Submitted:
02 October 2024
Posted:
04 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
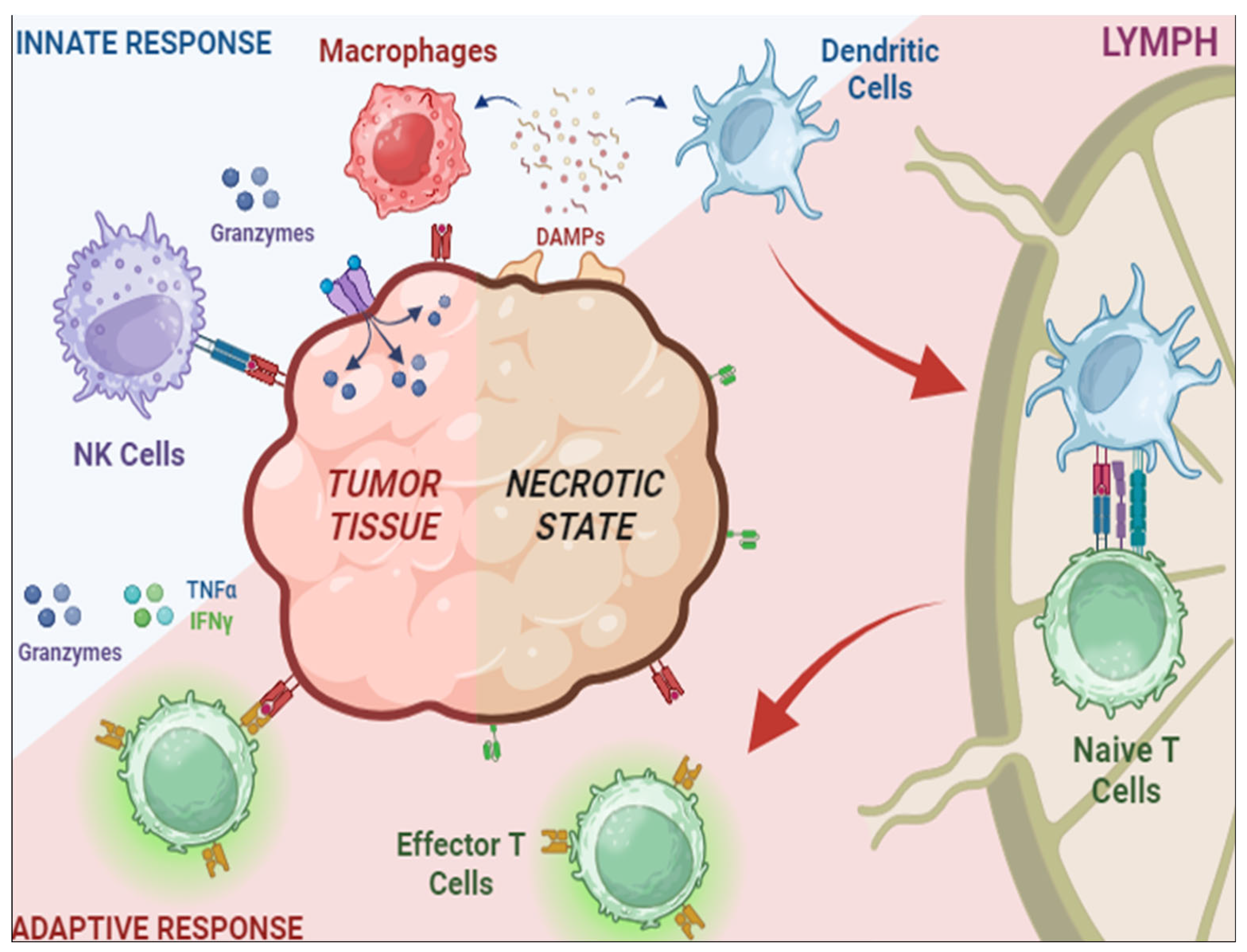
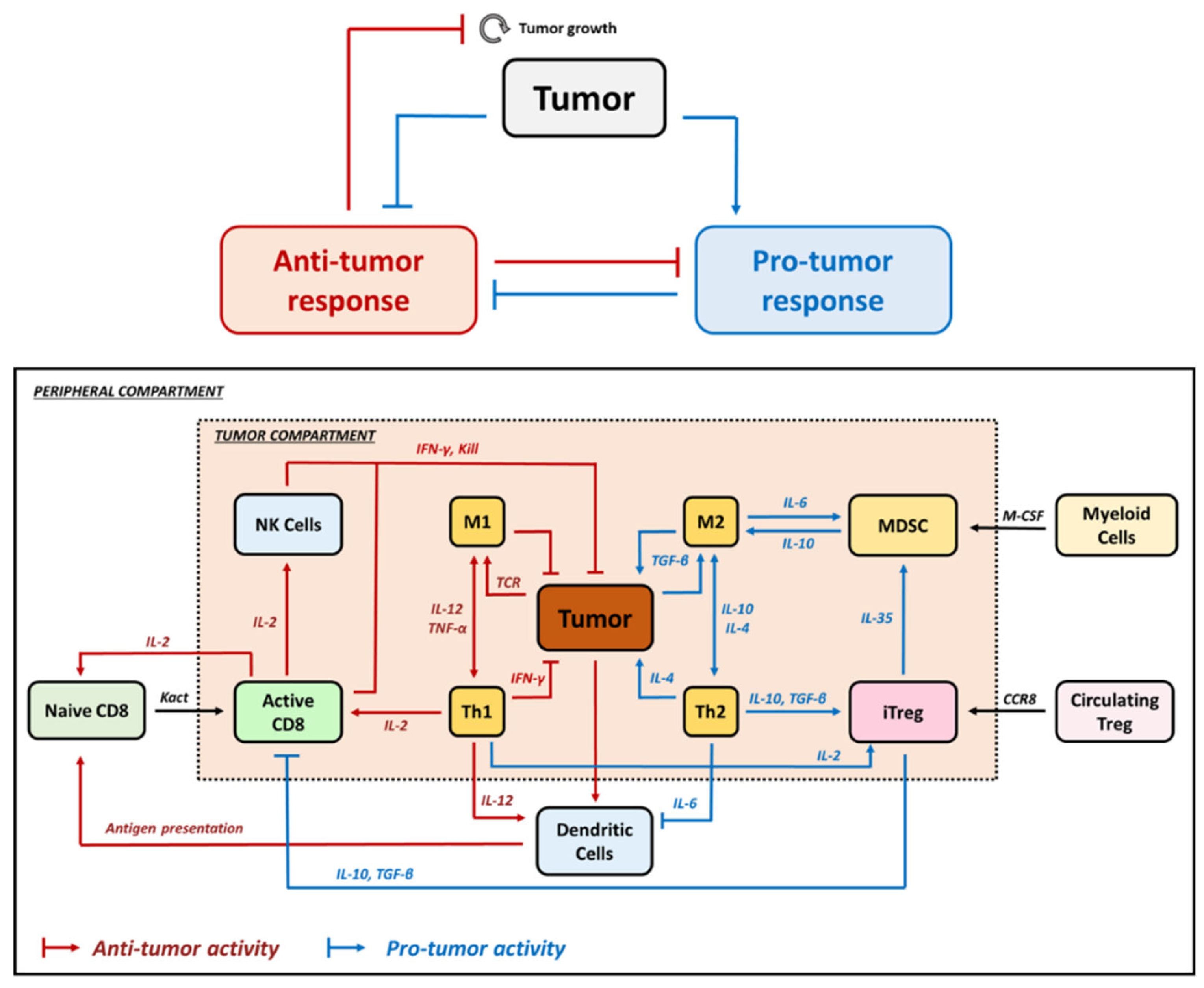
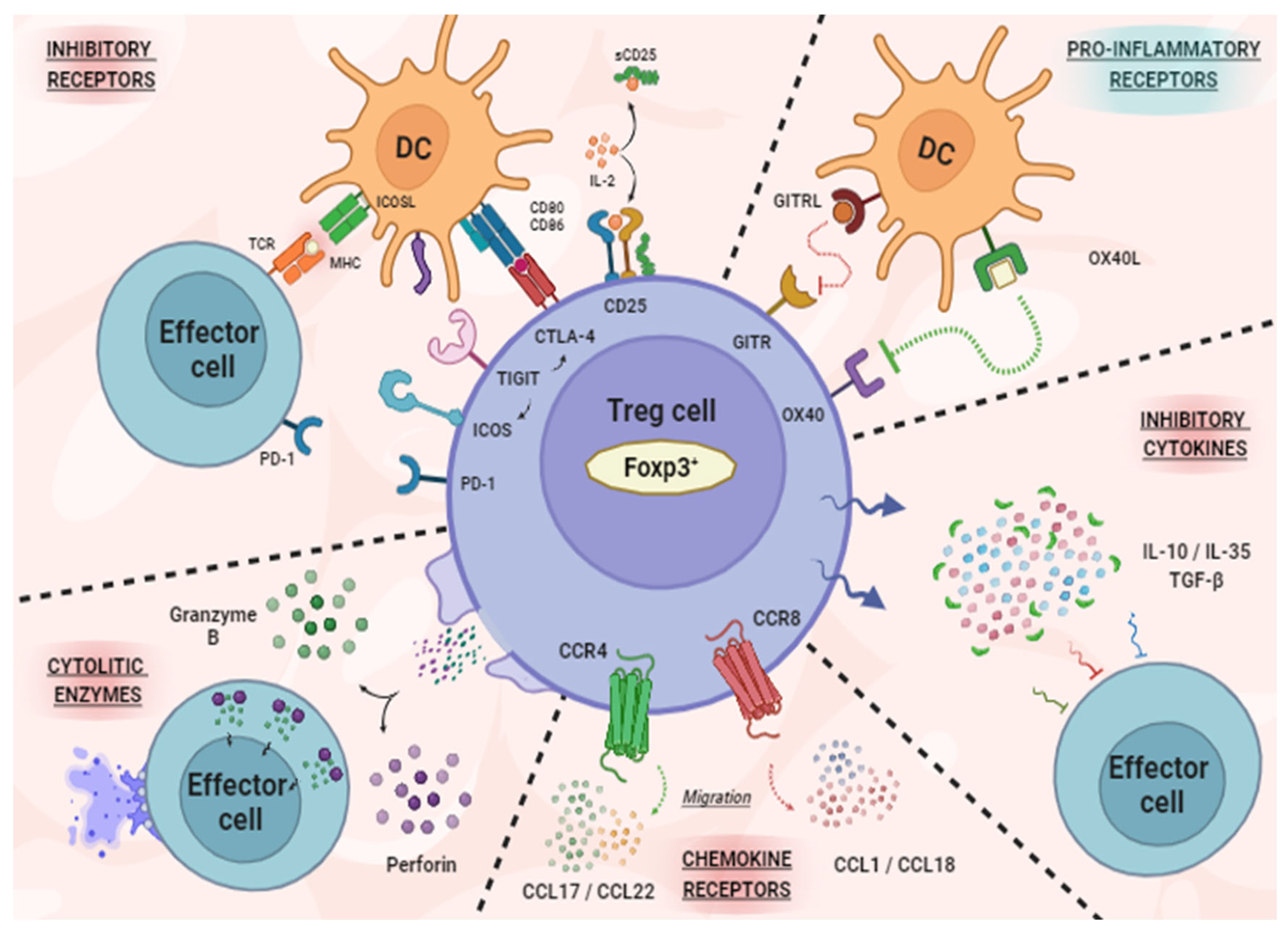
1. Regulatory T Cells in TME
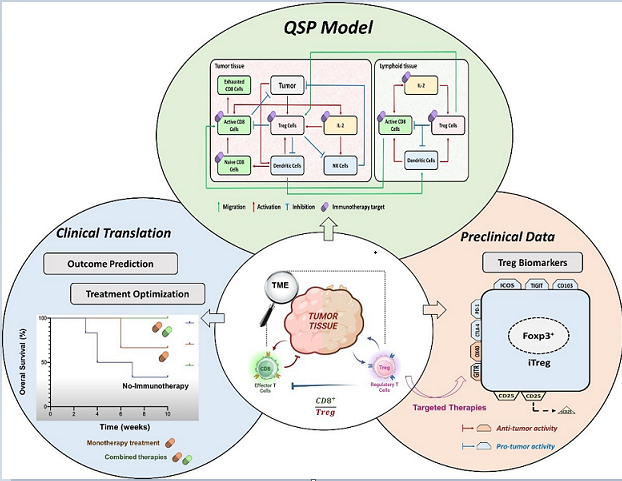
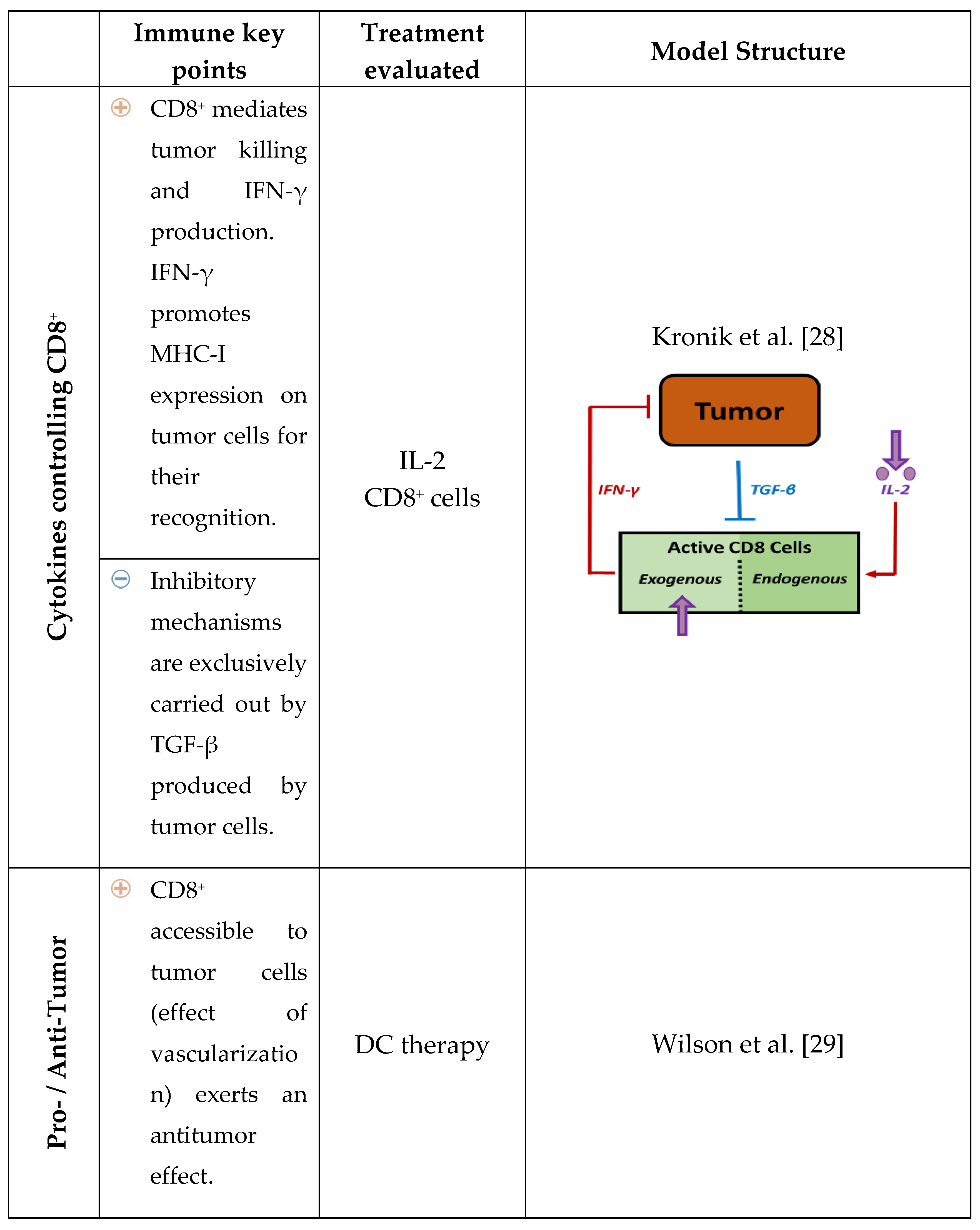
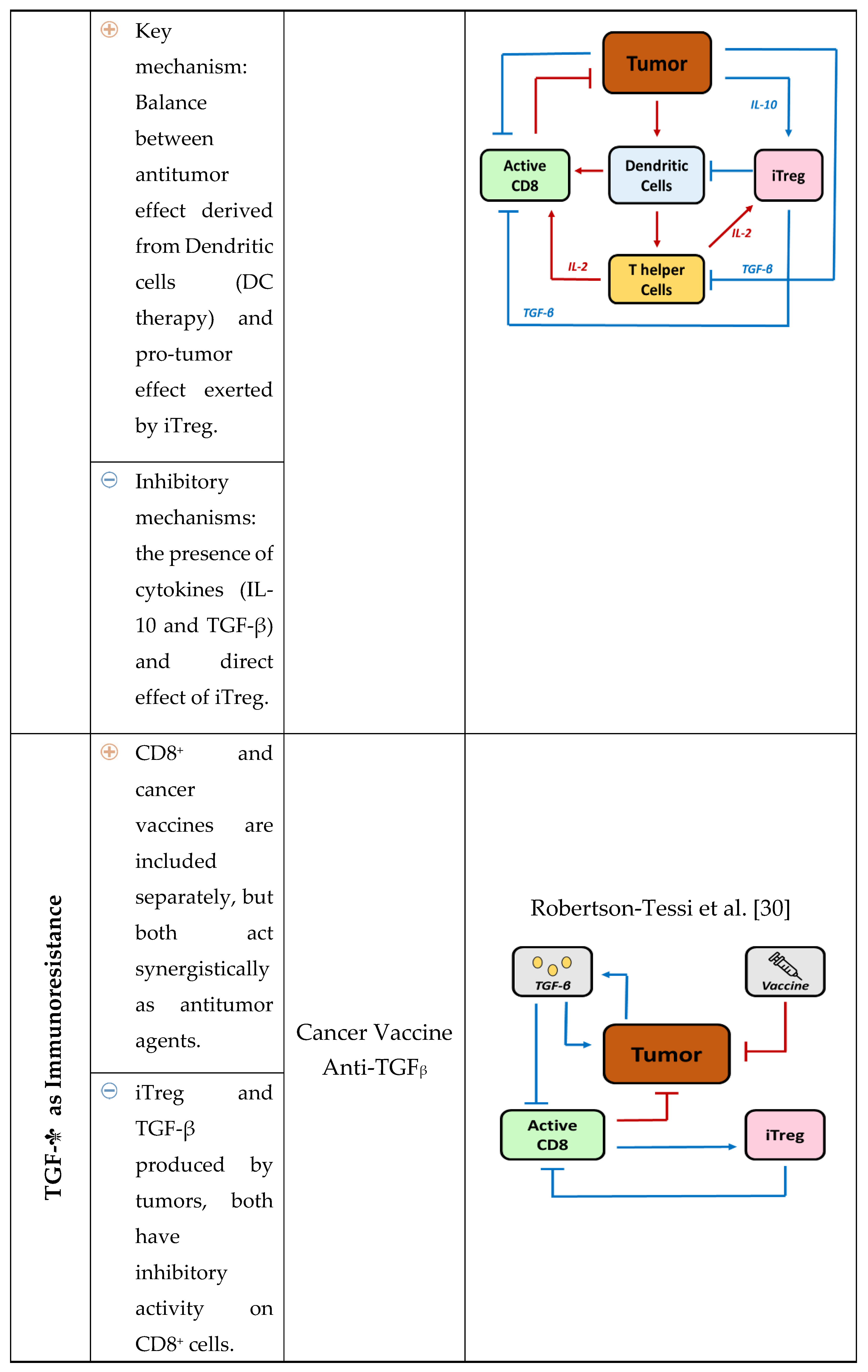
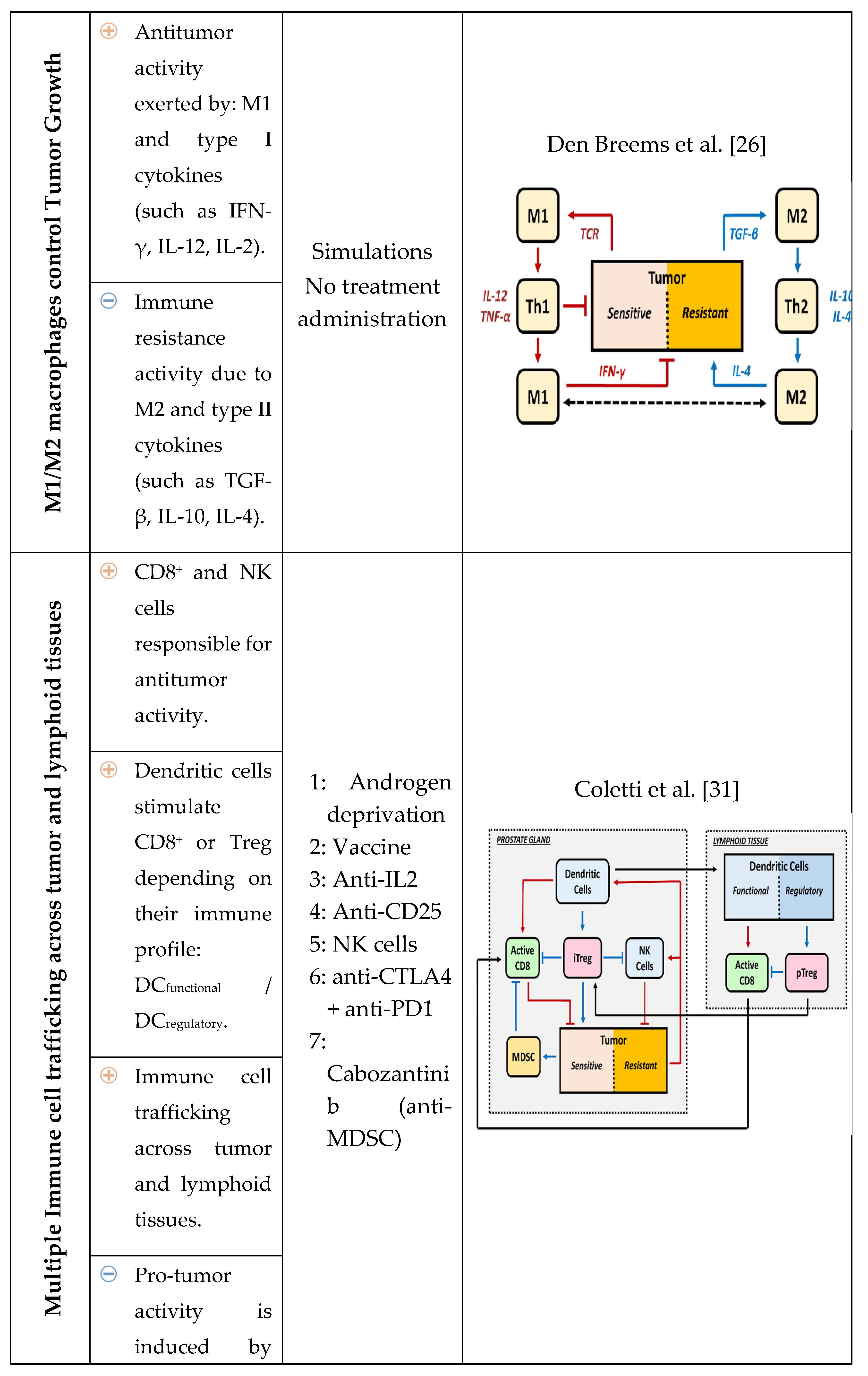
2. Relevance of Quantitative Mathematical Models to Explore Treg Activity in Tumor Growth
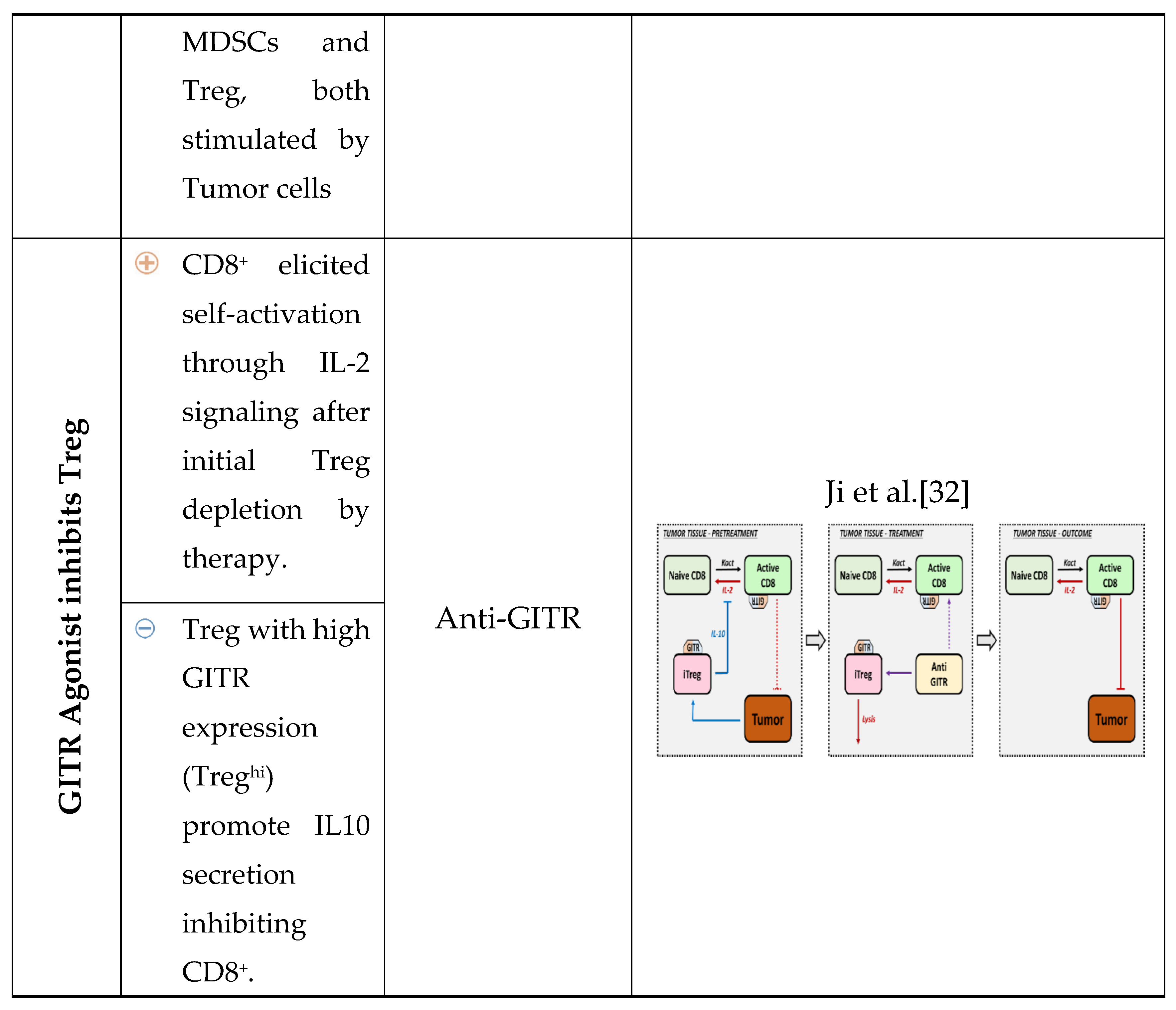
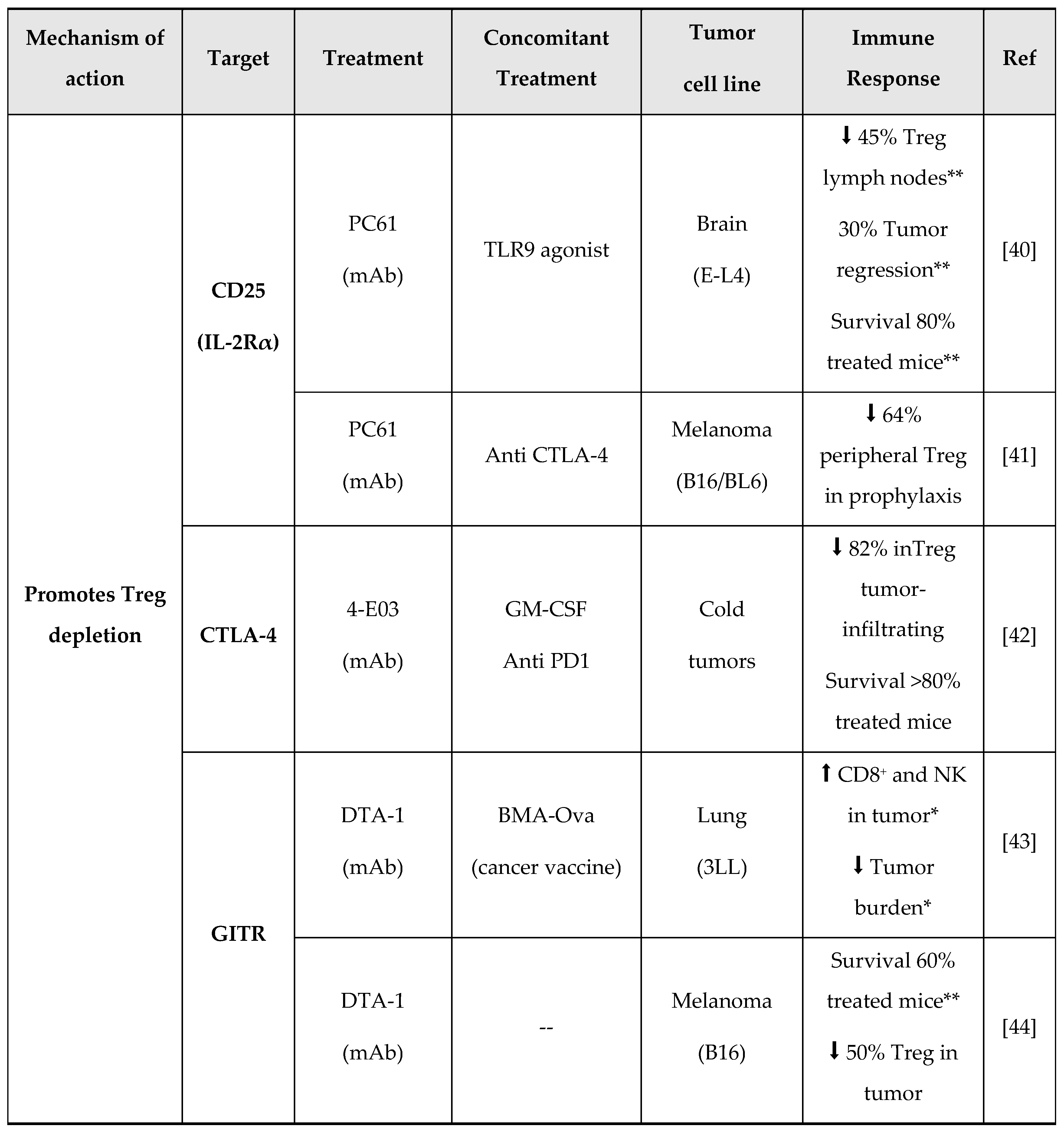
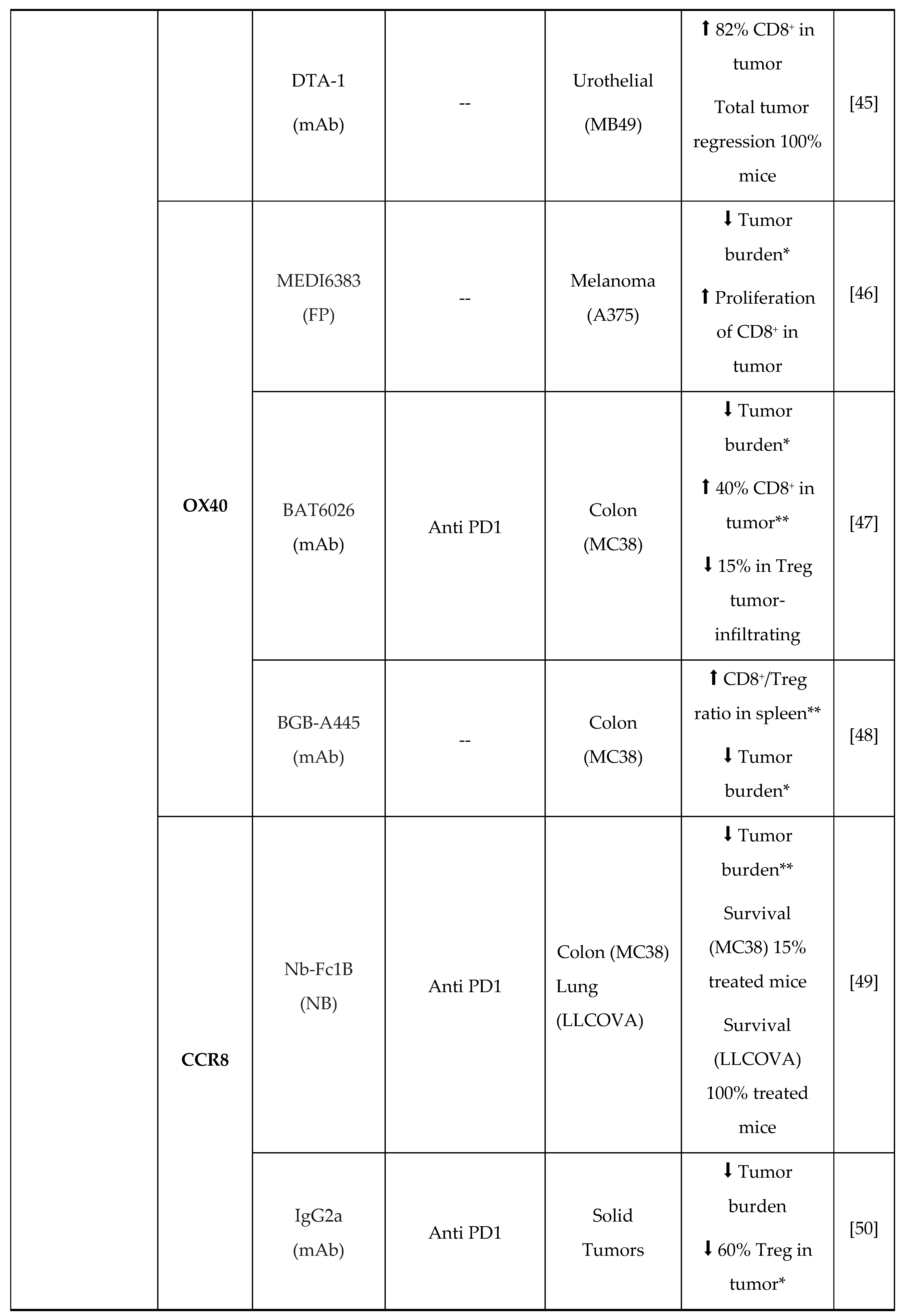
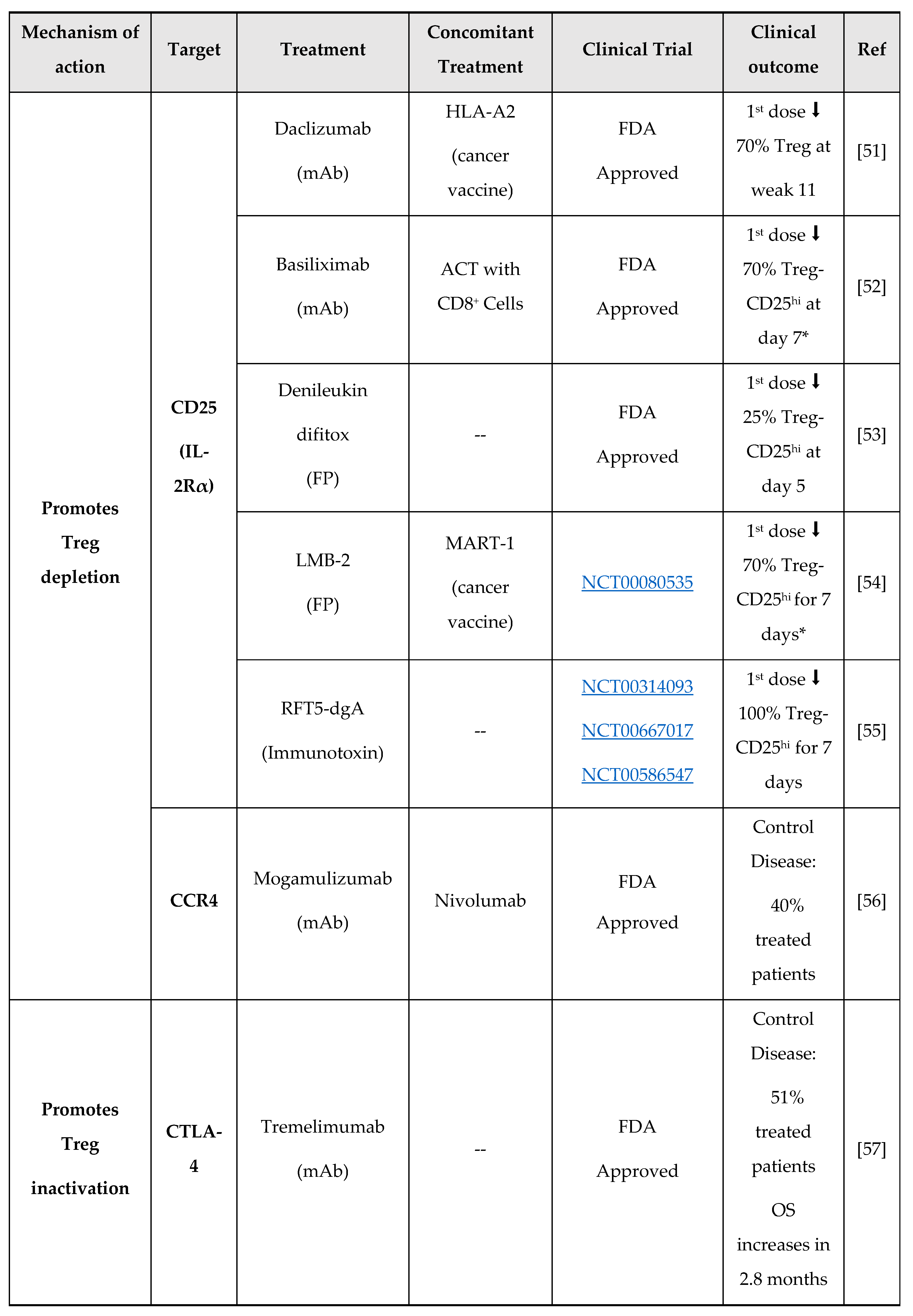
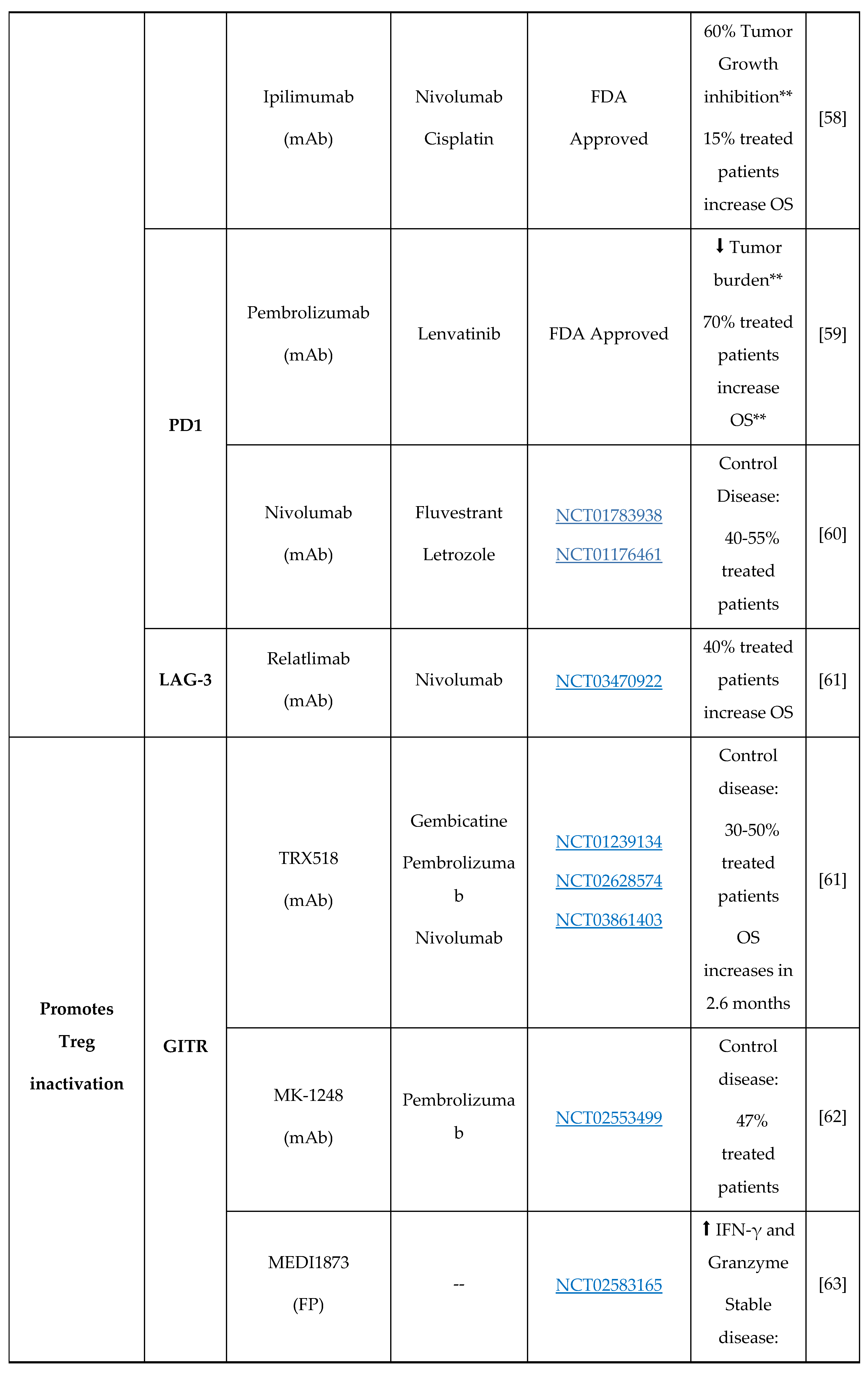
3. New Molecules for Treg Modulation
Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- J.C. Crispin, G.C. Tsokos, Cancer immunosurveillance by CD8 T cells, F100 Res. 9 (2020) 1–7. [CrossRef]
- T.K. Kim, E.N. Vandsemb, R.S. Herbst, L. Chen, Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities, Nat. Rev. Drug Discov. 21 (2022) 529–540. [CrossRef]
- S.A. Rosenberg, Progress in human tumour immunology and immunotherapy, Nature. 411 (2001) 380–384. [CrossRef]
- A. Ribas, L.H. Butterfield, J.A. Glaspy, J.S. Economou, Current developments in cancer vaccines and cellular immunotherapy, J. Clin. Oncol. 21 (2003) 2415–2432. [CrossRef]
- N. Mason, S. Dow, Cancer immunotherapy, Ther. Strateg. Vet. Oncol. 305 (2023) 121–152. [CrossRef]
- I. Mellman, D.S. Chen, T. Powles, S.J. Turley, The cancer-immunity cycle: Indication, genotype, and immunotype, Immunity. 56 (2023) 2188–2205. [CrossRef]
- S.T. Ferris, V. Durai, R. Wu, D.J. Theisen, J.P. Ward, M.D. Bern, J.T.D. Iv, P. Bagadia, T. Liu, G. Carlos, L. Li, W.E. Gillanders, G.F. Wu, W.M. Yokoyama, T.L. Murphy, R.D. Schreiber, K.M. Murphy, cDC1 prime and are licensed by CD4 T cells to induce antitumour immunity, 584 (2021) 624–629. [CrossRef]
- Y. Pan, Y. Yu, X. Wang, T. Zhang, Tumor-Associated Macrophages in Tumor Immunity, Front. Immunol. 11 (2020). [CrossRef]
- F. Castellino, R.N. Germain, Cooperation between CD4+ and CD8+ T cells: When, where, and how, Annu. Rev. Immunol. 24 (2006) 519–540. [CrossRef]
- S. Tang, Q. Ning, L. Yang, Z. Mo, S. Tang, Mechanisms of immune escape in the cancer immune cycle, Int. Immunopharmacol. 86 (2020) 106700. [CrossRef]
- M. Sengupta, R. Pal, A. Nath, B. Chakraborty, L.M. Singh, B. Das, S.K. Ghosh, Anticancer efficacy of noble metal nanoparticles relies on reprogramming tumor-associated macrophages through redox pathways and pro-inflammatory cytokine cascades, Cell. Mol. Immunol. 15 (2018) 1088–1090. [CrossRef]
- D.I. Gabrilovich, Myeloid-derived suppressor cells, Cancer Immunol Res. 5 (2018) 3–8. [CrossRef]
- A. Tanaka, S. Sakaguchi, Regulatory T cells in cancer immunotherapy, Cell Res. 27 (2017) 109–118. [CrossRef]
- M. V. Goldberg, C.G. Drake, LAG-3 in Cancer Immunotherapy, Curr Top Microbiol Immunol. 344 (2011) 269–278. [CrossRef]
- V. Gouirand, I. Habrylo, M.D. Rosenblum, Regulatory T Cells and Inflammatory Mediators in Autoimmune Disease, J. Invest. Dermatol. 142 (2022) 774–780. [CrossRef]
- E.M. Shevach, A.M. Thornton, tTregs, pTregs, and iTregs: similarities and differences, Immunol. Rev. 259 (2014) 88–102. [CrossRef]
- O. Yoshie, Ccr4 as a therapeutic target for cancer immunotherapy, Cancers (Basel). 13 (2021) 1–28. [CrossRef]
- O. Yoshie, K. Matsushima, CCR4 and its ligands: From bench to bedside, Int. Immunol. 27 (2015) 11–20. [CrossRef]
- S.K. Whiteside, F.M. Grant, D.S. Gyori, A.G. Conti, C.J. Imianowski, P. Kuo, R. Nasrallah, F. Sadiyah, S.A. Lira, F. Tacke, R.L. Eil, O.T. Burton, J. Dooley, A. Liston, K. Okkenhaug, J. Yang, R. Roychoudhuri, CCR8 marks highly suppressive Treg cells within tumours but is dispensable for their accumulation and suppressive function, Immunology. 163 (2021) 512–520. [CrossRef]
- D. Noyes, A. Bag, S. Oseni, J. Semidey-Hurtado, L. Cen, A.A. Sarnaik, V.K. Sondak, D. Adeegbe, Tumor-associated Tregs obstruct antitumor immunity by promoting T cell dysfunction and restricting clonal diversity in tumor-infiltrating CD8+ T cells, J. Immunother. Cancer. 10 (2022) e004605. [CrossRef]
- K. Peskov, I. Azarov, L. Chu, V. Voronova, Y. Kosinsky, G. Helmlinger, Quantitative mechanistic modeling in support of pharmacological therapeutics development in immuno-oncology, Front. Immunol. 10 (2019) 1–11. [CrossRef]
- M. Simeoni, P. Magni, C. Cammia, G. De Nicolao, V. Croci, E. Pesenti, M. Germani, I. Poggesi, M. Rocchetti, Predictive Pharmacokinetic-Pharmacodynamic Modeling of Tumor Growth Kinetics in Xenograft Models after Administration of Anticancer Agents, Cancer Res. 64 (2004) 1094–1101. [CrossRef]
- Z.P. Parra-Guillen, P. Berraondo, E. Grenier, B. Ribba, I.F. Troconiz, Mathematical model approach to describe tumour response in mice after vaccine administration and its applicability to immune-stimulatory cytokine-based strategies, AAPS J. 15 (2013) 797–807. [CrossRef]
- Y. Bulliard, R. Jolicoeur, M. Windman, S.M. Rue, S. Ettenberg, D.A. Knee, N.S. Wilson, G. Dranoff, J.L. Brogdon, Activating fc γ receptors contribute to the antitumor activities of immunoregulatory receptor-targeting antibodies, J. Exp. Med. 210 (2013) 1685–1693. [CrossRef]
- B. Chaudhary, E. Elkord, Regulatory T cells in the tumor microenvironment and cancer progression: Role and therapeutic targeting, Vaccines. 4 (2016) 1–25. [CrossRef]
- N.Y. den Breems, R. Eftimie, The re-polarisation of M2 and M1 macrophages and its role on cancer outcomes, J. Theor. Biol. 390 (2016) 23–39. [CrossRef]
- A. Arabameri, D. Asemani, J. Hadjati, A structural methodology for modeling immune-tumor interactions including pro- and anti-tumor factors for clinical applications, Math. Biosci. 304 (2018) 48–61. [CrossRef]
- N. Kronik, Y. Kogan, P.G. Schlegel, M. Wölflz, Erratum: Improving T-cell immunotherapy for melanoma through a mathematically motivated strategy: Efficacy in numbers (Journal of Immunotherapy (2012) (116)), J. Immunother. 35 (2012) 292. [CrossRef]
- S. Wilson, D. Levy, A Mathematical Model of the Enhancement of Tumor Vaccine Efficacy by Immunotherapy, Bull. Math. Biol. 74 (2012) 1485–1500. [CrossRef]
- M. Robertson-Tessi, A. El-Kareh, A. Goriely, A mathematical model of tumor-immune interactions, J. Theor. Biol. 294 (2012) 56–73. [CrossRef]
- R. Coletti, L. Leonardelli, S. Parolo, L. Marchetti, A QSP model of prostate cancer immunotherapy to identify effective combination therapies, Sci. Rep. 10 (2020) 1–18. [CrossRef]
- M. Movassaghi, R. Chung, C.B. Anderson, M. Stein, Y. Saenger, I. Faiena, Overcoming immune resistance in prostate cancer: Challenges and advances, Cancers (Basel). 13 (2021) 1–16. [CrossRef]
- Y. Ji, K. Madrasi, D.A. Knee, L. Gruenbaum, J.F. Apgar, J.M. Burke, B. Gomes, Quantitative systems pharmacology model of GITR-mediated T cell dynamics in tumor microenvironment, CPT Pharmacometrics Syst. Pharmacol. 12 (2023) 413–424. [CrossRef]
- D. Qin, Y. Zhang, P. Shu, Y. Lei, X. Li, Y. Wang, Targeting tumor-infiltrating tregs for improved antitumor responses, Front. Immunol. 15 (2024) 1–15. [CrossRef]
- P. Moussa, Transcriptomic analysis of GITR and GITR ligand reveals cancer immune heterogeneity with implications for GITR targeting, Am. J. Cancer Res. 14 (2024) 1634–1648. [CrossRef]
- Y. Dong, C. Yang, F. Pan, Post-Translational Regulations of Foxp3 in Treg Cells and Their Therapeutic Applications, Front. Immunol. 12 (2021) 1–16. [CrossRef]
- A. Serrano, N. Casares, I.F. Trocóniz, T. Lozano, J.J. Lasarte, S. Zalba, M.J. Garrido, Foxp3 inhibitory peptide encapsulated in a novel CD25-targeted nanoliposome promotes efficient tumor regression in mice, Acta Pharmacol. Sin. (2024) 1–13. [CrossRef]
- S. Mirzaei, M.H. Gholami, H.L. Ang, F. Hashemi, A. Zarrabi, A. Zabolian, K. Hushmandi, M. Delfi, H. Khan, M. Ashrafizadeh, G. Sethi, A.P. Kumar, Pre-clinical and clinical applications of small interfering rnas (Sirna) and co-delivery systems for pancreatic cancer therapy, Cells. 10 (2021). [CrossRef]
- S. Yonezawa, H. Koide, T. Asai, Recent advances in siRNA delivery mediated by lipid-based nanoparticles, Adv. Drug Deliv. Rev. 154–155 (2020) 64–78. [CrossRef]
- U. Jarry, S. Donnou, M. Vincent, P. Jeannin, L. Pineau, I. Fremaux, Y. Delneste, D. Couez, Treg depletion followed by intracerebral CpG-ODN injection induce brain tumor rejection, J. Neuroimmunol. 267 (2014) 35–42. [CrossRef]
- S.A. Quezada, K.S. Peggs, T.R. Simpson, Y. Shen, D.R. Littman, J.P. Allison, Limited tumor infiltration by activated T effector cells restricts the therapeutic activity of regulatory T cell depletion against established melanoma, J. Exp. Med. 205 (2008) 2125–2138. [CrossRef]
- M. Semmrich, J.B. Marchand, L. Fend, M. Rehn, C. Remy, P. Holmkvist, N. Silvestre, C. Svensson, P. Kleinpeter, J. Deforges, F. Junghus, K.L. Cleary, M. Bodén, L. Mårtensson, J. Foloppe, I. Teige, E. Quéméneur, B. Frendéus, Vectorized Treg-depleting αcTLA-4 elicits antigen cross-presentation and CD8 + T cell immunity to reject cold’ tumors, J. Immunother. Cancer. 10 (2022) 1–14. [CrossRef]
- L.X. Zhu, M. Davoodi, M.K. Srivastava, P. Kachroo, J.M. Lee, M.S. John, M. Harris-White, M. Huang, R.M. Strieter, S. Dubinett, S. Sharma, GITR agonist enhances vaccination responses in lung cancer, Oncoimmunology. 4 (2015) 1–12. [CrossRef]
- A.D. Cohen, D.A. Schaer, C. Liu, Y. Li, D. Hirschhorn-Cymmerman, S.C. Kim, A. Diab, G. Rizzuto, F. Duan, M.A. Perales, T. Merghoub, A.N. Houghton, J.D. Wolchok, Agonist anti-GITR monoclonal antibody induces melanoma tumor immunity in mice by altering regulatory T cell stability and intra-tumor accumulation, PLoS One. 5 (2010). [CrossRef]
- D. Coe, S. Begom, C. Addey, M. White, J. Dyson, J.G. Chai, Depletion of regulatory T cells by anti-GITR mAb as a novel mechanism for cancer immunotherapy, Cancer Immunol. Immunother. 59 (2010) 1367–1377. [CrossRef]
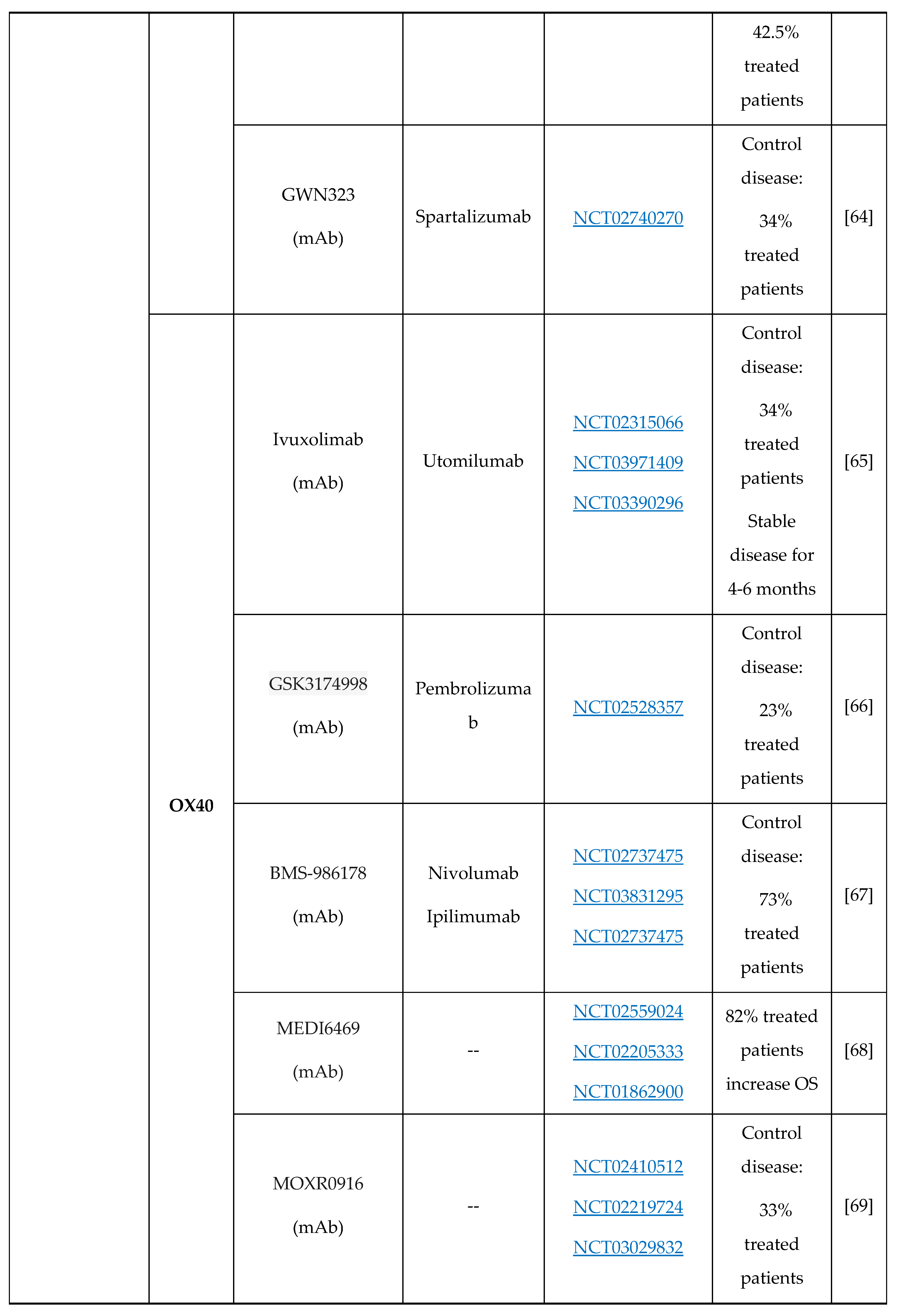
- M.D. Oberst, C. Augé, C. Morris, S. Kentner, K. Mulgrew, K. McGlinchey, J. Hair, S. Hanabuchi, Q. Du, M. Damschroder, H. Feng, S. Eck, N. Buss, L. de Haan, A.J. Pierce, H. Park, A. Sylwester, M.K. Axthelm, L. Picker, N.P. Morris, A. Weinberg, S.A. Hammond, Potent Immune Modulation by MEDI6383, an Engineered Human OX40 Ligand IgG4P Fc Fusion Protein, Mol. Cancer Ther. 17 (2018) 1024–1038. [CrossRef]
- S. Liang, D. Zheng, X. Liu, X. Mei, C. Zhou, C. Xiao, C. Qin, H. Yue, J. Lin, C. Liu, S. Li, J.C. Yu, BAT6026, a novel anti-OX40 antibody with enhanced antibody dependent cellular cytotoxicity effect for cancer immunotherapy, Front. Oncol. 13 (2023) 1–11. [CrossRef]
- B. Jiang, T. Zhang, M. Deng, W. Jin, Y. Hong, X. Chen, X. Chen, J. Wang, H. Hou, Y. Gao, W. Gong, X. Wang, H. Li, X. Zhou, Y. Feng, B. Zhang, B. Jiang, X. Lu, L. Zhang, Y. Li, W. Song, H. Sun, Z. Wang, X. Song, Z. Shen, X. Liu, K. Li, L. Wang, Y. Liu, BGB-A445, a novel non-ligand-blocking agonistic anti-OX40 antibody, exhibits superior immune activation and antitumor effects in preclinical models, Front. Med. 17 (2023) 1170–1185. [CrossRef]
- H. Van Damme, B. Dombrecht, M. Kiss, H. Roose, E. Allen, E. Van Overmeire, D. Kancheva, L. Martens, A. Murgaski, P.M.R. Bardet, G. Blancke, M. Jans, E. Bolli, M.S. Martins, Y. Elkrim, J. Dooley, L. Boon, J.K. Schwarze, F. Tacke, K. Movahedi, N. Vandamme, B. Neyns, S. Ocak, I. Scheyltjens, L. Vereecke, F.A. Nana, P. Merchiers, D. Laoui, J.A. Van Ginderachter, Therapeutic depletion of CCR8 + tumor-infiltrating regulatory T cells elicits antitumor immunity and synergizes with anti-PD-1 therapy, J. Immunother. Cancer. 9 (2021) 1–16. [CrossRef]
- J.R. Campbell, B.R. McDonald, P.B. Mesko, N.O. Siemers, P.B. Singh, M. Selby, T.W. Sproul, A.J. Korman, L.M. Vlach, J. Houser, S. Sambanthamoorthy, K. Lu, S. V. Hatcher, J. Lohre, R. Jain, R.Y. Lan, Fc-optimized anti-CCR8 antibody depletes regulatory t cells in human tumor models, Cancer Res. 81 (2021) 2983–2994. [CrossRef]
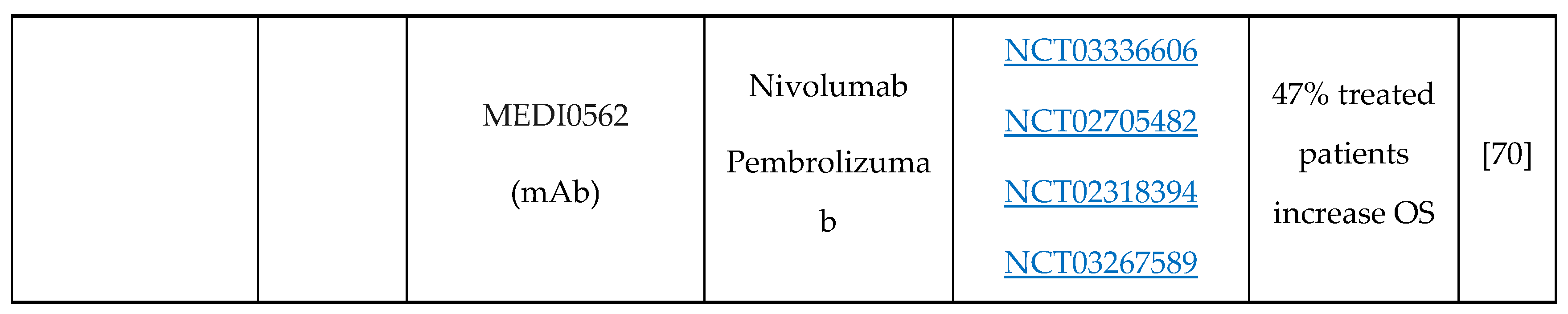
- A.J. Rech, R.H. Vonderheide, Clinical use of anti-CD25 antibody daclizumab to enhance immune responses to tumor antigen vaccination by targeting regulatory T cells, Ann. N. Y. Acad. Sci. 1174 (2009) 99–106. [CrossRef]
- R. Okita, Y. Yamaguchi, M. Ohara, K. Hironaka, M. Okawaki, I. Nagamine, T. Ikeda, A. Emi, J. Hihara, M. Okada, Targeting of CD4+CD25high cells while preserving CD4+CD25low cells with low-dose chimeric anti-CD25 antibody in adoptive immunotherapy of cancer., Int. J. Oncol. 34 (2009) 563–72. [CrossRef]
- M.T. Litzinger, R. Fernando, T.J. Curiel, D.W. Grosenbach, J. Schlom, C. Palena, IL-2 immunotoxin denileukin diftitox reduces regulatory T cells and enhances vaccine-mediated T-cell immunity, Blood. 110 (2007) 3192–3201. [CrossRef]
- D.J. Powell, A. Felipe-Silva, M.J. Merino, M. Ahmadzadeh, T. Allen, C. Levy, D.E. White, S. Mavroukakis, R.J. Kreitman, S.A. Rosenberg, I. Pastan, Administration of a CD25-Directed Immunotoxin, LMB-2, to Patients with Metastatic Melanoma Induces a Selective Partial Reduction in Regulatory T Cells In Vivo, J. Immunol. 179 (2007) 4919–4928. [CrossRef]
- D.J. Powell, P. Attia, V. Ghetie, J. Schindlerv, E.S. Vitetta, S.A. Rosenberg, Partial reduction of human FOXP3+ CD4 T cells in vivo after CD25-directed recombinant immunotoxin administration, J. Immunother. 31 (2008) 189–198. [CrossRef]
- D.S. Hong, O. Rixe, V.K. Chiu, P.M. Forde, T. Dragovich, Y. Lou, A. Nayak-Kapoor, R. Leidner, J.N. Atkins, A. Collaku, F.E. Fox, M.A. Marshall, A.J. Olszanski, Mogamulizumab in Combination with Nivolumab in a Phase I/II Study of Patients with Locally Advanced or Metastatic Solid Tumors, Clin. Cancer Res. 28 (2022) 479–488. [CrossRef]
- J.M. Kirkwood, P. Lorigan, P. Hersey, A. Hauschild, C. Robert, D. McDermott, M.A. Marshall, J. Gomez-Navarro, J.Q. Liang, C.A. Bulanhagui, Phase II trial of tremelimumab (CP-675,206) in patients with advanced refractory or relapsed melanoma, Clin. Cancer Res. 16 (2010) 1042–1048. [CrossRef]
- W. Chi, L. Zhang, X. Wang, J. Li, F. Li, Y. Ma, Q. Zhang, Effects of Nivolumab and Ipilimumab on the suppression of cisplatin resistant small cell lung cancer cells, Invest. New Drugs. 40 (2022) 709–717. [CrossRef]
- C. Yi, L. Chen, Z. Lin, L. Liu, W. Shao, R. Zhang, J. Lin, J. Zhang, W. Zhu, H. Jia, L. Qin, L. Lu, J. Chen, Lenvatinib Targets FGF Receptor 4 to Enhance Antitumor Immune Response of Anti–Programmed Cell Death-1 in HCC, Hepatology. 74 (2021) 2544–2560. [CrossRef]
- J. Koh, J.Y. Hur, K.Y. Lee, M.S. Kim, J.Y. Heo, B.M. Ku, J.M. Sun, S.H. Lee, J.S. Ahn, K. Park, M.J. Ahn, Regulatory (FoxP3+) T cells and TGF-β predict the response to anti-PD-1 immunotherapy in patients with non-small cell lung cancer, Sci. Rep. 10 (2020) 1–10. [CrossRef]
- D. Davar, R. Zappasodi, H. Wang, G.S. Naik, T. Sato, T. Bauer, D. Bajor, O. Rixe, W. Newman, J. Qi, A. Holland, P. Wong, L. Sifferlen, D. Piper, C.A. Sirard, T. Merghoub, J.D. Wolchok, J.J. Luke, Phase IB Study of GITR Agonist Antibody TRX518 Singly and in Combination with Gemcitabine, Pembrolizumab, or Nivolumab in Patients with Advanced Solid Tumors, Clin. Cancer Res. 28 (2022) 3990–4002. [CrossRef]
- R. Geva, M. Voskoboynik, K. Dobrenkov, K. Mayawala, J. Gwo, R. Wnek, E. Chartash, G. V. Long, First-in-human phase 1 study of MK-1248, an anti–glucocorticoid-induced tumor necrosis factor receptor agonist monoclonal antibody, as monotherapy or with pembrolizumab in patients with advanced solid tumors, Cancer. 126 (2020) 4926–4935. [CrossRef]
- A.S. Balmanoukian, J.R. Infante, R. Aljumaily, A. Naing, A. V. Chintakuntlawar, N.A. Rizvi, H.J. Ross, M. Gordon, P.R. Mallinder, N. Elgeioushi, I. Gonzalez-García, N. Standifer, J. Cann, N. Durham, S. Rahimian, R. Kumar, C.S. Denlinger, Safety and clinical activity of MEDI1873, a novel GITR agonist, in advanced solid tumors, Clin. Cancer Res. 26 (2020) 6196–6203. [CrossRef]
- S.A. Piha-Paul, R. Geva, T.J. Tan, D.W.T. Lim, C. Hierro, T. Doi, O. Rahma, A. Lesokhin, J.J. Luke, J. Otero, L. Nardi, A. Singh, A. Xyrafas, X. Chen, J. Mataraza, P.L. Bedard, First-in-human phase I/Ib open-label dose-escalation study of GWN323 (anti-GITR) as a single agent and in combination with spartalizumab (anti-PD-1) in patients with advanced solid tumors and lymphomas, J. Immunother. Cancer. 9 (2021). [CrossRef]
- O. Hamid, A.A. Chiappori, J.A. Thompson, T. Doi, S. Hu-Lieskovan, F.A.L.M. Eskens, W. Ros, A. Diab, J.P. Spano, N.A. Rizvi, J.S. Wasser, E. Angevin, P.A. Ott, A. Forgie, W. Yang, C. Guo, J. Chou, A.B. El-Khoueiry, First-in-human study of an OX40 (ivuxolimab) and 4-1BB (utomilumab) agonistic antibody combination in patients with advanced solid tumors, J. Immunother. Cancer. 10 (2022) 1–12. [CrossRef]
- S. Postel-Vinay, V.K. Lam, W. Ros, T.M. Bauer, A.R. Hansen, D.C. Cho, F. Stephen Hodi, J.H.M. Schellens, J.K. Litton, S. Aspeslagh, K.A. Autio, F.L. Opdam, M. McKean, N. Somaiah, S. Champiat, M. Altan, A. Spreafico, O. Rahma, E.M. Paul, C.M. Ahlers, H. Zhou, H. Struemper, S.A. Gorman, M. Watmuff, K.M. Yablonski, N. Yanamandra, M.J. Chisamore, E. V. Schmidt, A. Hoos, A. Marabelle, J.S. Weber, J. V. Heymach, First-in-human phase I study of the OX40 agonist GSK3174998 with or without pembrolizumab in patients with selected advanced solid tumors (ENGAGE-1), J. Immunother. Cancer. 11 (2023) e005301. [CrossRef]
- M. Gutierrez, V. Moreno, K.M. Heinhuis, A.J. Olszanski, A. Spreafico, M. Ong, Q. Chu, R.D. Carvajal, J. Trigo, M.O. de Olza, M. Provencio, F.Y. de Vos, F. de Braud, S. Leong, D. Lathers, R. Wang, P. Ravindran, Y. Feng, P. Aanur, I. Melero, OX40 agonist BMS-986178 alone or in combination with nivolumab and/or ipilimumab in patients with advanced solid tumors, Clin. Cancer Res. 27 (2021) 460–472. [CrossRef]
- R. Duhen, C. Ballesteros-Merino, A.K. Frye, E. Tran, V. Rajamanickam, S.C. Chang, Y. Koguchi, C.B. Bifulco, B. Bernard, R.S. Leidner, B.D. Curti, B.A. Fox, W.J. Urba, R.B. Bell, A.D. Weinberg, Neoadjuvant anti-OX40 (MEDI6469) therapy in patients with head and neck squamous cell carcinoma activates and expands antigen-specific tumor-infiltrating T cells, Nat. Commun. 12 (2021). [CrossRef]
- T.W. Kim, H.A. Burris, M.J. de Miguel Luken, M.J. Pishvaian, Y.-J. Bang, M. Gordon, A. Awada, D.R. Camidge, F.S. Hodi, G.A. McArthur, W.H. Miller, A. Cervantes, L.Q. Chow, A.M. Lesokhin, A. Rutten, M. Sznol, D. Rishipathak, S.-C. Chen, E. Stefanich, T. Pourmohamad, M. Anderson, J. Kim, M. Huseni, I. Rhee, L.L. Siu, First-In-Human Phase I Study of the OX40 Agonist MOXR0916 in Patients with Advanced Solid Tumors., Clin. Cancer Res. 28 (2022) 3452–3463. [CrossRef]
- B.S. Glisson, R.S. Leidner, R.L. Ferris, J. Powderly, N.A. Rizvi, B. Keam, R. Schneider, S. Goel, J.P. Ohr, J. Burton, Y. Zheng, S. Eck, M. Gribbin, K. Streicher, D.M. Townsley, S.P. Patel, Safety and clinical activity of MEDI0562, a humanized OX40 agonist monoclonal antibody, in adult patients with advanced solid tumors, Clin. Cancer Res. 26 (2020) 5358–5367. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





