MRI images. There were also other related studies [
30]. Noise reduction is another task that has benefitted from generative features of the autoencoder framework. The fusion of Bayes shrinkage fused wavelet transform (BSbFWT) was proposed for noise removal and an auto encoder block for generative a noiseless variant of an MRI image of prostate cancer [
31]. As MRI images can be prone to Gaussian and Rician noise which is introduced during image capturing by the MRI device and the imaging environment. The effectiveness of their noise reduced generated images is measured using several parameters like values of peak signal to noise ratio (PSNR), mean squared error (MSE), structural similarity index metric (SSIM) and mean absolute error (MAE) which outperforms numerous traditional filtering approaches presented in their work. Auto encoders can also be used in classification tasks [
29,
31,
33,
35]. Autoencoders were used to mesh a fully convoluted network for a classification task [
32]. They have proposed a Convolutional Mesh Autoencoder (CMA) framework for the classification of syndromic craniosynostosis (SC) of three known SC variants including Muenke, Crouzon and Apert disease in infants and adults using 3D computed tomography data (CT) images and others with very good percentage on evaluation metrices such as sensitivity, specificity and accuracy when compared to the human counterpart. These tasks were very critical as late and inaccurate diagnosis might prove irreversible, causing permanent damage to the brain. As indicated, an autoencoder which is infused with four convolutional layers for encoding and decoding was used for the construction of the face models from the CT images. Autoencoder were also used to detect complex anomalies presented in medical imaging [
32,
34,
36]. Autoencoders were applied to chest X-ray and digital pathology images [
35]. Abnormalities that are barely visible such as metastases in lymph nodes always proved difficult to detect as they resemble normal images in pathological slides. They proposed a deep perpetual autoencoder that learnt the shared patterns of normal images and content similarities with abnormal ones and restores them correctly. For evaluation of their mode, they used the receiver operating characteristics (ROC) as it integrates the classification performance of the normal and the abnormal class. Their models were also evaluated on non-medical related images. Their model performed well with medical images when compared to non-medical images making autoencoder suitable in medical imaging climes.
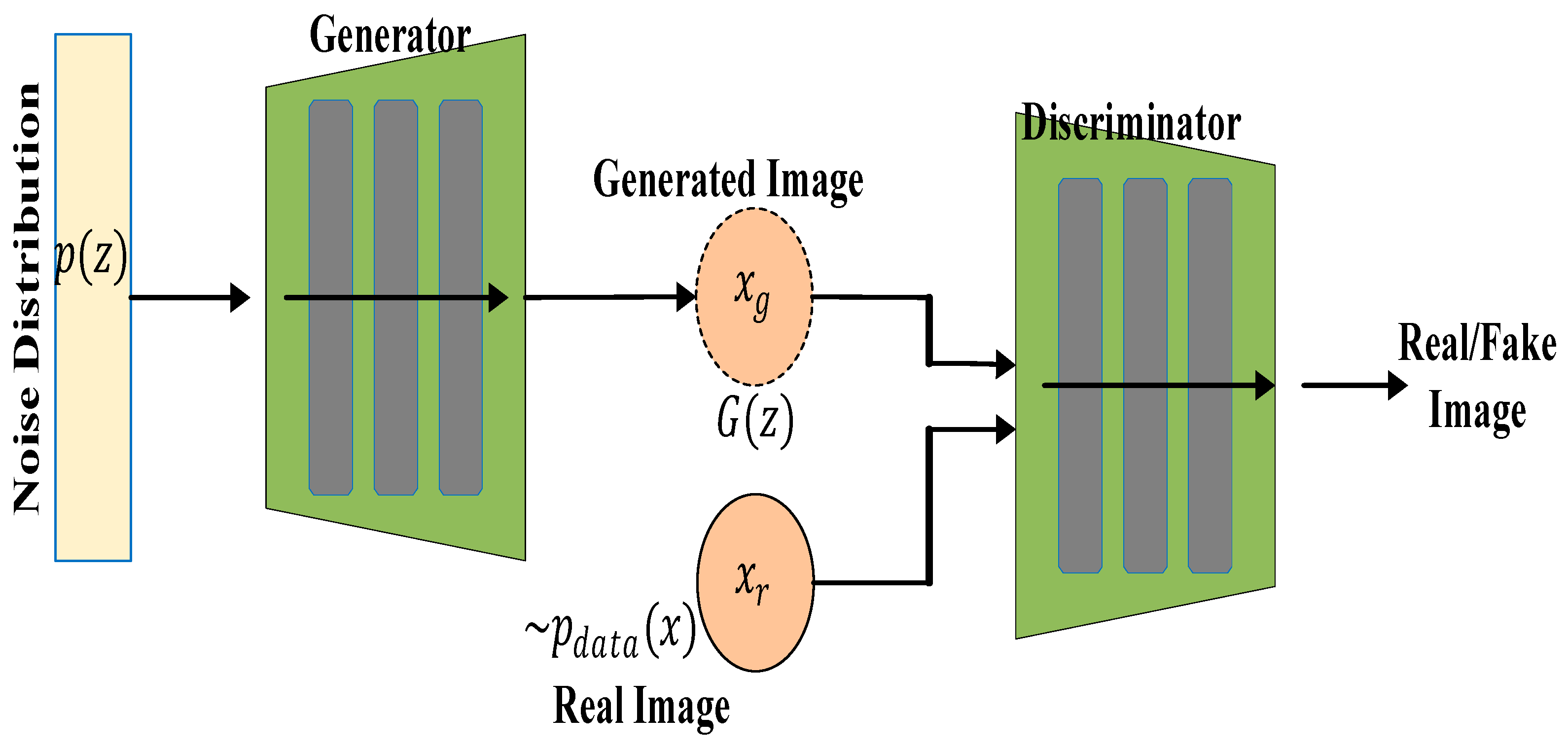
3.4. Generative Adversarial Network
Generative adversarial networks (GANs) are groups of deep learning artificial neural networks that can be used for generating synthetic medical images, data augmentation, and improving image resolution. They are valuable for enhancing small datasets, which are common in medical imaging, and can also be used to create better training data for improving model performance. GAN uses an unsupervised learning algorithm and can be used for mostly semi-supervised and unsupervised learning [
37,
38]. A GAN network consists of two main parts, namely the generator (G) and the discriminator (D). The generator, which comprises of a multi-layer perceptron (LP), learns the data distribution of the input image and produces a similar image to the input, also known as “fake data”. The job of the discriminator, which is also an MLP, is to discriminate between the generated image from the generator network and the input image. The result of this discrimination, which constitutes an error between the original input image and the generated image is fed back to the generator input to make the generated image more realistic i.e., closer to the original image. During training, the weights of the generator and discriminator are alternately updated, and the weights updates of the generator come from the discrimination error. Both networks are engaged in a competing optimization process. This process continues until there is an equilibrium between the generator and the discriminator networks [
1,
39,
40]. The architecture of the GAN network is shown in
Figure 7.
The Generator (G) consists of an input noise vector
such that
where
is a prior distribution which could be Gaussian. The output of the generator, which could be an image is represented by
and the objective is to maximize
. The input to the discriminator (D) is the image data sample
and the output of the generator network. The output to the discriminator is the probability that
is real or fake. This is represented by
where 0 represents fake and 1 represents real. The goal of the discriminator network is to maximize
. The discriminator loss
is given by
where
maximises the probability of correctly classifying the real data and
minimises the probability of misclassifying the generated data. When both generator and discriminator are at a stable equilibrium, the combined minimax game function is defined as
Table 3.
GAN based techniques.
Table 3.
GAN based techniques.
| Article |
Image Modality |
Task |
Disease/body part |
Variant used |
| [37] |
MRI/Retina fundus |
Image synthesis |
- |
- |
| [41] |
MRI |
Image resolution |
Brain |
Cycle-GAN |
| [42] |
CT |
Image synthesis |
Covid |
Enhanced vanilla |
| [43] |
X-Ray/CT |
Image Denoising |
Chest/Thorax |
CGAN |
| [44] |
Various |
Image resolution |
Various |
Enhanced vanilla |
| [45] |
- |
Image synthesis |
Skin cancer |
DCGAN |
| [46] |
MRI/CT |
Image synthesis |
Head/Neck |
Vanilla GAN |
| [47] |
MRI/CT |
Image resolution |
Bladder cancer |
Enhanced Vanilla |
| [48] |
Retina Fundus/MRI |
Image resolution |
Various |
Vanilla GAN |
| [49] |
CT/MRI |
Translation |
Thorax/brain |
CGAN |
3.4.1. Literature Review Findings for GAN
Deep learning technique such as GANs can be used in medical imaging in two different ways. The generative network in GANs has been used for image synthesis purposes to generate synthetic datasets for tasks where there are limited annotated datasets, and using the discriminating networks of GANs for anomaly detection [
38]. The quality of the generated synthetic images can be measured by the use of some qualitative metrics such as Fréchet inception distance (FID) which is a measure of similarities between the representations of the generated and the real input images, structural similarity index measure (SSIM), which indicates the similarities of the structures (usually image contrast and brightness) and peak signal to noise ratio (PSNR), which are used to analyse the sensitivity of the generated image. PSNR is the most important metrics when dealing with medical images. Quantitative metrics used for generated image quality includes number of parameters (NoP), to represent the total number of trainable parameters in the GAN network, and floating points of operations (FLoPs), which measures the cost of computation of the network [
37]. Most studies that have used GANs have however been used for imaged synthesis, image resolution, image translation, and image denoising. For example, a lightweight GAN (LEGAN) was proposed to generate high fidelity images from MRIs and retina fundus images [
40]. Their technique boasts of fewer parameters used in the training process and lower FID when compared to other variants of the GAN networks such as CGAN, DCGAN, Pix2Pix and so on. To achieve this, they used a two-stage GAN to create a coarse-to-fine paradigm which is necessary in generating images with a high sensitivity to the fine patterns of the original image. To lower the NoP, redundancy in the convolutional kernel was eliminated by using the principal components of a normally fully ranked convolutional kernel for feature extraction. The resolution of MRI images of the brain was improved by increasing the contrast using a variant of the GAN network known as Cycle-GAN to aid the segmentation of the MRI images [
44]. To achieve this, they used the image-to-image translation technique to create a high tissue contrast (HTC) of the real image. The attention block of the Cycle-GAN used in this study helped in focussing on a single tissue and increasing the contrast within the tissue other works that have used GANs for medical image resolution includes [
44]. For denoising tasks, GANs was used for denoising X-ray images using the CGAN variant of the GAN architectures [
46]. They purposed to deal with the spatially varying noise, which is often overlooked when dealing with medical images. To achieve this, the gradient of original image was merged with the noisy image to obtain the conditional information for the CGAN network thereby enhancing the contrast. The convolutional layers of the generator were used in full for better feature extraction. For improved consistency between the real and fake images, the reconstruction loss was combined with WGAN loss to create an objective loss for the network. They obtained remarkable PSNR and SSIM performance when compared to other state-of-the-art (SOTA) GAN architectures.
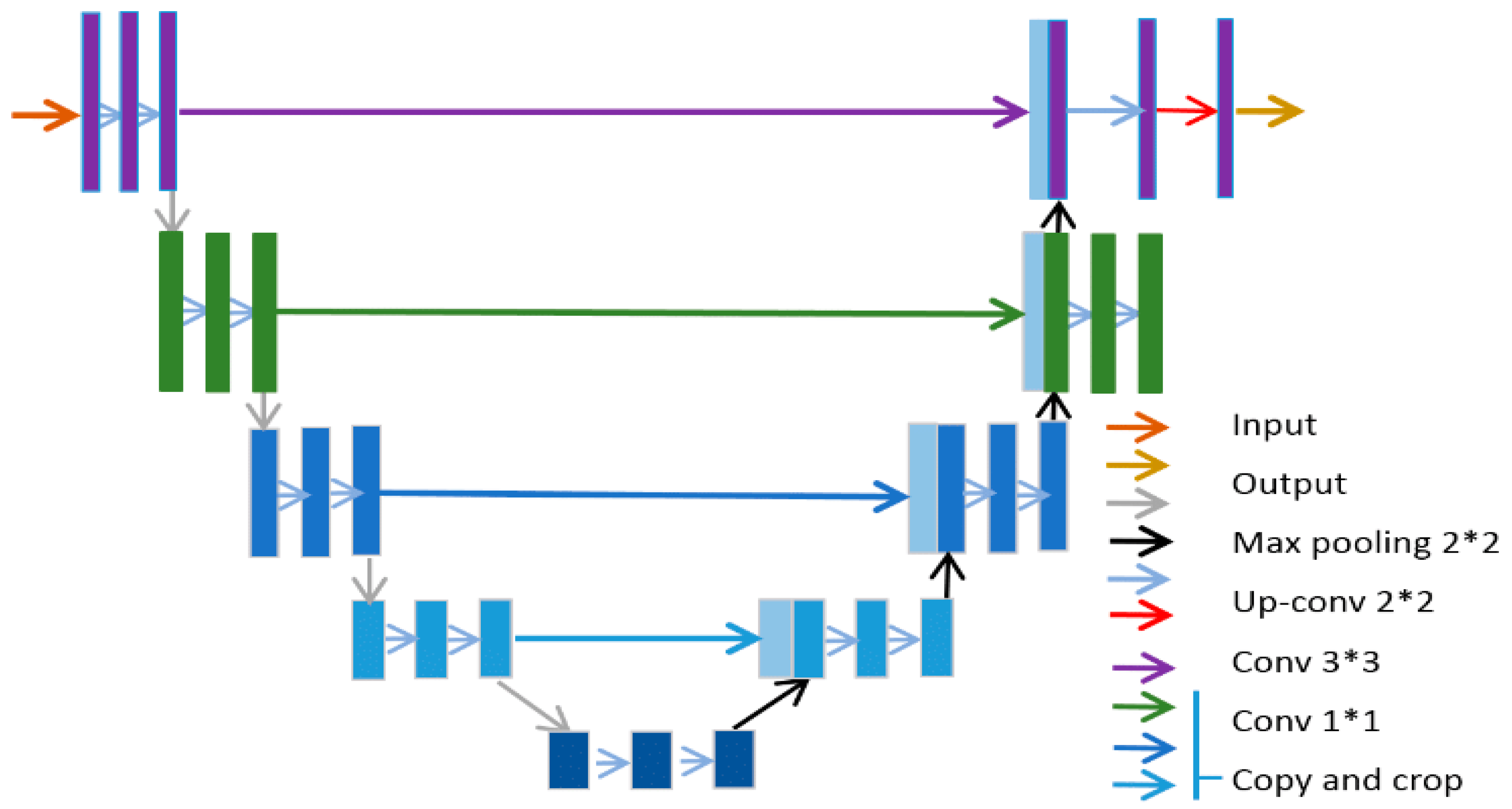
3.5. U-Net
U-Net architecture combines the best of CNN and encoder-decoder models, specifically for the purposes of segmenting medical images [
50,
51]. They have found applications in major medical image modalities such as CT, X-ray radiograph and MRI. The U-Net’s ability to exploit small, annotated data samples (based on its fully connected layers) by leveraging data augmentation and improved feature extraction made it a valuable technique for medical image segmentation [
51]. The U-shaped architectures with skip connections help to delineate objects in images, making it highly effective in medical image analysis, particularly in tasks like tumors detection, organ delineation, and segmentation of medical images from various modalities and they have been widely embraced variants among the many different deep learning networks [
52].
U-Net architecture is mainly composed of two paths. The first path is referred to as the contracting or encoder path. It uses a down sampling module that consists of several repeating convolutional blocks for semantic and contextual feature extraction. Each convolution block has two successive 3x3 convolutions, ReLU activation functions and the pooling layer. The pooling layer serves to increase the receptive field of the convolution network with no extra burden of computing resources that might be introduced by an additional convolution block. The second path of the U-net is the expansive path, or the decoder path and it is saddled with the task of up sampling spatial resolutions of the feature maps from the contracting path, usually by a factor of two. During this operation, the dimensions of the features are reduced, and a pixel-wise classification/resolution score is produced. The expansive path is made up of a 2x2 transposed convolution layer (reversing the operation in the contracting path), which is followed by a 3x3 convolutional layer and a ReLU activation function. There is also a bottleneck layer which serves as a connection between the two paths. It is also comprised of two blocks of 3x3 convolution layers and a ReLU activation function. The embedded skip connections in the bottleneck copy the output of each stage of the paths, helping to learn contextual and semantic representations in the deep and shallow layers respectively [
51,
53]. The U-Net architecture is shown in
Figure 8.
The encoders/contracting paths shown on the left-hand side of
Figure 8 comprise several layers. Each layer
in the encoder is represented by the function
where
represents input features from a previous layer,
,
is the convolutional weights and biases and
represents the activation function which is ReLU in most cases.
The output of the encoder is normally max-pooled and can be represented as
The output is then passed through the bottleneck layer which is another convolution function represented by
With
representing the number of layers in the encoder/contracting path. The up sampling taking place in the decoder is performed via a skip connection at every layer on the encoder. So, each layer in the decoder up samples the feature map, concatenates the corresponding encoder features and applies convolution with the given function as
At the output layer, there is a 1x1 convolution layer that uses SoftMax activation to map the desired output segments and is represented below:
During training of the U-Net, the loss function used is usually the cross-entropy loss and it is normally applied pixel-wise.
Table 4.
U-Net Segmentation techniques.
Table 4.
U-Net Segmentation techniques.
| Article |
Imaging Modality |
Disease/Body Part |
Variant Used |
| [55] |
CT |
Liver/ Lung |
Attention U-Net |
| [56] |
CT |
hepatocellular carcinoma |
Enhanced U-Net |
| [57] |
CT |
Liver |
Enhanced U-Net |
| [58] |
MRI |
Brain Tumour |
None |
| [54] |
MRI |
Brain Tumour |
Enhanced U-Net |
| [59] |
Colour Fundus |
Diabetic retinopathy |
Enhanced U-Net |
| [60] |
MRI |
Various/Musculoskeletal |
Enhanced U-Net |
| [61] |
MRI |
Lower limb muscle |
Attention U-Net/SCU-Net |
| [62] |
MRI |
Musculoskeletal |
Various |
| [63] |
Various |
Various |
Enhanced U-Net (U-Net++) |
3.5.1. Literature Review Findings for the U-Net
The U-Net is mostly used for segmentation tasks especially for segmentation of cancer for various image modalities. Over time, there have been a lot of modifications to the vanilla U-Net model as different researcher has tried to enhance the different facets of the U-Net, ranging from the skip connections to modifying the convolutional layers with attention networks in a bid to increase segmentation quality or reduce computational resource. A multi-level feature assembly MFLA U-Net was reported which is integrated with multi-scale information attention MSIA and pixel-vanishing attention mechanism [
64]. This enhanced U-Net model was designed to boost segmentation performance. Their model was tested on different medical imaging datasets with different modalities such as colonoscopy and dermoscopic images. They have used the dice index coefficients as a metrics to evaluate the effectiveness of their developed model in segmenting these images. Their model outperformed many state-of-the-art U-Net models on the datasets used for testing. A lightweight U-Net architecture was applied to on a publicly available brain tumour datasets (BraTs) to segment brain tumor [
58]. The focus of the study was mainly developing a low resource U-Net framework which had a multimodal CNN encoder-decoder. They also excluded augmentation to reduce the computational demand of their network. Their model achieved a remarkable performance with dice coefficient values of up to 0.93 for specific classes they segmented when compared to other U-Net models. An enhanced U-Net model with minimal parameter was reported [
54]. The authors achieved this by developing a framework known as Stack Multi-Connection Simple Reducing Net, otherwise known as SRNet. This network used fewer convolution operation in the down sampling and up sampling processes, which in turn helped to reduce the total parameter of the vanilla U-Net algorithm by 20%. They also modified the original architecture by ensuring the convolutional layers were not stacked, helping to reduce information loss. Their model was also tested with the BraTs dataset. They obtained matching results with popular variants of the U-Net model, also using the Dice coefficient parameters as an evaluation tool. The vanilla U-Net was modified for the purpose of improving the accuracy of segmentation [
58]. They explored the weakness of the U-Nets models which only focues on contextual information and neglects other useful features of the channel. The developed HDA-ResUNet which combined the best of attentions mechanisms, U-Net and dilated convolution. They evaluated their model on ISI and LiTS segmentation datasets and achieved a good performance in terms of the dice coefficient. This model also used fewer parameters compared to the conventional U-Net.
3.6. Transfer Learning
Deep learning techniques are known to be computationally intensive, owing to the large number of trainable parameters available in their networks. These parameters increase substantially as the network deepens. Also, the availability of large amounts of annotated medical image datasets is scarce and it is a very important element in the use of deep learning for medical image analysis [
65,
66]. Transfer learning algorithms was developed to help solve these problems by providing a means to reparametrize an already trained large deep learning network. These networks are CNN based networks, trained on millions of images with different classes. The resulting learnt parameters are saved to be reused on other datasets. Some aspects of these networks are modified to suit the new dataset. Transfer learning involves using pre-trained deep learning models (e.g., ResNet, VGG, DenseNet, GogleNet, XceptionNet, AlexNet, Inception V3 and SqueezeNet) and fine-tuning them on medical images. Developing models when using transfer learning is performed in two stages: Initializing the weights and finetuning. During initialization of the weights, the weights of a previously trained model with a different dataset, i.e. (AlexNet – trained on ImageNet) as shown in
Figure 9, are copied. When a new training dataset such as medical image in this case is used to train this model, the weights are updated. In the finetuning process, some of the CNN layers are frozen thereby the weights are not being updated. Another method of finetuning is freezing all the CNN layers barring the classification layer that is adjusted according to the requirements of the medical image data. This technique is highly effective when the available dataset is small or specific to a particular medical condition. It reduces training time and improves diagnostic accuracy [
67]. Transfer learning can be categorized into inductive, transductive and unsupervised learning based on data labels. They can also be categorized as homogenous and heterogeneous based on how consistent the dataset features and label between the source and target domain [
68,
69].
The concept of transfer learning can be modelled mathematically as described next. The source domain be represented as
where
is the input data from the source domain and
represents the corresponding labels and
is the number of samples. The target domain be represented as
where
is the input data from the target domain and
represents the corresponding labels
is the number of samples, with the assumption that
or
The objective of the transfer learning function is to find a model
that minimizes the loss in the target domain. If we represent the loss function by
, then
can be defined as:
where
is the specific loss algorithm, which can be cross-entropy or mean squared error and
is the target domains model parameters. The transfer learning process between the source and target domain takes two steps. We pretrain the model on the source domain, using the objective loss function given by the equation:
The learned parameters from the source domain are transferred to the target domain by freezing the base layers, where fixed or by fine-tuning specific layers i.e., .
The target domain overall model becomes:
The learning process is then optimized by regularizing the losses. Therefore, a combined loss function
given by
where
is the regularisation weight and
can be any term used for smooth transfer i.e.,
-norm.
3.6.1. Findings for Transfer Learning
Transfer learning (TL) techniques are mostly applied for the classification of medical images as summarised by the studies listed in
Table 5.
This is because the networks they are learning from have been pretrained for classification as well. Several transfer learning models such as VGG16, DenseNet 121 and ResNet 50 were used for the binary classification tasks of X-ray radiographs and CT images of Covid-19 patients [
74]. Most of the model parameters were frozen and the weights were not initialised. However, they were able to obtain very good results in terms of classification accuracy with VGG 16 model performing best with an accuracy of 99%. Transfer learning models like the models in [
74] were compared and also a custom CNN was built from scratch to compare the efficacy of the TL process [
79]. The models were however trained on different publicly available datasets with varying modalities, ranging from X-ray radiographs to CT of different diseases including lung cancer and brain tumour. The initial convolutional layers of the TL networks used in their work was frozen and the weights of the top layers were updated. The models were trained on all the datasets, and they obtained improved accuracy with the TL models with ResNet 50 showing the highest accuracy of 90% for the histopathological images. Transfer learning techniques were trained on the ImageNet datasets [
80]. They pre-trained a novel hybrid DCNN model which combined convolutional layers with global average pooling for each layer with a skip connection in each of the convolution layers. The 200,000 augmented unlabelled data were sourced from different repository of biopsy breast cancer image datasets, and this was used to train their DCNN model. The pretrained DCNN model was then used to classify annotated skin lesion cancer as benign or malignant based on the weights from their novel pretrained model. They obtained an improved accuracy when compared to other models that tested on similar datasets. A transfer learning model (ResNet 18) was used for the diagnosis of different stages of the Alzheimer's disease (AD) [
78]. The ResNet model was fine-tuned by unfreezing all the layers, allowing for the ResNet 18 model to update all it weights based on the DICOM MRI publicly available datasets used in their study. They obtained high accuracies for the three different classes in the range of 99% and their model outperformed other related works that classified AD.
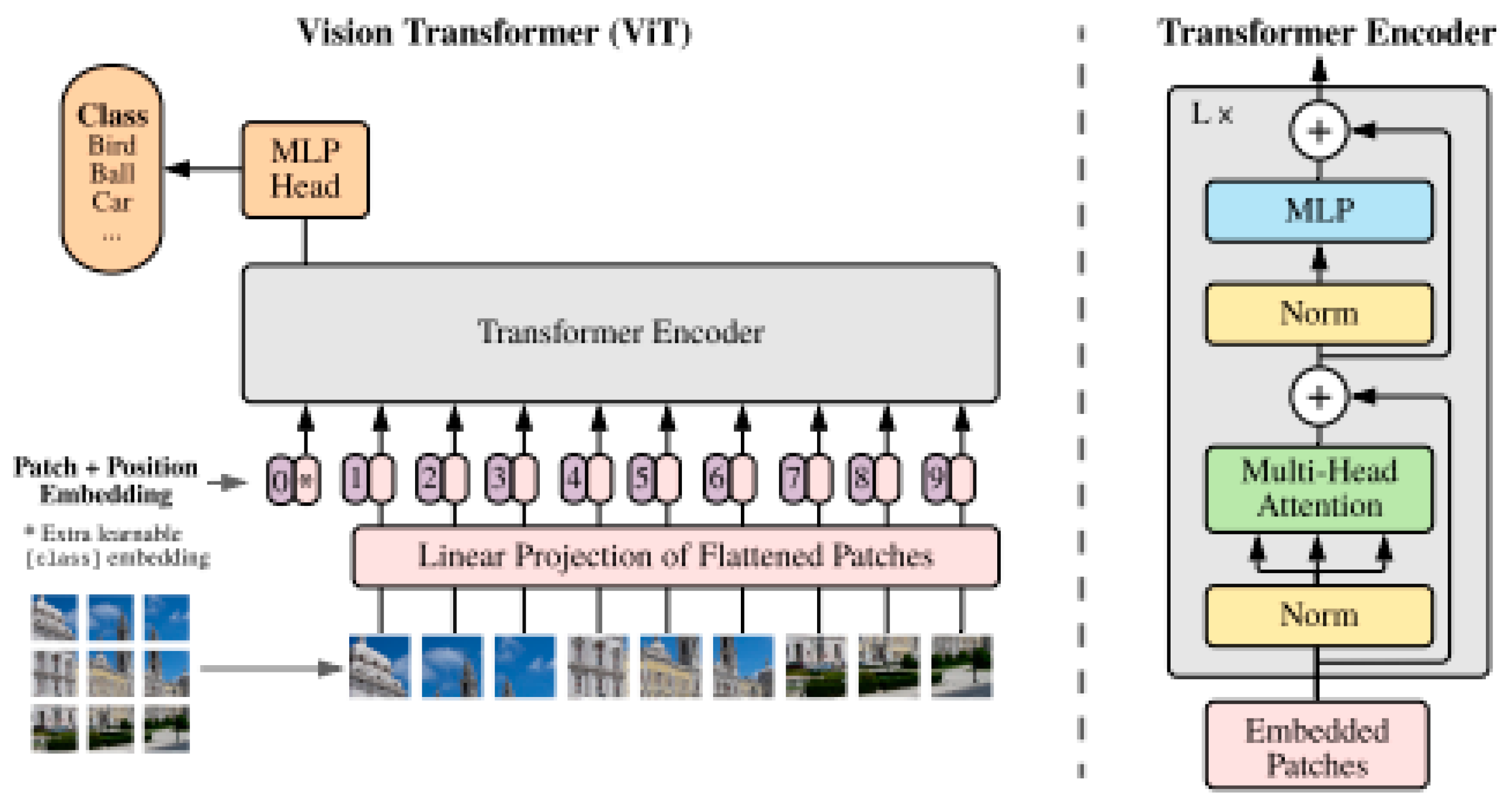
3.7. Vision Transformers
Despite the significant successes recorded in the enhancement of the diagnostic accuracy of deep learning models such as CNN, RNN and U-Net in the classification and segmentation of medical images, there have remained some limitations. Their reliance on localized feature extraction, leading to inductive bias and sequential operation, make them fall short when the medical imaging tasks requiring long range dependency and global feature extraction [
76]. Although initially designed for natural language processing tasks such as sentiment analysis [
77], machine translation and text summarization [
78], their ability to capture long range dependencies in image pixels helps to build a more robust segmentation/ classification model. Vision transformer is a relatively new deep learning architecture that is increasingly being applied to medical imaging. Developed by Google in 2020, it performs segmentation or classification using the transformer architecture. The ViT creates a partition of the input images into multiple patches of 16x16 pixels and linearly embeds them. For the pixels to be suitable for the transformer architecture, they must be transformed into fixed-length vectors [
79]. The self-attention mechanism represents the main feature of the ViT architecture as this forms the basis on the interaction between the pixel patches. It also uses positional encoding to represent the spatial location of the image patches. Feedforward layers, placed after the self-attention layers, are generally used in for making final decisions by the model [
80]. The original architecture of the vision transformer is shown in
Figure 10.
If we represent the input to the ViT model by an image
, then
can be defined such that
where
and
represent the height and width of the image respectively, and
is the number of channels which is usually 3 for RGB images. The input image is divided into non-overlapping patches of size
, such that
where
is the total number of patches and each patch is flatten as
The patches are sent through the transformer encoder layers consisting of the multi-head attention layer, which computes the relationships between patches using the
matrices (Keys, Values and Queries), which are computed as
where
is the positional patch embedding for each patch and
is the learnable weights projection matrices, which have the same size as embedding space. This is then passed to the feed forward neural network (FFN) and a classification head for a classification task [
82].
3.7.1. Literature Review Findings for Vision Transformers
Several studies used ViT and its variants including TNT, Swin, DeiT and PVT [
82] for classification, registration and segmentation of medical images. Just like the convolution-based transfer learning models, learnable parameters from transformer models such the ones listed above can also be used for specific DL tasks. For example, a pretrained Swin transformer was used for the classification of breast cancer using publicly available breast X-ray images [
88]. The dataset was resized to fit the Swin transformer input size and augmentation was also performed to improve generalization. They achieved very high classification rates based on their selected metrics for evaluation with an accuracy of 99.9% and a precision of 99.8%. They compared their results with convolution-based TL algorithms (ResNet50 and VGG16) and found that the Swin transformer had a superior performance. MRI images of Ischemic strokes were classified using ViT by [
83]. The vanilla ViT or ViT base used in the study also had limited fine-tuning and hyperparameters were adjusted to fit the datasets to be classified. They also augmented and resized the MRI images as preprocessing steps before deploying the ViT model. They achieved an impressive accuracy score of 97.59% when compared to VGG16 model used in a similar study, demonstrating the superiority of transformer models for classification tasks. Transformers were used for image registration tasks [
90]. In their study, they developed the TransMorph algorithm which is a hybrid transformer and convolution network. The network leveraged on the encoder/decoder architecture of transformers but instead of the attention mechanisms at the decoder, this was replaced with a convolution network. For the transformer network, they used the Swin variant due to its ability to extract feature maps at different resolutions by merging patch layers, making it suitable for the image registration task. The algorithm was tested on different image pair datasets comprising mainly MRI and CT modalities for registration purposes. They obtained very competitive results based on the Dice score evaluation when compared to both traditional and other DL methods used for similar tasks.
3.8. Hybrid Models
Most deployed deep learning methods use CNN as described in the preceding sections, with variants such as VGG, ResNet, LSTM, RNN, GAN and GRU all being used for different medical image analysis. However, different authors have tried to combine the strength of some of the models to deal with the weaknesses of the others by combining them together and this forms the basis for hybrid models [
3]. Some works have also combined different deep learning methodologies by focusing on the strengths of the methodologies. For example, a convolutional network’s strong local feature extraction has been combined with the long-range dependencies of transformers when performing medical analysis.
3.8.1. Convolution-Based Hybrid Models
A hybrid model of convolution algorithms was developed comprising SegNet, MultResUNet and Krill Herd Optimisation algorithm (KHO) to improve the segmentation of CT scans of liver lesion and RNA genome sequencing [
91]. The SegNet framework provided the segmentation capacity of their model, utilizing pixel-wise classification through the Softmax layer. A CNN based architecture was employed with the MultiResUNet to handle the lesion segmentation, together with the SegNet framework. The hyperparameters of the models, when optimized through the KHO algorithm, helped to improve the segmentation process. They tested their algorithm with a publicly available LiTs datasets and used evaluation metrics such as the Dice coefficient, F1-score and accuracy and obtained better results with F1-score comparatively higher than the models used for comparison. A hybrid model of convolutional methods involving RestNet and UNet model (ResUNet) was developed for the segmentation of liver and tumors using CT images [
92]. Their ResUnet model focused on improving the available models by providing improved image contrast and segmenting irregular tumor shapes and small tumor sizes. The combination uses the best of ResNet’s residual connection and the UNet’s encoder and decoder structure to enhance feature learning, segmentation precision and efficiency. They also implemented various augmentation techniques such as rotation and reflection to increase the variability in their dataset. They achieved an accuracy of 99.6% and a dice coefficient of 99.2%.
3.8.2. Convolution-Transformer Based Hybrid Models
A hybrid model called TBConvl-NET that combines CNN, LSTM and ViT for the segmentation of several diseases was developed using publicly available datasets of different modalities such as ultrasound and MRI [
93]. The hybrid model targeted some well-known challenges in segmenting medical images such as scale, texture and shape of pathology. Due to high computational resources required, they used a depth wise separable convolution, thereby reducing computational overhead. Swin transformer blocks were used in the skip connections to help deal with the varying scales of the data and help preserve semantic information. They used Dice index, accuracy and Jaccard index to evaluate their model and compared the developed model with other hybrid segmentation models and obtained improved results. A hybrid classification model that combines the transformer and the convolution model to improve the classification of skin lesion was developed using publicly available datasets [
94]. They used the Swin-Unet architecture to perform image segmentation, leveraging self-attention of the Swin transformer and robust hierarchical analysis of the UNet. They combined this with the Xception and ResNet 18 models for feature extraction to further improve on the image analysis. For hyperparameter tuning, a Hybrid Salp Swam Algorithm (HSSA) was used to obtain the optimal parameters, hence avoiding local minima during training. A gated recurrent unit (GRU) network was used for the eventual classification. They achieved an accuracy of 94.51% and 95.38% on both datasets used in the study. Their model performed better when compared to TL models like AlexNet and ResNet18.
4. Discussion
In this article, an in-depth discussion of various deep learning methodologies used for medical imaging analysis was provided. These included Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), Autoencoders, Generative Adversarial Networks (GANs), U-Net architectures, Vision Transformers, and hybrid models. For each of these methodologies, some of the key findings were presented.
CNNs are extensively used for medical imaging tasks like disease detection, classification, and segmentation and the most important contribution of the algorithm is the unrivalled feature extraction through convolutional layers, making them effective for tasks requiring spatial hierarchy. CNN models can be lightweighted in terms of computing resources as inspired by ResNet while also balancing the model performance. Deeper CNN architectures can be computationally expensive but can be very useful in multi-disease detection, for example in the classification and detection of COVID-19 and viral pneumonia, achieving high accuracy due to advanced hardware. They can also be effective when the task includes a pixel-level feature extraction. Examples can be seen in its use in the classification of ultrasound image classification of fatty liver. RNNs, particularly their variants like Long Short-Term Memory (LSTM) and Gated Recurrent Units (GRU), are suitable for sequential data tasks. Their ability to retain temporal dependencies is utilized in dynamic imaging and disease progression monitoring. RNNs which usually have convolution layers are effective for classification tasks, with different imaging modalities, with MRI dominating. They were applied to detecting several diseases such as Alzheimer’s disease and breast cancer. LSTM variants of the RNNs algorithm handle tasks like segmentation of temporal MRI images and noise removal in diagnostic images. Some modifications have also been applied to the RNN algorithm training, such as using the Honey Badger Algorithm (HBA) for optimisation of the model training parameters.
Autoencoders are unsupervised models used for dimensionality reduction, anomaly detection, and data augmentation. Their encoder-decoder structure helps compress and reconstruct medical images with minimal distortion. Autoencoders are employed for image analysis tasks like segmentation and denoising and have been used with several image modalities for the analysis of different diseases. Enhancing autoencoders by fusing with wavelet transform can also help improve noise removal from medical images. They can also be combined with convolutional networks to improve disease classification. GANs are used for generating synthetic medical images, data augmentation, and improving image resolution or quality. The main components of a GAN network comprise: a generator for creating synthetic images and a discriminator for distinguishing between real and generated images. GANs can be very computationally intensive due to the dual deep convolutional networks. Research around this area is focussed on reducing models’ parameters and maintaining performance. Different variant of GANs have employed different image analysis tasks with each variant tuned to fit the specific tasks.
U-Nets are specialised for medical image segmentation and have been widely adopted for tasks like tumour detection and organ delineation. The architecture combines encoder-decoder pathways with skip connections for improved feature representation. Different variants of the U-Net architecture also exist to enhance the vanilla model, making them suitable for specific image modality while performing its main task of segmentation. Also, enhancements have been created to reduce model operating costs, while retaining the segmentation process. More recent studies have been adding transformers to U-Nets to further enhance segmentation accuracy. Transfer learning leverages pretrained models (e.g., ResNet, VGG, DenseNet) on large datasets and fine-tunes them for medical imaging tasks. They are mostly used for classification tasks models like VGG16 and ResNet50 perform well for COVID-19 classification, achieving very high accuracy on different image modalities. Some transfer learning models are quite deep with millions of parameters, making them very computationally intensive. Custom hybrid models pretrained on unlabelled medical datasets have also been used to improve classification accuracy for skin lesions and breast cancer images, mimicking the transfer learning ideology. They provide an option in terms of computing resources and domain specific training and learning. ViTs are a relatively newer deep learning architecture, adapted for medical imaging tasks. They require global feature extraction and long-range dependencies. They divide images into patches and process them using self-attention mechanisms. ViT models can also be pre-trained like most CNN models and the learning transfer for a newer task. Several variants have also been developed depending on the task. With larger training datasets, ViTs have shown to outperform CNN models when used for tasks with fewer annotated data.
Hybrid models combine the strengths of multiple architectures (e.g., CNNs with RNNs, or CNNs with transformers) to improve medical image analysis. CNN-based hybrids like ResUNet combine ResNet’s feature extraction with U-Net’s segmentation capabilities. They were used widely for cancer related image segmentation. Convo-transformer hybrids, like TBConvl-NET integrate CNN, LSTM, and ViT for improved segmentation and classification of diseases. Hybrid models can also be optimised. The algorithm such as hybrid salp swarm algorithm (HSSA) could be used to obtain the optimal parameters thereby increasing model performance.
5. Conclusions
This review explored the transformative impact of deep learning algorithms on medical image analysis. Convolutional neural networks (CNNs) are widely used for feature extraction and disease classification across medical imaging modalities, achieving high accuracy with optimized computational costs. Recurrent neural networks (RNNs), including LSTM and GRU, enhance temporal analysis for disease progression monitoring. Autoencoders and generative adversarial networks (GANs) assist in data augmentation, denoising, and synthetic image generation. U-Net architectures improve segmentation for tumor detection and organ delineation. Vision transformers (ViTs) leverage attention mechanisms for superior classification and registration. Hybrid models combining CNNs, transformers, and optimization techniques enhance performance, while transfer learning mitigates data scarcity, ensuring robust results across imaging applications. Together, these advancements underscore the versatility and efficiency of deep learning in medical diagnostics, paving the way for improved clinical outcomes and personalized healthcare solutions. Future advancements may focus on computational efficiency, integrating multimodal data, and enhancing interpretability for clinical adoption.
Author Contributions
Conceptualization, O.S. and R.S.; methodology, O.S. and R.S.; investigation, , O.S. and R.S.; resources, , O.S. and R.S.; data curation, , O.S. and R.S.; writing—original draft preparation, , O.S. and R.S.; writing—review and editing, , O.S. and R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jeong, J.J.; Tariq, A.; Adejumo, T.; Trivedi, H.; Gichoya, J.W.; Banerjee, I. Systematic Review of Generative Adversarial Networks (GANs) for Medical Image Classification and Segmentation. J Digit Imaging 2022, 35, 137. [CrossRef]
- Yu-Jen Chen, Y.; Hua, K.; Hsu, C.; Cheng, W.; Hidayati, S.C. Computer-aided Classification of Lung Nodules on Computed Tomography Images via deep Learning Technique. OTT 2015. [CrossRef]
- Zhu, Z. Advancements in Automated Classification of Chronic Obstructive Pulmonary Disease based on Computed Tomography Imaging Features Through Deep Learning Approaches. Respiratory Medicine 2024, 234. [CrossRef]
- Sarmadi, A.; Razavi, Z.S.; Van Wijnen, A.J.; Soltani, M. Comparative Analysis of Vision Transformers and Convolutional Neural Networks in Osteoporosis Detection from X-ray Images. Sci Rep 2024, 14. [CrossRef]
- Takahashi, S.; Sakaguchi, Y.; Kouno, N.; Takasawa, K.; Ishizu, K.; Akagi, Y.; Aoyama, R.; Teraya, N.; Bolatkan, A.; Shinkai, N.; Machino, H.; Kobayashi, K.; Asada, K.; Komatsu, M.; Kaneko, S.; Sugiyama, M.; Hamamoto, R. Comparison of Vision Transformers and Convolutional Neural Networks in Medical Image Analysis: A Systematic Review. J Med Syst 2024, 48. [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Journal of Machine Learning Research 2014, 15, 1929-1958.
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. Journal of Big Data 2019, 6(60), 1-48.
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J Med Imag Rad Onc 2021, 65, 545.
- Masumoto, R.; Eguchi, Y.; Takeuchi, H.; Inage, K.; Narita, M.; Shiga, Y.; Inoue, M.; Toshi, N.; Tokeshi, S.; Okuyama, K.; Ohyama, S.; Suzuki, N.; Maki, S.; Furuya, T.; Ohtori, S.; Orita, S. Automatic Generation of Diffusion Tensor imaging for the Lumbar Nerve using Convolutional Neural Networks. Magnetic Resonance Imaging 2024, 114. [CrossRef]
- Shobayo, O.; Saatchi, R.; Ramlakhan, S. Convolutional Neural Network to Classify Infrared Thermal Images of Fractured Wrists in Pediatrics. Healthcare 2024, 12. [CrossRef]
- Chauhan, S.; Edla, D.R.; Boddu, V.; Rao, M.J.; Cheruku, R.; Nayak, S.R.; Martha, S.; Lavanya, K.; Nigat, T.D. Detection of COVID-19 Using Edge Devices by a Light-Weight Convolutional Neural Network from Chest X-ray Images. BMC Med Imaging 2024, 24. [CrossRef]
- Wu, Z.; Shen, C.; Van Den Hengel, A. Wider or Deeper: Revisiting the ResNet Model for Visual Recognition. Pattern Recognition 2019, 90, 119. [CrossRef]
- Abdulahi, A.T.; Ogundokun, R.O.; Adenike, A.R.; Shah, M.A.; Ahmed, Y.K. PulmoNet: A Novel Deep learning based Pulmonary Diseases Detection Model. BMC Med Imaging 2024, 24.
- Zhang, K.; Sun, M.; Han, T.X.; Yuan, X.; Guo, L.; Liu, T. Residual networks of residual networks: Multilevel residual networks. IEEE Transactions on Circuits and Systems for Video Technology 2017, 28, 1303–1314.
- Zhu, H.; Liu, Y.; Gao, X.; Zhang, L. Combined CNN and Pixel Feature Image for Fatty Liver Ultrasound Image Classification. Computational and Mathematical Methods in Medicine 2022, 2022, 1. [CrossRef]
- Kim, J.; Hong, J.; Park, H. Prospects of Deep Learning for Medical Imaging. Precis Future Med 2018, 2, 37. [CrossRef]
- Zhang, H.; Qie, Y. Applying Deep Learning to Medical Imaging: A Review. Applied Sciences 2023, 13. [CrossRef]
- Rajeev, R.; Samath, J.A.; Karthikeyan, N.K. An Intelligent Recurrent Neural Network with Long Short-Term Memory (LSTM) BASED Batch Normalization for Medical Image Denoising. J Med Syst 2019, 43. [CrossRef]
- Yao, W.; Bai, J.; Liao, W.; Chen, Y.; Liu, M.; Xie, Y. From CNN to Transformer: A Review of Medical Image Segmentation Models. J Digit Imaging. Inform. med. 2024, 37, 1529. [CrossRef]
- Cui, R.; Liu, M. RNN-based Longitudinal Analysis for Diagnosis of Alzheimer’s Disease. Computerized Medical Imaging and Graphics 2019, 73, 1. [CrossRef]
- Anbalagan, V.; Balasubramanian, V. HBO-GMRNN: Honey Badger Optimization Based Gain Modulated Recurrent Neural Network for Classification of Breast Cancer. Biomedical Signal Processing and Control 2024, 91. [CrossRef]
- Amarneni, S.; Valarmathi, D.R.S. Diagnosing the MRI Brain Tumour Images Through RNN-LSTM. e-Prime - Advances in Electrical Engineering, Electronics and Energy 2024, 9. [CrossRef]
- Zhu, K.; Chen, Y.; Ouyang, X.; White, G.; Agam, G. Fully RNN for Knee Ligament Tear Classification and Localization in MRI scans. ei 2022, 34. [CrossRef]
- Gulshan; Arora, A.S. Automated Prediction of Diabetes Mellitus Using Infrared Thermal Foot Images: Recurrent Neural Network Approach. Biomed. Phys. Eng. Express 2024, 10. [CrossRef]
- Ayub, S.; Kannan, R.J.; Alsini, R.; Hasanin, T.; Sasidhar, C. LSTM-Based RNN Framework to Remove Motion Artifacts in Dynamic Multicontrast MR Images with Registration Model. Wireless Communications and Mobile Computing 2022, 2022, 1. [CrossRef]
- Das, T.; Saha, G. Addressing Big Data Issues Using RNN Based Techniques. Journal of Information and Optimization Sciences 2020, 40, 1773. [CrossRef]
- Zhang, Y. In In A better autoencoder for image: Convolutional autoencoder; ICONIP17-DCEC. Available online: http://users.cecs.anu.edu.au/Tom.Gedeon/conf/ABCs2018/paper/ABCs2018_paper_58.pdf. 2018; pp 11-21. (accessed on October 2024).
- Baldi, P. In In Autoencoders, unsupervised learning, and deep architectures; Proceedings of ICML workshop on unsupervised and transfer learning; JMLR Workshop and Conference Proceedings: 2012; , pp 37–49. (accessed on November 2024).
- Vorontsov, E.; Molchanov, P.; Gazda, M.; Beckham, C.; Kautz, J.; Kadoury, S. Towards Annotation-Efficient Segmentation Via Image-to-Image Translation. Medical Image Analysis 2022, 82. [CrossRef]
- Wang, W.; Huang, Y.; Wang, Y.; Wang, L. In In Generalized autoencoder: A neural network framework for dimensionality reduction; Proceedings of the IEEE conference on computer vision and pattern recognition workshops; 2014; , pp 490–497.
- Juneja, M.; Kaur Saini, S.; Kaul, S.; Acharjee, R.; Thakur, N.; Jindal, P. Denoising of Magnetic Resonance Imaging Using Bayes Shrinkage Based Fused Wavelet Transform and Autoencoder Based Deep Learning Approach. Biomedical Signal Processing and Control 2021, 69. [CrossRef]
- O’ Sullivan, E.; Van De Lande, L.S.; Papaioannou, A.; Breakey, R.W.F.; Jeelani, N.O.; Ponniah, A.; Duncan, C.; Schievano, S.; Khonsari, R.H.; Zafeiriou, S.; Dunaway, D.J. Convolutional Mesh Autoencoders for the 3-Dimensional Identification of FGFR-Related Craniosynostosis. Sci Rep 2022, 12. [CrossRef]
- Wolf, D.; Payer, T.; Lisson, C.S.; Lisson, C.G.; Beer, M.; Götz, M.; Ropinski, T. Self-supervised Pre-training with Contrastive and Masked Autoencoder Methods for Dealing with Small Datasets in Deep Learning for Medical Imaging. Sci Rep 2023, 13. [CrossRef]
- Chen, R.; Song, Y.; Huang, J.; Wang, J.; Sun, H.; Wang, H. Rapid Diagnosis and Continuous Monitoring of Intracerebral Hemorrhage with Magnetic Induction Tomography Based on Stacked Autoencoder. Review of Scientific Instruments 2021, 92.
- Shvetsova, N.; Bakker, B.; Fedulova, I.; Schulz, H.; Dylov, D.V. Anomaly Detection in Medical Imaging With Deep Perceptual Autoencoders. IEEE Access 2021, 9, 118571. [CrossRef]
- Elhassan, T.A.; Mohd Rahim, M.S.; Siti Zaiton, M.H.; Swee, T.T.; Alhaj, T.A.; Ali, A.; Aljurf, M. Classification of Atypical White Blood Cells in Acute Myeloid Leukemia Using a Two-Stage Hybrid Model Based on Deep Convolutional Autoencoder and Deep Convolutional Neural Network. Diagnostics 2023, 13.
- Zhang, H.; Guo, W.; Zhang, S.; Lu, H.; Zhao, X. Unsupervised Deep Anomaly Detection for Medical Images Using an Improved Adversarial Autoencoder. J Digit Imaging 2022, 35, 153. [CrossRef]
- Li, D.; Fu, Z.; Xu, J. Stacked-Autoencoder-Based Model for COVID-19 Diagnosis on CT Images. Appl Intell 2020, 51, 2805. [CrossRef]
- Zhang, H.; Chen, J.; Liao, B.; Wu, F.; Bi, X. Deep Canonical Correlation Fusion Algorithm Based on Denoising Autoencoder for ASD Diagnosis and Pathogenic Brain Region Identification. Interdiscip Sci Comput Life Sci 2024, 16, 455. [CrossRef]
- Gao, J.; Zhao, W.; Li, P.; Huang, W.; Chen, Z. LEGAN: A Light and Effective Generative Adversarial Network for medical image synthesis. Computers in Biology and Medicine 2022, 148. [CrossRef]
- Singh, N.K.; Raza, K. In Medical Image Generation Using Generative Adversarial Networks: A Review; Springer Singapore: 2021; pp 77.
- Xun, S.; Li, D.; Zhu, H.; Chen, M.; Wang, J.; Li, J.; Chen, M.; Wu, B.; Zhang, H.; Chai, X.; Jiang, Z.; Zhang, Y.; Huang, P. Generative adversarial networks in medical image segmentation: A review. Computers in Biology and Medicine 2021, 140. [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Medical Image Analysis 2019, 58. [CrossRef]
- Hamghalam, M.; Wang, T.; Lei, B. High tissue contrast image synthesis via multistage attention-GAN: Application to segmenting brain MR scans. Neural Networks 2020, 132, 43. [CrossRef]
- Zhu, Q.; Ye, H.; Sun, L.; Li, Z.; Wang, R.; Shi, F.; Shen, D.; Zhang, D. GACDN: generative adversarial feature completion and diagnosis network for COVID-19. BMC Med Imaging 2021, 21. [CrossRef]
- Li, Y.; Zhang, K.; Shi, W.; Miao, Y.; Jiang, Z. A Novel Medical Image Denoising Method Based on Conditional Generative Adversarial Network. Computational and Mathematical Methods in Medicine 2021, 2021, 1. [CrossRef]
- Ahmad, W.; Ali, H.; Shah, Z.; Azmat, S. A new generative adversarial network for medical images super resolution. Sci Rep 2022, 12. [CrossRef]
- Mutepfe, F.; Kalejahi, B.K.; Meshgini, S.; Danishvar, S. Generative Adversarial Network Image Synthesis Method for Skin Lesion Generation and Classification. Journal of Medical Signals & Sensors 2021, 11, 237. [CrossRef]
- Touati, R.; Le, W.T.; Kadoury, S. A feature invariant generative adversarial network for head and neck MRI/CT image synthesis. Phys. Med. Biol. 2021, 66.
- Xiao, Y.; Chen, C.; Wang, L.; Yu, J.; Fu, X.; Zou, Y.; Lin, Z.; Wang, K. A novel hybrid generative adversarial network for CT and MRI super-resolution reconstruction. Phys. Med. Biol. 2023, 68. [CrossRef]
- Mahapatra, D.; Bozorgtabar, B.; Garnavi, R. Image super-resolution using progressive generative adversarial networks for medical image analysis. Computerized Medical Imaging and Graphics 2019, 71, 30. [CrossRef]
- Uzunova, H.; Ehrhardt, J.; Handels, H. Memory-efficient GAN-based domain translation of high resolution 3D medical images. Computerized Medical Imaging and Graphics 2020, 86. [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lecture Notes in Computer Science 2015, 234. [CrossRef]
- Azad, R.; Aghdam, K.; Rauland, A.; Jia, Y.; Avval, H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. .
- Krithika Alias Anbudevi, M.; Suganthi, K. Review of Semantic Segmentation of Medical Images Using Modified Architectures of UNET. Diagnostics 2022, 12. [CrossRef]
- Ehab, W.; Li, Y. Performance Analysis of UNet and Variants for Medical Image Segmentation. .
- Ding, Y.; Chen, F.; Zhao, Y.; Wu, Z.; Zhang, C.; Wu, D. A Stacked Multi-Connection Simple Reducing Net for Brain Tumor Segmentation. IEEE Access 2019, 7, 104011. [CrossRef]
- Wang, Z.; Zou, Y.; Liu, P.X. Hybrid dilation and attention residual U-Net for medical image segmentation. Computers in Biology and Medicine 2021, 134. [CrossRef]
- Khan, R.A.; Luo, Y.; Wu, F. RMS-UNet: Residual multi-scale UNet for liver and lesion segmentation. Artificial Intelligence in Medicine 2022, 124. [CrossRef]
- Kong, Z.; Zhang, M.; Zhu, W.; Yi, Y.; Wang, T.; Zhang, B. Data enhancement based on M2-Unet for liver segmentation in Computed Tomography. Biomedical Signal Processing and Control 2022, 79. [CrossRef]
- Chetty, G.; Yamin, M.; White, M. A low resource 3D U-Net based deep learning model for medical image analysis. Int. j. inf. tecnol. 2022, 14, 95. [CrossRef]
- Huang, K.; Yang, Y.; Huang, Z.; Liu, Y.; Lee, S. Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module. Bioengineering 2023, 10.
- Lin, Z.; Dall’ara, E.; Guo, L. A novel mean shape based post-processing method for enhancing deep learning lower-limb muscle segmentation accuracy. PLoS ONE 2024, 19.
- Henson, W.H.; Li, X.; Lin, Z.; Guo, L.; Mazzá, C.; Dall’ara, E. Automatic segmentation of lower limb muscles from MR images of post-menopausal women based on deep learning and data augmentation. PLoS ONE 2024, 19.
- Lin, Z.; Henson, W.H.; Dowling, L.; Walsh, J.; Dall’ara, E.; Guo, L. Automatic segmentation of skeletal muscles from MR images using modified U-Net and a novel data augmentation approach. Front. Bioeng. Biotechnol. 2024, 12.
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. In UNet++: A Nested U-Net Architecture for Medical Image Segmentation; Springer International Publishing: 2020; pp 3.
- Garbaz, A.; Oukdach, Y.; Charfi, S.; El Ansari, M.; Koutti, L.; Salihoun, M. MLFA-UNet: A multi-level feature assembly UNet for medical image segmentation. Methods 2024, 232, 52. [CrossRef]
- Kora, P.; Ooi, C.P.; Faust, O.; Raghavendra, U.; Gudigar, A.; Chan, W.Y.; Meenakshi, K.; Swaraja, K.; Plawiak, P.; Rajendra Acharya, U. Transfer learning techniques for medical image analysis: A review. Biocybernetics and Biomedical Engineering 2021, 42, 79. [CrossRef]
- Yu, X.; Wang, J.; Hong, Q.; Teku, R.; Wang, S.; Zhang, Y. Transfer learning for medical images analyses: A survey. Neurocomputing 2022, 489, 230. [CrossRef]
- Ayana, G.; Dese, K.; Abagaro, A.M.; Jeong, K.C.; Yoon, S.; Choe, S. Multistage Transfer Learning for Medical Images. Artif Intell Rev 2024, 57. [CrossRef]
- Atasever, S.; Azginoglu, N.; Terzi, D.S.; Terzi, R. A comprehensive survey of deep learning research on medical image analysis with focus on transfer learning. Clinical Imaging 2022, 94, 18. [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: a literature review. BMC Med Imaging 2022, 22.
- Santana, M.A.d.; Pereira, J.M.S.; Silva, F.L.d.; Lima, N.M.d.; Sousa, F.N.d.; Arruda, G.M.S.d.; Lima, R.d.C.F.d.; Silva, W.W.A.d.; Santos, W.P.d. Breast cancer diagnosis based on mammary thermography and extreme learning machines. Research on biomedical engineering 2018, 34, 45–53. [CrossRef]
- Kumar, S.; Choudhary, S.; Jain, A.; Singh, K.; Ahmadian, A.; Bajuri, M.Y. Brain Tumor Classification Using Deep Neural Network and Transfer Learning. Brain Topogr 2023, 36, 305. [CrossRef]
- B., A.; Kalirajan, K. An Intelligent Magnetic Resonance Imagining-Based Multistage Alzheimer’s Disease Classification using Swish-Convolutional Neural Networks. Med Biol Eng Comput 2024. [CrossRef]
- Saied, M.; Raafat, M.; Yehia, S.; Khalil, M.M. Efficient Pulmonary Nodules Classification Using Radiomics and Different Artificial Intelligence Strategies. Insights Imaging 2023, 14. [CrossRef]
- Yang, D.; Martinez, C.; Visuña, L.; Khandhar, H.; Bhatt, C.; Carretero, J. Detection and analysis of COVID-19 in medical images using deep learning techniques. Sci Rep 2021, 11. [CrossRef]
- Odusami, M.; Maskeliūnas, R.; Damaševičius, R.; Krilavičius, T. Analysis of Features of Alzheimer’s Disease: Detection of Early Stage from Functional Brain Changes in Magnetic Resonance Images Using a Finetuned ResNet18 Network. Diagnostics 2021, 11. [CrossRef]
- Kalusivalingam, A.K.; Sharma, A.; Patel, N.; Singh, V. Enhancing Diagnostic Accuracy in Medical Imaging through Convolutional Neural Networks and Transfer Learning Algorithms.
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13.
- Tian, D.; Jiang, S.; Zhang, L.; Lu, X.; Xu, Y. The role of large language models in medical image processing: a narrative review. Quant Imaging Med Surg 2023, 14, 1108. [CrossRef]
- Ogunleye, B.; Sharma, H.; Shobayo, O. Sentiment Informed Sentence BERT-Ensemble Algorithm for Depression Detection. BDCC 2024, 8. [CrossRef]
- Bora, A.; Cuayáhuitl, H. Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. MAKE 2024, 6, 2355. [CrossRef]
- Pu, Q.; Xi, Z.; Yin, S.; Zhao, Z.; Zhao, L. Advantages of transformer and its application for medical image segmentation: a survey. BioMed Eng OnLine 2024, 23.
- Berroukham, A.; Housni, K.; Lahraichi, M. In In Vision Transformers: A Review of Architecture, Applications, and Future Directions; IEEE: 2023-12-16; , pp 205.
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; Uszkoreit, J.; Houlsby, N. AN IMAGE IS WORTH 16X16 WORDS: TRANSFORMERS FOR IMAGE RECOGNITION AT SCALE. 2021.
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; Yang, Z.; Zhang, Y.; Tao, D. A Survey on Vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 87. [CrossRef]
- Tanimola, O.; Shobayo, O.; Popoola, O.; Okoyeigbo, O. Breast Cancer Classification Using Fine-Tuned SWIN Transformer Model on Mammographic Images. Analytics 2024, 3, 461. [CrossRef]
- Abbaoui, W.; Retal, S.; Ziti, S.; El Bhiri, B. Automated Ischemic Stroke Classification from MRI Scans: Using a Vision Transformer Approach. JCM 2024, 13.
- Chen, J.; Frey, E.C.; He, Y.; Segars, W.P.; Li, Y.; Du, Y. TransMorph: Transformer for unsupervised medical image registration. Medical Image Analysis 2022, 82. [CrossRef]
- Ramamurthy, M.; Krishnamurthi, I.; Vimal, S.; Robinson, Y.H. Deep Learning Based Genome Analysis and NGS-RNA LL Identification with a Novel Hybrid Model. Biosystems 2020, 197. [CrossRef]
- Rahman, H.; Bukht, T.F.N.; Imran, A.; Tariq, J.; Tu, S.; Alzahrani, A. A Deep Learning Approach for Liver and Tumor Segmentation in CT Images Using ResUNet. Bioengineering 2022, 9. [CrossRef]
- Iqbal, S.; Khan, T.M.; Naqvi, S.S.; Naveed, A.; Meijering, E. TBConvL-Net: A hybrid deep learning architecture for robust medical image segmentation. Pattern Recognition 2025, 158. [CrossRef]
- Obayya, M.; Saeed, M.K.; Alruwais, N.; Alotaibi, S.S.; Assiri, M.; Salama, A.S. Hybrid Metaheuristics With Deep Learning-Based Fusion Model for Biomedical Image Analysis. IEEE Access 2023, 11, 117149. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).