Submitted:
03 February 2025
Posted:
04 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. General Molecular Features
3. Glioblastoma Management
4. Clinical Relevance of the Molecular Profile
- Astrocytoma, IDH-mutant: previously, IDH-mutant tumors were classified as diffuse astrocytoma, anaplastic astrocytoma, or GBM. The latest classification now consolidates these into a single type of IDH-mutant astrocytoma, graded as 2, 3, or 4.
- Grading criteria: the grading of IDH-mutant diffuse astrocytic tumors is no longer based solely on histology. It also considers the presence of the CDKN2A/B homozygous deletion mutation, which results in a CNS WHO grade of 4, even if microvascular proliferation or necrosis is absent.
- GBM, IDH-wildtype: this classification identifies specific molecular markers for this tumor, such as the presence of a TERT promoter mutation (associated with increased telomerase activity, crucial for tumor cell immortalization) or EGFR gene amplification, leading to overexpression of the receptor, and the combined gain of chromosome 7 and loss of chromosome 10 (+7/−10). If any of these markers are found in an IDH-wildtype diffuse astrocytic glioma in adults, the diagnosis should be GBM, IDH-wildtype.
- Pediatric patients: the diagnostic criteria for IDH-wildtype diffuse astrocytomas differ in pediatric patients, who are diagnosed using different categories of pediatric-type gliomas [2].
- Proneural Group: characterized by proneural gene expression patterns and RTK I/LGm6 DNA methylation profiles. This subgroup often shows amplifications of genes such as cyclin-dependent kinase 4 (CDK4) and platelet-derived growth factor alpha (PDGFRA). It is more prevalent among younger adults.
- Classical Group: exhibits classical gene expression patterns and classic-like RTK II DNA methylation profiles. It is marked by frequent EGFR amplifications and the loss of CDKN2A/B genes.
- Mesenchymal Group: enriched for tumors with neurofibromatosis type 1 (NF1) loss and increased infiltration by macrophages [22]. This subgroup is associated with a mesenchymal or mesenchymal-like subtype.
5. microRNAs (miRNAs) and GBM
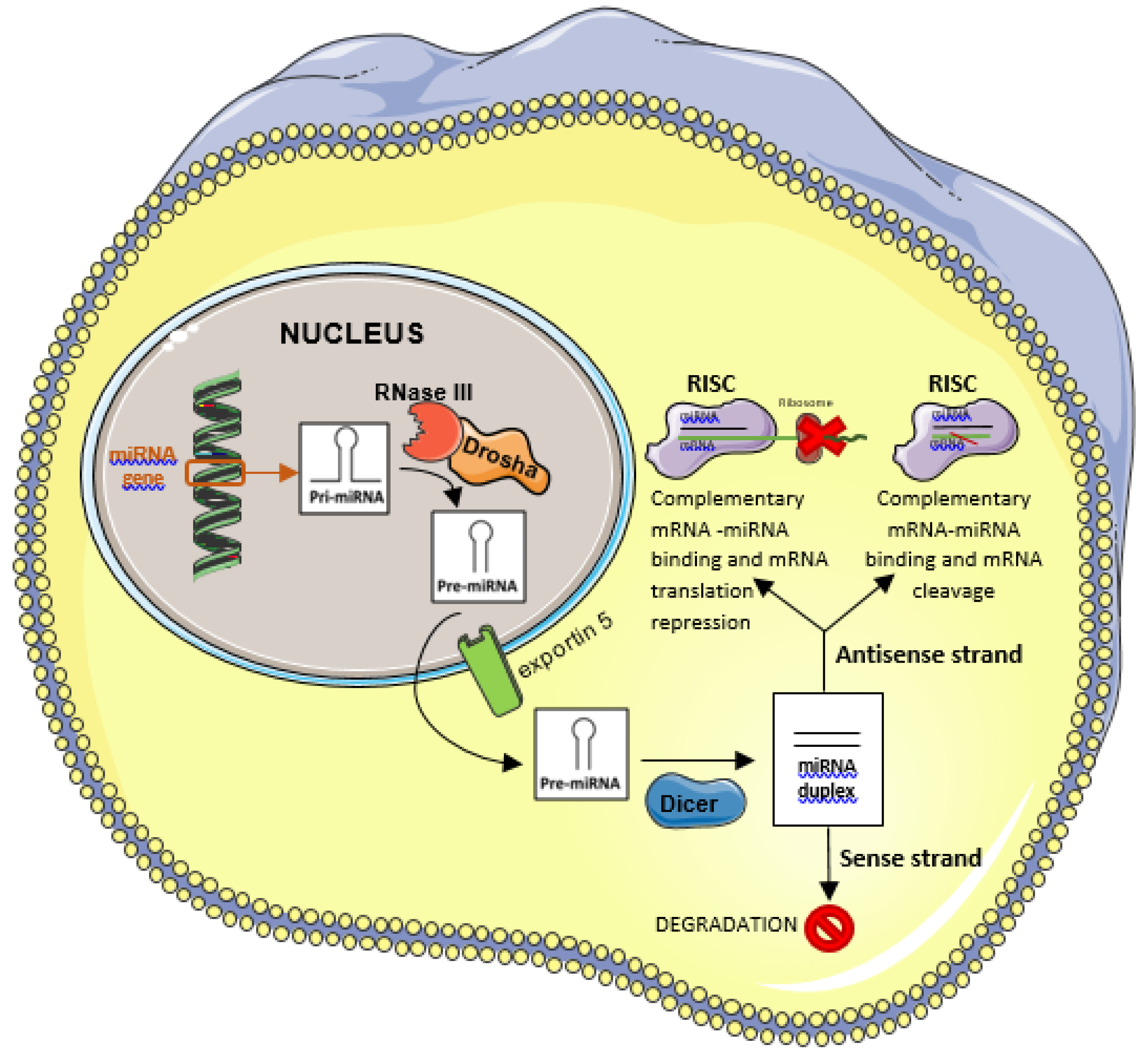
5.1. miRNAs Function and Biogenesis
5.2. miRNAs in Cancer
5.3. miRNAs in GBM
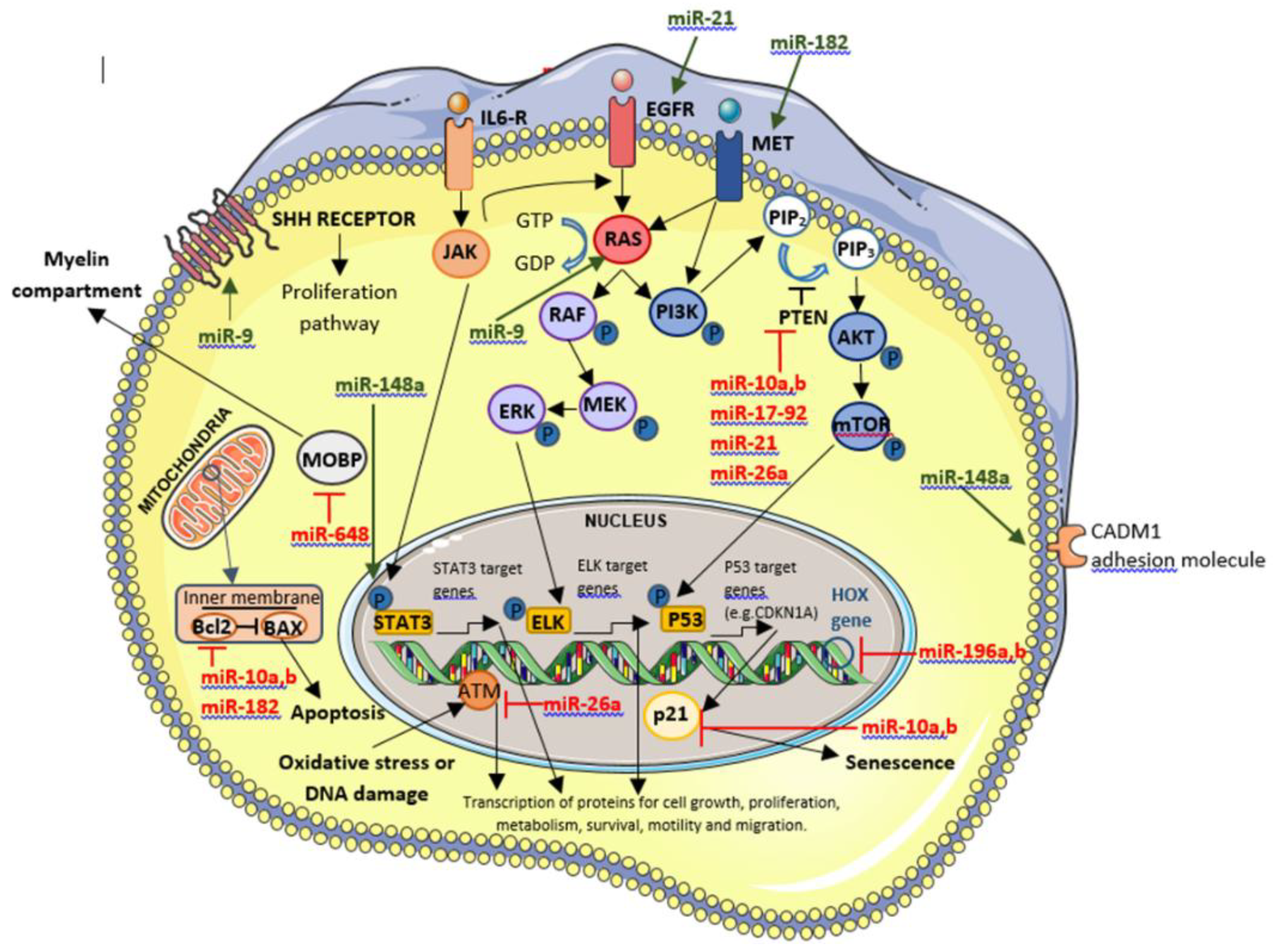
5.3.1. OncomiR-Upregulated miRNAs
5.3.1.1. OncomiR Involved in GBM Biogenesis
- miR-17-92 cluster
5.3.1.2. OncomiR Involved in GBM prognosis
- miR-9
- miR-10a and miR-10b
- miR-148a
- miR-182
- miR-196a and miR-196b
5.3.1.3. OncomiR with Effect on Therapy Efficacy
- miR-26a
- miR-648
5.3.1.4. OncomiR Involved in GBM Biogenesis that in the Future Will Have a Clinical Role Through Their Inhibition
- miR-21
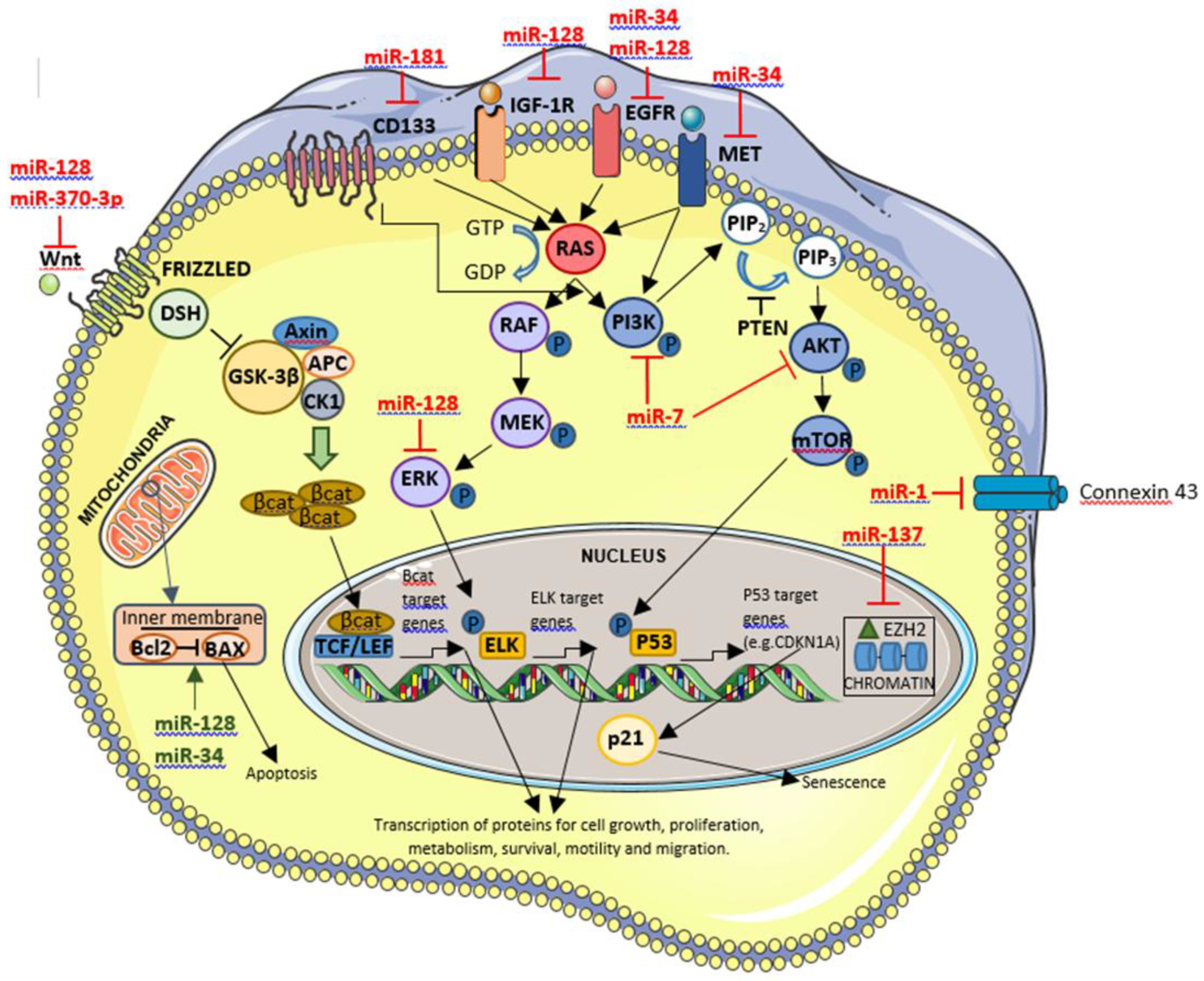
5.3.2. Tumor Suppressor miRNAs-Downregulated miRNAs
5.3.2.1. Tumor Suppressor miRNAs Involved in GBM Biogenesis
- miR-1
5.3.2.2. Tumor Suppressor miRNAs Involved in GBM Prognosis
- miR-128
- miR-137
- miR-181 family
5.3.2.3. Tumor Suppressor miRNAs with Effect on Therapy Efficacy
- miR-370-3p
5.3.2.4. Tumor Suppressor miRNAs Involved in GBM Biogenesis that in the Future Will Have a Clinical Role Through Their Inhibition
- miR-7
- miR-34
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schaff, L.R. , Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329(7), 574-587. [CrossRef]
- Louis, D.N. , Perry, A., Wesseling, P., Brat, D.J., Cree, I.A., Figarella-Branger, D., Hawkins, C., Ng, H.K., Pfister, S.M., Reifenberger, G., Soffietti, R., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol, 1: 23(8), 1231-1251. [Google Scholar] [CrossRef]
- Wen, P.Y. , Weller, M., Lee, E.Q., Alexander, B.M., Barnholtz-Sloan, J.S., Barthel, F.P., Batchelor, T.T., Bindra, R.S., Chang, S.M., Chiocca, E.A., et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol, 2020, 22(8), 1073-1113. [Google Scholar] [CrossRef]
- Ostrom, Q.T. , Cioffi, G., Waite, K., Kruchko, C., Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014-2018. Neuro Oncol, 2021: 23 (12 Suppl 2):iii1-iii105. [CrossRef]
- Vijapura, C. , Saad, Aldin. E., Capizzano, A.A., Policeni, B., Sato, Y., Moritani, T. Genetic Syndromes Associated with Central Nervous System Tumors. Radiographics. 2017, 37(1), 258–280. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P. , Lin, A.L., Young, R.J., DiStefano, N.M., Hyman, D.M., Li, B.T., Berger, M.F., Zehir, A., Ladanyi, M., Solit, D.B. et al. Genomic Correlates of Disease Progression and Treatment Response in Prospectively Characterized Gliomas. Clin Cancer Res, 5: 25(18), 5537-5547. [Google Scholar] [CrossRef]
- Stupp, R. , Mason, W.P., van den Bent, M.J., Weller, M., Fisher, B., Taphoorn, M.J., Belanger, K., Brandes, A.A., Marosi, C., Bogdahn, U. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med, 2005: 352(10):987-96. [CrossRef]
- Hanna, C. , Kurian, K.M., Williams, K., Watts, C., Jackson, A., Carruthers, R., Strathdee, K., Cruickshank, G., Dunn, L,, Erridge, S. et al. Pharmacokinetics, safety, and tolerability of olaparib and temozolomide for recurrent glioblastoma: results of the phase I OPARATIC trial. Neuro Oncol, 2020, 22(12): 1840-1850. [Google Scholar] [CrossRef]
- Hau, E. , Shen, H., Clark, C., Graham, P.H., Koh, E.S., L, McDonald, K. The evolving roles and controversies of radiotherapy in the treatment of glioblastoma. J Med Radiat Sci. 2016, 63(2), 114–23. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R. , Taillibert, S., Kanner, A., Read, W., Steinberg, D., Lhermitte, B., Toms, S., Idbaih, A., Ahluwalia, M.S., Fink, K., et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA. 2017, 318(23), 2306-2316; Erratum in: JAMA. 2018 May 1;319(17):1824. 10.1001/jama.2018.3431. [Google Scholar] [CrossRef]
- Brennan, C.W. , Verhaak, R.G., McKenna, A., Campos, B., Noushmehr, H., Salama, S.R., Zheng, S., Chakravarty, D., Sanborn, J.Z., Berman, S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013, 155(2), 462–77. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J. , Mashouf, L.A., Lim, M. CAR T Cell Therapy in Primary Brain Tumors: Current Investigations and the Future. Front Immunol, 2022, 13, 817296. [Google Scholar] [CrossRef]
- Wick, W. , Gorlia, T., Bendszus, M., Taphoorn, M., Sahm, F., Harting, I., Brandes, A.A., Taal, W,, Domont, J., Idbaih, A., et al. Lomustine and Bevacizumab in Progressive Glioblastoma. N Engl J Med. 2017, 377(20), 1954–1963. [Google Scholar] [CrossRef]
- Louis, D.N. , Aldape, K., Brat, D.J., Capper, D., Ellison, D.W., Hawkins, C., Paulus, W., Perry, A., Reifenberger, G, et al. Announcing cIMPACT-NOW: the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy. Acta Neuropathol 2017, 133(1), 1-3. [CrossRef]
- Leske, H. , Dalgleish, R., Lazar, A.J., Reifenberger, G., Cree, I.A. A common classification framework for histone sequence alterations in tumours: an expert consensus proposal. J Pathol 2021, 254(2), 109-120. [CrossRef]
- Louis, D.N. , Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W.K., Ohgaki, H., Wiestler, O.D., Kleihues, P., Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016, 131(6), 803-20. [CrossRef]
- Chen, L. , Voronovich, Z., Clark, K., Hands, I., Mannas, J., Walsh, M., Nikiforova, M.N., Durbin, E.B., Weiss, H., Horbinski, C. Predicting the likelihood of an isocitrate dehydrogenase 1 or 2 mutation in diagnoses of infiltrative glioma. Neuro Oncol. 2014, 16(11), 1478–83. [Google Scholar] [CrossRef]
- Capper, D. , Jones, D.T.W., Sill, M., Hovestadt, V., Schrimpf, D., Sturm, D., Koelsche, C., Sahm, F., Chavez, L., Reuss, D.E., et al. DNA methylation-based classification of central nervous system tumours. Nature, 2018: 22, 555(7697): 469-474. [CrossRef]
- Louis, D.N. , Wesseling, P., Aldape, K., Brat, D.J., Capper, D., Cree, I.A., Eberhart, C., Figarella-Branger, D., Fouladi, M., Fuller, G,N., et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol 2020, 30(4), 844-856. [CrossRef]
- Capper, D. , Stichel, D., Sahm, F., Jones, D.T.W., Schrimpf, D., Sill, M., Schmid, S., Hovestadt, V., Reuss, D.E., Koelsche, C., et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol 2018, 136(2), 181-210. [CrossRef]
- Korshunov, A. , Schrimpf, D., Ryzhova, M., Sturm, D., Chavez, L., Hovestadt, V., Sharma, T., Habel, A., Burford, A., Jones, C., et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol 2017, 134(3): 507-516. [CrossRef]
- Wang, Q. , Hu, B., Hu, X., Kim, H., Squatrito, M., Scarpace, L., deCarvalho, A.C., Lyu, S., Li, P., Li, Y., et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32(1), 42-56.e6, Erratum in: Cancer Cell. 2018 Jan 8;33(1):152. [CrossRef]
- Stupp, R. , Hegi, M.E., Mason, W.P., van den Bent, M.J., Taphoorn, M.J., Janzer, R.C., Ludwin, S.K., Allgeier, A., Fisher, B., Belanger, K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009, 10(5), 459-66. [CrossRef]
- Reifenberger, G. , Weber, R.G., Riehmer, V., Kaulich, K., Willscher, E., Wirth, H., Gietzelt, J., Hentschel, B., Westphal, M., Simon, M., Schackert, G., et al. Molecular characterization of long-term survivors of glioblastoma using genome- and transcriptome-wide profiling. Int J Cancer, 2014, 135(8), 1822-31. [Google Scholar] [CrossRef]
- Weller, M. , Felsberg, J., Hartmann, C., Berger, H., Steinbach, J.P., Schramm, J., Westphal, M., Schackert, G., Simon, M., Tonn, J.C., et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009, 27(34), 5743-50. [Google Scholar] [CrossRef]
- Hegi, M.E. , Diserens, A.C., Gorlia, T., Hamou, M.F., de Tribolet, N., Weller, M., Kros, J.M., Hainfellner, J.A., Mason, W., Mariani, L., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med, 2005, 352(10), 997-1003. [Google Scholar] [CrossRef]
- Weller, M. , Tabatabai, G., Kästner, B., Felsberg, J., Steinbach, J.P., Wick, A., Schnell, O., Hau, P., Herrlinger, U., Sabel, M.C., et al. MGMT Promoter Methylation Is a Strong Prognostic Biomarker for Benefit from Dose-Intensified Temozolomide Rechallenge in Progressive Glioblastoma: The DIRECTOR Trial. Clin Cancer Res, 2015, 21(9), 2057-64. [Google Scholar] [CrossRef]
- Malmström, A. , Grønberg, B.H., Marosi, C., Stupp, R., Frappaz, D., Schultz, H., Abacioglu, U., Tavelin, B., Lhermitte, B., Hegi, M.E., et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol 2012, 13(9), 916-26. [CrossRef]
- Tandel, S. G,, Biswas, M., Kakde, G.O., Tiwari, A., Suri, S.H., Turk, M., Laird, J.R., Asare, C.K., Ankrah, A.A., Khanna, N.N. A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers (Basel). 2019, 11(1), 111. [Google Scholar] [CrossRef]
- DeWitt, J.C. , Jordan, J.T., Frosch, M.P., Samore, W.R., Iafrate, A.J., Louis, D.N., Lennerz, J.K. Cost-effectiveness of IDH testing in diffuse gliomas according to the 2016 WHO classification of tumors of the central nervous system recommendations. Neuro Oncol. 2017, 19(12), 1640–1650. [Google Scholar] [CrossRef]
- Korshunov, A. , Chavez, L., Sharma, T., Ryzhova, M., Schrimpf, D., Stichel, D., Capper, D., Sturm, D., Kool, M., Habel, A., et al. Epithelioid glioblastomas stratify into established diagnostic subsets upon integrated molecular analysis. Brain Pathol. 2018, 28(5), 656–662. [Google Scholar] [CrossRef]
- Körber, V. , Yang, J., Barah, P., Wu, Y., Stichel, D., Gu, Z., Fletcher, M.N.C., Jones, D., Hentschel, B., Lamszus, K., et al. Evolutionary Trajectories of IDHWT Glioblastomas Reveal a Common Path of Early Tumorigenesis Instigated Years ahead of Initial Diagnosis. Cancer Cell 2019, 35(4), 692-704.e12. [CrossRef]
- Touat, M. , Li, Y.Y., Boynton, A.N., Spurr, L.F., Iorgulescu, J.B., Bohrson, C.L., Cortes-Ciriano, I., Birzu, C., Geduldig, J.E., Pelton, K., Lim-Fat, M.J., et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature, . 2020, 580(7804), 517-523. [Google Scholar] [CrossRef]
- Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro KD, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019, 576(7785), 112–120. [Google Scholar] [CrossRef]
- Hasan, H.; Afzal, M.; Castresanam, J.S.; Shahi, M.H. A Comprehensive Review of miRNAs and Their Epigenetic Effects in Glioblastoma. Cells. 2023, 12(12), 1578. [Google Scholar] [CrossRef]
- Lewis, B.P. , Burge, C.B., Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120(1), 15-20. [CrossRef]
- Ouellet, D.L.; Perron, M.P.; Gobeil, L.A.; Plante, P.; Provost, P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotechnol. 2006, 2006(4), 69616. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014, 15(8), 509–524. [Google Scholar] [CrossRef] [PubMed]
- Leonov, G. , Shah, K., Yee, D., Timmis, J., Sharp, T.V., Lagos, D. Suppression of AGO2 by miR-132 as a determinant of miRNA-mediated silencing in human primary endothelial cells. Int J Biochem Cell Biol, 2015, 69, 75-84. [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007, 302(1),1-12. [Google Scholar] [CrossRef]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA, 2021; 12(6), e1662. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, W. Regulation of angiogenesis by microRNAs in cancer. Mol Med Rep. 2021, 24(2), 583. [Google Scholar] [CrossRef]
- Uzuner, E.; Ulu, G.T.; Gürler, S.B.; Baran, Y. The Role of MiRNA in Cancer: Pathogenesis, Diagnosis, and Treatment. Methods Mol Biol. 2022, 2257, 375–422. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002, 99(24), 15524–15529. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Horton, S.; Saldivar, J.C.; Miuma, S.; Stampfer, M. R.; Heerema, N. A.; & Huebner, K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability [published correction appears in Genes Chromosomes Cancer. 2019 Nov;58(11):824. https://doi.org/10.1002/gcc.22797]. . Genes Chromosomes Cancer. 2013, 52(11), 1017-1029. 10.1002/gcc.22097. [CrossRef]
- Behrooz, A.B.; Latifi-Navid, H.; Nezhadi, A. , Świat, M.; Los, M.; Jamalpoor, Z.; & Ghavami, S. Molecular mechanisms of microRNAs in glioblastoma pathogenesis. Biochim Biophys Acta Mol Cell Res. 2023, 1870(6), 119482. [Google Scholar] [CrossRef]
- Makowska, M.; Smolarz, B.; Romanowicz, H. microRNAs (miRNAs) in Glioblastoma Multiforme (GBM)-Recent Literature Review. Int J Mol Sci. 2023, 24(4), 3521. [Google Scholar] [CrossRef]
- Barciszewska A., M. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia neuropathologica. 2016, 54(4), 369–374. [Google Scholar] [CrossRef]
- Coolen, M. , Katz, S., & Bally-Cuif, L. miR-9: a versatile regulator of neurogenesis. Frontiers in cellular neuroscience. 2013, 7, 220. [Google Scholar] [CrossRef]
- Dong, Z. , & Cui, H. Epigenetic modulation of metabolism in glioblastoma. Seminars in cancer biology. 2019, 57, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J. L. , Rodriguez-Cruz, V., & Rameshwar, P. High expression of miR-9 in CD133+ glioblastoma cells in chemoresistance to temozolomide. Journal of cancer stem cell research, 2015, 3, e1003. [Google Scholar] [CrossRef]
- Yuan, G. Q. , Wei, N. L., Mu, L. Y., Wang, X. Q., Zhang, Y. N., Zhou, W. N., & Pan, Y. W.. A 4-miRNAs signature predicts survival in glioblastoma multiforme patients. Cancer biomarkers : section A of Disease markers. 2017, , 20(4), 443–452. [CrossRef]
- Rezaei, O. , Honarmand, K., Nateghinia, S., Taheri, M., & Ghafouri-Fard, S. miRNA signature in glioblastoma: Potential biomarkers and therapeutic targets. Experimental and molecular pathology, . 2020, 117, 104550. [Google Scholar] [CrossRef]
- Santolini, E. , Puri, C., Salcini, A. E., Gagliani, M. C., Pelicci, P. G., Tacchetti, C., & Di Fiore, P. P. Numb is an endocytic protein. The Journal of cell biology.. 2000, 151(6), 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- de Menezes, M. R. , Acioli, M. E. A., da Trindade, A. C. L., da Silva, S. P., de Lima, R. E., da Silva Teixeira, V. G., & Vasconcelos, L. R. S. Potential role of microRNAs as biomarkers in human glioblastoma: a mini systematic review from 2015 to 2020. Molecular biology reports, 2021, 48(5), 4647–4658. [Google Scholar] [CrossRef]
- Junior, L. G. D. , Baroni, M., Lira, R. C. P., Teixeira, S., Fedatto, P. F., Silveira, V. S., Suazo, V. K., Veronez, L. C., Panepucci, R. A., Antônio, D. S. M. et al. High-throughput microRNA profile in adult and pediatric primary glioblastomas: the role of miR-10b-5p and miR-630 in the tumor aggressiveness. Molecular biology reports, 2020, 47(9), 6949–6959. [Google Scholar] [CrossRef]
- Gabriely, G. , Yi, M., Narayan, R. S., Niers, J. M., Wurdinger, T., Imitola, J., Ligon, K. L., Kesari, S., Esau, C., Stephens, R. M., Tannous, B. A., & Krichevsky, A. M. Human glioma growth is controlled by microRNA-10b. Cancer research, 2011, 71(10), 3563–3572. [Google Scholar] [CrossRef]
- Sun, L. , Yan, W., Wang, Y., Sun, G., Luo, H., Zhang, J., Wang, X., You, Y., Yang, Z., & Liu, N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain research. 2011, 1389, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, B. , Zhao, X., Ming, J., Liu, X., Liu, D., & Jiang, C. Stepwise detection and evaluation reveal miR-10b and miR-222 as a remarkable prognostic pair for glioblastoma. Oncogene, . 2019, 38(33), 6142–6157. [Google Scholar] [CrossRef]
- Cai, Q. , Zhu, A., & Gong, L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bulletin du cancer 2018, 105(7-8), 643–651. [CrossRef]
- Srinivasan, S. , Patric, I. R., & Somasundaram, K. A ten-microRNA expression signature predicts survival in glioblastoma. PloS one, . 2011, 6(3), e17438. [Google Scholar] [CrossRef]
- Vo, D. T. , Qiao, M., Smith, A. D., Burns, S. C., Brenner, A. J., & Penalva, L. O. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA biology 2011, 8(5), 817–828. [CrossRef]
- Shea, A. , Harish, V., Afzal, Z., Chijioke, J., Kedir, H., Dusmatova, S., Roy, A., Ramalinga, M., Harris, B., Blancato, J., et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer medicine. 2016, 5(8), 1917–1946. [Google Scholar] [CrossRef]
- Dambal, S. , Shah, M., Mihelich, B., & Nonn, L. The microRNA-183 cluster: the family that plays together stays together. Nucleic acids research. 2015, 43(15), 7173–7188. [CrossRef]
- Schneider, B. , William, D., Lamp, N., Zimpfer, A., Henker, C., Classen, C. F., & Erbersdobler, A. The miR-183/96/182 cluster is upregulated in glioblastoma carrying EGFR amplification. Molecular and cellular biochemistry, 2022, 477(9), 2297–2307. [Google Scholar] [CrossRef]
- Vilar, J. B. , Christmann, M., & Tomicic, M. T. Alterations in Molecular Profiles Affecting Glioblastoma Resistance to Radiochemotherapy: Where Does the Good Go? Cancers. 2022, 14(10), 2416. [Google Scholar] [CrossRef]
- Kouri, F. M. , Ritner, C., & Stegh, A. H. miRNA-182 and the regulation of the glioblastoma phenotype - toward miRNA-based precision therapeutics. Cell cycle (Georgetown, Tex.), 2015, 14(24), 3794–3800. [Google Scholar] [CrossRef]
- Zhao, H. , Shen, J., Hodges, T. R., Song, R., Fuller, G. N., & Heimberger, A. B. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Molecular cancer. 2017, 16(1), 59. [CrossRef]
- Xiao, Y. , Zhang, L., Song, Z., Guo, C., Zhu, J., Li, Z., & Zhu, S. Potential Diagnostic and Prognostic Value of Plasma Circulating MicroRNA-182 in Human Glioma. Medical science monitor: international medical journal of experimental and clinical research. [CrossRef]
- Jiang, L. , Mao, P. , Song, L., Wu, J., Huang, J., Lin, C., Yuan, J., Qu, L., Cheng, S. Y., & Li, J. miR-182 as a prognostic marker for glioma progression and patient survival. The American journal of pathology. 2010, 177(1), 29–38. [Google Scholar] [CrossRef]
- Chen, C, Zhang, Y, Zhang, L, Weakley, S.M., Yao, Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011, 15(1), 14-23. [CrossRef]
- Hassan, A. , Mosley, J., Singh, S., & Zinn, P. O. A Comprehensive Review of Genomics and Noncoding RNA in Gliomas. Topics in magnetic resonance imaging. 2017, 26(1), 3–14. [Google Scholar] [CrossRef]
- Lakomy, R. , Sana, J., Hankeova, S., Fadrus, P., Kren, L., Lzicarova, E., Svoboda, M., Dolezelova, H., Smrcka, M., Vyzula, R., et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer science. ( 2011, 102(12), 2186–2190. [CrossRef]
- Cardia A, Epistolio S, Zaed I, Sahnane N, Cerutti R, Cipriani D, Barizzi J, Spina P, Stefanini FM, Cerati M et al. Identification of MGMT Downregulation Induced by miRNA in Glioblastoma and Possible Effect on Temozolomide Sensitivity. J Clin Med. 2023, 12(5), 2061. [Google Scholar] [CrossRef]
- Huse, J. T. , Brennan, C., Hambardzumyan, D., Wee, B., Pena, J., Rouhanifard, S. H., Sohn-Lee, C., le Sage, C., Agami, R., Tuschl, T. et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & development, 1327. [Google Scholar] [CrossRef]
- Guo, P. , Lan, J., Ge, J., Nie, Q., Guo, L., Qiu, Y., & Mao, Q. MiR-26a enhances the radiosensitivity of glioblastoma multiforme cells through targeting of ataxia-telangiectasia mutated. Experimental cell research. 2014, 320(2), 200–208. [Google Scholar] [CrossRef]
- ParvizHamidi, M. , Haddad, G., Ostadrahimi, S., Ostadrahimi, N., Sadeghi, S., Fayaz, S., & Fard-Esfahani, P. Circulating miR-26a and miR-21 as biomarkers for glioblastoma multiform. Biotechnology and applied biochemistry 2019, 66(2), 261–265. [CrossRef]
- Siegel, S. R. , Mackenzie, J., Chaplin, G., Jablonski, N. G., & Griffiths, L. Circulating microRNAs involved in multiple sclerosis. Molecular biology reports. 2012, 39(5), 6219–6225. [Google Scholar] [CrossRef] [PubMed]
- Epistolio S, Dazio G, Zaed I, Sahnane N, Cipriani D, Polinelli F, Barizzi J, Spina P, Stefanini FM, Cerati M, Balbi S, Mazzucchelli L, Sessa F, Pesce GA, Reinert M, Cardia A, Marchi F, Frattini M. Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis. Cells. 2024, 13(3), 276. [Google Scholar] [CrossRef]
- Kreth, S. , Limbeck, E., Hinske, L. C., Schütz, S. V., Thon, N., Hoefig, K., Egensperger, R., & Kreth, F. W. In human glioblastomas transcript elongation by alternative polyadenylation and miRNA targeting is a potent mechanism of MGMT silencing. Acta neuropathological. 2013, 125(5), 671–681. [Google Scholar] [CrossRef]
- Kalkan, R. , & Atli, E. İ. The Impacts of miRNAs in Glioblastoma Progression. Critical reviews in eukaryotic gene expression. 2016, 26(2), 137–142. [Google Scholar] [CrossRef]
- Yang, C. H. , Wang, Y., Sims, M., Cai, C., & Pfeffer, L. M. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer letters. 2019, 465, 59–67. [CrossRef]
- Marumoto, T. , & Saya, H. Molecular biology of glioma. Advances in experimental medicine and biology. 2012, 746, 2–11. [Google Scholar] [CrossRef]
- Cardoso, A. M. , Morais, C. M., Pena, F., Marante, T., Cunha, P. P., Jurado, A. S., & Pedroso de Lima, M. C. Differentiation of glioblastoma stem cells promoted by miR-128 or miR-302a overexpression enhances senescence-associated cytotoxicity of axitinib. Human molecular genetics 2021, 30(3-4), 160–171. [CrossRef]
- Balandeh, E. , Mohammadshafie, K., Mahmoudi, Y., Hossein Pourhanifeh, M., Rajabi, A., Bahabadi, Z. R., Mohammadi, A. H., Rahimian, N., Hamblin, M. R., & Mirzaei, H. Roles of Non-coding RNAs and Angiogenesis in Glioblastoma. Frontiers in cell and developmental biology, 2021, 9, 716462. [Google Scholar] [CrossRef]
- Sun, J. , Zheng, G., Gu, Z., & Guo, Z. (2015). MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. Journal of neuro-oncology 2015. 122(3), 481–489. [CrossRef]
- Li, Y. , Fan, S., Xia, W., Qiao, B., Huang, K., Zhou, J., & Liang, M. MiR-181b suppresses angiogenesis by directly targeting cellular communication network factor 1. Laboratory investigation; a journal of technical methods and pathology, 2021, 101(8), 1026–1035. [Google Scholar] [CrossRef]
- Nadaradjane, A. , Briand, J., Bougras-Cartron, G., Disdero, V., Vallette, F. M., Frenel, J. S., & Cartron, P. F. miR-370-3p Is a Therapeutic Tool in Anti-glioblastoma Therapy but Is Not an Intratumoral or Cell-free Circulating Biomarker. Molecular therapy Nucleic acids. [CrossRef]
- Kirstein, A. , Schmid, T. E., & Combs, S. E. The Role of miRNA for the Treatment of MGMT Unmethylated Glioblastoma Multiforme. Cancers. 2020, 12(5), 1099. [Google Scholar] [CrossRef]
- Morales-Martínez, M. , & Vega, M. I. Role of MicroRNA-7 (MiR-7) in Cancer Physiopathology. International journal of molecular sciences, 2022, 23(16), 9091. [Google Scholar] [CrossRef]
- Alamdari-Palangi, V. , Amini, R., & Karami, H. MiRNA-7 enhances erlotinib sensitivity of glioblastoma cells by blocking the IRS-1 and IRS-2 expression. The Journal of pharmacy and pharmacology. 2020, 72(4), 531–538. [Google Scholar] [CrossRef]
- Li, Y. , Guessous, F., Zhang, Y., Dipierro, C., Kefas, B., Johnson, E., Marcinkiewicz, L., Jiang, J., Yang, Y., Schmittgen, T. D., et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer research, 2009, 69(19), 7569–7576. [Google Scholar] [CrossRef]
- Bazzoni, R. , & Bentivegna, A. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers. 2019, 11(3), 292. [Google Scholar] [CrossRef]
- Janaki Ramaiah, M. , Divyapriya, K., Kartik Kumar, S., & Rajesh, Y. B. R. D. Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme. Gene, 2020, 723, 144126. [Google Scholar] [CrossRef]
- Francipane, M. G. , Douradinha, B., Chinnici, C. M., Russelli, G., Conaldi, P. G., & Iannolo, G. Zika Virus: A New Therapeutic Candidate for Glioblastoma Treatment. International journal of molecular sciences, 2021, 22(20), 1099. [Google Scholar] [CrossRef]
- Zhu, Z.; Gorman, M.J.; McKenzie, L.D.; Chai, J.N.; Hubert, C.G.; Prager, B.C.; Fernandez, E.; Richner, J.M.; Zhang, R.; Shan, C.; et al. Zika virus has oncolytic activity against glioblastoma stem cells. J. Exp. Med. 2017, 214, 2843–2857. [Google Scholar] [CrossRef]
- Lubin, J.; Zhang, R.R.; Kuo, J. Zika Virus has Oncolytic Activity Against Glioblastoma Stem Cells. Neurosurgery. 2018, 82, E113–E114. [Google Scholar] [CrossRef]
- Kaid, C.; Goulart, E.; Caires-Júnior, L.C.; Araujo, B.H.S.; Schanoski, A.S.; Bueno, H.; Silva, K.A.T.; Astray, R.M.; Assoni, A.F.; Júnior, A.F.R.; et al. Zika Virus Selectively Kills Aggressive Human Embryonal CNS Tumor Cells In Vitro and In Vivo. Cancer Res. 2018, 78, 3363–3374. [Google Scholar] [CrossRef] [PubMed]
- Baronti, C.; Piorkowski, G.; Charrel, R.N.; Boubis, L.; Leparc-Goffart, I.; de Lamballerie, X. Complete Coding Sequence of Zika Virus from a French Polynesia Outbreak in 2013. Genome Announc. 2014, 2, e00500–14. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Target | Expression in GBM | Function/role in GBM if the miRNA is overexpressed | Clinical application | References |
|---|---|---|---|---|---|
| miR-9 | -RAS and MYC -PTCH1 |
↑ | Cancer cells proliferation↑ Tumor cells transformation↑ Inflammation↑ Angiogenesis↑ Apoptosis↓ |
In vitro: overexpression can enhance the resistance to TMZ in GBM cells. In patients: associated to short term survivors. |
[49,50,51,52] |
| miR-10a, b | -CDKN1A, BIM, BCL2, TEAP2C and PTEN -HOXD10 |
↑ | Cancer cells proliferation↑ Tumor cells migration↑ Invasion↑ EMT promotion↑ Apoptosis↓ GSC differentiation↑ |
In patients: the expression is higher in GBM than in other gliomas. |
[53,54,55,56,57,58] |
|
miR-17-92 cluster |
-Some cell-cycle inhibitors such as PTEN and CDKN1A | ↑ | Cancer cells proliferation↑ Apoptosis↓ GSC differentiation↑ |
In vitro: are associated with high aggressiveness, higher invasion and replication capability. |
[48] |
| miR-21 | -PTEN, p53 -EGFR, Cyclin D1 and AKT2 -SPOCK1 -RECK and TIMP3 |
↑ | Cancer cells proliferation↑ Tumor cells migration↑ Invasion↑ |
In patients: hypothesis that silencing of this miRNA can be used, in the future, as a therapy in the treatment of GBM. |
[35,48,53,55] |
| miR-26a | -PTEN, ATM | ↑ | Cancer cells proliferation↑ Invasion↑ |
In vitro: overexpression of this miRNA, reducing DNA repair ability, enhances radio sensitivity to radiotherapy. |
[75,76,77] |
| miR-148a | -CADM1 -FIH1 |
↑ | Angiogenesis↑ Invasion↑ |
In patients: -overexpression in the plasma from the serum of GBM patients if compared to healthy cases. -has been found to be expressed in the high risk group (i.e. patients characterized by low survival). |
[53,60,61,62] |
| miR-182 | -BCL2L12 -HIF2A -MET -CYLD -LRRC4 |
↑ | Uncontrolled cells proliferation↑ Apoptosis↑ GSC differentiation↑ |
In patients: -miR-182 correlates with better response to TMZ based chemotherapy and with better survival. |
[64,65,66] |
| miR-196a, b | -HOXB8, HOXC8, HOXD8, HOXA7, HOXB7 -ERG -HMGA2 -ANXA1 |
↑ | Cancer cells proliferation↑ Apoptosis↓ |
In vitro: -overexpression of this miRNA favours cells proliferation. In patients: -miR-196b expression correlated with OS. |
[71,72,73,74] |
| miR-648 | -MOBP | ↑ | Cancer cells proliferation↑ Invasion↑ |
In vitro: - the expression by transfection of miR-648 enhanced responsivity of TMZ in MGMT expressing T98G glioma cells. In patients: -correlation between OS and miR-648 expression. |
[74,78,79,80] |
| miRNA | Target | Expression in GBM | Function/role in GBM if the miRNA expression is inhibited | Clinical application | References |
|---|---|---|---|---|---|
| miR-1 | -Connexin-43 -G6PD | ↓ | Cancer cells proliferation↑ Tumor cells migration↑ Apoptosis↓ |
In vitro: inhibition can enhance the cells proliferation and the sensitivity of GBM cells towards TMZ. |
[53,81,82] |
|
miR-7 |
-EGFR, AKT/ PI3K pathway -PKM2 |
↓ | Cancer cells proliferation↑ GSC differentiation↑ |
In vitro: transfection of this miRNA in U373-MG GBM cell line resulted in significant suppression of EGFR mRNA and protein bringing to the inhibition of cells duplication. |
[35,90,91] |
|
miR-34 |
-Bcl2, NOTCH, NUMB -CDK6 -EGFR -c-Met |
↓ | Cancer cells proliferation↑ Apoptosis↓ Invasion↑ |
In vitro: -in G[92–99SC cultures the infection by ZIKV induced miR-34 expression inhibiting the anti-apoptotic protein Bcl-2 and Numb, involved in GSC invasion. - in mouse models ZIKV reduced brain tumor size and metastasis. |
[92,93,94,95,96,97,98,99] |
| miR-128 | -WNT -ERK -EGFR -IGF1R -Bcl2 -PDGFRA -caspase |
↓ | Cancer cells proliferation↑ Apoptosis↓ |
In vitro: its low expression can be associated to high-grade glioma cell lines and, consequently, to a worst prognosis. |
[53,83,84] |
|
miR-137 |
- EZH2 | ↓ | Cancer cells proliferation↑ Apoptosis↓ Angiogenesis↑ |
In vitro: expression level of miR-137 was down-regulated in GBM cells. In patients: the low level of this miRNA was related to poor prognosis in GBM patients. |
[53,85,86] |
| miR-181 family | -CD133 and BMI1 CCN1 |
↓ | Cancer cells proliferation↑ GSC differentiation↑ Apoptosis↓ Invasion↑ |
In patients: -the low level of this miRNA was related to poor prognosis in GBM patients. - low expression of miR-181c or low expression of miR-181d in combination with expression of miR-648, predict the worst prognosis. |
[73,79,81,87] |
| miR-370-3p | -WNT - FOX01, FOXM1 and TGFβ. |
↓ | Cancer cells proliferation↑ Invasion↑ |
In vitro: when miR-370-3p is upregulated, GBM growth is inhibited |
[88,89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





