Submitted:
12 June 2024
Posted:
18 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Post-Acute Sequelae Of SARS-CoV-2
1.2. Proposed Pathogenesis Of PASC
2. Coronavirus Replication & Persistence
2.1. Structure & Replication
2.2. Persistence Of SARS-CoV-2 Proteins & Genetic Material
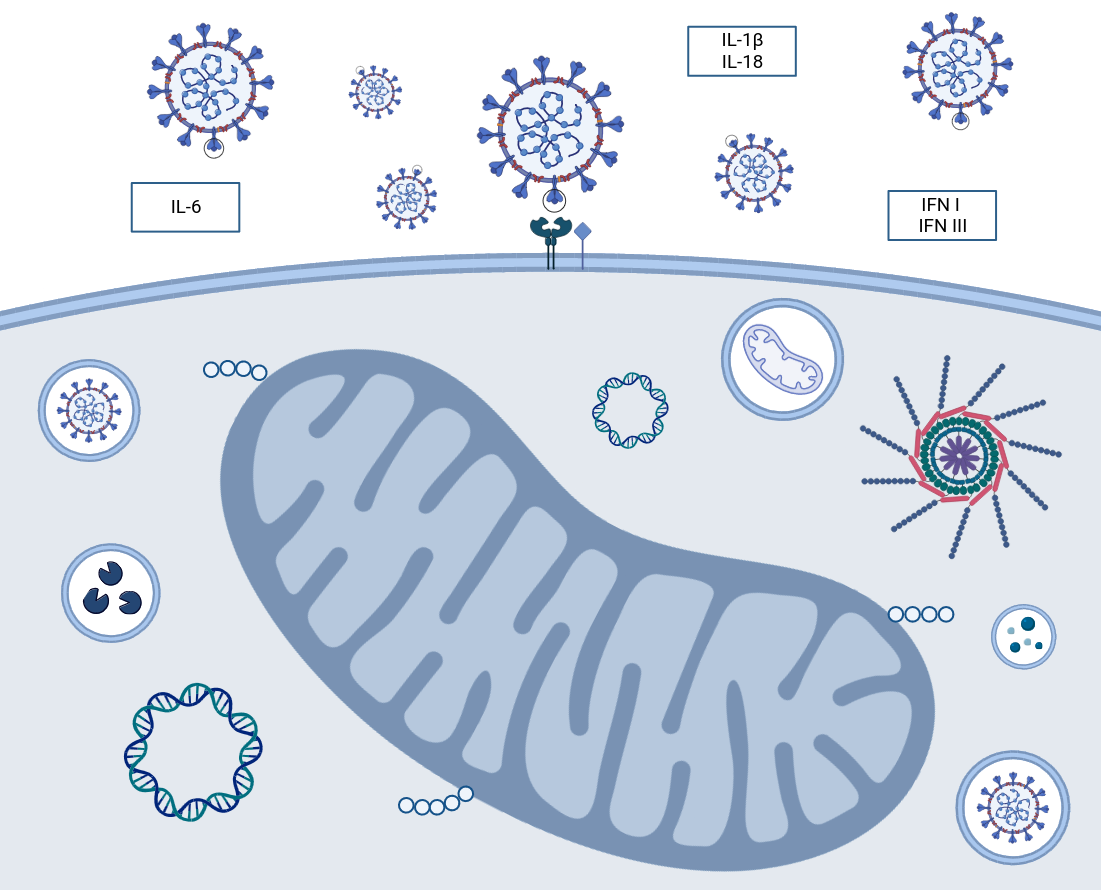
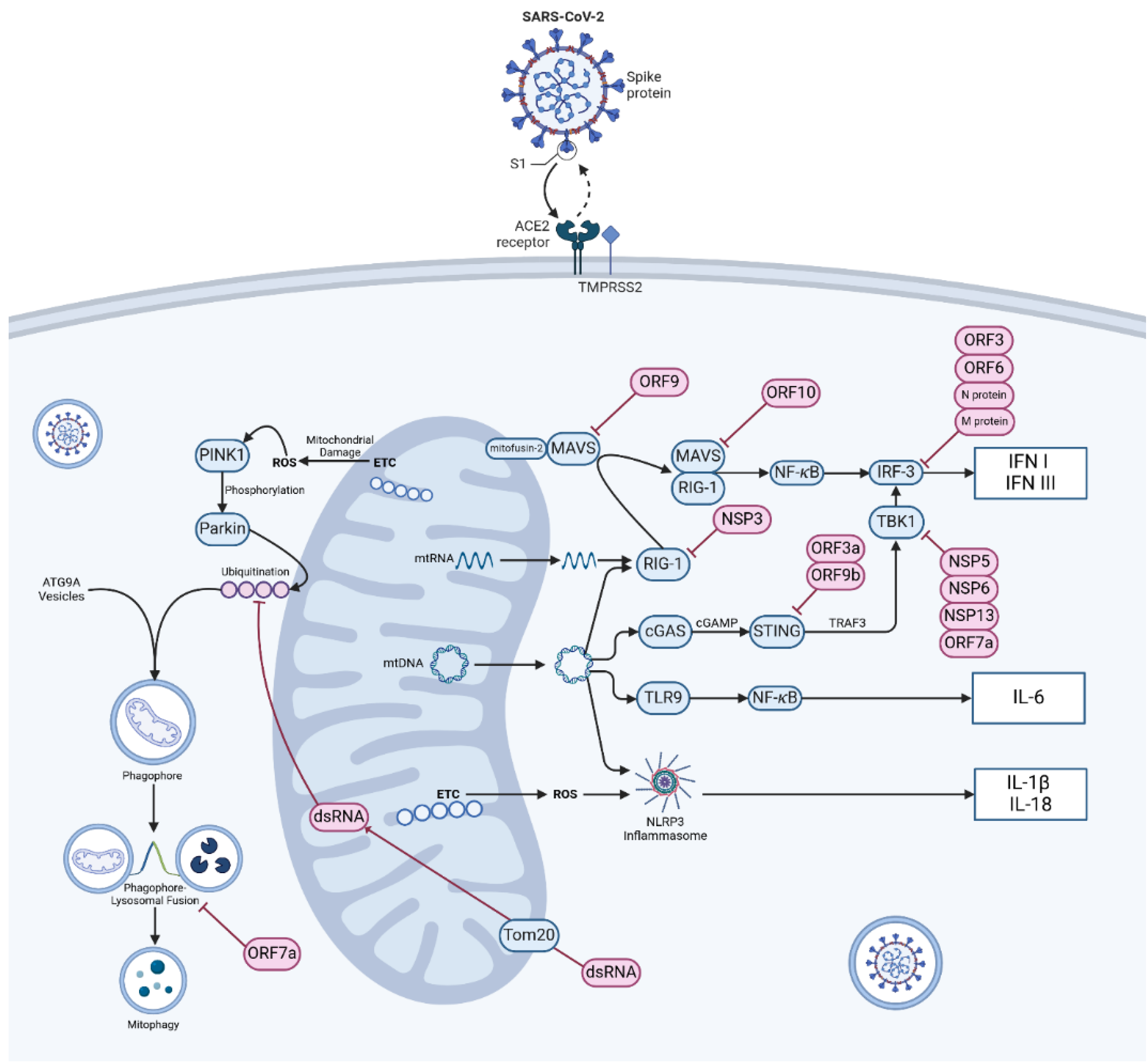
3. Autophagy, Mitochondrial Damage & Innate Immune Response
3.1. Autophagy & Mitophagy
3.2. Is Mitochondrial DNA A Warning Of An Ageing Immune System?
3.3. Mitochondrial Fusion & Fission
3.4. Mitochondrial Control of Inflammation
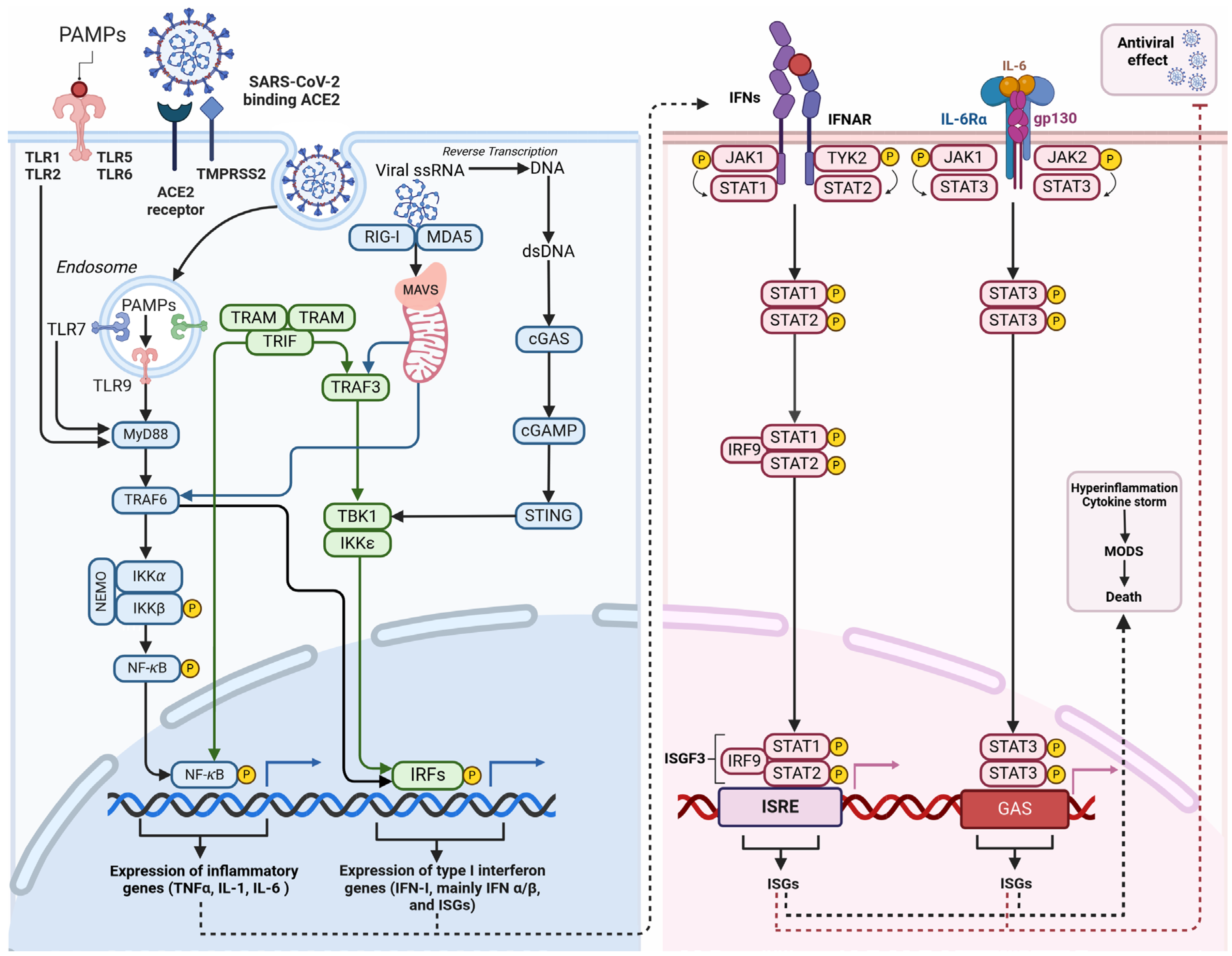
3.5. Viral Interference in Innate Immune Response
4. Alteration To Cellular Metabolism In SARS-CoV-2 Infection
4.1. Functional Change In Energy Production In Infected Cells
4.2. Immune System Impacts Of SARS-CoV-2 On Lipid Metabolism
4.3. Oxidative Stress
5. Mitochondrial Interference & mtDNA
5.1. Computational Modelling
5.2. Breakthroughs With Fluorescence Microscopy & Multi-Omics
5.3. mtDNA As A Prognostic Marker Of Acute COVID Infection?
5.4. Mitochondrial Dysfunction In COVID-19 Patients & Therapeutic Implications
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Post COVID-19 Condition (Long COVID) Available online:. Available online: https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition (accessed on 30 January 2024).
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-Acute COVID-19 Syndrome. Nat Med 2021, 27, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.L.; Catalfamo, C.J.; Farland, L.V.; Ernst, K.C.; Jacobs, E.T.; Klimentidis, Y.C.; Jehn, M.; Pogreba-Brown, K. Post-Acute Sequelae of COVID-19 in a Non-Hospitalized Cohort: Results from the Arizona CoVHORT. PLOS ONE 2021, 16, e0254347. [Google Scholar] [CrossRef] [PubMed]
- Overview | COVID-19 Rapid Guideline: Managing the Long-Term Effects of COVID-19 | Guidance | NICE Available online:. Available online: https://www.nice.org.uk/guidance/NG188 (accessed on 30 January 2024).
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major Findings, Mechanisms and Recommendations. Nat Rev Microbiol 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Lechner-Scott, J.; Levy, M.; Hawkes, C.; Yeh, A.; Giovannoni, G. Long COVID or Post COVID-19 Syndrome. Multiple Sclerosis and Related Disorders 2021, 55. [Google Scholar] [CrossRef] [PubMed]
- Tziolos, N.-R.; Ioannou, P.; Baliou, S.; Kofteridis, D.P. Long COVID-19 Pathophysiology: What Do We Know So Far? Microorganisms 2023, 11, 2458. [Google Scholar] [CrossRef] [PubMed]
- Haunhorst, S.; Bloch, W.; Wagner, H.; Ellert, C.; Krüger, K.; Vilser, D.C.; Finke, K.; Reuken, P.; Pletz, M.W.; Stallmach, A.; et al. Long COVID: A Narrative Review of the Clinical Aftermaths of COVID-19 with a Focus on the Putative Pathophysiology and Aspects of Physical Activity. Oxf Open Immunol 2022, 3. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Patel, K.; Greenwood, D.C.; Halpin, S.; Lewthwaite, P.; Salawu, A.; Eyre, L.; Breen, A.; O’Connor, R.; Jones, A.; et al. Long-Term Clinical Outcomes in Survivors of Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome Coronavirus Outbreaks after Hospitalisation or ICU Admission: A Systematic Review and Meta-Analysis. J Rehabil Med 2020, 52, jrm00063. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Joynt, G.M.; Wong, K.T.; Gomersall, C.D.; Li, T.S.; Antonio, G.; Ko, F.W.; Chan, M.C.; Chan, D.P.; Tong, M.W.; et al. Impact of Severe Acute Respiratory Syndrome (SARS) on Pulmonary Function, Functional Capacity and Quality of Life in a Cohort of Survivors. Thorax 2005, 60, 401–409. [Google Scholar] [CrossRef]
- Lam, M.H.-B.; Wing, Y.-K.; Yu, M.W.-M.; Leung, C.-M.; Ma, R.C.W.; Kong, A.P.S.; So, W.Y.; Fong, S.Y.-Y.; Lam, S.-P. Mental Morbidities and Chronic Fatigue in Severe Acute Respiratory Syndrome Survivors: Long-Term Follow-Up. Arch Intern Med 2009, 169, 2142–2147. [Google Scholar] [CrossRef]
- Lee, A.M.; Wong, J.G.W.S.; McAlonan, G.M.; Cheung, V.; Cheung, C.; Sham, P.C.; Chu, C.-M.; Wong, P.-C.; Tsang, K.W.T.; Chua, S.E. Stress and Psychological Distress among SARS Survivors 1 Year after the Outbreak. Can J Psychiatry 2007, 52, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Lancee, W.J.; Maunder, R.G.; Goldbloom, D.S. ; Coauthors for the Impact of SARS Study Prevalence of Psychiatric Disorders among Toronto Hospital Workers One to Two Years after the SARS Outbreak. Psychiatr Serv 2008, 59, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Shin, H.-S.; Park, H.Y.; Kim, J.L.; Lee, J.J.; Lee, H.; Won, S.-D.; Han, W. Depression as a Mediator of Chronic Fatigue and Post-Traumatic Stress Symptoms in Middle East Respiratory Syndrome Survivors. Psychiatry Investig 2019, 16, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Benatti, S.V.; Casati, M.; Binda, F.; Zuglian, G.; Imeri, G.; Conti, C.; Biffi, A.M.; Spada, M.S.; Bondi, E.; et al. Surviving COVID-19 in Bergamo Province: A Post-Acute Outpatient Re-Evaluation. Epidemiol Infect 2021, 149, e32. [Google Scholar] [CrossRef] [PubMed]
- Diem, L.; Fregolente-Gomes, L.; Warncke, J.D.; Hammer, H.; Friedli, C.; Kamber, N.; Jung, S.; Bigi, S.; Funke-Chambour, M.; Chan, A.; et al. Fatigue in Post-COVID-19 Syndrome: Clinical Phenomenology, Comorbidities and Association With Initial Course of COVID-19. J Cent Nerv Syst Dis 2022, 14, 11795735221102727. [Google Scholar] [CrossRef] [PubMed]
- Sahanic, S.; Tymoszuk, P.; Ausserhofer, D.; Rass, V.; Pizzini, A.; Nordmeyer, G.; Hüfner, K.; Kurz, K.; Weber, P.M.; Sonnweber, T.; et al. Phenotyping of Acute and Persistent Coronavirus Disease 2019 Features in the Outpatient Setting: Exploratory Analysis of an International Cross-Sectional Online Survey. Clin Infect Dis 2022, 75, e418–e431. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Ren, Y.; Lv, T. Encephalitis as a Clinical Manifestation of COVID-19. Brain Behav Immun 2020, 88, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Muccioli, L.; Pensato, U.; Cani, I.; Guarino, M.; Cortelli, P.; Bisulli, F. COVID-19-Associated Encephalopathy and Cytokine-Mediated Neuroinflammation. Ann Neurol 2020, 88, 860–861. [Google Scholar] [CrossRef] [PubMed]
- Maury, A.; Lyoubi, A.; Peiffer-Smadja, N.; de Broucker, T.; Meppiel, E. Neurological Manifestations Associated with SARS-CoV-2 and Other Coronaviruses: A Narrative Review for Clinicians. Rev Neurol (Paris) 2021, 177, 51–64. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat Rev Microbiol 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 Genome-Encoded Proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, R.; Lee, I.; Zhang, W.; Sun, J.; Wang, W.; Meng, X. Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Sci 2021, 30, 1114–1130. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; Zhao, Y.; Wang, W.; Wu, P.; Wang, Z.; Yu, Y.; Hou, Z.; Tan, G.; Liu, Q. SARS-CoV-2 Membrane Protein Inhibits Type I Interferon Production Through Ubiquitin-Mediated Degradation of TBK1. Front Immunol 2021, 12, 662989. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; De Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A Molecular Pore Spans the Double Membrane of the Coronavirus Replication Organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B.; Aleman, S.; Bach, K.; Boribong, B.P.; Buggert, M.; Cherry, S.; Chertow, D.S.; Davies, H.E.; Dupont, C.L.; et al. SARS-CoV-2 Reservoir in Post-Acute Sequelae of COVID-19 (PASC). Nature Immunology 2023, 24, 1616–1627. [Google Scholar] [CrossRef] [PubMed]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S.H. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res 2009, 16, 45–58. [Google Scholar] [CrossRef]
- Yang, E.; van Nimwegen, E.; Zavolan, M.; Rajewsky, N.; Schroeder, M.; Magnasco, M.; Darnell, J.E. Decay Rates of Human mRNAs: Correlation With Functional Characteristics and Sequence Attributes. Genome Res 2003, 13, 1863–1872. [Google Scholar] [CrossRef]
- Swank, Z.; Senussi, Y.; Manickas-Hill, Z.; Yu, X.G.; Li, J.Z.; Alter, G.; Walt, D.R. Persistent Circulating Severe Acute Respiratory Syndrome Coronavirus 2 Spike Is Associated With Post-Acute Coronavirus Disease 2019 Sequelae. Clin Infect Dis 2022, 76, e487–e490. [Google Scholar] [CrossRef] [PubMed]
- Craddock, V.; Mahajan, A.; Spikes, L.; Krishnamachary, B.; Ram, A.K.; Kumar, A.; Chen, L.; Chalise, P.; Dhillon, N.K. Persistent Circulation of Soluble and Extracellular Vesicle-linked Spike Protein in Individuals with Postacute Sequelae of COVID-19. J. Med. Virol. 2023, 95. [Google Scholar] [CrossRef]
- Chun Chau Lawrence Cheung; Denise Goh; Xinru Lim; Tracy Zhijun Tien; Jeffrey Chun Tatt Lim; Justina Nadia Lee; Benedict Tan; Zhi En Amos Tay; Wei Yee Wan; Eileen Xueqin Chen; et al. Residual SARS-CoV-2 Viral Antigens Detected in GI and Hepatic Tissues from Five Recovered Patients with COVID-19. Gut 2022, 71, 226. [Google Scholar] [CrossRef]
- Hany, M.; Zidan, A.; Gaballa, M.; Ibrahim, M.; Agayby, A.S.S.; Abouelnasr, A.A.; Sheta, E.; Torensma, B. Lingering SARS-CoV-2 in Gastric and Gallbladder Tissues of Patients with Previous COVID-19 Infection Undergoing Bariatric Surgery. Obes Surg 2023, 33, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Miura, C.S.; Lima, T.M.; Martins, R.B.; Jorge, D.M.M.; Tamashiro, E.; Anselmo-Lima, W.T.; Arruda, E.; Valera, F.C.P. Asymptomatic SARS-COV-2 Infection in Children’s Tonsils. Braz J Otorhinolaryngol 2022, 88, 9. [Google Scholar] [CrossRef]
- Stein, S.R.; Ramelli, S.C.; Grazioli, A.; Chung, J.-Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; et al. SARS-CoV-2 Infection and Persistence in the Human Body and Brain at Autopsy. Nature 2022, 612, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Mukerji, S.S.; Solomon, I.H. What Can We Learn from Brain Autopsies in COVID-19? Neurosci Lett 2021, 742, 135528. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Tan, B. What Can Cerebrospinal Fluid Testing and Brain Autopsies Tell Us about Viral Neuroinvasion of SARS-CoV-2. J Med Virol 2021, 93, 4247–4257. [Google Scholar] [CrossRef] [PubMed]
- Douaud, G.; Lee, S.; Alfaro-Almagro, F.; Arthofer, C.; Wang, C.; McCarthy, P.; Lange, F.; Andersson, J.L.R.; Griffanti, L.; Duff, E.; et al. SARS-CoV-2 Is Associated with Changes in Brain Structure in UK Biobank. Nature 2022, 604, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Roden, A.C.; Boland, J.M.; Johnson, T.F.; Aubry, M.C.; Lo, Y.-C.; Butt, Y.M.; Maleszewski, J.J.; Larsen, B.T.; Tazelaar, H.D.; Khoor, A.; et al. Late Complications of COVID-19A Morphologic, Imaging, and Droplet Digital Polymerase Chain Reaction Study of Lung Tissue. Arch Pathol Lab Med 2022, 146, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Zollner, A.; Koch, R.; Jukic, A.; Pfister, A.; Meyer, M.; Rössler, A.; Kimpel, J.; Adolph, T.E.; Tilg, H. Postacute COVID-19 Is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 2022, 163, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Goh, D.; Lim, J.C.T.; Fernaíndez, S.B.; Joseph, C.R.; Edwards, S.G.; Neo, Z.W.; Lee, J.N.; Caballero, S.G.; Lau, M.C.; Yeong, J.P.S. Case Report: Persistence of Residual Antigen and RNA of the SARS-CoV-2 Virus in Tissues of Two Patients with Long COVID. Frontiers in Immunology 2022, 13. [Google Scholar]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Huo, Y.; Sawant, A.; Tan, Y.; Mahdi, A.H.; Li, T.; Ma, H.; Bhatt, V.; Yan, R.; Coleman, J.; Dreyfus, C.F.; et al. Tumor Suppressor PALB2 Maintains Redox and Mitochondrial Homeostasis in the Brain and Cooperates with ATG7/Autophagy to Suppress Neurodegeneration. PLOS Genetics 2022, 18, e1010138. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.D.; Galle-Treger, L.; Akbari, O. Role of Autophagy in Lung Inflammation. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Mechanisms and Functions of Inflammasomes. Cell 2014, 157, 1013–1022. [Google Scholar] [CrossRef]
- Deretic, V.; Levine, B. Autophagy Balances Inflammation in Innate Immunity. Autophagy 2018, 14, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-C.; Jha, S.; Linhoff, M.W.; Ting, J.P.-Y. The NLR Gene Family: From Discovery to Present Day. Nat Rev Immunol 2023, 23, 635–654. [Google Scholar] [CrossRef]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.-J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy Proteins Regulate Innate Immune Response by Inhibiting NALP3 Inflammasome-Mediated Mitochondrial DNA Release. Nat Immunol 2011, 12, 222–230. [Google Scholar] [CrossRef]
- Poon, A.; Saini, H.; Sethi, S.; O’Sullivan, G.A.; Plun-Favreau, H.; Wray, S.; Dawson, L.A.; McCarthy, J.M. The Role of SQSTM1 (P62) in Mitochondrial Function and Clearance in Human Cortical Neurons. Stem Cell Reports 2021, 16, 1276–1289. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The Ubiquitin Kinase PINK1 Recruits Autophagy Receptors to Induce Mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- Yamano, K.; Wang, C.; Sarraf, S.A.; Münch, C.; Kikuchi, R.; Noda, N.N.; Hizukuri, Y.; Kanemaki, M.T.; Harper, W.; Tanaka, K.; et al. Endosomal Rab Cycles Regulate Parkin-Mediated Mitophagy. eLife 7. [CrossRef]
- Lim, J.; Lachenmayer, M.L.; Wu, S.; Liu, W.; Kundu, M.; Wang, R.; Komatsu, M.; Oh, Y.J.; Zhao, Y.; Yue, Z. Proteotoxic Stress Induces Phosphorylation of P62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates. PLoS Genet 2015, 11, e1004987. [Google Scholar] [CrossRef]
- Geisler, S.; Holmström, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-Mediated Mitophagy Is Dependent on VDAC1 and P62/SQSTM1. Nat Cell Biol 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Shadel, G.S. Mitochondrial DNA in Innate Immune Responses and Inflammatory Pathology. Nat Rev Immunol 2017, 17, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.C.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, K.; Hisata, S.; Choi, A.M.K. The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxid Redox Signal 2015, 23, 1329–1350. [Google Scholar] [CrossRef] [PubMed]
- Pinti, M.; Cevenini, E.; Nasi, M.; De Biasi, S.; Salvioli, S.; Monti, D.; Benatti, S.; Gibellini, L.; Cotichini, R.; Stazi, M.A.; et al. Circulating Mitochondrial DNA Increases with Age and Is a Familiar Trait: Implications for “Inflamm-Aging. ” Eur J Immunol 2014, 44, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Guerra Martinez, C.; Torres-Odio, S.; Bell, S.L.; Birdwell, C.E.; Bryant, J.D.; Tong, C.W.; Watson, R.O.; West, L.C.; West, A.P. Elevated Type I Interferon Responses Potentiate Metabolic Dysfunction, Inflammation, and Accelerated Aging in mtDNA Mutator Mice. Science Advances 2021, 7, eabe7548. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Otsu, K. Mitochondrial DNA as an Inflammatory Mediator in Cardiovascular Diseases. Biochem J 2018, 475, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Guilbaud, E.; Tait, S.W.G.; Yamazaki, T.; Galluzzi, L. Mitochondrial Control of Inflammation. Nat Rev Immunol 2023, 23, 159–173. [Google Scholar] [CrossRef]
- Orekhov, A.N.; Nikiforov, N.N.; Ivanova, E.A.; Sobenin, I.A. Possible Role of Mitochondrial DNA Mutations in Chronification of Inflammation: Focus on Atherosclerosis. J Clin Med 2020, 9, 978. [Google Scholar] [CrossRef]
- Peripheral Artery Disease, Calf Skeletal Muscle Mitochondrial DNA Copy Number, and Functional Performance - Mary M McDermott, Charlotte A Peterson, Robert Sufit, Luigi Ferrucci, Jack M Guralnik, Melina R Kibbe, Tamar S Polonsky, Lu Tian, Michael H Criqui, Lihui Zhao, James H Stein, Lingyu Li, Christiaan Leeuwenburgh, 2018 Available online:. Available online: https://journals.sagepub.com/doi/10.1177/1358863X18765667 (accessed on 7 May 2024).
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 Coordinately Regulate Mitochondrial Fusion and Are Essential for Embryonic Development. J Cell Biol 2003, 160, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, L.; Scorrano, L. A Cut Short to Death: Parl and Opa1 in the Regulation of Mitochondrial Morphology and Apoptosis. Cell Death Differ 2007, 14, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, E. OPA1 and PARL Keep a Lid on Apoptosis. Cell 2006, 126, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol Biol Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chan, D.C. Emerging Functions of Mammalian Mitochondrial Fusion and Fission. Hum Mol Genet 2005, 14 Spec No. 2, R283–289. [Google Scholar] [CrossRef]
- Griffin, E.E.; Detmer, S.A.; Chan, D.C. Molecular Mechanism of Mitochondrial Membrane Fusion. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2006, 1763, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Knott, A.B.; Perkins, G.; Schwarzenbacher, R.; Bossy-Wetzel, E. Mitochondrial Fragmentation in Neurodegeneration. Nat Rev Neurosci 2008, 9, 505–518. [Google Scholar] [CrossRef]
- Wakabayashi, T. Megamitochondria Formation - Physiology and Pathology. J Cell Mol Med 2002, 6, 497–538. [Google Scholar] [CrossRef] [PubMed]
- Principles of the Mitochondrial Fusion and Fission Cycle in Neurons - PubMed Available online:. Available online: https://pubmed.ncbi.nlm.nih.gov/23525002/ (accessed on 13 March 2024).
- Shang, Y.; Li, Z.; Cai, P.; Li, W.; Xu, Y.; Zhao, Y.; Xia, S.; Shao, Q.; Wang, H. Megamitochondria Plasticity: Function Transition from Adaption to Disease. Mitochondrion 2023, 71, 64–75. [Google Scholar] [CrossRef]
- Grel, H.; Woznica, D.; Ratajczak, K.; Kalwarczyk, E.; Anchimowicz, J.; Switlik, W.; Olejnik, P.; Zielonka, P.; Stobiecka, M.; Jakiela, S. Mitochondrial Dynamics in Neurodegenerative Diseases: Unraveling the Role of Fusion and Fission Processes. International Journal of Molecular Sciences 2023, 24, 13033. [Google Scholar] [CrossRef]
- Holder, K.; Reddy, P.H. The COVID-19 Effect on the Immune System and Mitochondrial Dynamics in Diabetes, Obesity, and Dementia. The Neuroscientist 2020. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Lee, W.; Ku, K.B.; Yoon, G.Y.; Moon, H.-W.; Kim, C.; Kim, M.-H.; Yi, Y.-S.; Jun, S.; Kim, B.-T.; et al. SARS-CoV-2 Aberrantly Elevates Mitochondrial Bioenergetics to Induce Robust Virus Propagation. Sig Transduct Target Ther 2024, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liu, J.-J.; Ma, S.-L. Analysis of Mitochondrial Function in Lymphocytes Obtained from COVID-19 Patients. Int J Immunopathol Pharmacol 2023, 37, 03946320231210736. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Chen, Y.-F.; Chen, Y.-F.; Chung, L.-C.; Tamilselvi, S.; Shen, C.-Y.; Day, C.H.; Chen, R.-J.; Viswanadha, V.P.; Kuo, W.-W.; et al. The Multifaceted Link between Inflammation and Human Diseases. Journal of Cellular Physiology 2018, 233, 6458–6471. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Marzetti, E. Cell Death and Inflammation: The Role of Mitochondria in Health and Disease. Cells 2021, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Bowie, A.G. Immune Sensing of DNA. Immunity 2013, 38, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating Mitochondrial DAMPs Cause Inflammatory Responses to Injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. Cytoplasmic DNA Innate Immune Pathways. Immunological Reviews 2011, 243, 99–108. [Google Scholar] [CrossRef] [PubMed]
- T, O.; S, H.; O, Y.; M, T.; T, T.; T, T.; J, O.; T, M.; H, N.; K, N.; et al. Mitochondrial DNA That Escapes from Autophagy Causes Inflammation and Heart Failure. Nature 2012, 485. [Google Scholar] [CrossRef]
- Hu, M.-M.; Shu, H.-B. Mitochondrial DNA-Triggered Innate Immune Response: Mechanisms and Diseases. Cell Mol Immunol 2023, 20, 1403–1412. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, M.; Yuan, C.; Ma, Z.; Li, W.; Zhang, Y.; Su, L.; Xu, J.; Liu, W. Progress of cGAS-STING Signaling in Response to SARS-CoV-2 Infection. Front Immunol 2022, 13, 1010911. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, C.; Liao, Z.; Xu, P. cGAS-STING Signaling in Cell Death: Mechanisms of Action and Implications in Pathologies. European Journal of Immunology 2023, 53, 2350386. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Chen, J.; Zhao, L.; Wu, S.; Chen, X.; Chen, H. Mitochondrial DNA Leakage Triggers Inflammation in Age-Related Cardiovascular Diseases. Front. Cell Dev. Biol. 2024, 12. [Google Scholar] [CrossRef] [PubMed]
- Jabir, M.S.; Hopkins, L.; Ritchie, N.D.; Ullah, I.; Bayes, H.K.; Li, D.; Tourlomousis, P.; Lupton, A.; Puleston, D.; Simon, A.K.; et al. Mitochondrial Damage Contributes to Pseudomonas Aeruginosa Activation of the Inflammasome and Is Downregulated by Autophagy. Autophagy 2015, 11, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wang, W.; Lu, L.; Wang, L.; Chen, X.; Guo, R.; Li, S.; Jiang, J. Inactivation of Beclin-1-Dependent Autophagy Promotes Ursolic Acid-Induced Apoptosis in Hypertrophic Scar Fibroblasts. Experimental Dermatology 2018, 27, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Scozzi, D.; Cano, M.; Ma, L.; Zhou, D.; Zhu, J.H.; O’Halloran, J.A.; Goss, C.; Rauseo, A.M.; Liu, Z.; Sahu, S.K.; et al. Circulating Mitochondrial DNA Is an Early Indicator of Severe Illness and Mortality from COVID-19. JCI Insight 2021, 6, e143299–143299. [Google Scholar] [CrossRef]
- Gilkerson, R.W.; De Vries, R.L.A.; Lebot, P.; Wikstrom, J.D.; Torgyekes, E.; Shirihai, O.S.; Przedborski, S.; Schon, E.A. Mitochondrial Autophagy in Cells with mtDNA Mutations Results from Synergistic Loss of Transmembrane Potential and mTORC1 Inhibition. Hum Mol Genet 2012, 21, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, J.; Zhang, X.; Li, X.; Wu, X.; Zhao, Y.; Ren, J. Circulating Mitochondrial DNA-Triggered Autophagy Dysfunction via STING Underlies Sepsis-Related Acute Lung Injury. Cell Death Dis 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Rodgers, M.A.; Bowman, J.W.; Liang, Q.; Jung, J.U. Regulation Where Autophagy Intersects the Inflammasome. Antioxid Redox Signal 2014, 20, 495–506. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, Y.; Wu, J.; Qi, N. MAVS: A Two-Sided CARD Mediating Antiviral Innate Immune Signaling and Regulating Immune Homeostasis. Front Microbiol 2021, 12, 744348. [Google Scholar] [CrossRef]
- Vazquez, C.; Horner, S.M. MAVS Coordination of Antiviral Innate Immunity. J Virol 2015, 89, 6974–6977. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Q.; Pan, Y.; Wang, C. Sensing and Responding to Cytosolic Viruses Invasions: An Orchestra of Kaleidoscopic Ubiquitinations. Cytokine & Growth Factor Reviews 2015, 26, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J Virol 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Duan, L.; Huang, B.; Xiong, Y.; Zhang, G.; Huang, H. The SARS-CoV-2 Papain-like Protease Suppresses Type I Interferon Responses by Deubiquitinating STING. Sci Signal 2023, 16, eadd0082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhang, M.; Yang, Z.; Zhou, Z.; Huang, J.; Zhao, B. Role of E3 Ubiquitin Ligases and Deubiquitinating Enzymes in SARS-CoV-2 Infection. Front Cell Infect Microbiol 2023, 13, 1217383. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, X.; Chang, M.; Xue, Z.; Wang, W.; Bai, L.; Zhao, S.; Liu, E. ORF3a Protein of Severe Acute Respiratory Syndrome Coronavirus 2 Inhibits Interferon-Activated Janus Kinase/Signal Transducer and Activator of Transcription Signaling via Elevating Suppressor of Cytokine Signaling 1. Front Microbiol 2021, 12, 752597. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, H.; Meng, Q.; Xie, J.; Li, Y.; Chen, H.; Zheng, Y.; Wang, X.; Qi, H.; Zhang, J.; et al. SARS-CoV-2 Orf9b Suppresses Type I Interferon Responses by Targeting TOM70. Cell Mol Immunol 2020, 17, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, P.C.; Behrmann, I.; Haan, S.; Hermanns, H.M.; Müller-Newen, G.; Schaper, F. Principles of Interleukin (IL)-6-Type Cytokine Signalling and Its Regulation. Biochem J 2003, 374, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Y.; Khan, N.; Close, B.J.; Goel, R.K.; Blum, B.; Tavares, A.H.; Kenney, D.; Conway, H.L.; Ewoldt, J.K.; Chitalia, V.C.; et al. SARS-CoV-2 Disrupts Proximal Elements in the JAK-STAT Pathway. J Virol 95. [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000. [Google Scholar] [CrossRef]
- Onoja, A.; von Gerichten, J.; Lewis, H.-M.; Bailey, M.J.; Skene, D.J.; Geifman, N.; Spick, M. Meta-Analysis of COVID-19 Metabolomics Identifies Variations in Robustness of Biomarkers. International Journal of Molecular Sciences 2023, 24, 14371. [Google Scholar] [CrossRef]
- Casari, I.; Manfredi, M.; Metharom, P.; Falasca, M. Dissecting Lipid Metabolism Alterations in SARS-CoV-2. Prog Lipid Res 2021, 82, 101092. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; García de Vicuña, A.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef] [PubMed]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics Analysis Reveals a Modified Amino Acid Metabolism That Correlates with Altered Oxygen Homeostasis in COVID-19 Patients. Sci Rep 2021, 11, 6350. [Google Scholar] [CrossRef] [PubMed]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L. de B.; Souza, G.F. de; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; Junior, C.A.O. de B.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metabolism 2020, 32, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Ajaz, S.; McPhail, M.J.; Singh, K.K.; Mujib, S.; Trovato, F.M.; Napoli, S.; Agarwal, K. Mitochondrial Metabolic Manipulation by SARS-CoV-2 in Peripheral Blood Mononuclear Cells of Patients with COVID-19. Am J Physiol Cell Physiol 2021, 320, C57–C65. [Google Scholar] [CrossRef] [PubMed]
- Langston, P.K.; Shibata, M.; Horng, T. Metabolism Supports Macrophage Activation. Front Immunol 2017, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D. M1 and M2 Macrophages: Oracles of Health and Disease. Crit Rev Immunol 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Lincet, H.; Wu, Z.; Coquerel, A.; Forgez, P.; Alifano, M.; Fournel, L. The Key Role of Warburg Effect in SARS-CoV-2 Replication and Associated Inflammatory Response. Biochimie 2021, 180, 169–177. [Google Scholar] [CrossRef]
- Moreno-Altamirano, M.M.B.; Kolstoe, S.E.; Sánchez-García, F.J. Virus Control of Cell Metabolism for Replication and Evasion of Host Immune Responses. Front Cell Infect Microbiol 2019, 9, 95. [Google Scholar] [CrossRef]
- Clayton, S.A.; Daley, K.K.; MacDonald, L.; Fernandez-Vizarra, E.; Bottegoni, G.; O’Neil, J.D.; Major, T.; Griffin, D.; Zhuang, Q.; Adewoye, A.B.; et al. Inflammation Causes Remodeling of Mitochondrial Cytochrome c Oxidase Mediated by the Bifunctional Gene C15orf48. Sci Adv 7. [CrossRef]
- Floyd, B.J.; Wilkerson, E.M.; Veling, M.T.; Minogue, C.E.; Xia, C.; Beebe, E.T.; Wrobel, R.L.; Cho, H.; Kremer, L.S.; Alston, C.L.; et al. Mitochondrial Protein Interaction Mapping Identifies New Regulators of Respiratory Chain Function. Mol Cell 2016, 63, 621–632. [Google Scholar] [CrossRef]
- Wise, D.R.; Ward, P.S.; Shay, J.E.S.; Cross, J.R.; Gruber, J.J.; Sachdeva, U.M.; Platt, J.M.; DeMatteo, R.G.; Simon, M.C.; Thompson, C.B. Hypoxia Promotes Isocitrate Dehydrogenase-Dependent Carboxylation of α-Ketoglutarate to Citrate to Support Cell Growth and Viability. Proc Natl Acad Sci U S A 2011, 108, 19611–19616. [Google Scholar] [CrossRef]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 Mediates Adaptation to Hypoxia by Actively Downregulating Mitochondrial Oxygen Consumption. Cell Metab 2006, 3, 187–197. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Soto Albrecht, Y.; Murdock, D.G.; Angelin, A.; Singh, L.N.; et al. Core Mitochondrial Genes Are Down-Regulated during SARS-CoV-2 Infection of Rodent and Human Hosts. Sci Transl Med 2023, 15, eabq1533. [Google Scholar] [CrossRef]
- Lu, H.; Koju, N.; Sheng, R. Mammalian Integrated Stress Responses in Stressed Organelles and Their Functions. Acta Pharmacol Sin 2024, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Costa-Mattioli, M.; Walter, P. The Integrated Stress Response: From Mechanism to Disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.-H.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial Dysfunction Is Signaled to the Integrated Stress Response by OMA1, DELE1 and HRI 2019, 715896.
- García, M.A.; Meurs, E.F.; Esteban, M. The dsRNA Protein Kinase PKR: Virus and Cell Control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, P.J.; Wang, F.; Du, Y.; Liu, Q.; Pickard, R.T.; Gonciarz, M.D.; Coskun, T.; Hamang, M.J.; Sindelar, D.K.; Ballman, K.K.; et al. The Metabolic Effects of GDF15 Are Mediated by the Orphan Receptor GFRAL. Nat Med 2017, 23, 1215–1219. [Google Scholar] [CrossRef]
- Johann, K.; Kleinert, M.; Klaus, S. The Role of GDF15 as a Myomitokine. Cells 2021, 10, 2990. [Google Scholar] [CrossRef]
- Song, J.-W.; Lam, S.M.; Fan, X.; Cao, W.-J.; Wang, S.-Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.-P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab 2020, 32, 188–202. [Google Scholar] [CrossRef]
- Hong, W. Combating COVID-19 with Chloroquine. J Mol Cell Biol 2020, 12, 249–250. [Google Scholar] [CrossRef]
- Kornhuber, J.; Hoertel, N.; Gulbins, E. The Acid Sphingomyelinase/Ceramide System in COVID-19. Mol Psychiatry 2022, 27, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Carpinteiro, A.; Gripp, B.; Hoffmann, M.; Pöhlmann, S.; Hoertel, N.; Edwards, M.J.; Kamler, M.; Kornhuber, J.; Becker, K.A.; Gulbins, E. Inhibition of Acid Sphingomyelinase by Ambroxol Prevents SARS-CoV-2 Entry into Epithelial Cells. Journal of Biological Chemistry 2021, 296. [Google Scholar] [CrossRef]
- Hu, X.; Chen, D.; Wu, L.; He, G.; Ye, W. Declined Serum High Density Lipoprotein Cholesterol Is Associated with the Severity of COVID-19 Infection. Clinica Chimica Acta 2020, 510, 105–110. [Google Scholar] [CrossRef]
- Masana, L.; Correig, E.; Ibarretxe, D.; Anoro, E.; Arroyo, J.A.; Jericó, C.; Guerrero, C.; Miret, M.; Näf, S.; Pardo, A.; et al. Low HDL and High Triglycerides Predict COVID-19 Severity. Sci Rep 2021, 11, 7217. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-H. The Current Status of Research on High-Density Lipoproteins (HDL): A Paradigm Shift from HDL Quantity to HDL Quality and HDL Functionality. Int J Mol Sci 2022, 23, 3967. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Kong, S.Y.; Ro, Y.S.; Ryu, H.H.; Shin, S.D. Serum Cholesterol Levels and Risk of Cardiovascular Death: A Systematic Review and a Dose-Response Meta-Analysis of Prospective Cohort Studies. Int J Environ Res Public Health 2022, 19, 8272. [Google Scholar] [CrossRef]
- Filippas-Ntekouan, S.; Liberopoulos, E.; Elisaf, M. Lipid Testing in Infectious Diseases: Possible Role in Diagnosis and Prognosis. Infection 2017, 45, 575–588. [Google Scholar] [CrossRef]
- Trinder, M.; Boyd, J.H.; Brunham, L.R. Molecular Regulation of Plasma Lipid Levels during Systemic Inflammation and Sepsis. Current Opinion in Lipidology 2019, 30, 108. [Google Scholar] [CrossRef]
- Zewinger, S.; Drechsler, C.; Kleber, M.E.; Dressel, A.; Riffel, J.; Triem, S.; Lehmann, M.; Kopecky, C.; Säemann, M.D.; Lepper, P.M.; et al. Serum Amyloid A: High-Density Lipoproteins Interaction and Cardiovascular Risk. Eur Heart J 2015, 36, 3007–3016. [Google Scholar] [CrossRef]
- Lee, W.; Ahn, J.H.; Park, H.H.; Kim, H.N.; Kim, H.; Yoo, Y.; Shin, H.; Hong, K.S.; Jang, J.G.; Park, C.G.; et al. COVID-19-Activated SREBP2 Disturbs Cholesterol Biosynthesis and Leads to Cytokine Storm. Signal Transduct Target Ther 2020, 5, 186. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zeng, W.; Su, J.; Wan, H.; Yu, X.; Cao, X.; Tan, W.; Wang, H. Hypolipidemia Is Associated with the Severity of COVID-19. J Clin Lipidol 2020, 14, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, J.; Sampieri, K.; Clohessy, J.G.; Mendez, L.; Gonzalez-Billalabeitia, E.; Liu, X.-S.; Lee, Y.-R.; Fung, J.; Katon, J.M.; et al. An Aberrant SREBP-Dependent Lipogenic Program Promotes Metastatic Prostate Cancer. Nat Genet 2018, 50, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Chi, Z.; Jiang, D.; Xu, T.; Yu, W.; Wang, Z.; Chen, S.; Zhang, L.; Liu, Q.; Guo, X.; et al. Cholesterol Homeostatic Regulator SCAP-SREBP2 Integrates NLRP3 Inflammasome Activation and Cholesterol Biosynthetic Signaling in Macrophages. Immunity 2018, 49, 842–856. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int J Mol Sci 2020, 21, 3544. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Czajkowsky, D.M. SARS-CoV-2 Infection and Oxidative Stress: Pathophysiological Insight into Thrombosis and Therapeutic Opportunities. Cytokine Growth Factor Rev 2022, 63, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Tekos, F.; Skaperda, Z.; Goutzourelas, N.; Phelps, D.S.; Floros, J.; Kouretas, D. The Importance of Redox Status in the Frame of Lifestyle Approaches and the Genetics of the Lung Innate Immune Molecules, SP-A1 and SP-A2, on Differential Outcomes of COVID-19 Infection. Antioxidants (Basel) 2020, 9, 784. [Google Scholar] [CrossRef] [PubMed]
- Gain, C.; Song, S.; Angtuaco, T.; Satta, S.; Kelesidis, T. The Role of Oxidative Stress in the Pathogenesis of Infections with Coronaviruses. Front. Microbiol. 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Forcados, G.E.; Muhammad, A.; Oladipo, O.O.; Makama, S.; Meseko, C.A. Metabolic Implications of Oxidative Stress and Inflammatory Process in SARS-CoV-2 Pathogenesis: Therapeutic Potential of Natural Antioxidants. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Meyer, K.; Patra, T.; Vijayamahantesh, null; Ray, R. SARS-CoV-2 Spike Protein Induces Paracrine Senescence and Leukocyte Adhesion in Endothelial Cells. J Virol 2021, 95, e0079421. [Google Scholar] [CrossRef]
- Muhammad, Y.; Kani, Y.A.; Iliya, S.; Muhammad, J.B.; Binji, A.; El-Fulaty Ahmad, A.; Kabir, M.B.; Umar Bindawa, K.; Ahmed, A. Deficiency of Antioxidants and Increased Oxidative Stress in COVID-19 Patients: A Cross-Sectional Comparative Study in Jigawa, Northwestern Nigeria. SAGE Open Med 2021, 9, 2050312121991246. [Google Scholar] [CrossRef] [PubMed]
- Duca, L.; Ottolenghi, S.; Coppola, S.; Rinaldo, R.; Dei Cas, M.; Rubino, F.M.; Paroni, R.; Samaja, M.; Chiumello, D.A.; Motta, I. Differential Redox State and Iron Regulation in Chronic Obstructive Pulmonary Disease, Acute Respiratory Distress Syndrome and Coronavirus Disease 2019. Antioxidants (Basel) 2021, 10, 1460. [Google Scholar] [CrossRef] [PubMed]
- McCracken, I.R.; Saginc, G.; He, L.; Huseynov, A.; Daniels, A.; Fletcher, S.; Peghaire, C.; Kalna, V.; Andaloussi-Mäe, M.; Muhl, L.; et al. Lack of Evidence of Angiotensin-Converting Enzyme 2 Expression and Replicative Infection by SARS-CoV-2 in Human Endothelial Cells. Circulation 2021, 143, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.M.; Levy, J.H. COVID-19 and Its Implications for Thrombosis and Anticoagulation. Blood 2020, 135, 2033–2040. [Google Scholar] [CrossRef]
- Vollbracht, C.; Kraft, K. Oxidative Stress and Hyper-Inflammation as Major Drivers of Severe COVID-19 and Long COVID: Implications for the Benefit of High-Dose Intravenous Vitamin C. Front Pharmacol 2022, 13, 899198. [Google Scholar] [CrossRef] [PubMed]
- Mikerov, A.N.; Umstead, T.M.; Gan, X.; Huang, W.; Guo, X.; Wang, G.; Phelps, D.S.; Floros, J. Impact of Ozone Exposure on the Phagocytic Activity of Human Surfactant Protein A (SP-A) and SP-A Variants. Am J Physiol Lung Cell Mol Physiol 2008, 294, L121–L130. [Google Scholar] [CrossRef] [PubMed]
- George, P.M.; Wells, A.U.; Jenkins, R.G. Pulmonary Fibrosis and COVID-19: The Potential Role for Antifibrotic Therapy. The Lancet Respiratory Medicine 2020, 8, 807–815. [Google Scholar] [CrossRef]
- Wu, K.E.; Fazal, F.M.; Parker, K.R.; Zou, J.; Chang, H.Y. RNA-GPS Predicts SARS-CoV-2 RNA Residency to Host Mitochondria and Nucleolus. Cell Syst 2020, 11, 102–108. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug-Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Shi, C.-S.; Qi, H.-Y.; Boularan, C.; Huang, N.-N.; Abu-Asab, M.; Shelhamer, J.H.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. J Immunol 2014, 193, 3080–3089. [Google Scholar] [CrossRef]
- Medini, H.; Zirman, A.; Mishmar, D. Immune System Cells from COVID-19 Patients Display Compromised Mitochondrial-Nuclear Expression Co-Regulation and Rewiring toward Glycolysis. iScience 2021, 24, 103471. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.; Silverstein, A.; Flores, M.; Cao, K.; Kumagai, H.; Mehta, H.H.; Yen, K.; Kim, S.-J.; Cohen, P. Host Mitochondrial Transcriptome Response to SARS-CoV-2 in Multiple Cell Models and Clinical Samples. Sci Rep 2021, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hou, P.; Ma, W.; Wang, X.; Wang, H.; Yu, Z.; Chang, H.; Wang, T.; Jin, S.; Wang, X.; et al. SARS-CoV-2 ORF10 Suppresses the Antiviral Innate Immune Response by Degrading MAVS through Mitophagy. Cell Mol Immunol 2022, 19, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front Microbiol 2021, 12, 780768. [Google Scholar] [CrossRef] [PubMed]
- Budzińska, M.; Gałgańska, H.; Karachitos, A.; Wojtkowska, M.; Kmita, H. The TOM Complex Is Involved in the Release of Superoxide Anion from Mitochondria. J Bioenerg Biomembr 2009, 41, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, A.; Oldani, M.; Forcella, M.E.; Vantaggiato, C.; Cappelletti, G.; Pontremoli, C.; Valenti, F.; Forni, D.; Saresella, M.; Biasin, M.; et al. SARS-CoV-2 ORF3c Impairs Mitochondrial Respiratory Metabolism, Oxidative Stress, and Autophagic Flux. iScience 2023, 26, 107118. [Google Scholar] [CrossRef] [PubMed]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Schneck, E.; Edinger, F.; Hecker, M.; Sommer, N.; Pak, O.; Weissmann, N.; Hecker, A.; Reichert, M.; Markmann, M.; Sander, M.; et al. Blood Levels of Free-Circulating Mitochondrial DNA in Septic Shock and Postsurgical Systemic Inflammation and Its Influence on Coagulation: A Secondary Analysis of a Prospective Observational Study. J Clin Med 2020, 9, 2056. [Google Scholar] [CrossRef] [PubMed]
- Edinger, F.; Edinger, S.; Koch, C.; Markmann, M.; Hecker, M.; Sander, M.; Schneck, E. Peak Plasma Levels of mtDNA Serve as a Predictive Biomarker for COVID-19 in-Hospital Mortality. J Clin Med 2022, 11, 7161. [Google Scholar] [CrossRef]
- Denning, N.-L.; Aziz, M.; Gurien, S.D.; Wang, P. DAMPs and NETs in Sepsis. Front Immunol 2019, 10, 2536. [Google Scholar] [CrossRef]
- Valdés-Aguayo, J.J.; Garza-Veloz, I.; Vargas-Rodríguez, J.R.; Martinez-Vazquez, M.C.; Avila-Carrasco, L.; Bernal-Silva, S.; González-Fuentes, C.; Comas-García, A.; Alvarado-Hernández, D.E.; Centeno-Ramirez, A.S.H.; et al. Peripheral Blood Mitochondrial DNA Levels Were Modulated by SARS-CoV-2 Infection Severity and Its Lessening Was Associated With Mortality Among Hospitalized Patients With COVID-19. Front Cell Infect Microbiol 2021, 11, 754708. [Google Scholar] [CrossRef] [PubMed]
- Bľandová, G.; Janoštiaková, N.; Kodada, D.; Pastorek, M.; Lipták, R.; Hodosy, J.; Šebeková, K.; Celec, P.; Krasňanská, G.; Eliaš, V.; et al. Mitochondrial DNA Variability and Covid-19 in the Slovak Population. Mitochondrion 2024, 75, 101827. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.-H.; Li, X.-H.; Song, L.-Y.; Yu, S.-L.; Fang, Z.-C.; Liu, Y.-Q.; Yuan, L.-Y.; Peng, C.-Y.; Zhang, S.-Y.; et al. Common mtDNA Variations at C5178a and A249d/T6392C/G10310A Decrease the Risk of Severe COVID-19 in a Han Chinese Population from Central China. Military Med Res 2021, 8, 57. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Chu, P.-Y. Advances in Understanding Mitochondrial MicroRNAs (mitomiRs) on the Pathogenesis of Triple-Negative Breast Cancer (TNBC). Oxid Med Cell Longev 2021, 2021, 5517777. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Research 2019, 79, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lu, Y.; Zhang, H.; Zhang, J.; Fang, X.; Wang, J.; Li, M. Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules 2022, 12, 1863. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.T.; Enguita, F.J.; Taylor, D.; Griffin, R.J.; Priebe, W.; Emmett, M.R.; Sajadi, M.M.; Harris, A.D.; Clement, J.; Dybas, J.M.; et al. Role of miR-2392 in Driving SARS-CoV-2 Infection. Cell Rep 2021, 37, 109839. [Google Scholar] [CrossRef]
- Olivieri, F.; Prattichizzo, F.; Giuliani, A.; Matacchione, G.; Rippo, M.R.; Sabbatinelli, J.; Bonafè, M. miR-21 and miR-146a: The microRNAs of Inflammaging and Age-Related Diseases. Ageing Research Reviews 2021, 70, 101374. [Google Scholar] [CrossRef]
- Ballinas-Verdugo, M.A.; Jiménez-Ortega, R.F.; Martínez-Martínez, E.; Rivas, N.; Contreras-López, E.A.; Carbó, R.; Sánchez, F.; Bojalil, R.; Márquez-Velasco, R.; Sánchez-Muñoz, F.; et al. Circulating miR-146a as a Possible Candidate Biomarker in the Indeterminate Phase of Chagas Disease. Biological Research 2021, 54. [Google Scholar] [CrossRef]
- Ouyang, L.; Gong, J. Mitochondrial-Targeted Ubiquinone: A Potential Treatment for COVID-19. Medical Hypotheses 2020, 144, 110161. [Google Scholar] [CrossRef]
- Finnigan, L.E.M.; Cassar, M.P.; Koziel, M.J.; Pradines, J.; Lamlum, H.; Azer, K.; Kirby, D.; Montgomery, H.; Neubauer, S.; Valkovič, L.; et al. Efficacy and Tolerability of an Endogenous Metabolic Modulator (AXA1125) in Fatigue-Predominant Long COVID: A Single-Centre, Double-Blind, Randomised Controlled Phase 2a Pilot Study. eClinicalMedicine 2023, 59. [Google Scholar] [CrossRef] [PubMed]
- Gendrot, M.; Andreani, J.; Duflot, I.; Boxberger, M.; Le Bideau, M.; Mosnier, J.; Jardot, P.; Fonta, I.; Rolland, C.; Bogreau, H.; et al. Methylene Blue Inhibits Replication of SARS-CoV-2 in Vitro. Int J Antimicrob Agents 2020, 56, 106202. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.J. da S.; Leão, A.H.F.F.; Erustes, A.G.; Morais, I.B. de M.; Vrechi, T.A. de M.; Zamarioli, L. dos S.; Pereira, C.A.S.; Marchioro, L. de O.; Sperandio, L.P.; Lins, Í.V.F.; et al. Pharmacological Modulators of Autophagy as a Potential Strategy for the Treatment of COVID-19. Int J Mol Sci 2021, 22, 4067. [Google Scholar] [CrossRef] [PubMed]
- Babajani, A.; Hosseini-Monfared, P.; Abbaspour, S.; Jamshidi, E.; Niknejad, H. Targeted Mitochondrial Therapy With Over-Expressed MAVS Protein From Mesenchymal Stem Cells: A New Therapeutic Approach for COVID-19. Front Cell Dev Biol 2021, 9, 695362. [Google Scholar] [CrossRef]
- Bramante, C.T.; Buse, J.B.; Liebovitz, D.M.; Nicklas, J.M.; Puskarich, M.A.; Cohen, K.; Belani, H.K.; Anderson, B.J.; Huling, J.D.; Tignanelli, C.J.; et al. Outpatient Treatment of COVID-19 and Incidence of Post-COVID-19 Condition over 10 Months (COVID-OUT): A Multicentre, Randomised, Quadruple-Blind, Parallel-Group, Phase 3 Trial. Lancet Infect Dis 2023, 23, 1119–1129. [Google Scholar] [CrossRef]
| Target/strategy | Effect/hypothesis | Intervention/treatment | ClinicalTrials.gov identifier (NCT number) | Ref. | |
| Antioxidants | N-acetylcysteine (NAC) | By supplementing with glycine and cysteine amino acids (in the form of N-acetylcysteine), it is possible to enhance GSH levels and improve mitochondrial function. As a result, this approach may help to lower oxidative stress, inflammation, and endothelial dysfunction. | Glycine N-acetylcysteine Alanine (placebo) |
NCT04703036 | |
| Quercetin Bromelain Zinc Vitamin C |
Zinc ionophore may act as antiviral agent, Bromelain is an anti-inflammatory agent, Vitamin C and Quercetin are antioxidants that also stimulate mitochondrial biogenesis |
Quercetin Bromelain Zinc Vitamin C |
NCT04468139 | ||
| Ubiquinol | Unique spa rehabilitation programmes in the High Tatras mountains can help patients with post-COVID 19 syndrome to restore impaired mitochondrial metabolism. The study has the potential to enhance physical and mental activity, boost immunity, reduce oxidative stress, and expedite the recovery process. | Ubiquinol (reduced coenzyme Q10) Mountain spa rehabilitation |
NCT05178225 | ||
| Mitochondrial-targeted ubiquinone (MitoQ) | MitoQ may help treat COVID-19 by reducing cytokine storms and restoring T cell function through improving mitochondrial dysfunction, which is linked to severe COVID-19 cases. Using MitoQ in the early stages could effectively slow down or postpone the progression of the disease in elderly COVID-19 patients or those with other comorbidities. |
MitoQ | - | [178] | |
| Mitoquinone | The overall objective of this study is to determine whether the daily administration of mito-MES at a dosage of 20 mg is effective in preventing confirmed SARS-CoV-2 infection. The study aims to compare the treatment to a placebo over a 14-day period and will focus on high-risk individuals who have had close contact with confirmed COVID-19 cases. | Mitoquinone/mitoquinol mesylate (mito-MES) Placebo |
NCT05886816 | ||
| Vitamin C Vitamin E Melatonin N-acetyl cysteine Pentoxifylline |
Inclusion of antioxidants like N-acetylcysteine (NAC), vitamin C, melatonin, and vitamin E in the treatment helps enhance intracellular GSH levels, sequester ROS, safeguard cell membrane lipids, cytosol proteins, nuclear DNA, and mitochondria. | Vitamin C Vitamin E Melatonin N-acetyl cysteine Pentoxifylline |
NCT04570254 | ||
| α-Lipoic acid | ALA has both antioxidant properties and the ability to suppress the NF-kB transcription factor, resulting in the inhibition of cytokine and pro-inflammatory factor production. | NAC (N-acetyl cysteine) α-lipoic acid (ALA) Liposomal glutathione (GSH) |
NCT05371288 | ||
| Mitochondria-targeted therapies | Biogenesis enhancers | Both adults and children with severe COVID-19 and multisystem inflammatory syndrome in children (MIS-C) have been found to have a significant lack of arginine. The limited availability of arginine in the plasma has been suggested as a factor contributing to problems with endothelial function, immune regulation, and excessive blood clotting. |
Arginine Hydrochloride | NCT05855330 | |
| The hypothesis proposes that supplementing with L-citrulline (CIT) is superior to ARG administration in correcting hypoargininemia, alleviating lymphocyte dysfunction, rectifying immunosuppression, and improving organ function in septic patients admitted to intensive care. | L-citrulline Placebo (water) |
NCT04404426 | |||
| Assessment of the effect of drug AXA1135 on improving bioenergetic function (measured via phosphocreatine recovery rate) in patients with fatigue-predominant PASC | AXA1125 | NCT05152849 | [179] | ||
| Mitochondria-protective agents | The study assesses the effect of methylene blue as a broad-spectrum antiviral agent and its stabilising impact on mitochondria. | Methylene mlue | - | [180] | |
| Autophagy modulation | Blocking autophagy in the early phase of COVID-19 infection could potentially control the body’s antiviral IFN response and suppress viral reproduction. |
Lysosomotropic agents (e.g., chloroquine, hydroxychloroquine, azithromycin, artemisinin, and imatinib),
Protease inhibitors/activating agents (camostat mesylate, lopinavir, ritonavir, umifenovir and teicoplanin), PI3K/AKT/mTOR modulators (e.g., rapamycin, wortmannin) |
[181] | ||
| Modulation of the mitochondrial function | The study builds on previous research suggesting that exogenous ketone supplementation can increase mitochondrial respiration in various tissues, including skeletal muscle and adipose tissue | Agilent Seahorse XF Cell Mito Stress Test Ketoneaid (Ketone monoester) |
NCT05798260 | ||
| MicroRNA targeting | miR-2392 | SBCov207 aims to mitigate the negative impacts of miR-2392 upregulation observed in COVID-19 patients, including mitochondrial dysfunction, heightened inflammation, increased glycolysis, and hypoxia, by inhibiting miR-2392. | SBCov207- antisense-based therapeutic against human miR-2392 | - | [175] |
| Enhancing the MAVS pathway | Overexpression of MAVS | The use of mesenchymal stem cells to deliver targeted mitochondrial therapy, specifically with an over-expressed MAVS protein, is being explored as a promising new treatment approach for COVID-19. The aim of this approach is to selectively enhance interferon (IFN) production and innate immune responses against SARS-CoV-2. |
Mesenchymal stem cells | - | [182] |
| Immune boosting action | Upregulation of TLR 3 | The presence of 13-cis retinoic acid led to a time-dependent upregulation of TLR3, MAVS, and IFN regulatory factor 1. Isotretinoin as “the Immunity passport" |
Isotretinoin (13 cis retinoic acid) |
NCT04353180 | |
| Drug repurposing | Metformin | By modulating mitochondrial function and reducing oxidative stress, metformin may offer potential relief for the cytokine storm and hyperinflammatory response observed in severe cases of COVID-19. Metformin can enhance the function of immune cells by optimizing their energy metabolism. | Metformin Placebo Fluvoxamine Ivermectin |
NCT04510194 | [183] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





