Submitted:
19 February 2025
Posted:
19 February 2025
You are already at the latest version
Abstract
Keywords:
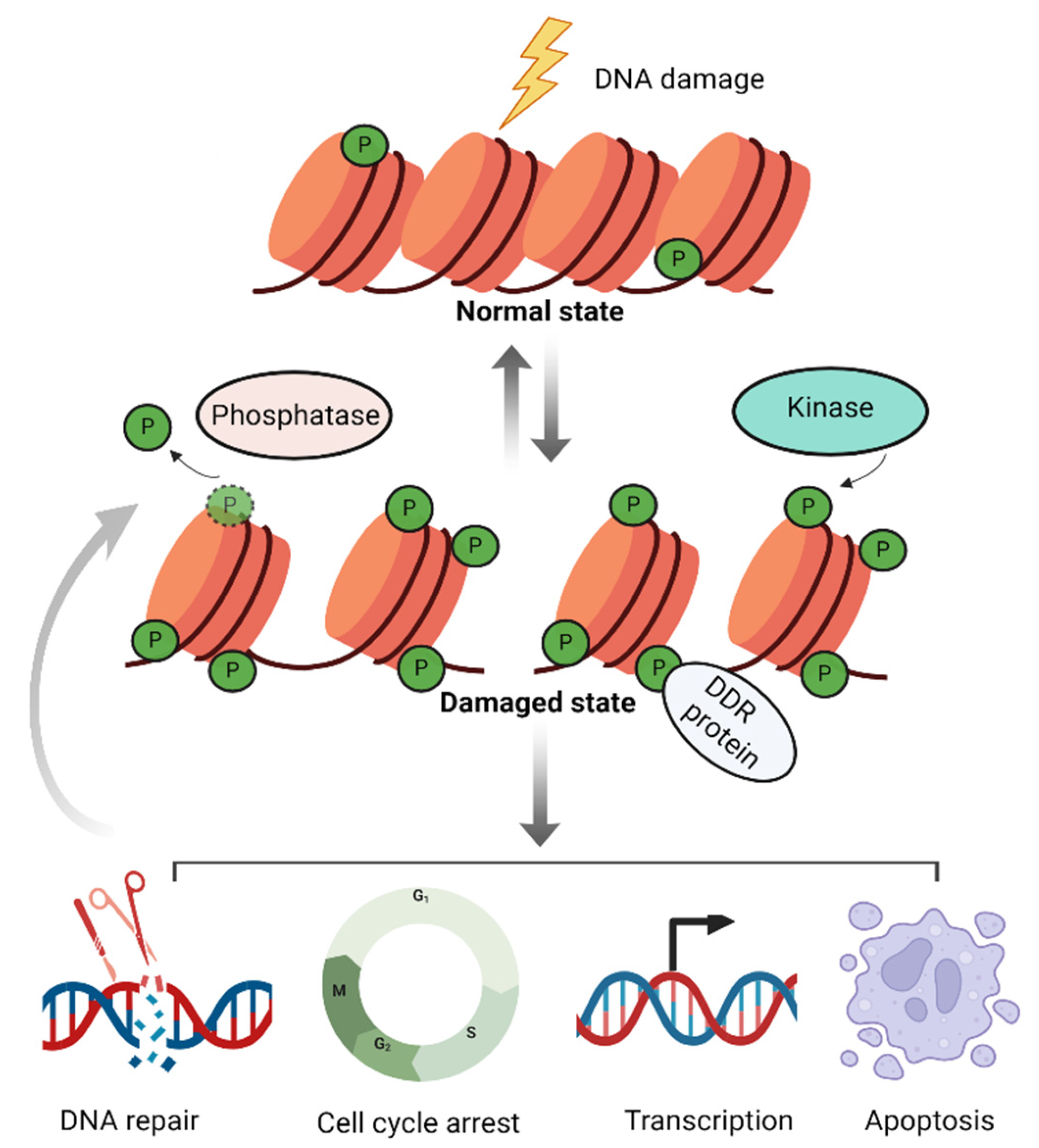
1. Introduction
2. Histone Phosphorylation in the DNA Damage Response
2.1. H2A Phosphorylation in the DNA Damage Response
2.1.1. γH2AX
2.1.2. Other H2A sites
2.2. H3
2.3. H4
2.4. H2B and H1
3. Histone Phosphorylation in Cancer Research and Therapy
4. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DDR | DNA damage response |
| 53BP1 | p53-binding protein 1 |
| ATM | ataxia telangiectasia-mutated |
| ATR | ATM- and Rad3-related |
| BRCA1 | breast cancer 1 |
| CKII | casein kinase II |
| DNA-PK | DNA-dependent protein kinase |
| DSB | double-strand break |
| EGF | epidermal growth factor |
| HR | homologous recombination |
| HU | hydroxyurea |
| IR | ionizing radiation |
| JAK2 | janus kinase 2 |
| MMR | mismatch repair |
| MMS | methyl methane-sulphonate |
| MRN | Mre11-Rad50-Nbs1 |
| NHEJ | nonhomologous end-joining |
| PTM | posttranslational modification |
| RPA | replication protein A |
| SSA | single-strand annealing |
| ssDNA | single-stranded DNA |
| TOPK | T-LAK cell-originated protein kinases |
| UV | ultraviolet |
| DDR | DNA damage response |
| 53BP1 | p53-binding protein 1 |
References
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: making it safe to play with knives. Molecular cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. The DNA damage response and cancer therapy. Nature 2012, 481, 287–294. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—an evolving hallmark of cancer. Nature reviews Molecular cell biology 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Delint-Ramirez, I.; Madabhushi, R. DNA damage and its links to neuronal aging and degeneration. Neuron 2025, 113, 7–28. [Google Scholar] [CrossRef]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nature reviews Molecular cell biology 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Jiricny, J. The multifaceted mismatch-repair system. Nature reviews Molecular cell biology 2006, 7, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Mullenders, L.H. Solar UV damage to cellular DNA: from mechanisms to biological effects. Photochemical & Photobiological Sciences 2018, 17, 1842–1852. [Google Scholar]
- Kumar, N.; Raja, S.; Van Houten, B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Research 2020, 48, 11227–11243. [Google Scholar] [CrossRef]
- Ijsselsteijn, R.; Jansen, J.G.; de Wind, N. DNA mismatch repair-dependent DNA damage responses and cancer. DNA repair 2020, 93, 102923. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry 2004, 73, 39–85. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends in cell biology 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annual review of genetics 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S. Mechanism and regulation of DNA end resection in eukaryotes. Critical reviews in biochemistry and molecular biology 2016, 51, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nature cell biology 2017, 19, 1–9. [Google Scholar] [CrossRef]
- Ui, A.; Chiba, N.; Yasui, A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer science 2020, 111, 1443–1451. [Google Scholar] [CrossRef]
- Katsuki, Y.; Jeggo, P.A.; Uchihara, Y.; Takata, M.; Shibata, A. DNA double-strand break end resection: a critical relay point for determining the pathway of repair and signaling. Genome Instability & Disease 2020, 1, 155–171. [Google Scholar]
- Wang, K.; Li, L.; Zhang, Y.; Gao, D. Crosstalk between signaling pathways and DNA damage response. Genome Instability & Disease 2020, 1, 81–91. [Google Scholar]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nature reviews Molecular cell biology 2017, 18, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of histone modification. Histone Mutations and Cancer 2021, 1–16. [Google Scholar]
- Audia, J.E.; Campbell, R.M. Histone modifications and cancer. Cold Spring Harbor perspectives in biology 2016, 8, a019521. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: how histone modifications regulate gene expression. Trends in Genetics 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Van, H.T.; Santos, M.A. Histone modifications and the DNA double-strand break response. Cell Cycle 2018, 17, 2399–2410. [Google Scholar] [CrossRef] [PubMed]
- Arnaudo, A.M.; Garcia, B.A. Proteomic characterization of novel histone post-translational modifications. Epigenetics & chromatin 2013, 6, 1–7. [Google Scholar]
- Campos, E.I.; Reinberg, D. Histones: annotating chromatin. Annual review of genetics 2009, 43, 559–599. [Google Scholar] [CrossRef]
- Lai, P.M.; Chan, K.M. Roles of Histone H2A Variants in Cancer Development, Prognosis, and Treatment. International Journal of Molecular Sciences 2024, 25, 3144. [Google Scholar] [CrossRef]
- Stope, M.B. Phosphorylation of histone H2A. X as a DNA-associated biomarker. World Academy of Sciences Journal 2021, 3, 1–5. [Google Scholar] [CrossRef]
- Yao, S.; Feng, Y.; Zhang, Y.; Feng, J. DNA damage checkpoint and repair: From the budding yeast Saccharomyces cerevisiae to the pathogenic fungus Candida albicans. Computational and Structural Biotechnology Journal 2021, 19, 6343–6354. [Google Scholar] [CrossRef]
- Merighi, A.; Gionchiglia, N.; Granato, A.; Lossi, L. The phosphorylated form of the histone H2AX (γH2AX) in the brain from embryonic life to old age. Molecules 2021, 26, 7198. [Google Scholar] [CrossRef] [PubMed]
- Rahmanian, N.; Shokrzadeh, M.; Eskandani, M. Recent advances in γH2AX biomarker-based genotoxicity assays: A marker of DNA damage and repair. DNA repair 2021, 108, 103243. [Google Scholar] [CrossRef]
- Zhao, S.; Allis, C.D.; Wang, G.G. The language of chromatin modification in human cancers. Nature Reviews Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Salzano, M.; Sanz-García, M.; Monsalve, D.M.; Moura, D.S.; Lazo, P.A. VRK1 chromatin kinase phosphorylates H2AX and is required for foci formation induced by DNA damage. Epigenetics 2015, 10, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, K.S.; Kuttikrishnan, S.; Ahmad, N.; Habeeba, U.; Mariyam, Z.; Suleman, M.; Bhat, A.A.; Uddin, S. H2AX: A key player in DNA damage response and a promising target for cancer therapy. Biomedicine & Pharmacotherapy 2024, 175, 116663. [Google Scholar]
- Song, H.; Shen, R.; Liu, X.; Yang, X.; Xie, K.; Guo, Z.; Wang, D. Histone post-translational modification and the DNA damage response. Genes & Diseases 2023, 10, 1429–1444. [Google Scholar]
- Oberdoerffer, P.; Miller, K.M. Histone H2A variants: Diversifying chromatin to ensure genome integrity. Seminars in cell & developmental biology 2023, 135, 59–72. [Google Scholar]
- Xie, A.; Puget, N.; Shim, I.; Odate, S.; Jarzyna, I.; Bassing, C.H.; Alt, F.W.; Scully, R. Control of sister chromatid recombination by histone H2AX. Molecular cell 2004, 16, 1017–1025. [Google Scholar] [CrossRef]
- Oh, J.-M.; Myung, K. Crosstalk between different DNA repair pathways for DNA double strand break repairs. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 2022, 873, 503438. [Google Scholar] [CrossRef]
- Guha, S.; Bhaumik, S.R. Transcription-coupled DNA double-strand break repair. DNA repair 2022, 109, 103211. [Google Scholar] [CrossRef]
- Lee, J.-H.; Paull, T.T. Cellular functions of the protein kinase ATM and their relevance to human disease. Nature Reviews Molecular Cell Biology 2021, 22, 796–814. [Google Scholar] [CrossRef] [PubMed]
- Rass, E.; Willaume, S.; Bertrand, P. 53BP1: keeping it under control, even at a distance from DNA damage. Genes 2022, 13, 2390. [Google Scholar] [CrossRef]
- Kciuk, M.; Gielecińska, A.; Mujwar, S.; Mojzych, M.; Kontek, R. Cyclin-dependent kinases in DNA damage response. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 2022, 1877, 188716. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. ATM’s role in the repair of DNA double-strand breaks. Genes 2021, 12, 1370. [Google Scholar] [CrossRef]
- Danovski, G.; Panova, G.; Keister, B.; Georgiev, G.; Atemin, A.; Uzunova, S.; Stamatov, R.; Kanev, P.-B.; Aleksandrov, R.; Blagoev, K.B. Diffusion of activated ATM explains γH2AX and MDC1 spread beyond the DNA damage site. Iscience 2024, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Ho, T.L.; Hariharan, A.; Goh, H.C.; Wong, Y.L.; Verkaik, N.S.; Lee, M.Y.; Tam, W.L.; van Gent, D.C.; Venkitaraman, A.R. Rapid recruitment of p53 to DNA damage sites directs DNA repair choice and integrity. Proceedings of the National Academy of Sciences 2022, 119, e2113233119. [Google Scholar] [CrossRef]
- Salguero, I.; Belotserkovskaya, R.; Coates, J.; Sczaniecka-Clift, M.; Demir, M.; Jhujh, S.; Wilson, M.D.; Jackson, S.P. MDC1 PST-repeat region promotes histone H2AX-independent chromatin association and DNA damage tolerance. Nature communications 2019, 10, 5191. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Zhu, Q.; Wani, G.; He, J.; Wang, Q.-e.; Wani, A.A. USP3 counteracts RNF168 via deubiquitinating H2A and γH2AX at lysine 13 and 15. Cell cycle 2014, 13, 106–114. [Google Scholar] [CrossRef]
- Kocyłowski, M.K.; Rey, A.J.; Stewart, G.S.; Halazonetis, T.D. Ubiquitin-H2AX fusions render 53BP1 recruitment to DNA damage sites independent of RNF8 or RNF168. Cell Cycle 2015, 14, 1748–1758. [Google Scholar] [CrossRef]
- Krishnan, R.; Lapierre, M.; Gautreau, B.; Nixon, K.C.; El Ghamrasni, S.; Patel, P.S.; Hao, J.; Yerlici, V.T.; Guturi, K.K.N.; St-Germain, J. RNF8 ubiquitylation of XRN2 facilitates R-loop resolution and restrains genomic instability in BRCA1 mutant cells. Nucleic Acids Research 2023, 51, 10484–10505. [Google Scholar] [CrossRef]
- Sadoughi, F.; Hallajzadeh, J.; Asemi, Z.; Mansournia, M.A.; Alemi, F.; Yousefi, B. Signaling pathways involved in cell cycle arrest during the DNA breaks. DNA repair 2021, 98, 103047. [Google Scholar] [CrossRef] [PubMed]
- Siler, J.; Guo, N.; Liu, Z.; Qin, Y.; Bi, X. γH2A/γH2AX Mediates DNA Damage-Specific Control of Checkpoint Signaling in Saccharomyces cerevisiae. International Journal of Molecular Sciences 2024, 25, 2462. [Google Scholar] [CrossRef] [PubMed]
- Hammet, A.; Magill, C.; Heierhorst, J.; Jackson, S.P. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO reports 2007, 8, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Aricthota, S.; Rana, P.P.; Haldar, D. Histone acetylation dynamics in repair of DNA double-strand breaks. Frontiers in Genetics 2022, 13, 926577. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.W.; Yu, D.Y.; Pray-Grant, M.G.; Qiu, Q.; Harmon, K.E.; Megee, P.C.; Grant, P.A.; Smith, M.M.; Christman, M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 2002, 419, 411–415. [Google Scholar] [CrossRef]
- van Attikum, H.; Fritsch, O.; Hohn, B.; Gasser, S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004, 119, 777–788. [Google Scholar] [CrossRef]
- Horigome, C.; Oma, Y.; Konishi, T.; Schmid, R.; Marcomini, I.; Hauer, M.H.; Dion, V.; Harata, M.; Gasser, S.M. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Molecular cell 2014, 55, 626–639. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Krebs, J.E.; Peterson, C.L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptationin response to DNA damage. Genes & development 2006, 20, 2437–2449. [Google Scholar]
- Gerhold, C.B.; Gasser, S.M. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends in cell biology 2014, 24, 619–631. [Google Scholar] [CrossRef]
- Keogh, M.-C.; Kim, J.-A.; Downey, M.; Fillingham, J.; Chowdhury, D.; Harrison, J.C.; Onishi, M.; Datta, N.; Galicia, S.; Emili, A. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 2006, 439, 497–501. [Google Scholar] [CrossRef]
- Chen, L.; Lai, Y.; Zhu, X.; Ma, L.; Bai, Q.; Vazquez, I.; Xiao, Y.; Liu, C.; Li, D.; Gao, C. The role of specific PP2A complexes in the dephosphorylation of γ-H2AX. Journal of Cell Science 2015, 128, 421. [Google Scholar] [CrossRef]
- Li, X.; Nan, A.; Xiao, Y.; Chen, Y.; Lai, Y. PP2A–B56ϵ complex is involved in dephosphorylation of γ-H2AX in the repair process of CPT-induced DNA double-strand breaks. Toxicology 2015, 331, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Cao, P.; Greer, P.; Nagengast, E.; Kolb, R.; Mumby, M.; Cowan, K. Protein phosphatase 2A has an essential role in the activation of γ-irradiation-induced G2/M checkpoint response. Oncogene 2010, 29, 4317–4329. [Google Scholar] [CrossRef] [PubMed]
- Nakada, S.; Chen, G.I.; Gingras, A.C.; Durocher, D. PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. The EMBO Reports 2008, 9, 1019–1026. [Google Scholar] [CrossRef]
- Moon, S.-H.; Nguyen, T.-A.; Darlington, Y.; Lu, X.; Donehower, L.A. Dephosphorylation of γ-H2AX by WIP1: an important homeostatic regulatory event in DNA repair and cell cycle control. Cell cycle 2010, 9, 2092–2096. [Google Scholar] [CrossRef]
- Zhong, J.; Liao, J.; Liu, X.; Wang, P.; Liu, J.; Hou, W.; Zhu, B.; Yao, L.; Wang, J.; Li, J. Protein phosphatase PP6 is required for homology-directed repair of DNA double-strand breaks. Cell cycle 2011, 10, 1411–1419. [Google Scholar] [CrossRef]
- Dziegielewski, J.; Bońkowska, M.A.; Poniecka, E.A.; Heo, J.; Du, K.; Crittenden, R.B.; Bender, T.P.; Brautigan, D.L.; Larner, J.M. Deletion of the SAPS1 subunit of protein phosphatase 6 in mice increases radiosensitivity and impairs the cellular DNA damage response. DNA repair 2020, 85, 102737. [Google Scholar] [CrossRef]
- Xiao, A.; Li, H.; Shechter, D.; Ahn, S.H.; Fabrizio, L.A.; Erdjument-Bromage, H.; Ishibe-Murakami, S.; Wang, B.; Tempst, P.; Hofmann, K. WSTF regulates the H2A. X DNA damage response via a novel tyrosine kinase activity. Nature 2009, 457, 57–62. [Google Scholar] [CrossRef]
- Cook, P.J.; Ju, B.G.; Telese, F.; Wang, X.; Glass, C.K.; Rosenfeld, M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 2009, 458, 591–596. [Google Scholar] [CrossRef]
- Harvey, A.C.; Jackson, S.P.; Downs, J.A. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics 2005, 170, 543–553. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Moore, J.D.; Yazgan, O.; Ataian, Y.; Krebs, J.E. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics 2007, 176, 15–25. [Google Scholar] [CrossRef]
- Kozmin, S.G.; Dominska, M.; Kokoska, R.J.; Petes, T.D. A tale of two serines: the effects of histone H2A mutations S122A and S129A on chromosome nondisjunction in Saccharomyces cerevisiae. Genetics 2025, 229, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Côté, V.; Côté, J. DNA damage-induced phosphorylation of histone H2A at serine 15 is linked to DNA end resection. Molecular and Cellular Biology 2021, 41, e00056–00021. [Google Scholar] [CrossRef]
- Xie, A.; Odate, S.; Chandramouly, G.; Scully, R.A. H2AX post-translational modifications in the IR response and homologous recombination. Cell cycle 2010, 9, 3602–3610. [Google Scholar] [CrossRef] [PubMed]
- House, N.C.; Polleys, E.J.; Quasem, I.; De la Rosa Mejia, M.; Joyce, C.E.; Takacsi-Nagy, O.; Krebs, J.E.; Fuchs, S.M.; Freudenreich, C.H. Distinct roles for S. cerevisiae H2A copies in recombination and repeat stability, with a role for H2A. 1 threonine 126. Elife 2019, 8, e53362. [Google Scholar] [CrossRef]
- Sawicka, A.; Seiser, C. Histone H3 phosphorylation–a versatile chromatin modification for different occasions. Biochimie 2012, 94, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Niida, H.; Zineldeen, D.H.; Tagami, H.; Tanaka, M.; Saito, H.; Nakanishi, M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 2008, 132, 221–232. [Google Scholar] [CrossRef]
- Sharma, A.K.; Bhattacharya, S.; Khan, S.A.; Khade, B.; Gupta, S. Dynamic alteration in H3 serine 10 phosphorylation is G1-phase specific during ionization radiation induced DNA damage response in human cells. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2015, 773, 83–91. [Google Scholar] [CrossRef]
- Ozawa, K. Reduction of phosphorylated histone H3 serine 10 and serine 28 cell cycle marker intensities after DNA damage. Cytometry Part A: The Journal of the International Society for Analytical Cytology 2008, 73, 517–527. [Google Scholar] [CrossRef]
- Monte-Serrano, E.; Morejón-García, P.; Campillo-Marcos, I.; Campos-Díaz, A.; Navarro-Carrasco, E.; Lazo, P.A. The pattern of histone H3 epigenetic PTMs is regulated by the VRK1 chromatin kinase. Epigenetics & Chromatin 2023, 16, 18. [Google Scholar]
- Monaco, L.; Kolthur-Seetharam, U.; Loury, R.; Murcia, J.M.-d.; de Murcia, G.; Sassone-Corsi, P. Inhibition of Aurora-B kinase activity by poly (ADP-ribosyl) ation in response to DNA damage. Proceedings of the National Academy of Sciences 2005, 102, 14244–14248. [Google Scholar] [CrossRef] [PubMed]
- Tjeertes, J.V.; Miller, K.M.; Jackson, S.P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. The EMBO journal 2009, 28, 1878–1889. [Google Scholar] [CrossRef]
- Lan, J.; Lepikhov, K.; Giehr, P.; Walter, J. Histone and DNA methylation control by H3 serine 10/threonine 11 phosphorylation in the mouse zygote. Epigenetics & chromatin 2017, 10, 1–19. [Google Scholar]
- Tian, Y.; Zhang, C.; Tian, X.; Zhang, L.; Yin, T.; Dang, Y.; Liu, Y.; Lou, H.; He, Q. H3T11 phosphorylation by CKII is required for heterochromatin formation in Neurospora. Nucleic Acids Research 2024, 52, 9536–9550. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kang, B.-H.; Jang, H.; Kim, T.W.; Choi, J.; Kwak, S.; Han, J.; Cho, E.-J.; Youn, H.-D. AKT phosphorylates H3-threonine 45 to facilitate termination of gene transcription in response to DNA damage. Nucleic acids research 2015, 43, 4505–4516. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.L.; Turner, F.B.; Krishnamoorthy, T.; Wolner, B.; Ahn, S.-H.; Foley, M.; Dorsey, J.A.; Peterson, C.L.; Berger, S.L.; Allis, C.D. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Current biology 2005, 15, 656–660. [Google Scholar] [CrossRef]
- Utley, R.T.; Lacoste, N.; Jobin-Robitaille, O.; Allard, S.; Côté, J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Molecular and cellular biology 2005. [Google Scholar] [CrossRef]
- Clouaire, T.; Legube, G. A snapshot on the cis chromatin response to DNA double-strand breaks. Trends in Genetics 2019, 35, 330–345. [Google Scholar] [CrossRef]
- Hossain, M.B.; Shifat, R.; Johnson, D.G.; Bedford, M.T.; Gabrusiewicz, K.R.; Cortes-Santiago, N.; Luo, X.; Lu, Z.; Ezhilarasan, R.; Sulman, E.P. TIE2-mediated tyrosine phosphorylation of H4 regulates DNA damage response by recruiting ABL1. Science advances 2016, 2, e1501290. [Google Scholar] [CrossRef]
- Millan-Zambrano, G.; Santos-Rosa, H.; Puddu, F.; Robson, S.C.; Jackson, S.P.; Kouzarides, T. Phosphorylation of histone H4T80 triggers DNA damage checkpoint recovery. Molecular cell 2018, 72, 625–635. [Google Scholar] [CrossRef]
- Lee, C.-S.; Lee, K.; Legube, G.; Haber, J.E. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nature structural & molecular biology 2014, 21, 103–109. [Google Scholar]
- Fernandez-Capetillo, O.; Allis, C.D.; Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. The Journal of experimental medicine 2004, 199, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.L.; Ajiro, K.; Samejima, K.; Kloc, M.; Cheung, P.; Mizzen, C.A.; Beeser, A.; Etkin, L.D.; Chernoff, J.; Earnshaw, W.C. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 2003, 113, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Andrés, M.; García-Gomis, D.; Ponte, I.; Suau, P.; Roque, A. Histone H1 post-translational modifications: update and future perspectives. International journal of molecular sciences 2020, 21, 5941. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jeong, K.W.; Kim, H.; Choi, J.; Lu, W.; Stallcup, M.R.; An, W. Functional interplay between p53 acetylation and H1. 2 phosphorylation in p53-regulated transcription. Oncogene 2012, 31, 4290–4301. [Google Scholar] [CrossRef]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. γH2AX and cancer. Nature Reviews Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef]
- Kawashima, S.; Kawaguchi, N.; Taniguchi, K.; Tashiro, K.; Komura, K.; Tanaka, T.; Inomata, Y.; Imai, Y.; Tanaka, R.; Yamamoto, M. γ-H2AX as a potential indicator of radiosensitivity in colorectal cancer cells. Oncology Letters 2020, 20, 2331–2337. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Yin, T.C.; Chen, Y.-T.; Chai, C.-Y.; Wang, J.Y.; Liu, M.-C.; Lin, Y.-C.; Kan, J.Y. High expression of phospho-H2AX predicts a poor prognosis in colorectal cancer. Anticancer Research 2015, 35, 2447–2453. [Google Scholar]
- Xiao, J.; Duan, Q.; Wang, Z.; Yan, W.; Sun, H.; Xue, P.; Fan, X.; Zeng, X.; Chen, J.; Shao, C. Phosphorylation of TOPK at Y74, Y272 by Src increases the stability of TOPK and promotes tumorigenesis of colon. Oncotarget 2016, 7, 24483. [Google Scholar] [CrossRef]
- Hsu, J.-Y.; Sun, Z.-W.; Li, X.; Reuben, M.; Tatchell, K.; Bishop, D.K.; Grushcow, J.M.; Brame, C.J.; Caldwell, J.A.; Hunt, D.F. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 2000, 102, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Imhof, A.; Patel, D.; Kahl, P.; Hoffmeyer, K.; Friedrichs, N.; Müller, J.M.; Greschik, H.; Kirfel, J.; Ji, S. Phosphorylation of histone H3T6 by PKCβI controls demethylation at histone H3K4. Nature 2010, 464, 792–796. [Google Scholar] [CrossRef]
- Metzger, E.; Wissmann, M.; Yin, N.; Müller, J.M.; Schneider, R.; Peters, A.H.; Günther, T.; Buettner, R.; Schüle, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef]
- Wen, W.; Zhu, F.; Zhang, J.; Keum, Y.-S.; Zykova, T.; Yao, K.; Peng, C.; Zheng, D.; Cho, Y.-Y.; Ma, W.-y. MST1 promotes apoptosis through phosphorylation of histone H2AX. Journal of Biological Chemistry 2010, 285, 39108–39116. [Google Scholar] [CrossRef]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—cause and consequence of genome function. Nature Reviews Genetics 2022, 23, 563–580. [Google Scholar] [CrossRef]
- Dawson, M.A.; Bannister, A.J.; Göttgens, B.; Foster, S.D.; Bartke, T.; Green, A.R.; Kouzarides, T. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 2009, 461, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, M.K.; Sinha, S.; Mendez, M.T.; Sanyal, T.; Mahmud, H.; Kay, N.E.; Gupta, M.; Xu, C.; Vesely, S.K.; Mukherjee, P. Aberrantly Expressed Mitochondrial Lipid Kinase, AGK, Activates JAK2–Histone H3 Axis and BCR Signal: A Mechanistic Study with Implication in CLL Therapy. Clinical Cancer Research 2024, OF1–OF15. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Choi, B.Y.; Cho, Y.-Y.; Mizuno, H.; Kang, B.S.; Bode, A.M.; Dong, Z. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer research 2005, 65, 5818–5827. [Google Scholar] [CrossRef]
- Komar, D.; Juszczynski, P. Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clinical Epigenetics 2020, 12, 1–14. [Google Scholar] [CrossRef]
- Lau, P.N.I.; Cheung, P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proceedings of the National Academy of Sciences 2011, 108, 2801–2806. [Google Scholar] [CrossRef]
- Cho, Y.-Y. RSK2 and its binding partners in cell proliferation, transformation and cancer development. Archives of pharmacal research 2017, 40, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Widjaja, L.; Werner, R.A.; Krischke, E.; Christiansen, H.; Bengel, F.M.; Bogdanova, N.; Derlin, T. Individual radiosensitivity reflected by γ-H2AX and 53BP1 foci predicts outcome in PSMA-targeted radioligand therapy. European Journal of Nuclear Medicine and Molecular Imaging 2023, 50, 602–612. [Google Scholar] [CrossRef]
- Banjarnahor, C.T.U.; Hardiany, N.S.; Wahjoepramono, E.J.; Hariyanto, A.D.; Sadikin, M. High concentration of γ-H2AX correlates with a marker of apoptotic suppression and PI3K/Akt pathway upregulation in glioblastoma multiforme. Oncology Letters 2023, 25, 1–7. [Google Scholar] [CrossRef]
- Hosking, H.; Pederick, W.; Neilsen, P.; Fenning, A. Considerations for the Use of the DNA Damage Marker γ-H2AX in Disease Modeling, Detection, Diagnosis, and Prognosis. Aging and Cancer 2024, 5, 62–69. [Google Scholar] [CrossRef]
- Aitmagambetova, M.; Smagulova, G.; Sakhanova, S.; Kereyeva, N.; Koishybaev, A.; Amanzholkyzy, A.; Tulyaeva, A.; Zholmukhamedova, D.; Kandygulova, G.; Imanbaev, N. The γ-H2AX foci as an indicator for double-stranded DNA breaks and response to ongoing chemotherapy in breast cancer women: a pilot study. European Review for Medical & Pharmacological Sciences 2023, 27. [Google Scholar]
- Zhao, H.; Qu, M.; Li, Y.; Wen, K.; Xu, H.; Song, M.; Xie, D.; Ao, X.; Gong, Y.; Sui, L. An estimate assay for low-level exposure to IR based on mass spectrometry quantification of γ-H2AX in human peripheral blood lymphocytes. Frontiers in Public Health 2022, 10, 1031743. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, K.; Chen, Y.; Liu, J.; Zhang, X.; Zhou, Y.; Liu, Q.; Wang, B.; Chen, T.; Cao, X. RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal transduction and targeted therapy 2022, 7, 240. [Google Scholar] [CrossRef] [PubMed]
- Llavanera, M.; Delgado-Bermudez, A.; Ribas-Maynou, J.; Salas-Huetos, A.; Yeste, M. A systematic review identifying fertility biomarkers in semen: a clinical approach through omics to diagnose male infertility. Fertility and Sterility 2022, 118, 291–313. [Google Scholar] [CrossRef]
- Zorzompokou, C.; Ipeirotis, M.; Martzoukos, M.K.; Marangos, P. Detection of DNA Double-Stranded Breaks in Mouse Oocytes. J. Vis. Exp 2023, 196, e65494. [Google Scholar] [CrossRef]
- Wu, S.; Cao, R.; Tao, B.; Wu, P.; Peng, C.; Gao, H.; Liang, J.; Yang, W. Pyruvate Facilitates FACT-Mediated γH2AX Loading to Chromatin and Promotes the Radiation Resistance of Glioblastoma. Advanced Science 2022, 9, 2104055. [Google Scholar] [CrossRef]
- Kono, T.; Ozawa, H. A comprehensive review of current therapeutic strategies in cancers targeting DNA damage response mechanisms in head and neck squamous cell cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 2024, 189255. [Google Scholar]
- Moon, J.; Kitty, I.; Renata, K.; Qin, S.; Zhao, F.; Kim, W. DNA damage and its role in cancer therapeutics. International Journal of Molecular Sciences 2023, 24, 4741. [Google Scholar] [CrossRef] [PubMed]
| Histone phosphorylation sites | Kinases | Function | Ref. |
|---|---|---|---|
| H1.2-T145 | DNA-PK | chromatin remodel; p53 transcription | [95,96] |
| H2A.1-T126 | unknown | affects the stability and repair of fragile DNA regions | [76] |
| H2A-S122 | Bub1 | DNA repair; chromosome segregation | [70,73] |
| H2A-S15 | Mec1 | influencing chromatin dynamics and DNA end resection | [74] |
| H2AX-S139 (H2A-S129 in yeast) | ATM, ATR, DNA-PK | DNA repair; damage signal transduction; transcription; checkpoint regulation; apoptosis | [35,39,40,47,53,56,57,104] |
| H2AX-T101 | unknown | reduce cells’ sensitivity to IR | [75] |
| H2AX-Y142 | WSTF | DNA repair | [68,69] |
| H2B-S14 | MST1 | chromatin remodeling and apoptosis | [93,94] |
| H2B-T129 | Mec1/Tel1 | unclear, possibly coordinated with the function of γH2AX | [92] |
| H3-S10 | Aurora-B | transcription; modulate chromatin structure | [79,80,84,108] |
| H3-S28 | MSK1 | modulate chromatin structure; transcription | [80,110] |
| H3-T11 | CHK1, CKII | DNA repair; transcription; maintenance of heterochromatin | [78,84,85] |
| H3-T45 | AKT | transcription | [86] |
| H4-S1 | CKII | DNA repair | [87,88] |
| H4-T80 | Cla4 | checkpoint regulation | [91] |
| H4-Y51 | TIE2 | DNA repair | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





