Submitted:
25 March 2025
Posted:
26 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Escherichia coli Collection
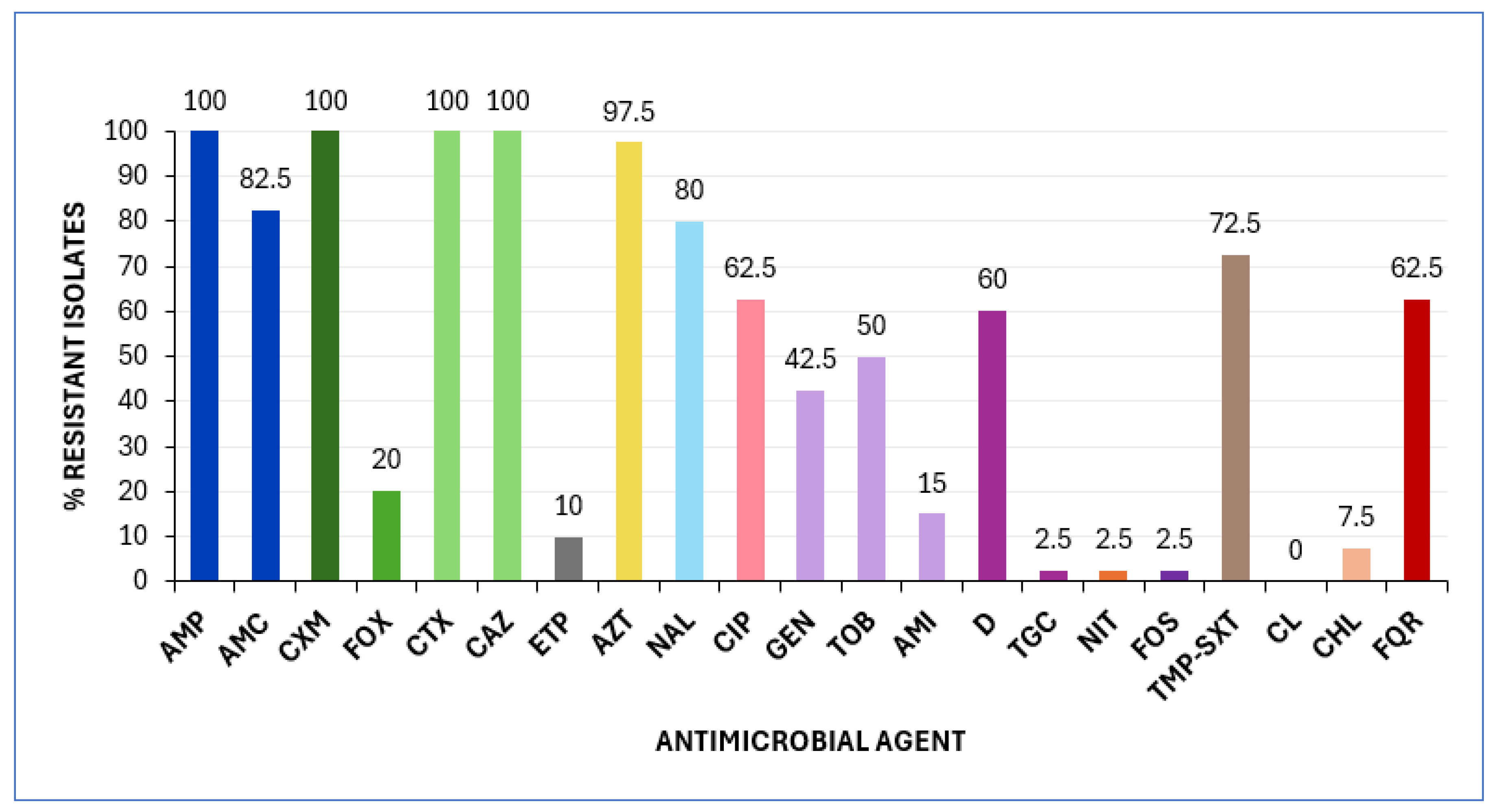
2.1.1. Antimicrobial Susceptibility Testing (AST) and Genotypic Characterization of bla Genes
2.1.2. Virulence Traits
2.1.3. Clonal Groups
2.1.3.1. Characterization of Clonal Complex (CC) 131 Isolates
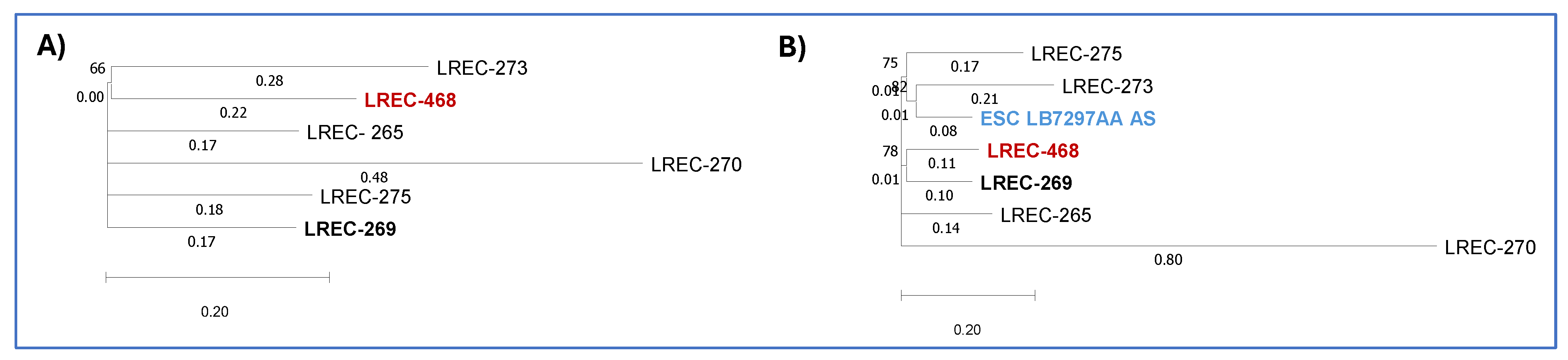
2.1.3.2. In silico characterization of ST1193 isolate
- (A)
- Phylogenetic dendrogram based on SNP counts per substitution within the core genome of the Algerian and Spanish ST1193 isolates. The WGS comparison resulted in a core genome covering 95.65% of the reference genome LREC-269 (5.4Mb).
- (B)
- Phylogenetic dendrogram incorporating the Algerian, Spanish and ESC_RA5887AA ST1193 isolates, showing SNP counts per substitution. The WGS comparison resulted in a core genome covering 82.6% of the reference genome LREC-269 (5.4Mb).
2.2. Klebsiella Pneumoniae Collection
2.2.1. AST and Genotypic Characterization of bla Genes
2.1.2. Virulence Traits
3. Discussion
4. Materials and Methods
4.1. E. coli and K. pneumoniae Collections
4.2. Antimicrobial Susceptibility Testing (AST)
4.3. Detection and Typing of Antimicrobial Resistance Genes
4.4. Molecular Characterization of E. coli: Virulence Traits, Phylogroup, Clonotype and Virotype Assignment
4.5. Phenotypic and Genotypic Detection of Hypervirulent K. pneumoniae
4.6. Whole Genome Sequencing (WGS) and Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foxman, B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28(1):1-13. [CrossRef]
- Flores-Mireles, A.L., Walker, J.N., Caparon, M., Hultgren, S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269-84. [CrossRef]
- Yang, X., Chen, H., Zheng, Y., Qu, S., Wang, H., Yi, F. Disease burden and long-term trends of urinary tract infections: A worldwide report. Front Public Health. 2022;10:888205. [CrossRef]
- Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-55. [CrossRef]
- Manges, A.R., Geum, H.M., Guo, A., Edens, T.J., Fibke, C.D., Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32(3). [CrossRef]
- García-Meniño, I., Lumbreras, P., Lestón, L., Álvarez-Álvarez, M., García, V., Hammerl, J.A., et al. Occurrence and Genomic Characterization of Clone ST1193 Clonotype 14-64 in Uncomplicated Urinary Tract Infections Caused by Escherichia coli in Spain. Microbiol Spectr. 2022;10(3):e0004122. [CrossRef]
- García-Meniño, I., García, V., Lumbreras-Iglesias, P., Fernández, J., Mora, A. Fluoroquinolone resistance in complicated urinary tract infections: association with the increased occurrence and diversity of clonal complex 131, together with ST1193. Front Cell Infect Microbiol. 2024;14:1351618. [CrossRef]
- Pitout, J.D.D., Peirano, G., Chen, L., DeVinney, R., Matsumura, Y. Escherichia coli ST1193: Following in the Footsteps of E. coli ST131. Antimicrob Agents Chemother. 2022;66(7):e0051122. [CrossRef]
- Cummins, E.A., Snaith, A.E., McNally, A., Hall, R.J. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis. 2021. [CrossRef]
- Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D.L., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318-27. [CrossRef]
- Ait-Mimoune, N., Hassaine, H., Boulanoir, M. Bacteriological profile of urinary tract infections and antibiotic susceptibility of Escherichia coli in Algeria. Iran J Microbiol. 2022;14(2):156-60. [CrossRef]
- Bakour, S., Sahli, F., Touati, A., Rolain, J.M. Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbes New Infect. 2015;3:34-6. [CrossRef]
- Brahmia, S., Lalaoui, R., Nedjai, S., Djahmi, N., Chettibi, S., Rolain, J.M., et al. First Clinical Cases of KPC-2-Producing Klebsiella pneumoniae ST258 in Algeria and Outbreak of Klebsiella penumoniae ST101 Harboring blaOXA-48 Gene in the Urology Department of Annaba Hospital. Microb Drug Resist. 2021;27(5):652-9. [CrossRef]
- Benbrahim, C., Barka, M.S., Benmahdi, L., Zatout, A., Khadir, A. Klebsiella pneumoniae producing extended spectrum β-lactamase in Regional Military University Hospital of Oran, Algeria: antibiotic resistance, biofilm formation, and detection of blaCTX-M and blaTEM genes. Afr J Clin Exper Microbiol. 2021;22 (1):28-37. 2: (1). [CrossRef]
- Gómez-Duarte, O.G., Arzuza, O,. Urbina, D., Bai, J., Guerra, J., Montes, O., et al. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children's diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog Dis. 2010;7(2):199-206. [CrossRef]
- Clermont, O., Christenson, J.K., Denamur, E., Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58-65. [CrossRef]
- Clermont, O., Dixit, O.V.A., Vangchhia, B., Condamine, B., Dion, S., Bridier-Nahmias, A., et al. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ Microbiol. 2019;21(8):3107-17. [CrossRef]
- Weissman, S.J., Johnson, J.R., Tchesnokova, V., Billig, M., Dykhuizen, D., Riddell, K., et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol. 2012;78(5):1353-60. [CrossRef]
- Dahbi, G., Mora, A., Mamani, R., López, C., Alonso, M.P., Marzoa, J., et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int J Med Microbiol. 2014;304(8):1247-57. [CrossRef]
- Spurbeck, R.R., Dinh, P.C., Walk, S.T., Stapleton, A.E., Hooton, T.M., Nolan, L.K., et al. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun. 2012;80(12):4115-22. [CrossRef]
- Johnson, J.R., Porter, S., Johnston, B., Kuskowski, M.A., Spurbeck, R.R., Mobley, H.L., et al. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates From Patients With Urosepsis. Open Forum Infect Dis. 2015;2(3):ofv083. [CrossRef]
- Jaureguy, F., Landraud, L., Passet, V., Diancourt, L., Frapy, E., Guigon, G., et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics. 2008;9:560. [CrossRef]
- Shon, A.S., Bajwa, R.P., Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107-18. [CrossRef]
- Russo, T.A., Olson, R., Fang, C.T., Stoesser, N., Miller, M., MacDonald, U., et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56(9). [CrossRef]
- Institute for Health Metrics and Evaluation (IHME) UoOM. [Available from: https://vizhub.healthdata.org/microbe].
- Peirano, G., Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs. 2019;79(14):1529-41. [CrossRef]
- Peirano, G., Pitout, J.D.D. Rapidly spreading Enterobacterales with OXA-48-like carbapenemases. J Clin Microbiol. 2025;63(2):e0151524. [CrossRef]
- Nabti, L.Z., Sahli, F., Radji, N., Mezaghcha, W., Semara, L., Aberkane, S., et al. High Prevalence of Multidrug-Resistant Escherichia coli in Urine Samples from Inpatients and Outpatients at a Tertiary Care Hospital in Sétif, Algeria. Microb Drug Resist. 2019;25(3):386-93. [CrossRef]
- Zenati, F., Barguigua, A., Nayme, K., Benbelaïd, F., Khadir, A., Bellahsene, C., et al. Characterization of uropathogenic ESBL-producing Escherichia coli isolated from hospitalized patients in western Algeria. J Infect Dev Ctries. 2019;13(4):291-302. [CrossRef]
- Touati, A., Mairi, A. Carbapenemase-Producing Enterobacterales in Algeria: A Systematic Review. Microb Drug Resist. 2020;26(5):475-82. [CrossRef]
- López-Cerero, L., Salamanca, E., Delgado-Valverde, M., Rodríguez-Martínez, J.M., Rodríguez-Baño, J., Pascual, Á. Higher prevalence of CTX-M-27-producing Escherichia coli belonging to ST131 clade C1 among residents of two long-term care facilities in Southern Spain. Eur J Clin Microbiol Infect Dis. 2022;41(2):335-8. [CrossRef]
- Spurbeck, R.R., Mobley, H.L.T. Chapter 9—Uropathogenic Escherichia coli. In Escherichia coli, Pathotypes and Principles of Pathogenesis, 2nd ed.; Editor Michael Donnenberg; Elsevier Inc., Academic Press: Amsterdam, The Netherlands; 2013. p. 275-304.
- Wang, M.C., Fan, Y.H., Zhang, Y.Z., Bregente, C.J.B., Lin, W.H., Chen, C.A., et al. Characterization of uropathogenic Escherichia coli phylogroups associated with antimicrobial resistance, virulence factor distribution, and virulence-related phenotypes. Infect Genet Evol. 2023;114:105493. [CrossRef]
- Colpan, A., Johnston, B., Porter, S., Clabots, C., Anway, R., Thao, L., et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis. 2013;57(9):1256-65. [CrossRef]
- Banerjee, R., Johnson, J.R. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2014;58(9):4997-5004. [CrossRef]
- Pitout, J.D., DeVinney, R. ST131: a multidrug-resistant clone primed for global domination. F1000Res. 2017;6. [CrossRef]
- Baba Ahmed-Kazi Tani, Z., Decré, D., Genel, N., Boucherit-Otmani, Z., Arlet, G., Drissi, M. Molecular and epidemiological characterization of enterobacterial multidrug-resistant strains in Tlemcen Hospital (Algeria) (2008-2010). Microb Drug Resist. 2013;19(3):185-90. [CrossRef]
- Agabou, A., Pantel, A., Ouchenane, Z., Lezzar, N., Khemissi, S., Satta, D., et al. First description of OXA-48-producing Escherichia coli and the pandemic clone ST131 from patients hospitalised at a military hospital in Algeria. Eur J Clin Microbiol Infect Dis. 2014;33(9):1641-6. [CrossRef]
- Yahiaoui, M., Robin, F., Bakour, R., Hamidi, M., Bonnet, R., Messai, Y. Antibiotic Resistance, Virulence, and Genetic Background of Community-Acquired Uropathogenic Escherichia coli from Algeria. Microb Drug Resist. 2015;21(5):516-26. [CrossRef]
- Brahmi, S., Dunyach-Rémy, C., Touati, A., Lavigne, J.P. CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin Microbiol Infect. 2015;21(3):e18-20. [CrossRef]
- Chenouf, N.S., Carvalho, I., Messaï, C.R., Ruiz-Ripa, L., Mama, O.M., Titouche, Y., et al. Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Brolier Liver in the Center of Algeria, with Detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli. Microb Drug Resist. 2021;27(2):268-76. [CrossRef]
- Bachiri, T., Bakour, S., Ladjouzi, R., Thongpan, L., Rolain, J.M., Touati, A. High rates of CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in wild boars and Barbary macaques in Algeria. J Glob Antimicrob Resist. 2017;8:35-40. [CrossRef]
- Johnson, T.J., Elnekave, E., Miller, E.A., Munoz-Aguayo, J., Flores Figueroa, C., Johnston, B., et al. Phylogenomic Analysis of Extraintestinal Pathogenic Escherichia coli Sequence Type 1193, an Emerging Multidrug-Resistant Clonal Group. Antimicrob Agents Chemother. 2019;63(1). [CrossRef]
- Wyrsch, E.R., Bushell, R.N., Marenda, M.S., Browning, G.F., Djordjevic, S.P. Global Phylogeny and F Virulence Plasmid Carriage in Pandemic Escherichia coli ST1193. Microbiol Spectr. 2022;10(6):e0255422. [CrossRef]
- Birgy, A., Madhi, F., Jung, C., Levy, C., Cointe, A., Bidet, P., et al. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J Antimicrob Chemother. 2020;75(1):96-105. [CrossRef]
- Valenza, G., Werner, M., Eisenberger, D., Nickel, S., Lehner-Reindl, V., Höller, C., et al. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J Glob Antimicrob Resist. 2019;17:305-8. [CrossRef]
- Ding, Y., Zhang, J., Yao, K., Gao, W., Wang, Y. Molecular characteristics of the new emerging global clone ST1193 among clinical isolates of Escherichia coli from neonatal invasive infections in China. Eur J Clin Microbiol Infect Dis. 2021;40(4):833-40. [CrossRef]
- Kim, Y., Oh, T., Nam, Y.S., Cho, S.Y., Lee, H.J. Prevalence of ST131 and ST1193 Among Bloodstream Isolates of Escherichia coli not Susceptible to Ciprofloxacin in a Tertiary Care University Hospital in Korea, 2013 - 2014. Clin Lab. 2017;63(9):1541-3. [CrossRef]
- Nguyen, Q., Nguyen, T.T.N., Pham, P., Chau, V., Nguyen, L.P.H., Nguyen, T.D., et al. Genomic insights into the circulation of pandemic fluoroquinolone-resistant extra-intestinal pathogenic Escherichia coli ST1193 in Vietnam. Microb Genom. 2021;7(12). [CrossRef]
- Tchesnokova, V.L., Rechkina, E., Larson, L., Ferrier, K., Weaver, J.L., Schroeder, D.W., et al. Rapid and Extensive Expansion in the United States of a New Multidrug-resistant Escherichia coli Clonal Group, Sequence Type 1193. Clin Infect Dis. 2019;68(2):334-7. [CrossRef]
- Yehouenou, C.L., Bogaerts, B., De Keersmaecker, S.C.J., Roosens, N.H.C., Marchal, K., Tchiakpe, E., et al. Whole-Genome Sequencing-Based Antimicrobial Resistance Characterization and Phylogenomic Investigation of 19 Multidrug-Resistant and Extended-Spectrum Beta-Lactamase-Positive Escherichia coli Strains Collected From Hospital Patients in Benin in 2019. Front Microbiol. 2021;12:752883. [CrossRef]
- Byarugaba, D.K., Erima, B., Wokorach, G., Alafi, S., Kibuuka, H., Mworozi, E., et al. Resistome and virulome of high-risk pandemic clones of multidrug-resistant extra-intestinal pathogenic Escherichia coli (ExPEC) isolated from tertiary healthcare settings in Uganda. PLoS One. 2023;18(11):e0294424. [CrossRef]
- Gomi, R., Matsumura, Y., Yamamoto, M., Tanaka, M., Komakech, A.J., Matsuda, T., et al. Genomic surveillance of antimicrobial-resistant Escherichia coli in fecal sludge and sewage in Uganda. Water Res. 2024;248:120830. [CrossRef]
- Achtman, M., Zhou, Z., Charlesworth, J., Baxter, L. EnteroBase: hierarchical clustering of 100 000s of bacterial genomes into species/subspecies and populations. Philos Trans R Soc Lond B Biol Sci. 2022;377(1861):20210240. [CrossRef]
- Falgenhauer, L., Nordmann, P., Imirzalioglu, C., Yao, Y., Falgenhauer, J., Hauri, A.M., et al. Cross-border emergence of clonal lineages of ST38 Escherichia coli producing the OXA-48-like carbapenemase OXA-244 in Germany and Switzerland. Int J Antimicrob Agents. 2020;56(6):106157. [CrossRef]
- Grevskott, D.H., Radisic, V., Salvà-Serra, F., Moore, E.R.B, Akervold, K.S., Victor, M.P., et al. Emergence and dissemination of epidemic-causing OXA-244 carbapenemase-producing Escherichia coli ST38 through hospital sewage in Norway, 2020-2022. J Hosp Infect. 2024;145:165-73. [CrossRef]
- Roy Chowdhury, P., Hastak, P., DeMaere, M., Wyrsch, E., Li, D., Elankumaran, P., et al. Phylogenomic analysis of a global collection of Escherichia coli ST38: evidence of interspecies and environmental transmission? mSystems. 2023;8(5):e0123622. [CrossRef]
- Bouaziz, A., Loucif, L., Ayachi, A., Guehaz, K., Bendjama, E., Rolain, J.M. Migratory White Stork (Ciconia ciconia): A Potential Vector of the OXA-48-Producing Escherichia coli ST38 Clone in Algeria. Microb Drug Resist. 2018;24(4):461-8. [CrossRef]
- Tafoukt, R., Touati, A., Leangapichart, T., Bakour, S., Rolain, J.M. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017;120:185-9. [CrossRef]
- Yagoubat, M., Ould El-Hadj-Khelil, A., Malki, A., Bakour, S., Touati, A., Rolain, J.M. Genetic characterisation of carbapenem-resistant Gram-negative bacteria isolated from the University Hospital Mohamed Boudiaf in Ouargla, southern Algeria. J Glob Antimicrob Resist. 2017;8:55-9. [CrossRef]
- Belmahdi, M., Bakour, S., Al Bayssari, C., Touati, A., Rolain, J.M. Molecular characterisation of extended-spectrum β-lactamase- and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. J Glob Antimicrob Resist. 2016;6:108-12. [CrossRef]
- Abderrahim, A., Djahmi, N., Pujol, C., Nedjai, S., Bentakouk, M.C., Kirane-Gacemi, D., et al. First Case of NDM-1-Producing Klebsiella pneumoniae in Annaba University Hospital, Algeria. Microb Drug Resist. 2017;23(7):895-900. [CrossRef]
- Zemmour, A., Dali-Yahia, R., Maatallah, M., Saidi-Ouahrani, N., Rahmani, B., Benhamouche, N., et al. High-risk clones of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from the University Hospital Establishment of Oran, Algeria (2011-2012). PLoS One. 2021;16(7):e0254805. [CrossRef]
- Liu, C., Guo, J., Lu, M., Shen, N., Du, P. Dissemination of the mobilised RND efflux pump gene cluster tmexCD-toprJ among Klebsiella pneumoniae. Lancet Microbe. 2023;4(3):e135. [CrossRef]
- Russo, T.A., Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3). [CrossRef]
- Al Ismail, D., Campos-Madueno, E.I., Donà, V., Endimiani, A. Hypervirulent Klebsiella pneumoniae (hvKP): Overview, Epidemiology, and Laboratory Detection. Pathog Immun. 2025;10(1):80-119. [CrossRef]
- Li, W., Sun, G., Yu, Y., Li, N., Chen, M., Jin, R., et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225-32. [CrossRef]
- Russo, T.A., MacDonald, U. The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. mSphere. 2020;5(1). [CrossRef]
- Russo, T.A., Alvarado, C.L., Davies, C.J., Drayer, Z.J., Carlino-MacDonald, U., Hutson, A., et al. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. mBio. 2024;15(2):e0286723. [CrossRef]
- Tang, Y., Du, P., Du, C., Yang, P., Shen, N., Russo, T.A., et al. Genomically defined hypervirulent Klebsiella pneumoniae contributed to early-onset increased mortality. Nat Commun. 2025;16(1):2096. [CrossRef]
- European Centre for Disease Prevention and Control. Emergence of hypervirulent Klebsiella pneumoniae ST23 carrying carbapenemase genes in EU/EEA countries, first update. 14 February 2024. ECDC: Stockholm; 2024. ISBN 978-92-9498-691-7. Catalogue number: TQ-02-24-218-EN-N. [CrossRef]
- Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Nicolas-Chanoine, M.H., Decré, D., et al. Development of a multiplex PCR assay for identification of Klebsiella pneumoniae hypervirulent clones of capsular serotype K2. J Med Microbiol. 2014;63(Pt 12):1608-14. [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical breakpoints 2025 [Available from: https://www.eucast.org/clinical_breakpoints].
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA, USA, 2024.
- Magiorakos, A.P., Srinivasan, A., Carey, R.B., Carmeli, Y., Falagas, M.E., Giske, C.G., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-81. [CrossRef]
- García-Meniño, I., García, V., Alonso, M.P., Blanco, J.E., Blanco, J., Mora, A. Clones of enterotoxigenic and Shiga toxin-producing Escherichia coli implicated in swine enteric colibacillosis in Spain and rates of antibiotic resistance. Vet Microbiol. 2021;252:108924. [CrossRef]
- Poirel, L., Walsh, T.R., Cuvillier, V., Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-23. [CrossRef]
- Rebelo, A.R., Bortolaia, V., Kjeldgaard, J.S., Pedersen, S.K., Leekitcharoenphon, P., Hansen, I.M., et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6). [CrossRef]
- Borowiak, M., Fischer, J., Hammerl, J.A., Hendriksen, R.S., Szabo, I., Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317-24. [CrossRef]
- García, V., García-Meniño, I., Gómez, V., Jiménez-Orellana, M., Méndez, A., Aguarón, A., et al. Mobile colistin resistance (MCR), extended-spectrum beta-lactamase (ESBL) and multidrug resistance monitoring in Escherichia coli (commensal and pathogenic) in pig farming: need of harmonized guidelines and clinical breakpoints. Front Microbiol. 2022;13:1042612. [CrossRef]
- García-Meniño, I., Díaz-Jiménez, D., García, V., de Toro, M., Flament-Simon, S.C., Blanco, J., et al. Genomic Characterization of Prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli Within Swine Enteric Colibacillosis in Spain. Front Microbiol. 2019;10. [CrossRef]
- Kaas, R.S., Leekitcharoenphon, P., Aarestrup, F.M., Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9(8):e104984. [CrossRef]

| Phylogroup | Clonotype (CH)a | No isolates | UPEC status | ExPEC status | FQR |
| B2 | CH40-30 | 17 | 15 | 16 | 17 |
| CH14-64 | 1 | 1 | 1 | 1 | |
| CH108-Neg | 2 | 2 | 2 | 1 | |
| CH1012-Neg | 1 | 0 | 0 | 1 | |
| D | CH26-5 | 6 | 0 | 6 | 1 |
| CH26-Neg | 3 | 0 | 3 | 0 | |
| B1 | CH65-27 | 2 | 0 | 0 | 0 |
| CH27-54 | 1 | 0 | 0 | 0 | |
| CH65-32 | 1 | 0 | 0 | 0 | |
| CH7-604 | 1 | 0 | 0 | 0 | |
| A | CH11-54 | 1 | 0 | 1 | 1 |
| CH11-Neg | 1 | 0 | 0 | 1 | |
| CH7-94 | 1 | 0 | 0 | 0 | |
| F | CH88-Neg | 2 | 0 | 0 | 2 |
| Isolate | UPEC statusa | ExPEC statusb | Virotypec | Phenotypic resistanced | Virulence genes profile |
| Ec4 | + | + | NT | AMP, CXM, CTX, CAZ, AZT, NAL, CIP, TOB, AMI, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, papGII |
| Ec5 | + | + | NT | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, TOB, AMI, D, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, papGII |
| Ec8 | + | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, AMI, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec10 | + | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec19 a | + | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec20 a | + | + | E | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII, cnfI, hlyA |
| Ec22 | + | + | F | AMP, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec23 | + | - | NT | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, TOB, AMI, D, NIT, TMP-SXT | fyuA, yfcV, chuA, papAH, papGII |
| Ec28 | + | + | NT | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, sat, papGII |
| Ec30 | + | + | NT | AMP, AMC, CXM, CTX, FOX, CAZ, ETP, AZT, NAL, CIP, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, sat, papGII |
| Ec31 | + | + | NT | AMP, AMC, CXM, CTX, FOX, CAZ, AZT, NAL, CIP, GEN, TOB, D, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, sat, papGII |
| Ec41 | - | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, D, TMP-SXT | chuA, iutA, kpsMII-K5, sat, papGII |
| Ec43 | + | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, D TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec45 | + | + | NT | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, AMI, D, TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, sat, papGII |
| Ec52 | + | + | F | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB, D TMP-SXT | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII |
| Ec55 a | + | + | E | AMP, AMC, CXM, CTX, CAZ, AZT, NAL, CIP, GEN, TOB | fyuA, yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII, cnfI, hlyA |
| Ec57 | - | + | E | AMP, AMC, CXM, CTX, FOX, CAZ, AZT, NAL, CIP, GEN, TOB, D, TGC, FOS, TMP-SXT, CHL | yfcV, chuA, papAH, iutA, kpsMII-K5, sat, papGII, cnfI, hlyA |
| ID Code for isolate/genomea | Ec1a/ LREC-468 |
| O:H antigensb | O75:H5 |
| ST#1 / ST#2c | 1193/53 |
| cgMLSTd | 140226 |
|
Acquired resistances and point mutations (in bold)e |
aac(3)-IIa, aac(6')-Ib-cr, blaCTX-M-15, blaOXA-1, catB3 gyrA p.S83L, gyrA p.D87N, parC p.S80I, parE p.L416F |
| Plasmid content: Inc. group [pMLST]f | IncF [F-:A1:B10], IncI1-I [ST Unknown], ColBS512-like |
| Virulence genesg | aslA, chuA, cia, csgA, fdeC, fimH, fyuA, gad, iha, irp2, iucC, iutA, kpsE, kpsMII_K1, neuC, nlpI, ompT, papA_F43, sat, shiA, sitA, terC, tia, usp, vat, yehA, yehB, yehC, yehD, yfcV |
| Phenotypic resistanceh | AMP, AMC, CXM, CAZ, CTX, ATM, NAL, CIP, GEN |
|
Hypermucoviscous Phenotype (HMV) a |
|||||||
| Isolate | ML | MH | TSA | CA | Phenotypic resistanceb | Virulence genesc | blagenes |
| KP6b | + | - | + | - | AMP, CXM, CTX, CAZ, AZT, CIP, FOS | terB | CTX-M-15 |
| KP10c | + | + | + | + | AMP, AMC, CXM, FOX, CTX, CAZ, ETP, AZT, NAL, CIP, GEN, TOB, NIT, FOS | iucA, peg-344, rmpA | NDM, CTX-M-15 |
| KP16 | + | + | + | + | AMP, AMC, CXM, FOX, CTX, CAZ, ETP, AZT, NAL, CIP, GEN, TOB, TGC, NIT, FOS, TMP-SXT, CHL | iucA, peg-344, rmpA | NDM, CTX-M-15 |
| KP20a | + | + | + | - | AMP, CXM, CTX, CAZ, AZT, CIP, FOS, TMP-SXT | terB | CTX-M-15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





