1. Introduction
Innate immunity, as one of the mechanisms of vertebrate immunity, has a major role in identifying itself with pathogenic microorganisms and is extremely important both for resistance to invasion by pathogenic bacteria and for stimulating the development of acquired immunity [
1].The key to innate immunity is the immune recognition and interaction with pathogenic microorganisms [
2]. When a pathogenic microorganism invades an organism, immune cells recognize the invading pathogen or intracellular damage signal by means of Pattern Recognize Receptors (PRRs) expressed on the surface of the cell membrane or in the cytoplasmic matrix [
3]. The NLRs (nod-like receptor family) are the most widely studied pattern recognition receptors [
4], which is located in the cytoplasm and involved in the recognition of bacteria and viruses [
5]. Tbk1 is an important serine/threonine kinase that interacts with different junction molecules and mediates the phosphorylation of IRF3 (interferon regulatory factor3), thus playing a central regulatory role in interferon production.
Nod1 (
nucleotide-binding oligomerization domain-containing protein 1) and nod2 (
nucleotide-binding oligomerization domain-containing protein 2) were first NLR family members to be identified and have important pattern recognition receptors for bacteria and viruses [
6,
7]. Both nod1 and nod2 contain three conserved domains: the N-terminal Caspase activiting and recruitment domain (CARD), the central nucleotide-binding oligomerization domain (NOD), which mediates intermolecular oligomerisation, and the C-terminal leucine-rich repeat domains (LRRs), which have ligand recognition functions [
8,
9]. In mammals, nod1 specifically recognises
γ-D-Glu-mDAP (iE-DAP) in bacteria [
10], and nod2 specifically recognises the
Muramyl Dipeptide (MDP) in bacteria, thus triggers an immune response by activating the downstream NF-κB signalling pathway [
9,
11]. These two proteins are activated by stimulation of pathogenic components of bacteria, then recruit and activate the receptor-interacting serine-threonine kinase 2 (RIPK2) through the CARD domain, which further activates the IKK kinase complex, ultimately promoting the transcription of corresponding genes such as inflammatory cytokines [
12,
13]. At the same time, nod2 also induces the activation of mitogen-activated protein kinases (MAPK) signaling pathway and promotes cytokine production [
14]. In addition to recognizing MDP, nod2 was also found to recognize viral ssRNA and promote interferon regulatory factors to induce the production of type I interferon [
15]. Nod2 has an important function in the antiviral immune response by enabling cells to resist viral infection and activating acquired immunity. The nod1 and nod2 genes in some teleost fish have been cloned and functionally studied including
Channel Catfish (Ictalurus punctatus) [
8],
Grass carp (Ctenopharyngodon idella) [
16], miiuy croaker (
Miichthys Miiuy) [
17], zebrafish (
Danio rerio) [
18] and Nile tilapia (
Oreochromis niloticus) [
19].
Tbk1 is a serine/threonine kinase commonly expressed in the IKK family, also known as NFkB-activating kinase (NAK), which plays an important role in interferon production and antiviral innate immunity. Following viral infection, virus-specific features are recognized by the PRR and further induce a signaling cascade downstream of the PRR.Tbk1 is an important checkpoint that receives signals from the TLR pathway, the RLR pathway, and the cGAMP synthase (cGAS)) pathway, and is essential in the generation of type I IFN antiviral immune responses in the body. It was found that in addition to its interferon-induced function, tbk1, as a ubiquitously expressed protein, plays a role in the insulin signaling pathway, cellular autophagy and mitophagy, and antiviral and antibacterial immune responses in mammals. This suggests that tbk1 is not only a key molecule in the induction of interferon production, but also an important molecule in cell biological processes. Homologs of tbk1 have been cloned and characterized in a variety of scleractinian fishes, such as zebrafish (
Danio rerio) [
20], Atlantic salmon (
Salmo salar) [
21], common carp (
Cyprinus carpio) [
22], grass carp (
Ctenophyngodon idella) [
23], cyprinid fish (
Mylophyngodon piceus) [
24] and crucian carp (
Carassius auratus) [
25].
Spotted knifejaw is one of the valuable mariculture species in China. However, with the increasing level of aquaculture intensification, disease problems have become a problem that restricts the healthy culture and promotion of spotted knifejaw
(Mukherjee et al. 2019). There are three main types of diseases that affect spotted knifejaw aquaculture including viral, bacterial and parasitic diseases. Recently, RSIV-type megalocytivirus (SKIV-ZJ07), SKIV-TJ and ISKNV-type megalocytivirus (SKIV-SD) were isolated from spotted knifejaw [
26,
27,
28]. Bacterial diseases such as black body disease caused by
Vibrio harveyi can lead to ascites, enteritis and serious damage to internal organs, which greatly affect the aquaculture industry [
29]. Therefore, in this study, the CDS regions of
Opnod1,
Opnod2 and
Optbk1 genes were cloned, and their expression patterns in different tissues of spotted knifejaw were examined by quantitative real-time PCR (qRT-PCR), as well as the expression levels in different tissues after infection by SKIV-SD or V.
harveyi, with the aim of further investigating the role of these genes in the immune response of spotted knifejaw. The aim was to lay the groundwork for further studies on the role of this gene in the immune response of the sole.
2. Materials and Methods
2.1. Experimental Fish
The spotted knifejaw used in this experiment were obtained from Shandong Laizhou Mingbo Aquatic Co. The body weight of fish is about 150g, healthy without disease. Before the experiment, they were temporarily kept in a water tank for a week, and the water temperature was maintained at about 25 ℃.
2.2. Sample Processing and Collection
Eleven tissues of liver, spleen, kidney, head kidney, heart, intestine, gills, brain, skin, muscle and blood were collected from five spotted knifejaw randomly after anesthetised by MS-222. The tissues were quickly placed in liquid nitrogen and subsequently transferred to -80°C refrigerator for further experiments.
Twenty healthy spotted knifejaw were selected for the spotted knifejaw SKIV-SD infection experiment. The virus used in this experiment was provided by Professor Qiwei Qin from South China Agricultural University. The individuals were anaesthetised and injected intraperitoneally with 100 µl (109 copies) of virus solution per fish. Samples were taken at five time points: 0 d, 1 d, 4 d, 7 d and 10 d. Three individuals were randomly selected at each time point, and three tissues were taken from each fish: liver, spleen and kidney. Tissues were removed and rapidly frozen in pre-prepared liquid nitrogen, and the tissue samples were subsequently stored in a -80°C refrigerator for subsequent RNA extraction.
Fifty healthy spotted knifejaw were selected for the Vibrio harveyi infection experiment. The V. harveyi used in this experiment were kept in this laboratory. The V. harveyi solution was diluted to 1×109 cfu/mL with PBS, and the spotted knifejaw were anesthetized and injected intraperitoneally with 100 µL of the bacterial solution per fish. Samples were taken at eight time points: 0 h, 6 h, 12 h, 24 h, 48 h, 72 h, 96 h and 120 h. Three spotted knifejaw were sampled at each time point, and three tissues were taken from each fish: liver, spleen and kidney. The tissues were immediately frozen in liquid nitrogen and subsequently stored in a -80°C refrigerator for subsequent experiments.
2.3. Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from each tissue by using an RNA extraction kit (Invitrogen, USA), the integrity of the RNA was authenticated by 1% agarose gel electrophoresis, and the concentration and purity of the RNA was measured by spectrophotometer. The cDNA was synthesized using the Prime Script RT reagent kit with gDNA eraser (TaKaRa, Japan).
2.4. Amplification of the CDS Region of the Opnod1, Opnod2 and Optbk1 Genes
Based on the sequence information of Opnod1, Opnod2 and Optbk1 genes, primers were designed for common PCR amplification using the newly synthesized cDNA as template to verify the integrity of their ORF regions. The reaction system for PCR was KOD TM OneMaster Mix (Toyobo, Japan) 25 μL, ORF F/R 1 μL, cDNA template 2 μL, and The PCR amplification conditions were: 98 ℃ for 5 min; 98 ℃ for 10 s, 59 ℃ for 5 s, 72℃ for 30 s, 35 cycles; 72 ℃ for 7 min, then stored at 4 ℃. PCR products were detected by agarose gel electrophoresis, and target fragments with the correct band size were recovered by gelling using a DNA gel recovery kit (Vazyme, Nanjing). The recovered product was ligated to pEASY-E1 vector (Transgen, China) and transformed into Trans-T1 receptor cells (Transgen, China), which were coated and cultured overnight. Finally, the positive monoclonal clones were selected and sent to Qingdao Ruibiotech Company for sequencing to obtain the ORF region sequences of Opnod1, Opnod2 and Optbk1.
2.5. Sequence Analysis of Opnod1, Opnod2 and Optbk1 Genes
The
Opnod1,
Opnod2 and
Optbk1 genes sequences were analyzed using Editseq software to predict their amino acid sequences, molecular weights and isoelectric points. The sequences were predicted using an online software (
http://www.cbs.dtu.dk/services/SignalP/ ) to predict their signal peptides and use the online website (
http://www.cbs.dtu.dk/services/TMHMM/ ) for transmembrane analysis. The domain prediction was performed using the online software (
http://smart.embl-heidelberg.de/). Homology comparison of amino acid was performed by DNAMAN 8 software and evolutionary trees were constructed by MEGA X software.
2.6. Sequence Analysis of Opnod1, Opnod2 and Optbk1 Genes
Quantitative real-time PCR (qRT-PCR) was applied to detect the reletive expression levels of
Opnod1,
Opnod2 and
Optbk1 genes in different tissues of healthy spotted knifejaw, as well as in immune-related tissues after stimulation with SKIV-SD or
V.harveyi. β-actin was chosen as the internal reference gene and the primers used for qRT-PCR are listed in
Table 1. Quantification of
Opnod1,
Opnod2 and
Optbk1 genes was performed on an ABI 7500 Fast Real-time (Applied Biosystems, USA) instrument using the SYBR
® Premix Ex Taq TM kit (TaKaRa, Japan) according to the instructions. Three parallel replicates were set up for each sample and the relative expression of genes was calculated using 2
–ΔΔCt method (Livak and Schmittgen 2002). All experimental data were expressed as mean ± standard error (Mean±SE) and subjected to one-way ANOVA and Duncan’s multiple comparisons using SPSS16.0 software. Differences were considered significant when P<0.05 and highly significant when P<0.01.
2.7. In Vitro Stimulation of Kidney Cells of O. punctatus with Poly I:C and LPS
The monolayer cultured, well-grown spotted knifejaw kidney cell line was inoculated into a 12-well plate and left for 24h until the cell coverage reached about 90%. The medium of the 12-well plate was aspirated off and washed 3 times with 1×PBS and replaced with fresh L15 medium. Stimulation of spotted knifejaw kidney cells with different concentrations of poly I:C at final concentrations of 0 μg/mL, 50 μg/mL, 100 μg/mL, and 200 μg/mL was added to each well of 12-well plates, and an equal volume of PBS was added to control group. LPS at final concentrations of 0 μg/mL, 10 μg/mL, 50 μg/mL and 100 μg/mL were added to 12-well plates, and an equal volume of PBS was added to the control group. The above cell samples were cultured in a 24℃ incubator for 6 h. The cell samples were collected by Trizol respectively, and stored in a -80℃ refrigerator for use in the subsequent RNA extraction for the preparation of quantitative templates as Method 2.6.
4. Discussion
In this experiment, the full length of the CDS region of the
Opnod1 and
Opnod2 genes were cloned, and then analyzed for sequence homology and evolutionary analysis with the
nod1 and nod2 genes of other species. The results showed that the Opnod1 and Opnod2 possesses all the characteristic domains of the NLR family [
13]. The protein domains of Opnod1 and Opnod2 are highly conserved compared to those of other species, suggesting that they share similar functional properties in recognition of pathogenic microorganism. Opnod1 has 7 LRR domains, which are similar to those of Nile tilapia [
19] and orange-spotted grouper [
30], while Nod1 genes of grass carp, channel catfish and zebrafish have 6 LRR domains [
16,
31]. The C-terminus of Opnod2 has 6 LRR domains, which are identical to those of miiuy croaker [
17] and orange-spotted grouper [
30], while in grass carp and zebrafish the number of LRRs is 5. The differences of LRR domains between in vertebrates may be related to the various environment and the replication and differentiation of immune related genes. In the genome-wide identification, we found 34 genes with NACHT domain. Structure analysis demonstrated that these NLRs genes were divided into four subfamilies that will expand our understanding of the NLR family in marine fish.
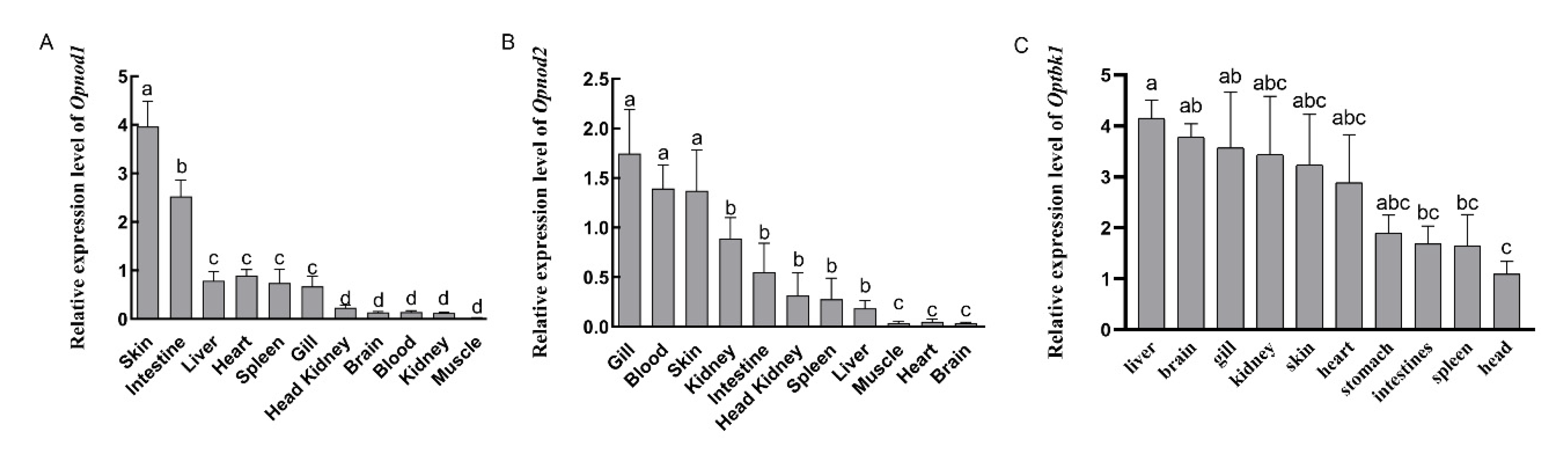
In this study, we detected that
Opnod1 and
Opnod2 were widely expressed in various tissues by using qRT-PCR.
Opnod1 gene was highly expressed in the skin, which was similar with orange-spotted grouper. In contrast, in healthy zebrafish, grass carp and channel catfish, the relative expression level of nod1 was highest in the spleen and lowest in the blood;
Opnod2 gene was highly expressed in gill and skin tissues of spotted knifejaw, and in Nile tilapia,
nod2 gene was highly expressed in the spleen, while in orange-spotted grouper
nod2 gene was most highly expressed in the kidney, followed by skin and gill. This is similar to the tissue expression pattern of the
nod2 gene in spotted knifejaw. Taken together, this suggests that nod1 and nod2 have an important role in the innate immunity of spotted knifejaw. The role of nod1 and nod2 in antibacterial and antiviral immune responses has been reported in a few species of fish such as Indian major carp (
Labeo rohita) [
32], Mrigalcarp (
Cirrhinus mrigala) [
33], orange-spotted grouper (
Epinephelus coioides) [
30], rainbow trout (
Oncorhynchus mykiss) [
11], grass carp (
Ctenopharyngodon idella) [
16], channel catfish (
Ictalurus punctatus) [
8] and zebrafish (
Danio rerio) [
18]. Nile tilapia showed significant changes in the expression levels of
nod1 and
nod2 in blood, spleen, kidney, intestine and gills following injection of
Streptococcus agalactiae [
19]. In Mrigalcarp the expression levels of
nod1 and
nod2 in the gills, liver, kidney and intestine were significantly increased after infection with either Streptococcus agalactiae or
Aeromonas hydrophila [
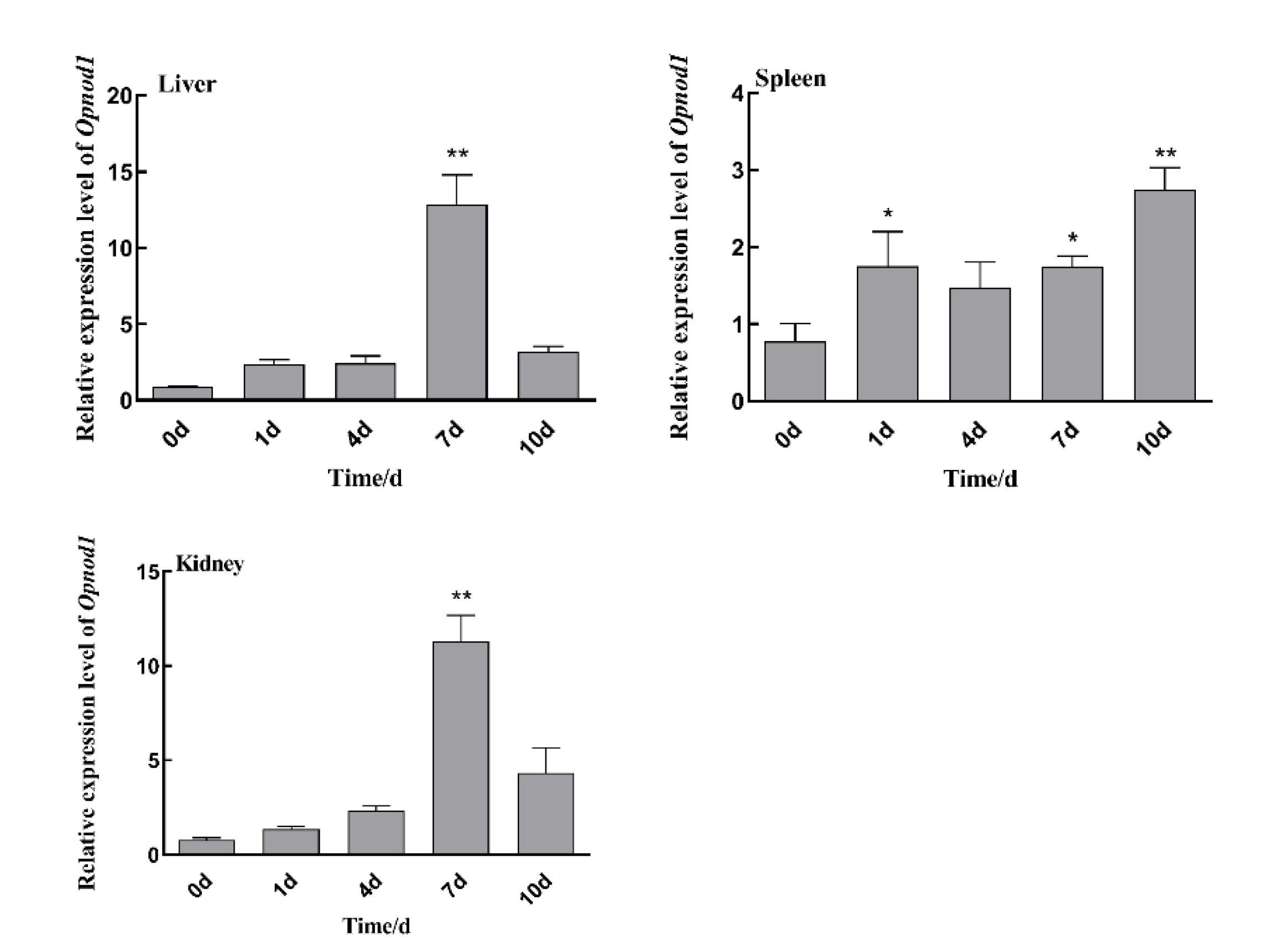
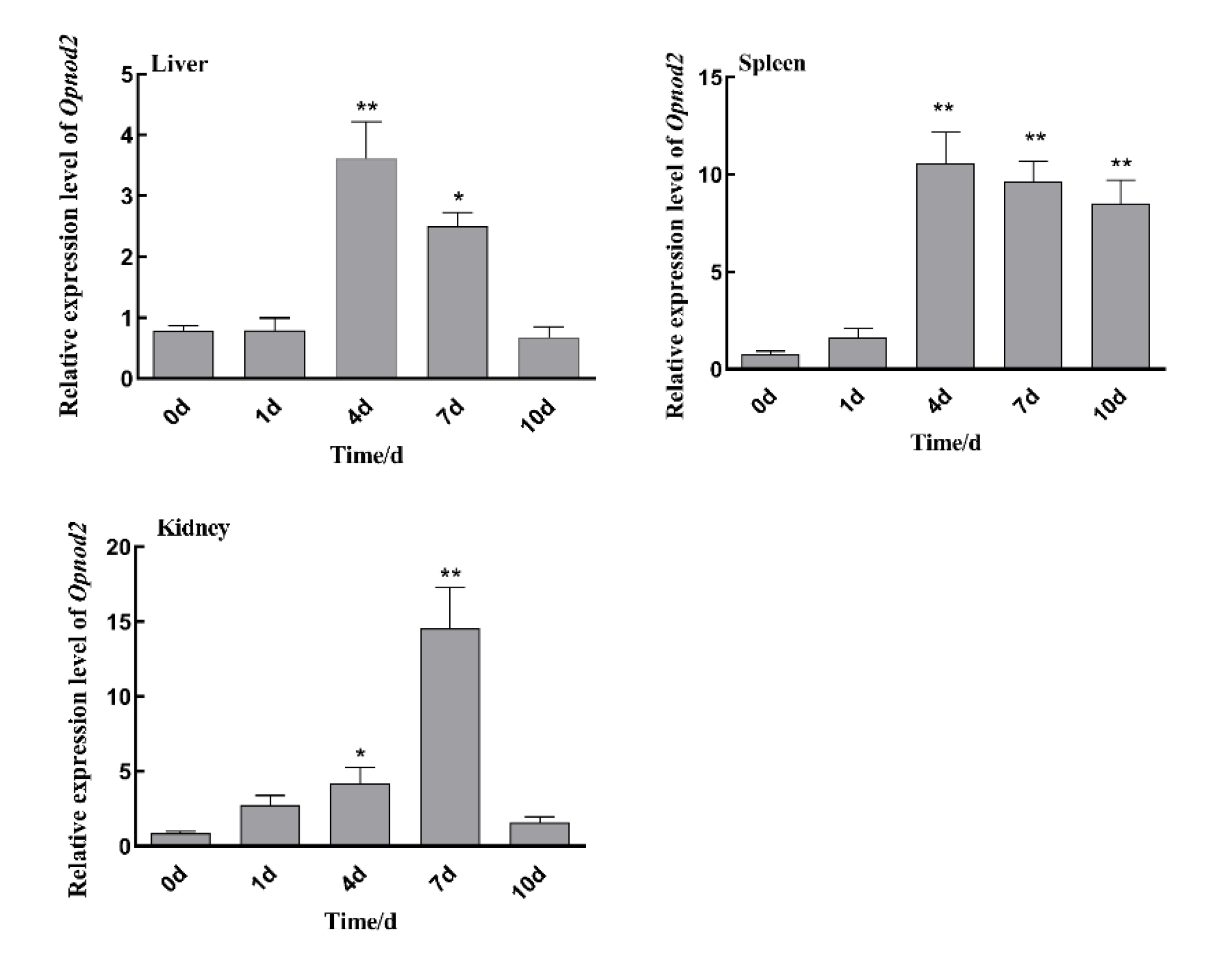
33]. In this study, we found that the relative expression levels of
Opnod1 and
Opnod2 genes in liver, spleen and kidney tissues of spotted knifejaw were significantly changed after infection by SKIV-SD or
V. harveyi, indicating that
Opnod1 and
Opnod2 play an important role in the immune response of spotted knifejaw.
Studies in mammals have shown that nod1 recognizes the peptidoglycan molecules of
γ-D-Glu-mDAP (iE-DAP) to sense the presence of bacterial pathogens. Mammalian nod2 detects
Muramyl Dipeptide (MDP) in peptidoglycans of Gram-positive and Gram-negative bacteria, and both receptors trigger immune responses by activating NF-κB [
34].Meanwhile, in teleost fishes, the detected ligands of nod1 and nod2 remains unclear. In zebrafish, nod2 responded significantly to MDP stimulation, but not to stimuli in Poly I: C and LPS [
18]. This suggests that zebrafish nod2 may be a receptor for MDP, which is consistent with Nile tilapia [
19]. In our research,
Opnod1 and
Opnod2 are terribly possible involved in antiviral and antibacterial infection process, while the activation mechanisms are unclear and need further study.
In mammals, tbk1 has been widely studied as an important molecular bridge connecting the signaling of TLRs (TLR3 and TLR4) and RLRs, which activates the transcription factors IRF3 and IRF7 to produce type I interferons. In this study, we identified the zebra seabream tbk1 gene, and after multiple sequence comparisons we found that the tbk1 sequence is highly conserved, and phylogenetic analysis of tbk1 showed that Optbk1 is more closely related to tbk1 from other teleost, and is likely to be a homologous gene of mammalian tbk1. Tissue expression analysis showed that Optbk1 was expressed in a variety of tissue, which was similar to that in other fish species, but the expression pattern of tbk1 was slightly different in different species. It was shown that grass carp (
Ctenopharyngodon idellus) tbk1 was mainly expressed in the spleen [
35], and Large yellow croaker (
Larimichthys crocea) tbk1 was highly expressed in the brain [
36]. The highest level of expression of Optbk1 was found in the liver of the spotted rock seabream, whereas the expression level was lower in the spleen and the head and kidneys, which was similar to that in the dark sandpiper (
Odontobutis obscurus) tbk1 tissue expression results were similar [
37], and it is hypothesized that Optbk1 is not required in the immune tissues of healthy spotted knifejaw. Black carp (
Mylophyngodon piceus) tbk1 exhibits antiviral activity against grass carp SVCV and GCRV [
24], large yellow croaker (
Larimichthys crocea) tbk1 can interact with the E3 ubiquitin ligase, Nrdp1, to defend against infections [
36].
In this study, the expression levels of Opnod1, Opnod2 and Optbk1 were up-regulated to different degrees in immune tissues liver, spleen and kidney after stimulation by SKIV-SD and V.harveyi, in addition spotted knifejaw kidney cell showed up-regulation to different degrees after stimulation by poly I:C and LPS in vitro, the above results suggest that NOD1/2-TBK1 signaling pathway plays an important role in immunity process of teleost, but the molecular mechanism in teleosts are still very limited.
In summary, the present study showed that Opnod1, Opnod2 and Optbk1 are involved in the immune response process of spotted knifejaw, which will provide evidence for further research on the role mechanism of NOD1/2-TBK1 signal pathway in the immunity of spotted knifejaw against diseases.
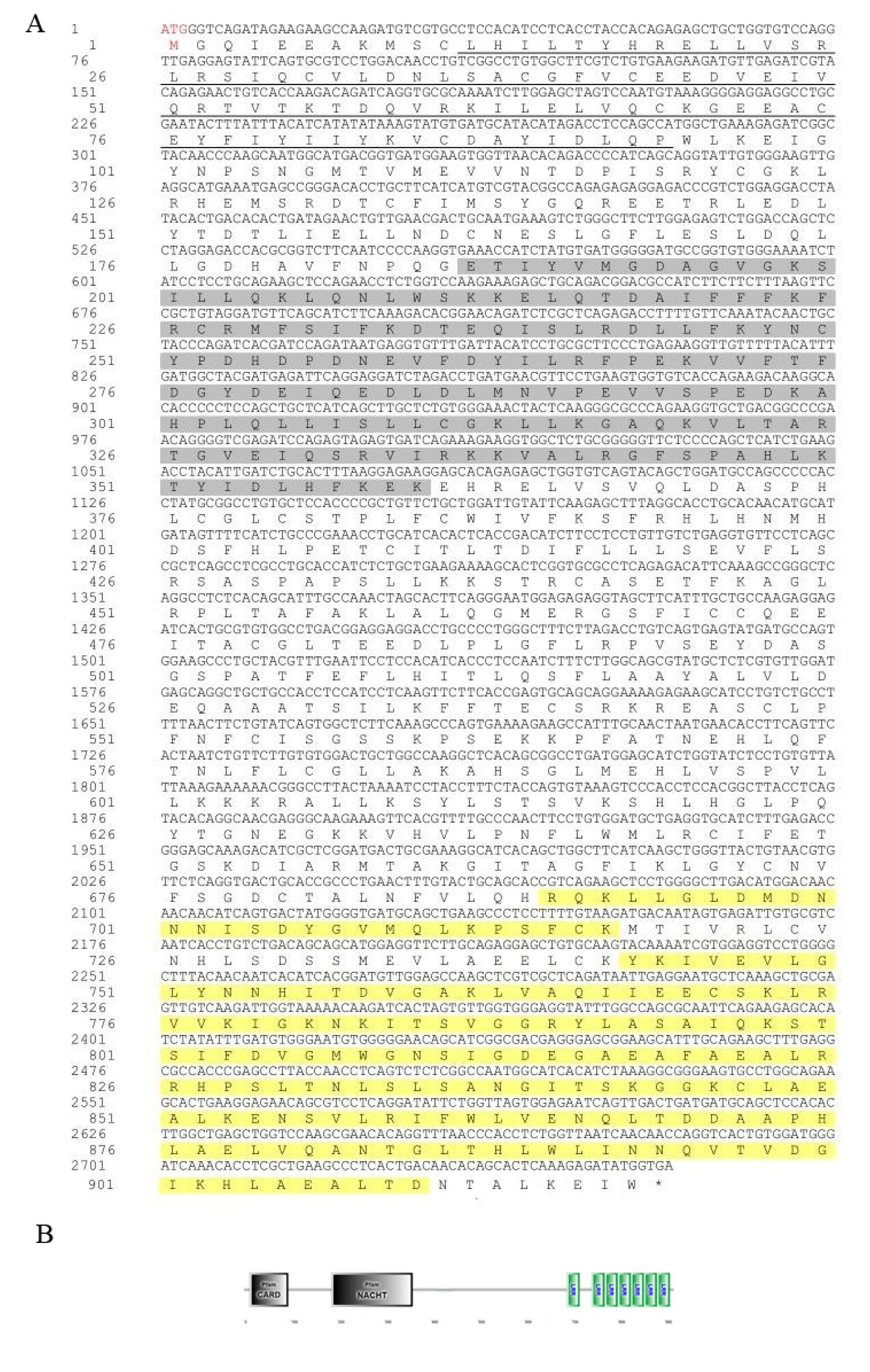
Figure 1.
Amino acid sequence and predicted domains of Opnod1 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence, the start codon “ATG” is marked with a red font, and the stop codon “TAG” is marked with an asterisk. The CARD domain is marked with an underscore “____”, the NOD domain is marked with gray shading, and the LRR domain is markedwith yellow shading. (B) The protein domain prediction of Opnod1. The CARD and NACHT domains are marked with gray shading, and the LRR domain is marked with green shading.
Figure 1.
Amino acid sequence and predicted domains of Opnod1 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence, the start codon “ATG” is marked with a red font, and the stop codon “TAG” is marked with an asterisk. The CARD domain is marked with an underscore “____”, the NOD domain is marked with gray shading, and the LRR domain is markedwith yellow shading. (B) The protein domain prediction of Opnod1. The CARD and NACHT domains are marked with gray shading, and the LRR domain is marked with green shading.
Figure 2.
Amino acid sequence and predicted domains of Opnod2 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence, the start codon “ATG” is marked with a red font, and the stop codon “TGA” is marked with an asterisk. (B) The protein domain prediction of Opnod2. The CARD domain is marked with an underscore, the NACHT domain is marked with gray shading, and the LRR domain is marked with green shading.
Figure 2.
Amino acid sequence and predicted domains of Opnod2 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence, the start codon “ATG” is marked with a red font, and the stop codon “TGA” is marked with an asterisk. (B) The protein domain prediction of Opnod2. The CARD domain is marked with an underscore, the NACHT domain is marked with gray shading, and the LRR domain is marked with green shading.
Figure 3.
Amino acid sequence and predicted domains of Optbk1 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence (B) The protein domain prediction of Optbk1. The S-TKc domain is marked with an asterisk.
Figure 3.
Amino acid sequence and predicted domains of Optbk1 protein. (A) The translated amino acid sequence is marked below the nucleotide sequence (B) The protein domain prediction of Optbk1. The S-TKc domain is marked with an asterisk.
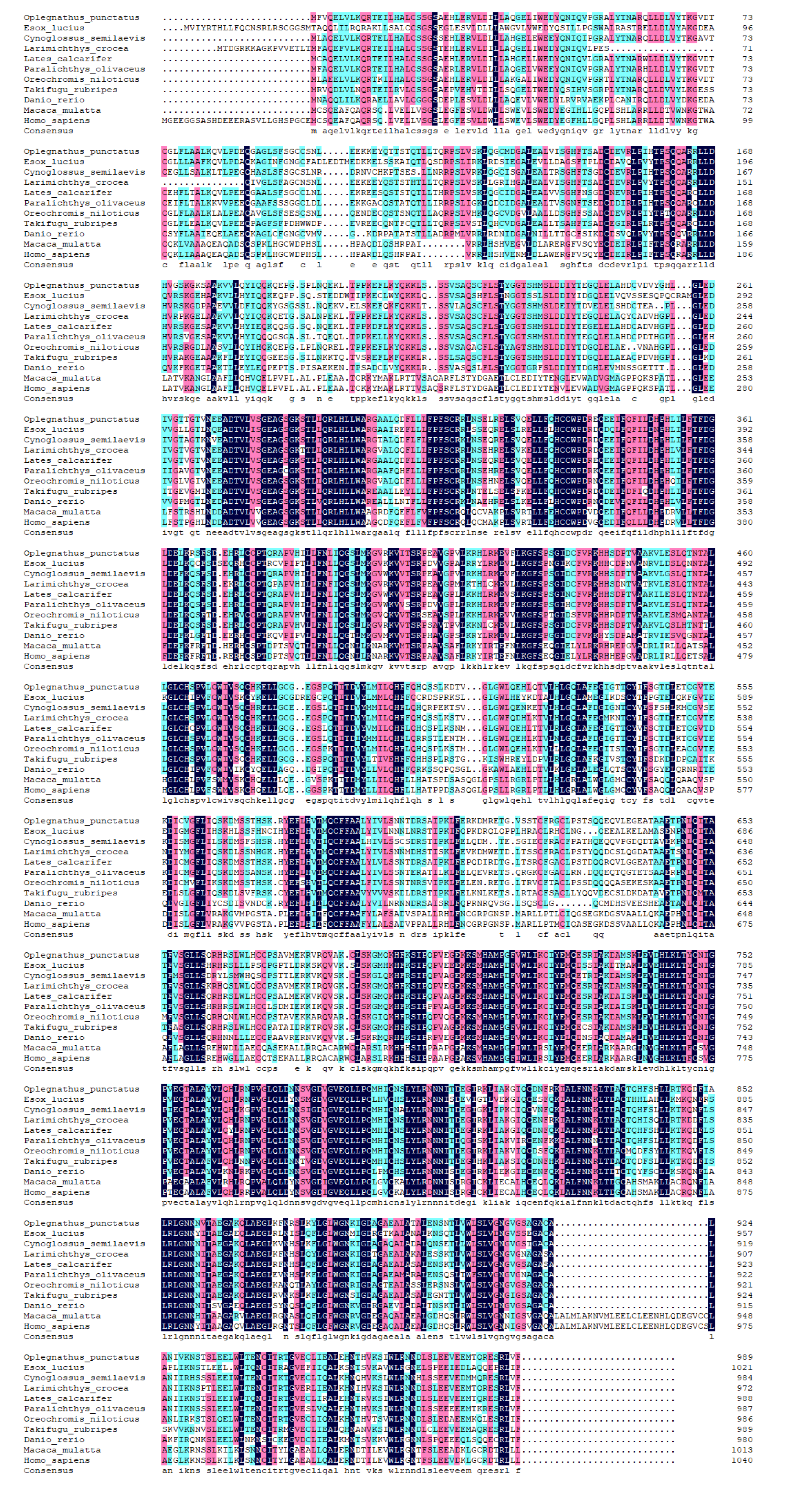
Figure 3.
Multiple alignment of the deduced amino acids of nod1 among Oplegnathus punctatus and other different species.
Figure 3.
Multiple alignment of the deduced amino acids of nod1 among Oplegnathus punctatus and other different species.
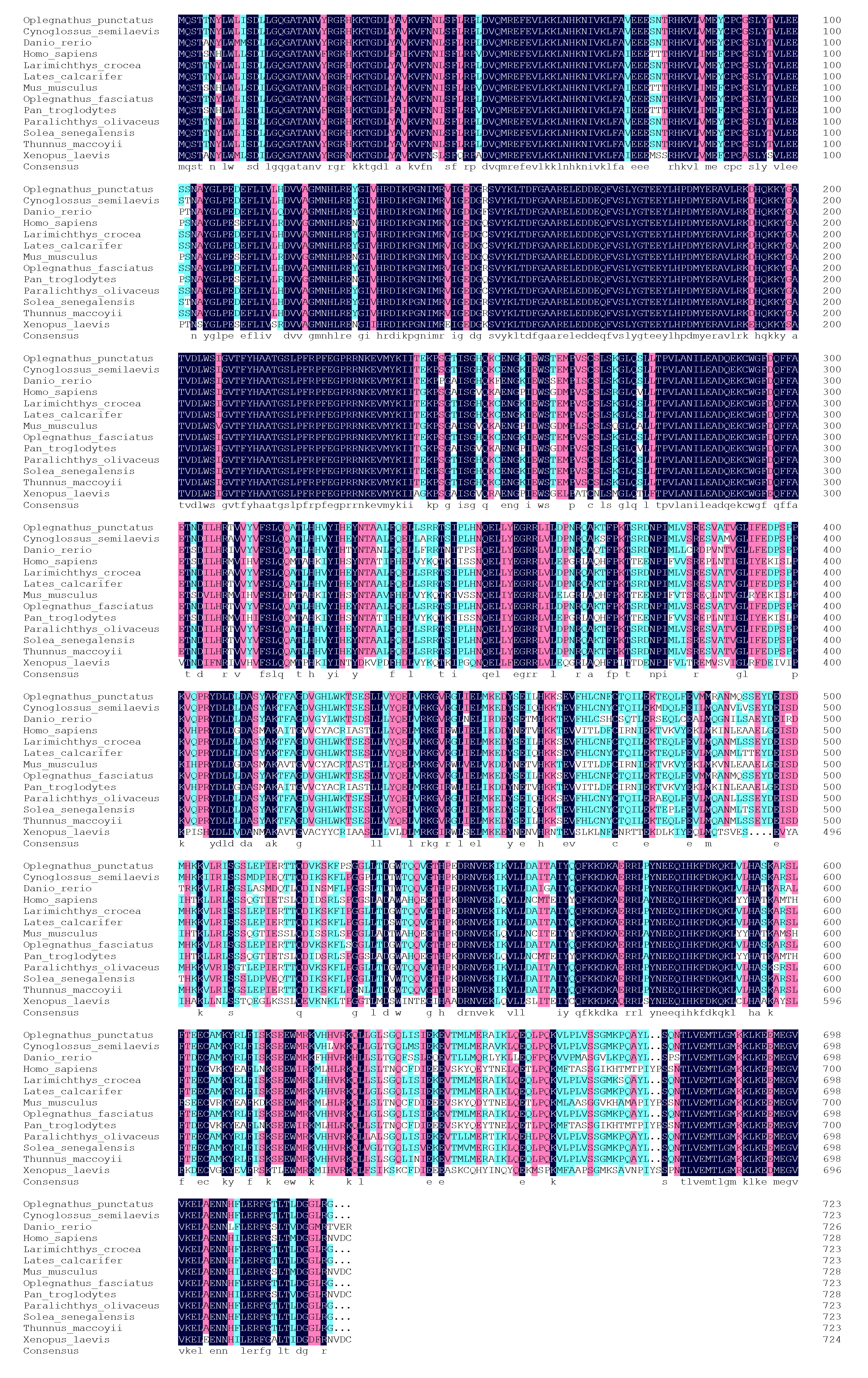
Figure 4.
Multiple alignment of the deduced amino acids of nod2 among Oplegnathus punctatus and other different species.
Figure 4.
Multiple alignment of the deduced amino acids of nod2 among Oplegnathus punctatus and other different species.
Figure 5.
Multiple alignment of the deduced amino acids of tbk1 among Oplegnathus punctatus and other different species.
Figure 5.
Multiple alignment of the deduced amino acids of tbk1 among Oplegnathus punctatus and other different species.
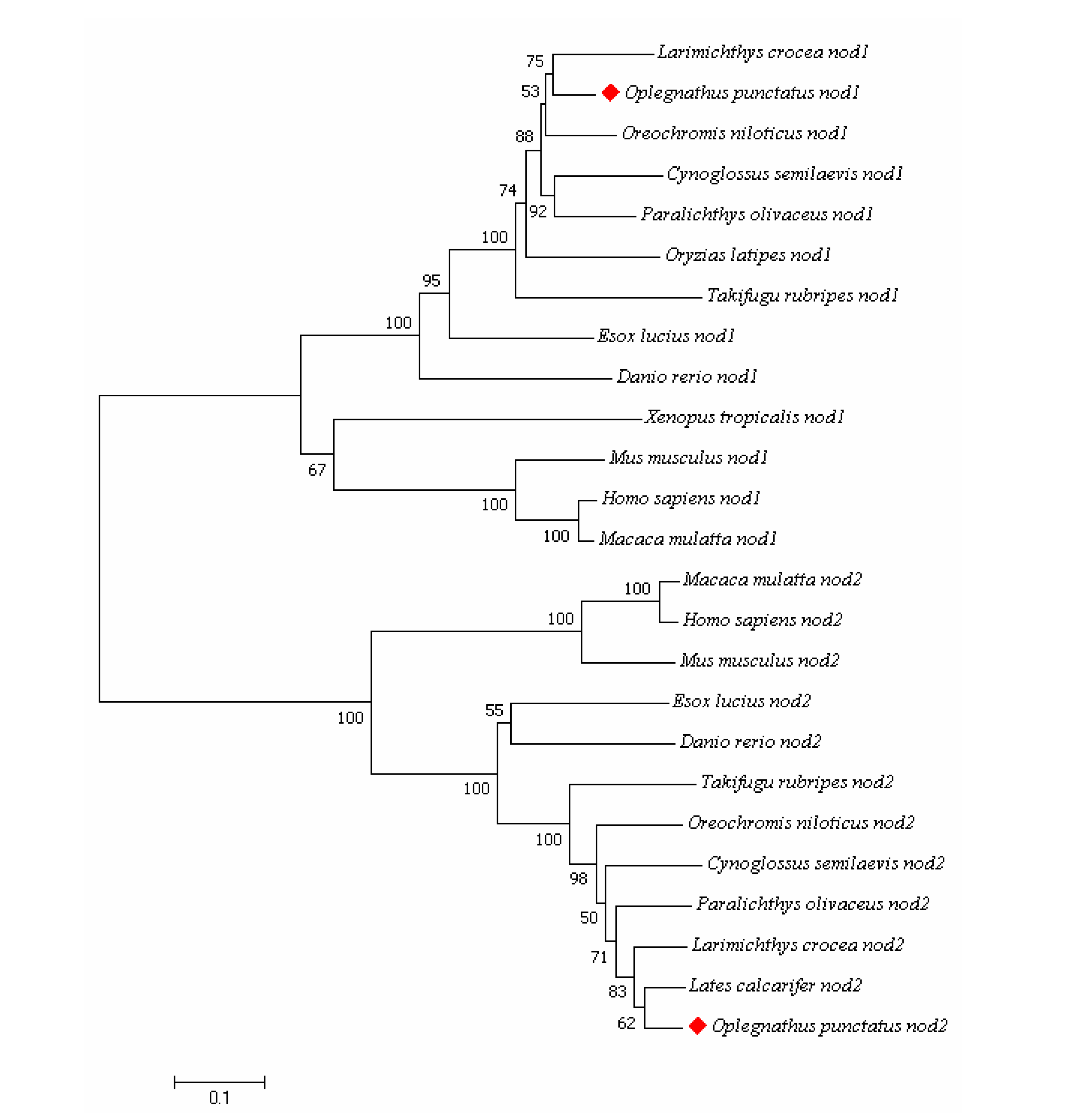
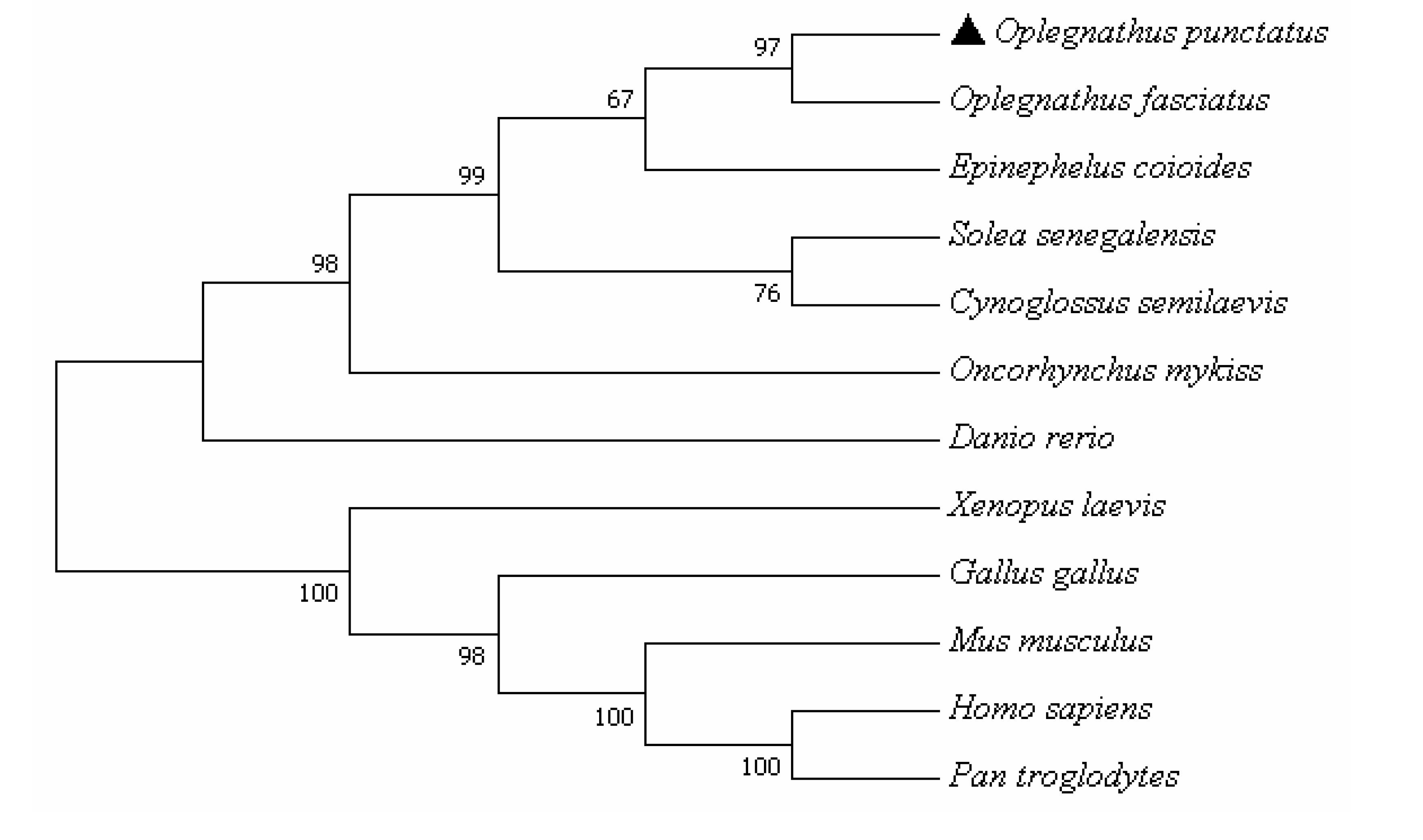
Figure 6.
Phylogenetic tree based on the amino acid sequences of nod1 and nod2 from various species.
Figure 6.
Phylogenetic tree based on the amino acid sequences of nod1 and nod2 from various species.
Figure 7.
Phylogenetic tree based on the amino acid sequences of tbk1 from various species.
Figure 7.
Phylogenetic tree based on the amino acid sequences of tbk1 from various species.
Figure 8.
Expression of Opnod1, Opnod2 and Optbk1 mRNA in different tissues of healthy O. punctatus.
Figure 8.
Expression of Opnod1, Opnod2 and Optbk1 mRNA in different tissues of healthy O. punctatus.
Figure 9.
Relative expression of Opnod1 gene in three tissues (liver, spleen, and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3).The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 9.
Relative expression of Opnod1 gene in three tissues (liver, spleen, and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3).The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 10.
Relative expression of Opnod2 gene in three tissues (liver, spleen and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 10.
Relative expression of Opnod2 gene in three tissues (liver, spleen and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
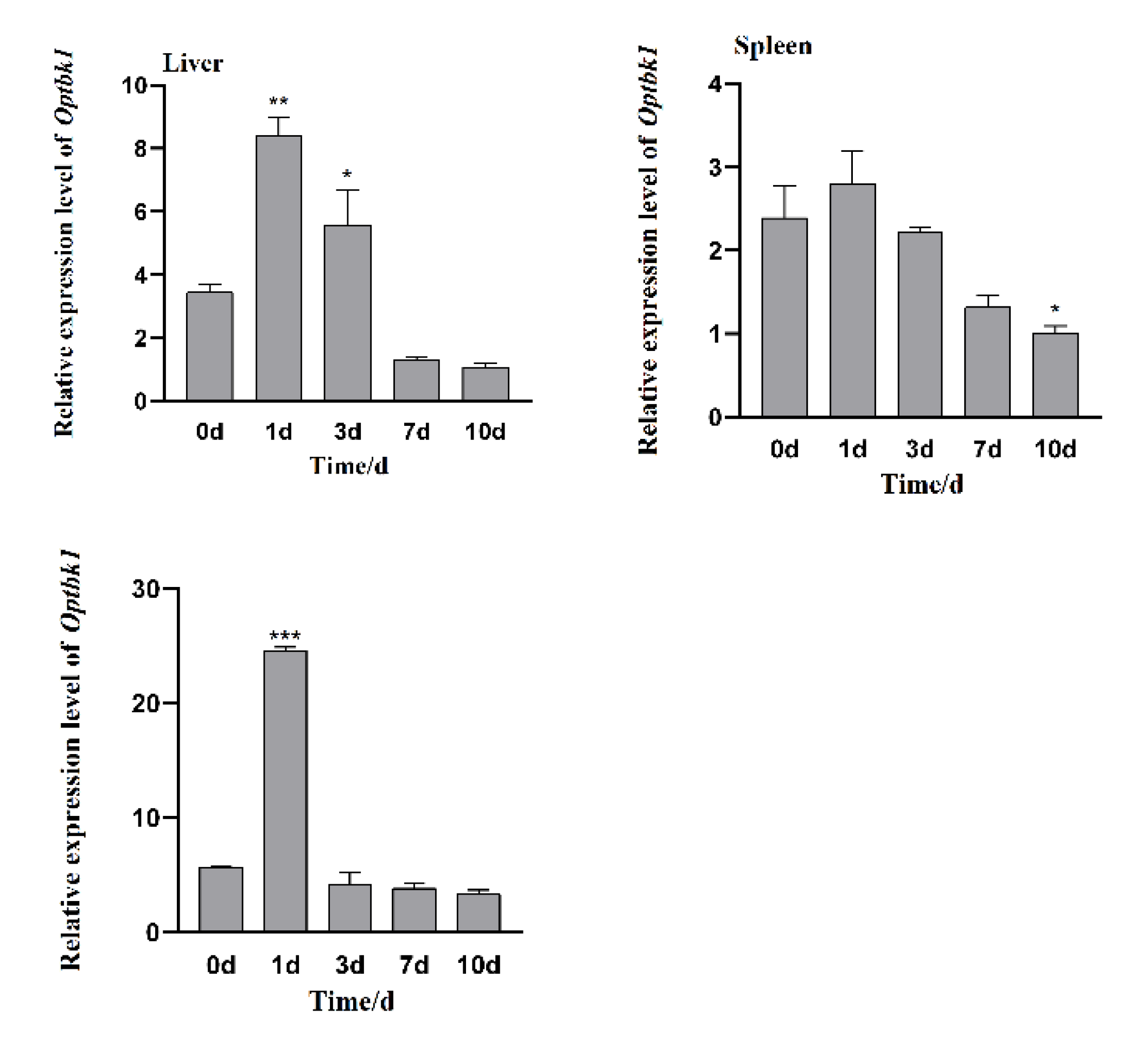
Figure 11.
Relative expression of Optbk1 gene in three tissues (liver, spleen, and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3).The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 11.
Relative expression of Optbk1 gene in three tissues (liver, spleen, and kidney) of Oplegnathus punctatus at different time points after SKIV-SD infection. All the data are shown as mean±SE (n=3).The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
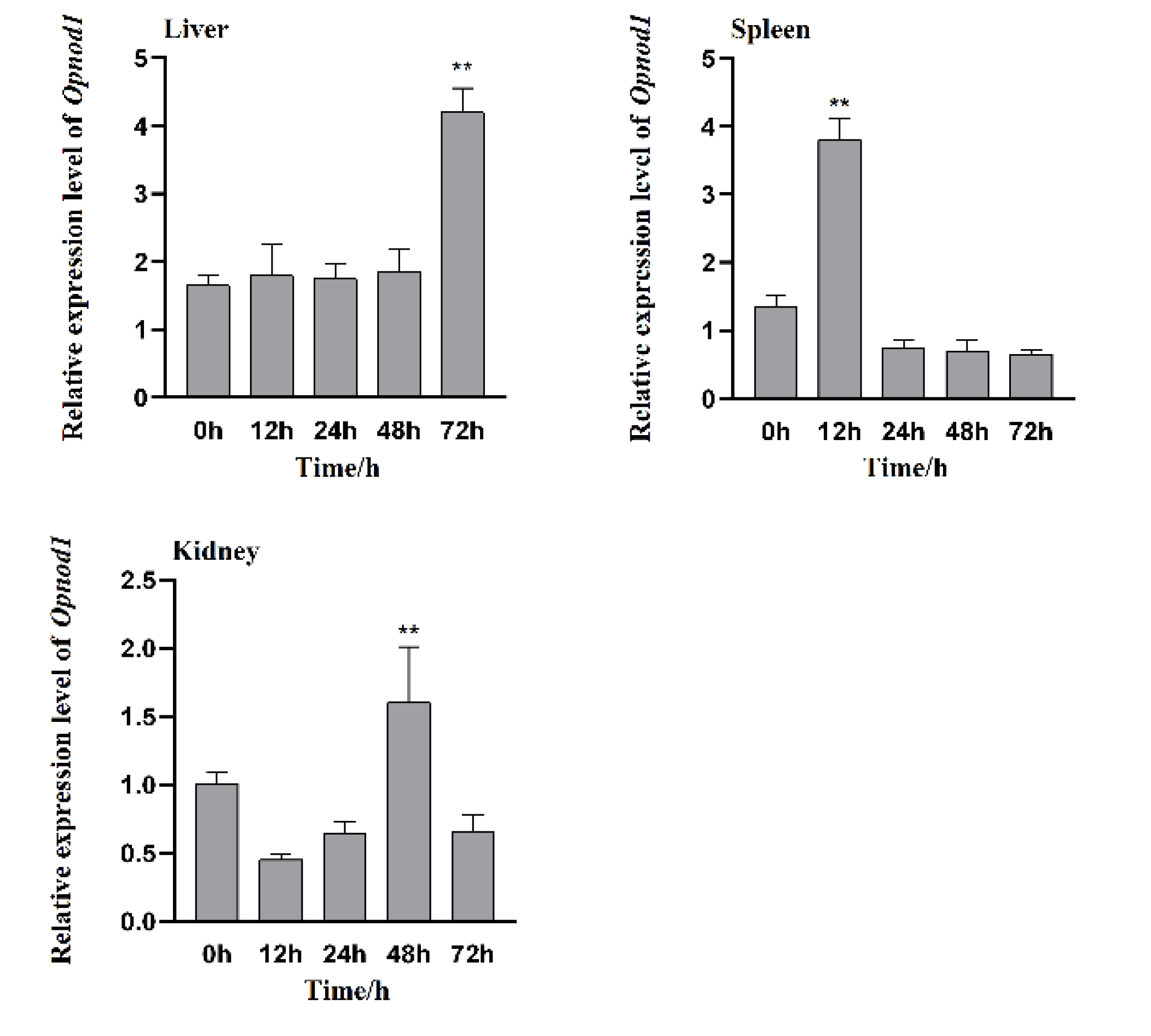
Figure 12.
Relative expression of Opnod1 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 12.
Relative expression of Opnod1 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
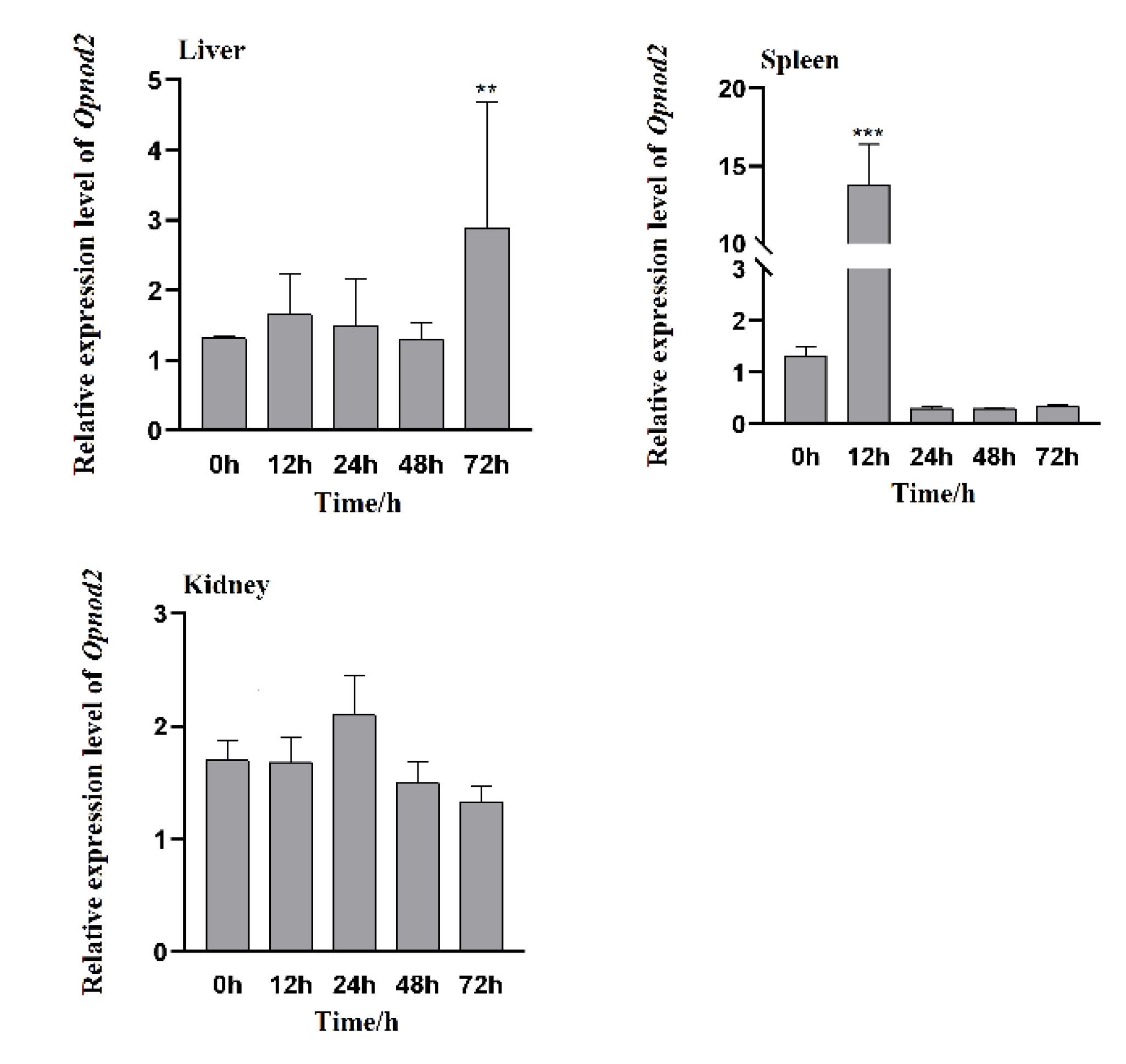
Figure 13.
Relative expression of Opnod2 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 13.
Relative expression of Opnod2 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
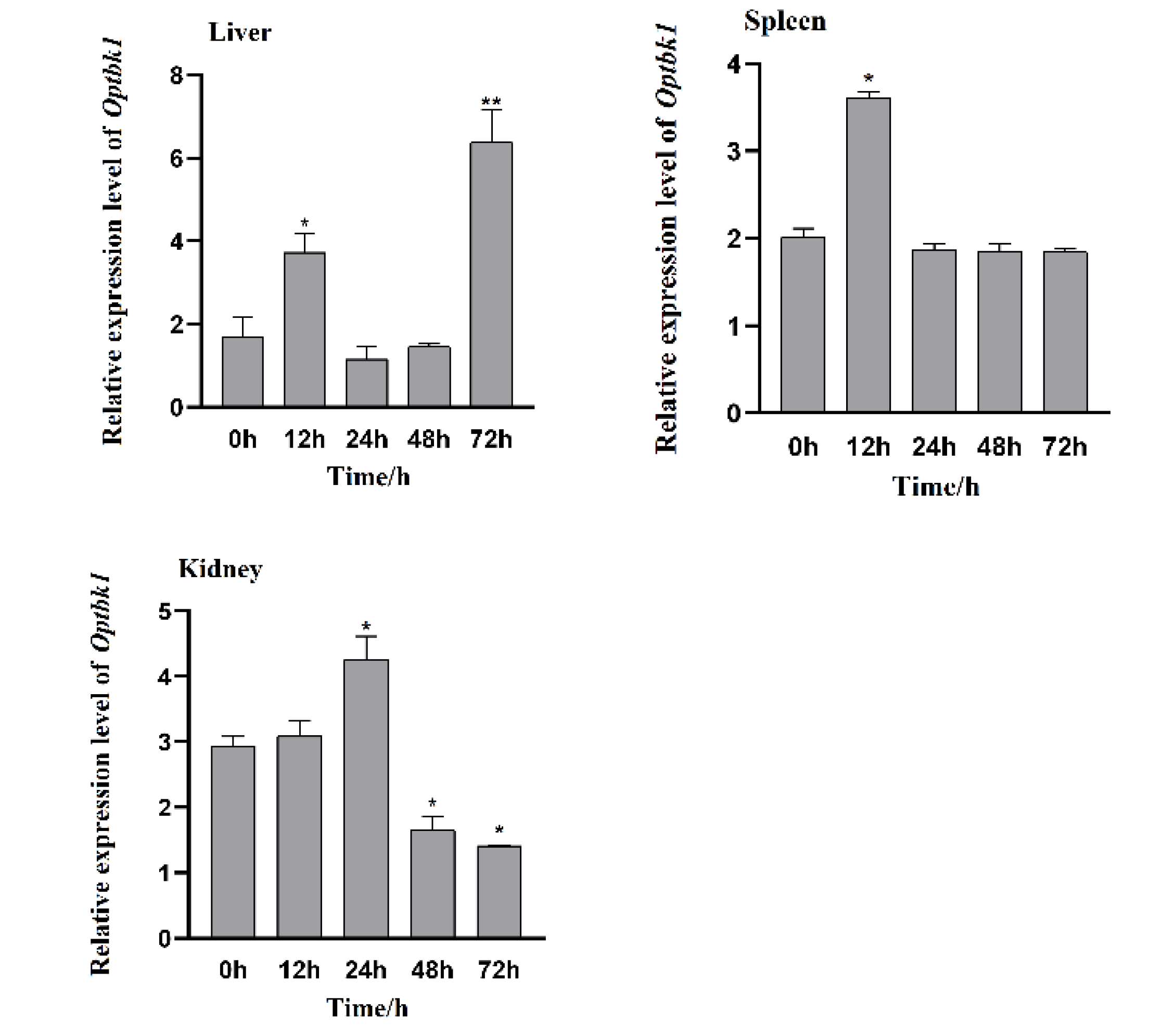
Figure 14.
Relative expression of Optbk1 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
Figure 14.
Relative expression of Optbk1 gene in three tissues (liver, spleen and kidney) of O. punctatus at different time points after V. harveyi infection. All the data are shown as mean±SE (n=3). The asterisk indicates a significant difference (*P<0.05, **P<0.01, ***P<0.001).
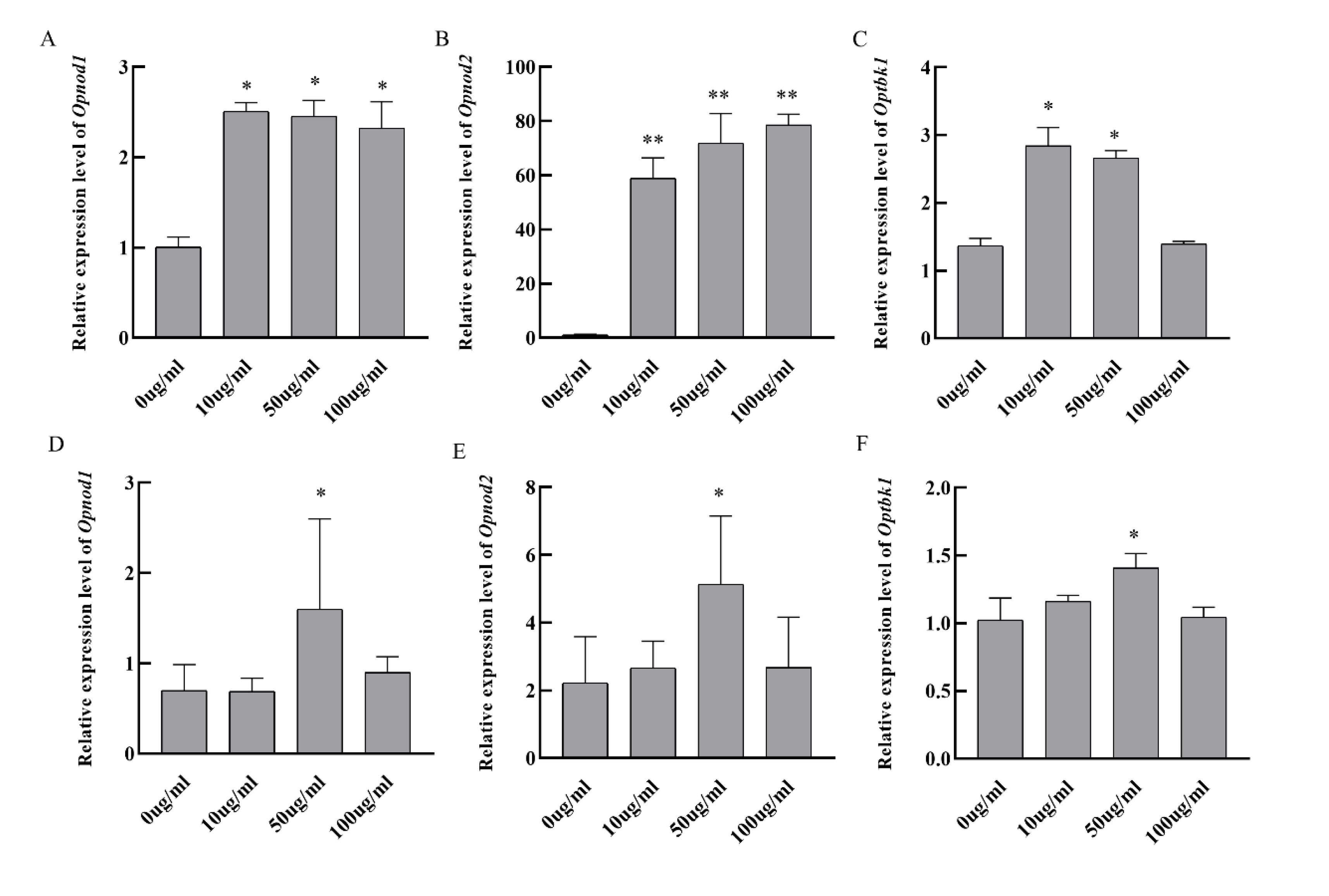
Figure 15.
Expression analysis of Opnod1, Opnod2 and Optbk1 in kidney cells at different concentrations of poly I:C and LPS stimulated. (A-C: different concentrations of poly I:C stimulation of kidney cells; D-E: different concentrations of LPS stimulated kidney cells).
Figure 15.
Expression analysis of Opnod1, Opnod2 and Optbk1 in kidney cells at different concentrations of poly I:C and LPS stimulated. (A-C: different concentrations of poly I:C stimulation of kidney cells; D-E: different concentrations of LPS stimulated kidney cells).
Table 1.
Primers used in this study.
Table 1.
Primers used in this study.
| Primers Primer |
Sequence(5′-3′) |
Use Application |
| Opnod1-ORF-F |
ATGGGTCAGATAGAAGAAGCCAAG |
ORF verification |
| Opnod1-ORF-R |
TCACCATATCTCTTTGAGTGCTGTG |
| Opnod2-ORF-F |
ATGTTTGTCCAGGAGCTTGTGCTG |
| Opnod2-ORF-R |
TCAGAAGACCAGTCTTGATTCACG |
| Optbk1-ORF-F |
ATGCAGAGCACCACTAACTACCTG |
| Optbk1-ORF-R |
TCAGCCTCTCAGACCTCCGTCCAG |
| Opnod1-qRT-F |
GTTGGTGGGAGGTATTTGG |
qRT-PCR |
| Opnod1-qRT-R |
GTTGGTAAGGCTCGGGTG |
| Opnod2-qRT-F |
GGGGCAATAAGATAGGCG |
| Opnod2-qRT-R |
TGACGATGTTGGCGAGGG |
| Optbk1-qRT-F |
AGGACGACGAGCACTTTGTG |
| Optbk1-qRT-R |
CGTATTTCTTCTGGTGGTCTTTT |
| β-actin-F |
GCTGTGCTGTCCCTGT |
| β-actin-R |
GAGTAGCCACGCTCTGTC |
Table 2.
Amino acid similarity of Nod1, Nod2 and TBK1 proteins among Oplegnathus punctatus and other vertebrates.
Table 2.
Amino acid similarity of Nod1, Nod2 and TBK1 proteins among Oplegnathus punctatus and other vertebrates.
| |
Species |
GenBank Access Number |
Similarity/% |
| Nod 1 |
Oreochromis niloticus |
XP_005472430.1 |
86.27 |
| |
Larimichthys crocea |
XP_019134818.2 |
85.07 |
| |
Paralichthys olivaceus |
XP_019946646.1 |
84.39 |
| |
Cynoglossus semilaevis |
XP_008322367.1 |
81.05 |
| |
Oryzias latipes |
XP_020565632.1 |
78.45 |
| |
Takifugu rubripes |
XP_003965935.3 |
74.4 |
| |
Esox lucius |
XP_010883447.1 |
71.35 |
| |
Danio rerio |
XP_002665106.3 |
65.25 |
| |
Macaca mulatta |
XP_028701734.1 |
50.71 |
| |
Mus musculus |
NP_001164478.1 |
50.33 |
| |
Homo sapiens |
XP_011513383.1 |
49.6 |
| |
Xenopus laevis |
XP_031759856.1 |
48.74 |
| Nod2 |
Lates calcarifer |
XP_018522174 |
89.59 |
| |
Larimichthys crocea |
XP_010727419.3 |
85.64 |
| |
Oreochromis niloticus |
XP_003437591.1 |
83.82 |
| |
Paralichthys olivaceus |
XP_019935411.1 |
83.22 |
| |
Cynoglossus semilaevis |
XP_008335431.1 |
79.27 |
| |
Takifugu rubripes |
XP_029701512.1 |
77.25 |
| |
Esox lucius |
XP_010894874.4 |
67.4 |
| |
Danio rerio |
NP_001314973.1 |
64.18 |
| |
Macaca mulatta |
XP_014981593.2 |
46.26 |
| |
Homo sapiens |
NP_071445.1 |
46.26 |
| |
Mus musculus |
AAN84594.1 |
45.54 |
| Tbk1 |
Oplegnathus fasciatus |
AHX37216.1 |
99.86 |
| |
Thunnus maccoyii |
XP_042259009.1 |
98.47 |
| |
Larimichthys crocea |
AKM77645.1 |
98.06 |
| |
Epinephelus coioides |
ATI15615.1 |
97.93 |
| |
Lates calcarifer |
XP_018530412.1 |
97.92 |
| |
Paralichthys olivaceus |
XP_019966450.1 |
97.09 |
| |
Solea senegalensis |
XP_043878151.1 |
96.26 |
| |
Cynoglossus semilaevis |
XP_008313509.1 |
95.29 |
| |
Danio rerio |
NP_001038213.2 |
85.48 |
| |
Mus musculus |
NP_062760.3 |
71.78 |
| |
Homo sapiens |
NP_037386.1 |
71.65 |
| |
Pan troglodytes |
XP_509194.2 |
71.64 |
| |
Xenopus laevis |
NP_001086516.1 |
64.03 |