Submitted:
17 June 2024
Posted:
20 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Definition of Amyloid
1.2. Historical Amyloid Studies
1.3. Amyloid Toxic Protein Aggregates Promote Neurodegeneration, Cognitive Decline, and Motor Dysfunction, However Functional Amyloids Have Beneficial Properties
1.4. The Morphology of Toxic Amyloid Deposits in Brain Tissues
2. Amyloid Precursor Protein and Its Bioactive Fragments
2.1. Proteolytic Processing of APP
2.2. The Enigmatic and Perplexing Story of APP Processing in Brain Tissues
2.3. Amyloid Deposition Can Impact on Tissue Pathology
2.4. Determining the Impact of Amyloid Fibrils on Normal Tissue Physiology Using Quantum Mechanics
3. Identification of Proteins with Amyloid Aggregative Potential
3.1. Determination of Amyloid Forming Peptide Sequences in Proteins
3.2. The Impact of Misfolded Proteins on Normal Tissue Functional Properties
4. The Attributes of Functional Amyloids
4.1. Bacterial Amyloids
4.2. Marine Amyloids
4.3. Insect Amyloids
4.4. Human Amyloids
4.5. Mammalian Amyloid-Related Proteins
4.6. Amyloids that Display Metallo-Enzyme Catalytic Activities
5. Amyloid Fibrils as Building Blocks for Innovative Biomaterials
5.1. Functional Amyloids and Their Application in Tissue Engineering
5.2. Engineered Amyloid Polymers
5.3. Amyloid Cell Attachment Matrices and Hydrogels for Cell Delivery in Tissue Repair Strategies
5.4. Application of Amyloid Fibrils in Nanobiology and Organic Microelectronics
5.5. Amyloid Fibrillar Assemblies and Neuromorphic Computing
6. Concluding Remarks
7. Conclusions
Author Contributions
Disclosures
Acknowledgments
References
- Benson:, M. : et al.,. (2018) Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 25, 215-219.
- Ke, P. , et al. (2020) Half a century of amyloids: past, present and future.. Chem Soc Rev. 5473; 49. [Google Scholar]
- Buxbaum, J.N.; Linke, R.P. A Molecular History of the Amyloidoses. J. Mol. Biol. 2012, 421, 142–159. [Google Scholar] [CrossRef]
- Puchtler, H.; Sweat, F. A REVIEW OF EARLY CONCEPTS OF AMYLOID IN CONTEXT WITH CONTEMPORARY CHEMICAL LITERATURE FROM 1839 TO 1859. J. Histochem. Cytochem. 1966, 14, 123–134. [Google Scholar] [CrossRef]
- Sipe, J. , Cohen, AS (2000) Review: history of the amyloid fibril.. J Struct Biol.
- Yakupova, E.I.; Bobyleva, L.G.; Vikhlyantsev, I.M.; Bobylev, A.G. Congo Red and amyloids: history and relationship. Biosci. Rep. 2019, 39, 1495–1506. [Google Scholar] [CrossRef]
- Walker, L. (2020) Aβ plaques. 1.
- Westermark, G. e. a. (2018) Noncerebral Amyloidoses: Aspects on Seeding, Cross-Seeding, and Transmission.. Cold Spring Harb Perspect Med. 0243; 8. [Google Scholar]
- Chiti, F. , Dobson, CM. . ( and human disease.. Annu Rev Biochem. 75, 333–366.
- Fändrich, M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007, 64, 2066–2078. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; E Balch, W.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLOS Biol. 2005, 4, e6. [Google Scholar] [CrossRef]
- Lamy, C.; Duyckaerts, C.; Delaere, P.; Payan, C.; Fermanian, J.; Poulain, V.; Hauw, J.J. COMPARISON OF SEVEN STAINING METHODS FOR SENILE PLAQUES AND NEUROFIBRILLARY TANGLES IN A PROSPECTIVE SERIES OF 15 ELDERLY PATIENTS. Neuropathol. Appl. Neurobiol. 1989, 15, 563–578. [Google Scholar] [CrossRef]
- Menter, T.; Bachmann, M.; Grieshaber, S.; Tzankov, A. A More Accurate Approach to Amyloid Detection and Subtyping: Combining in situ Congo Red Staining and Immunohistochemistry. Pathobiology 2016, 84, 49–55. [Google Scholar] [CrossRef]
- Li, D.; Liu, C. Structural Diversity of Amyloid Fibrils and Advances in Their Structure Determination. Biochemistry 2020, 59, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Auriemma Citarella, A. , Di Biasi, L, De Marco, F, Tortora, G.. (2022) ENTAIL: yEt aNoTher amyloid fIbrils cLassifier.. BMC Bioinformatics. 23, 517. [CrossRef]
- Griffith, D.; Holehouse, A.S. PARROT is a flexible recurrent neural network framework for analysis of large protein datasets. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, B.; Zhang, D.; Liu, Y.; Zhang, M.; Zhao, C.; Zheng, J. Design principles and fundamental understanding of biosensors for amyloid-β detection. J. Mater. Chem. B 2020, 8, 6179–6196. [Google Scholar] [CrossRef]
- Kaushik, A. , Jayant, RD, Tiwari, S, Vashist, A, Nair, M.. (2016) Nano-biosensors to detect beta-amyloid for Alzheimer's disease management.. Biosens Bioelectron. 80, 273-287. [CrossRef]
- Gao, H. , Chen, J, Huang, Y, Zhao, R.. (2024) Advances in targeted tracking and detection of soluble amyloid-β aggregates as a biomarker of Alzheimer's disease.. Talanta. 268, 125311. [CrossRef]
- Sharma, P. , Kim, ES, Mishra, S, Ganbold, E, Seong, RS, Kim, YM, Jahng, GH, Rhee, HY, Han, HS, Kim, DH, Kim, ST, Kim, NY.. (2022) Ultrasensitive probeless capacitive biosensor for amyloid beta (Aβ1-42) detection in human plasma using interdigitated electrodes.. Biosens Bioelectron. 212, 114365. [CrossRef]
- Zheng, Y.; Zhang, L.; Zhao, J.; Li, L.; Wang, M.; Gao, P.; Wang, Q.; Zhang, X.; Wang, W. Advances in aptamers against Aβ and applications in Aβ detection and regulation for Alzheimer's disease. Theranostics 2022, 12, 2095–2114. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, J.; Chae, M.-S.; Lee, W.; Yang, S.-H.; Kim, Y.; Kim, S.M.; Lee, J.S.; Lee, J.H.; Choi, J.; et al. Multifunctionalized Reduced Graphene Oxide Biosensors for Simultaneous Monitoring of Structural Changes in Amyloid-β 40. Sensors 2018, 18, 1738. [Google Scholar] [CrossRef]
- Wang, X. , Li, L, Gu, X, Yu, B, Jiang, M.. (2021) Switchable electrochemical aptasensor for amyloid-β oligomers detection based on triple helix switch coupling with AuNPs@CuMOF labeled signaling displaced-probe.. Mikrochim Acta. 188, 49. [CrossRef]
- Xing, Y. , Xia, N.. (2015) Biosensors for the Determination of Amyloid-Beta Peptides and their Aggregates with Application to Alzheimer's Disease. 48.
- Bu, X.-L. , Xiang, Y, Jin, W.-S.et al.. (2018) Blood-derived amyloid-β protein induces Alzheimer's disease pathologies. Molecular Psychiatry. 1948; 23. [Google Scholar]
- Heppner, F. , Ransohoff, RM, Becher, B (2015) Immune attack: the role of inflammation in Alzheimer disease Nature Reviews Neuroscience. 16.
- Jorfi, M. , Maaser-Hecker, A, Tanzi, RE.. (2023) The neuroimmune axis of Alzheimer's disease. Genome Medicine. 15.
- Brody, D. , Magnoni, S, Schwetye, KE, Spinner, ML, Esparza, TJ, Stocchetti, N, Zipfel, GJ, Holtzman, DM.. (2008) Amyloid-beta dynamics correlate with neurological status in the injured human brain.. Science. 1221. [Google Scholar]
- Serra-Batiste, M. , Ninot-Pedrosa, M, Bayoumi, M, Gairí, M, Maglia, G, Carulla, N.. (2016) Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments.. Proc Natl Acad Sci U S A. 1086. [Google Scholar]
- Kim, W. , Hecht, MH. . ( 2005) Sequence determinants of enhanced amyloidogenicity of Alzheimer A beta 42 peptide relative to A beta 40.. J Biol Chem. 280, 35069–35076.
- Itoh, S. , Yagi-Utsumi, M, Kato, K, Okumura, H.. (2022) Key Residue for Aggregation of Amyloid-β Peptides. 3139; 13. [Google Scholar]
- Hsu, F. , Park, G, Guo, Z.. (2018) Key Residues for the Formation of Aβ42 Amyloid Fibrils.. ACS Omega. 8401; 3. [Google Scholar]
- McLaurin, J. , Franklin, T, Zhang, X, Deng, J, Fraser, PE.. (1999) Interactions of Alzheimer amyloid-beta peptides with glycosaminoglycans effects on fibril nucleation and growth.. Eur J Biochem. 1101. [Google Scholar]
- Gu, L. , Guo, Z.. (2013) Alzheimer's Aβ42 and Aβ40 peptides form interlaced amyloid fibrils.. J Neurochem.
- Iwatsubo, T. , Mann, DMA, Odaka, A, Suzuki, N, Ihara, Y.. (1995) Amyloid β protein (Aβ) deposition: Aβ42(43) precedes Aβ40 in Down syndrome.. Ann Neurol. 37.
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Zhang, Y. , Chen, H, Li, R, Sterling, K, Song, W.. (2023) Amyloid β-based therapy for Alzheimer's disease: challenges, successes and future.. Signal Transduct Target Ther. 8.
- Dovidchenko, N.V.; Leonova, E.I.; Galzitskaya, O.V. Mechanisms of amyloid fibril formation. Biochem. (Moscow) 2014, 79, 1515–1527. [Google Scholar] [CrossRef]
- Siddiqi, M. , Majid, N, Malik, S, Alam, P, Khan, RH. . ( Protofibrils and Fibrils.. Subcell Biochem. 93, 471–503.
- Kyle, R. (2001) Amyloidosis: a convoluted story.. British J Haematol.
- Ow, S. , Dunstan, DE.. (2014) A brief overview of amyloids and Alzheimer's disease.. Protein Sci. 1315; 23. [Google Scholar]
- Citron, M. Strategies for disease modification in Alzheimer's disease. Nat. Rev. Neurosci. 2004, 5, 677–685. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer's disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Hardy, J. , Allsop, D.. (1991) Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 12.
- O'Brien, R.J.; Wong, P.C. Amyloid Precursor Protein Processing and Alzheimer's Disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef]
- Chiellini, G. Understanding Amyloid Structures and Disease: A Continuing Challenge in Health Research. Int. J. Mol. Sci. 2021, 22, 6620. [Google Scholar] [CrossRef]
- dos Santos, H.M.; Bertollo, A.G.; Mingoti, M.E.D.; Grolli, R.E.; Kreuz, K.M.; Ignácio, Z.M. Dementia and depression: Biological connections with amyloid β protein. Basic Clin. Pharmacol. Toxicol. 2024, 134, 563–573. [Google Scholar] [CrossRef]
- Jackson, M.P.; Hewitt, E.W. Why are Functional Amyloids Non-Toxic in Humans? Biomolecules 2017, 7, 71. [Google Scholar] [CrossRef]
- Rubel, M.S.; Fedotov, S.A.; Grizel, A.V.; Sopova, J.V.; Malikova, O.A.; Chernoff, Y.O.; Rubel, A.A. Functional Mammalian Amyloids and Amyloid-Like Proteins. Life 2020, 10, 156. [Google Scholar] [CrossRef]
- Reynolds, N.P. Amyloid-like peptide nanofibrils as scaffolds for tissue engineering: Progress and challenges (Review). Biointerphases 2019, 14, 040801. [Google Scholar] [CrossRef]
- Mankar, S.; Anoop, A.; Sen, S.; Maji, S.K. Nanomaterials: amyloids reflect their brighter side. Nano Rev. 2011, 2. [Google Scholar] [CrossRef]
- Das, S.; Jacob, R.S.; Patel, K.; Singh, N.; Maji, S.K. Amyloid Fibrils: Versatile Biomaterials for Cell Adhesion and Tissue Engineering Applications. Biomacromolecules 2018, 19, 1826–1839. [Google Scholar] [CrossRef]
- Binaymotlagh, R.; Chronopoulou, L.; Palocci, C. Peptide-Based Hydrogels: Template Materials for Tissue Engineering. J. Funct. Biomater. 2023, 14, 233. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef]
- Sweers, K.K.M.; Bennink, M.L.; Subramaniam, V. Nanomechanical properties of single amyloid fibrils. J. Physics: Condens. Matter 2012, 24, 243101. [Google Scholar] [CrossRef]
- Li, J. , Zhang, F.. (2021) Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges.. Int J Mol Sci. 1096; 22. [Google Scholar]
- Fukuma, T.; Mostaert, A.S.; Jarvis, S.P. Explanation for the mechanical strength of amyloid fibrils. Tribol. Lett. 2006, 22, 233–237. [Google Scholar] [CrossRef]
- Sawaya, M. , Hughes, MP, Rodriguez, JA, Riek, R, Eisenberg, DS.. (2021) The expanding amyloid family: Structure, stability, function, and pathogenesis.. Cell. 4857. [Google Scholar]
- Boon, B. , Bulk, M, Jonker, AJ, Morrema, THJ, van den Berg, E, Popovic, M, Walter, J, Kumar, S, van der Lee, SJ, Holstege, H, Zhu, X, Van Nostrand, WE, Natté, R, van der Weerd, L, Bouwman, FH, van de Berg, WDJ, Rozemuller, AJM, Hoozemans, JJM.. (2020) The coarse-grained plaque: a divergent Aβ plaque-type in early-onset Alzheimer's disease.. Acta Neuropathol.
- Kim, K. , et al. ( 1988) Production and characterization of monoclonal antibodies reactive to synthetic cerebrovascular amyloid peptide.. Neurosci Res Commun. 2, 121–130.
- Tsuchida, K.; Shioi, J.; Yamada, S.; Boghosian, G.; Wu, A.; Cai, H.; Sugahara, K.; Robakis, N.K. Appican, the Proteoglycan Form of the Amyloid Precursor Protein, Contains Chondroitin Sulfate E in the Repeating Disaccharide Region and 4-O-Sulfated Galactose in the Linkage Region. J. Biol. Chem. 2001, 276, 37155–37160. [Google Scholar] [CrossRef]
- Pangalos, M.N.; Shioi, J.; Efthimiopoulos, S.; Wu, A.; Robakis, N.K. Characterization of Appican, the Chondroitin Sulfate Proteoglycan Form of the Alzheimer Amyloid Precursor Protein. Neurodegeneration 1996, 5, 445–451. [Google Scholar] [CrossRef]
- Müller, U.C.; Deller, T.; Korte, M. Not just amyloid: physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 2017, 18, 281–298. [Google Scholar] [CrossRef]
- Hefter, D. , Ludewig, S, Draguhn, A, Korte, M. . ( and Electrical Activity of the Brain.. Neuroscientist. 26, 231–251.
- Turner, P. , O'Connor, K, Tate, WP, Abraham, WC. . ( plasticity and memory.. Prog Neurobiol. 70, 1–32.
- Korte, M. , Herrmann, U, Zhang, X, Draguhn, A.. (2012) The role of APP and APLP for synaptic transmission, plasticity, and network function: lessons from genetic mouse models.. Exp Brain Res.
- Russell, C.L.; Semerdjieva, S.; Empson, R.M.; Austen, B.M.; Beesley, P.W.; Alifragis, P. Amyloid-β Acts as a Regulator of Neurotransmitter Release Disrupting the Interaction between Synaptophysin and VAMP2. PLOS ONE 2012, 7, e43201. [Google Scholar] [CrossRef]
- Allinson, T. , Parkin, ET, Turner, AJ, Hooper, NM.. (2003) ADAMs family members as amyloid precursor proteinα-secretases. J Neurosci Res. 74.
- Willem, M.; Tahirovic, S.; Busche, M.A.; Ovsepian, S.V.; Chafai, M.; Kootar, S.; Hornburg, D.; Evans, L.D.B.; Moore, S.C.; Daria, A.; et al. η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 2015, 526, 443–447. [Google Scholar] [CrossRef]
- Andrew, R.J.; Kellett, K.A.; Thinakaran, G.; Hooper, N.M. A Greek Tragedy: The Growing Complexity of Alzheimer Amyloid Precursor Protein Proteolysis. J. Biol. Chem. 2016, 291, 19235–19244. [Google Scholar] [CrossRef]
- Gabriele, R.M.C.; Abel, E.; Fox, N.C.; Wray, S.; Arber, C. Knockdown of Amyloid Precursor Protein: Biological Consequences and Clinical Opportunities. Front. Neurosci. 2022, 16, 835645. [Google Scholar] [CrossRef]
- Hampel, H. , Hu, Y, Hardy, J, Blennow, K, Chen, C, Perry, G, Kim, SH, Villemagne, VL, Aisen, P, Vendruscolo, M, Iwatsubo, T, Masters, CL, Cho, M, Lannfelt,L, Cummings, JL, Vergallo, A. T. (2023) he amyloid-β pathway in Alzheimer's disease: a plain language summary.. Neurodegener Dis Manag. 13.
- Chen, G. , Xu, TH, Yan, Y, Zhou, YR, Jiang, Y, Melcher, K, Xu, HE. (2017) Amyloid beta: structure, biology y and structure-based therapeutic development.. Acta Pharmacol Sin. 1205; 38. [Google Scholar]
- Rambaran, R. , Serpell, LC.. (2008) Amyloid fibrils: abnormal protein assembly.. Prion. 2.
- Sosa, L.J.; Cáceres, A.; Dupraz, S.; Oksdath, M.; Quiroga, S.; Lorenzo, A. The physiological role of the amyloid precursor protein as an adhesion molecule in the developing nervous system. J. Neurochem. 2017, 143, 11–29. [Google Scholar] [CrossRef]
- Al-Kuraishy, H. , Jabir,MS, Al-Gareeb, AI, Albuhadily, AK, Albukhaty, S, Sulaiman, GM, Batiha, GE.. (2023) Evaluation and targeting of amyloid precursor protein (APP)/amyloid beta (Aβ) axis in amyloidogenic and non-amyloidogenic pathways: A time outside the tunnel.. Ageing Res Rev. 92, 102119. [CrossRef]
- Orobets, K. , Karamyshev, AL.. (2023) Amyloid Precursor Protein and Alzheimer's Disease.. Int J Mol Sci. 24, 14794. [CrossRef]
- Kuo, C.-C.; Chiang, A.W.T.; Baghdassarian, H.M.; Lewis, N.E. Dysregulation of the secretory pathway connects Alzheimer’s disease genetics to aggregate formation. Cell Syst. 2021. [Google Scholar] [CrossRef]
- Sun, J.; Roy, S. The physical approximation of APP and BACE-1: A key event in alzheimer's disease pathogenesis. Dev. Neurobiol. 2017, 78, 340–347. [Google Scholar] [CrossRef]
- Tackenberg, C. , Nitsch, RM.. (2019) The secreted APP ectodomain sAPPα, but not sAPPβ, protects neurons against Aβ oligomer-induced dendritic spine loss and increased tau phosphorylation.. Mol Brain. 12, 27. [CrossRef]
- Baratchi, S.; Evans, J.; Tate, W.P.; Abraham, W.C.; Connor, B. Secreted amyloid precursor proteins promote proliferation and glial differentiation of adult hippocampal neural progenitor cells. Hippocampus 2011, 22, 1517–1527. [Google Scholar] [CrossRef]
- Dar, N.J.; Glazner, G.W. Deciphering the neuroprotective and neurogenic potential of soluble amyloid precursor protein alpha (sAPPα). Cell. Mol. Life Sci. 2020, 77, 2315–2330. [Google Scholar] [CrossRef]
- Habib, A.; Sawmiller, D.; Tan, J. Restoring Soluble Amyloid Precursor Protein α Functions as a Potential Treatment for Alzheimer's Disease. J. Neurosci. Res. 2016, 95, 973–991. [Google Scholar] [CrossRef]
- Castro, M.A.; Hadziselimovic, A.; Sanders, C.R. The vexing complexity of the amyloidogenic pathway. Protein Sci. 2019, 28, 1177–1193. [Google Scholar] [CrossRef]
- Nalivaeva, N. , Turner, AJ.. (2013) The amyloid precursor protein: a biochemical enigma in brain development, function and disease.. FEBS Lett. 587, 2046-2054. [CrossRef]
- Chasseigneaux, S.; Allinquant, B. Functions of Aβ, sAPPα and sAPPβ: similarities and differences. J. Neurochem. 2011, 120, 99–108. [Google Scholar] [CrossRef]
- Proft, J. , Weiss, N.. (2012) Jekyll and Hide, The two faces of amyloid β. 5.
- Frohman, E.M.; Racke, M.K.; Raine, C.S. Multiple Sclerosis — The Plaque and Its Pathogenesis. New Engl. J. Med. 2006, 354, 942–955. [Google Scholar] [CrossRef]
- Grant, J. , Ghosn, EE, Axtell, RC, Herges, K, Kuipers, HF, Woodling, NS, et al.. (2012) Reversal of paralysis and reduced inflammation from peripheral administration of beta-amyloid in TH1 and TH17 versions of experimental autoimmune encephalomyelitis. 4.
- Trapp, B.D.; Peterson, J.; Ransohoff, R.M.; Rudick, R.; Mörk, S.; Bö, L. Axonal Transection in the Lesions of Multiple Sclerosis. New Engl. J. Med. 1998, 338, 278–285. [Google Scholar] [CrossRef]
- Ferguson, B.; Matyszak, M.K.; Esiri, M.M.; Perry, V.H. Axonal damage in acute multiple sclerosis lesions. Brain 1997, 120, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Simard, A.R.; Soulet, D.; Gowing, G.; Julien, J.-P.; Rivest, S. Bone Marrow-Derived Microglia Play a Critical Role in Restricting Senile Plaque Formation in Alzheimer's Disease. Neuron 2006, 49, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, S.; McGeer, P.; Akiyama, H.; Zhu, S.; Selkoe, D. Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. J. Neuroimmunol. 1989, 24, 173–182. [Google Scholar] [CrossRef]
- Li, X. , Feng, X, Sun, X, Hou, N, Han, F, Liu, Y.. (2022) Global, regional, and national burden of Alzheimer's disease and other dementias, 1990-2019. Front Aging Neurosci. 9: 14, 9374; 14. [Google Scholar]
- Gillet, J.-N. From molecular dynamics to quantum mechanics of misfolded proteins and amyloid-like macroaggregates applied to neurodegenerative diseases. J. Mol. Graph. Model. 2021, 110, 108046. [Google Scholar] [CrossRef]
- Sohail, A.; Ashiq, U. Quantum inspired improved AI computing for the sensors of cardiac mechano-biology. Sensors Int. 2023, 4. [Google Scholar] [CrossRef]
- De la Paz, M. , Serrano, L. . ( 2004) Sequence deteminants of amyloid fibril formation.. Proc Natl Acad Sci USA.. 101, 87–92.
- DuBay, K.F.; Pawar, A.P.; Chiti, F.; Zurdo, J.; Dobson, C.M.; Vendruscolo, M. Prediction of the Absolute Aggregation Rates of Amyloidogenic Polypeptide Chains. J. Mol. Biol. 2004, 341, 1317–1326. [Google Scholar] [CrossRef]
- Pawar, A. , Dubay, KF, Zurdo, J, et al.. (2005) Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative disease.. J Mol Biol.
- Zibaee, S. , Makin, OS, Goedert, M, et al.. (2007) A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta and tau using the amino acid sequence alone. 16.
- Fernandez-Escamilla, A.-M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Palato, L.M.; Pilcher, S.; Oakes, A.; Lamba, A.; Torres, J.; Monjaraz, L.I.L.; Munoz, C.; Njoo, E.; Rinauro, D.J.; Menefee, K.A.; et al. Amyloidogenicity of naturally occurring full-length animal IAPP variants. J. Pept. Sci. 2019, 25, e3199–e3199. [Google Scholar] [CrossRef]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65–65. [Google Scholar] [CrossRef] [PubMed]
- Castillo, V.; Graña-Montes, R.; Sabate, R.; Ventura, S. Prediction of the aggregation propensity of proteins from the primary sequence: Aggregation properties of proteomes. Biotechnol. J. 2011, 6, 674–685. [Google Scholar] [CrossRef] [PubMed]
- de Groot, N. , Castillo, V, Graña-Montes, R, Ventura, S.. (2012) AGGRESCAN: method, application, and perspectives for drug design.. Methods Mol Biol.
- Tsolis, A. , Papandreou, NC, Iconomidou, VA, Hamodrakas, SJ.. (2013) A consensus method for the prediction of 'aggregation-prone' peptides in globular proteins.. PLoS One. 5417; 8. [Google Scholar]
- Cherny, I.; Gazit, E. Amyloids: Not Only Pathological Agents but Also Ordered Nanomaterials. Angew. Chem. Int. Ed. 2008, 47, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, H.; Lai, L. Identification of amyloid fibril-forming segments based on structure and residue-based statistical potential. Bioinformatics 2007, 23, 2218–2225. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, S.A.; Uspenskaya, M.V.; Leclercq, J.; Falgarone, T.; Zhouravleva, G.A.; Kajava, A.V. AmyloComp: a bioinformatic tool for prediction of amyloid co-aggregation. J. Mol. Biol. 2024, 168437. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, J.W.; Szczurek, W.; Szulc, N.; Szefczyk, M.; Kotulska, M. PACT - Prediction of amyloid cross-interaction by threading. Sci. Rep. 2023, 13, 1–10. [Google Scholar] [CrossRef]
- Emily, M.; Talvas, A.; Delamarche, C. MetAmyl: A METa-Predictor for AMYLoid Proteins. PLOS ONE 2013, 8, e79722. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P. , Kanthawong, S, Nantasenamat, C, Hasan, MM, Shoombuatong, W.. (2021) iAMY-SCM: Improved prediction and analysis of amyloid proteins using a scoring card method with propensity scores of dipeptides.. Genomics.
- Niu, M.; Li, Y.; Wang, C.; Han, K. RFAmyloid: A Web Server for Predicting Amyloid Proteins. Int. J. Mol. Sci. 2018, 19, 2071. [Google Scholar] [CrossRef] [PubMed]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65–65. [Google Scholar] [CrossRef]
- Hamodrakas, S.J. Protein aggregation and amyloid fibril formation prediction software from primary sequence: towards controlling the formation of bacterial inclusion bodies. FEBS J. 2011, 278, 2428–2435. [Google Scholar] [CrossRef]
- Charoenkwan, P. , Ahmed, S, Nantasenamat, C, Quinn, JMW, Moni, MA, Lio,' P, Shoombuatong, W.. (2022) AMYPred-FRL is a novel approach for accurate prediction of amyloid proteins by using feature representation learning.. Sci Rep. 7697; 12. [Google Scholar]
- Nair, S.S.K.; Reddy, N.S.; Hareesha, K. AmylPepPred: Amyloidogenic Peptide Prediction tool. Bioinformation 2012, 8, 994–995. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wu, N.; Guo, J.; Fan, Y. Prediction of amyloid fibril-forming segments based on a support vector machine. BMC Bioinform. 2009, 10, S45–S45. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Robang, A.S.; Sarma, S.; Le, J.V.; E Helmicki, M.; Lambert, M.J.; Guerrero-Ferreira, R.; Arboleda-Echavarria, J.; Paravastu, A.K.; Hall, C.K. Sequence patterns and signatures: Computational and experimental discovery of amyloid-forming peptides. PNAS Nexus 2022, 1, pgac263. [Google Scholar] [CrossRef]
- Morris, K. , Rodger, A, Hicks, MR, Debulpaep, M, Schymkowitz, J, Rousseau, F, Serpel, LC.. (2013) Exploring the sequence-structure relationship for amyloid peptides.
- López de la Paz, M. , Serrano, L. . ( 2004) Sequence determinants of amyloid fibril formation.. Proc Natl Acad Sci U S A. 101, 87–92.
- Al-Garawi, Z.S.; Morris, K.L.; Marshall, K.E.; Eichler, J.; Serpell, L.C. The diversity and utility of amyloid fibrils formed by short amyloidogenic peptides. Interface Focus 2017, 7, 20170027–20170027. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.J.; Sievers, S.A.; Karanicolas, J.; Ivanova, M.I.; Baker, D.; Eisenberg, D. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. 2006, 103, 4074–4078. [Google Scholar] [CrossRef]
- Galzitskaya, O. , Garbuzynskiy, SO, Lobanov, MY. . ( 2006) Prediction of amyloidogenic and disordered regions in protein chains.. PLoS Comput Biol. 2, e177.
- Família, C. , Dennison, SR, Quintas, A, Phoenix, DA. . ( 2015) Prediction of Peptide and Protein Propensity for Amyloid Formation.. PLoS One. 10, e0134679.
- Belli, M. , Ramazzotti, M, Chiti, F. . ( 2011) Prediction of amyloid aggregation in vivo.. EMBO Rep. 12, 657–663.
- Abdelrahman, S.; Alghrably, M.; Lachowicz, J.I.; Emwas, A.-H.; Hauser, C.A.E.; Jaremko, M. “What Doesn’t Kill You Makes You Stronger”: Future Applications of Amyloid Aggregates in Biomedicine. Molecules 2020, 25, 5245. [Google Scholar] [CrossRef]
- Xiao, X. , Wang, Y, Seroski, DT, Wong, KM, Liu, R, Paravastu, AK, Hudalla, GA, Hall, CK.. (2021) De novo design of peptides that coassemble into β sheet-based nanofibrils.. Sci Adv. 7668; 7. [Google Scholar]
- Cherny, I.; Gazit, E. Amyloids: Not Only Pathological Agents but Also Ordered Nanomaterials. Angew. Chem. Int. Ed. 2008, 47, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Herczenik, E.; Gebbink, M.F.B.G. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008, 22, 2115–2133. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.V.; E Bredesen, D. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr. Opin. Cell Biol. 2004, 16, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D. (2003) Folding proteins in fatal ways.. Nature.
- Taylor, J.P.; Hardy, J.; Fischbeck, K.H. Toxic Proteins in Neurodegenerative Disease. Science 2002, 296, 1991–1995. [Google Scholar] [CrossRef] [PubMed]
- Kopito, R. , Ron, D. . ( 2000) Conformational disease.. Nat Cell Biol. 2, E207–E209.
- Kopito, R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000, 10, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Ellis, R.J. Protein folding and misfolding inside and outside the cell. EMBO J. 1998, 17, 5251–5254. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C. (2003) Protein folding and misfolding.. Nature.
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef]
- Roan, N.R.; Müller, J.A.; Liu, H.; Chu, S.; Arnold, F.; Stürzel, C.M.; Walther, P.; Dong, M.; Witkowska, H.E.; Kirchhoff, F.; et al. Peptides Released by Physiological Cleavage of Semen Coagulum Proteins Form Amyloids that Enhance HIV Infection. Cell Host Microbe 2011, 10, 541–550. [Google Scholar] [CrossRef]
- Tennent, G. , Lovat, LB, Pepys, MB (1995) Serum amyloid P component prevents proteolysis of the amyloid fibrils of Alzheimer disease and systemic amyloidosis.. PNAS USA. 4299; 92. [Google Scholar]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Sitia, R.; Braakman, I. Quality control in the endoplasmic reticulum protein factory. Nature 2003, 426, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M. , Goldberg, AL.. (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases.. Neuron. 29.
- Mostaert, A.S.; Crockett, R.; Kearn, G.; Cherny, I.; Gazit, E.; Serpell, L.C.; Jarvis, S.P. Mechanically functional amyloid fibrils in the adhesive of a marine invertebrate as revealed by Raman spectroscopy and atomic force microscopy. Arch. Histol. Cytol. 2009, 72, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Guyonnet, B.; Egge, N.; Cornwall, G.A.; Ghule, P.N.; Xie, R.-L.; Medina, R.; Colby, J.L.; Jones, S.N.; Lian, J.B.; Stein, J.L.; et al. Functional Amyloids in the Mouse Sperm Acrosome. Mol. Cell. Biol. 2014, 34, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Whelly, S.; Johnson, S.; Powell, J.; Borchardt, C.; Hastert, M.C.; Cornwall, G.A. Nonpathological Extracellular Amyloid Is Present during Normal Epididymal Sperm Maturation. PLOS ONE 2012, 7, e36394. [Google Scholar] [CrossRef] [PubMed]
- Whelly, S.; Muthusubramanian, A.; Powell, J.; Johnson, S.; Hastert, M.C.; Cornwall, G.A. Cystatin-related epididymal spermatogenic subgroup members are part of an amyloid matrix and associated with extracellular vesicles in the mouse epididymal lumen. Mol. Hum. Reprod. 2016, 22, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Guyonnet, B.; Egge, N.; Cornwall, G.A.; Ghule, P.N.; Xie, R.-L.; Medina, R.; Colby, J.L.; Jones, S.N.; Lian, J.B.; Stein, J.L.; et al. Functional Amyloids in the Mouse Sperm Acrosome. Mol. Cell. Biol. 2014, 34, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Sood, R. , Domanov, Y, Pietiainen, M, Kontinen, VP, Kinnunen, PKJ.. (2008) Binding of LL-37 to model biomembranes: Insight into target vs. host cell recognition.. Biochem Biophys Acta. 1778. [Google Scholar]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional Amyloids As Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; E Balch, W.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLOS Biol. 2005, 4, e6. [Google Scholar] [CrossRef]
- Münch, J.; Rücker, E.; Ständker, L.; Adermann, K.; Goffinet, C.; Schindler, M.; Wildum, S.; Chinnadurai, R.; Rajan, D.; Specht, A.; et al. Semen-Derived Amyloid Fibrils Drastically Enhance HIV Infection. Cell 2007, 131, 1059–1071. [Google Scholar] [CrossRef]
- Jang, H.; Arce, F.T.; Mustata, M.; Ramachandran, S.; Capone, R.; Nussinov, R.; Lal, R. Antimicrobial Protegrin-1 Forms Amyloid-Like Fibrils with Rapid Kinetics Suggesting a Functional Link. Biophys. J. 2011, 100, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Li, J. , McQuade, T, Siemer, AB, Napetschnig, J, Moriwaki, K, Hsiao, YS, Damko, E, Moquin, D, Walz, T, McDermott, A, et al.. (2012) The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis.. Cell.
- Audas, T.E.; Audas, D.E.; Jacob, M.D.; Ho, J.D.; Khacho, M.; Wang, M.; Perera, J.K.; Gardiner, C.; Bennett, C.A.; Head, T.; et al. Adaptation to Stressors by Systemic Protein Amyloidogenesis. Dev. Cell 2016, 39, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J. High Performance Marine and Terrestrial Bioadhesives and the Biomedical Applications They Have Inspired. Molecules 2022, 27, 8982. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, J.A.; Varghese, N.R.; Johnston, C.L.; Sunde, M. Functional Amyloids: Where Supramolecular Amyloid Assembly Controls Biological Activity or Generates New Functionality. J. Mol. Biol. 2023, 435, 167919. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Molecular Self-Assembly of Peptide Nanostructures: Mechanism of Association and Potential Uses. Curr. Nanosci. 2006, 2, 105–111. [Google Scholar] [CrossRef]
- Adler-Abramovich, L.; Aronov, D.; Beker, P.; Yevnin, M.; Stempler, S.; Buzhansky, L.; Rosenman, G.; Gazit, E. Self-assembled arrays of peptide nanotubes by vapour deposition. Nat. Nanotechnol. 2009, 4, 849–854. [Google Scholar] [CrossRef]
- Carny, O.; Shalev, D.E.; Gazit, E. Fabrication of Coaxial Metal Nanocables Using a Self-Assembled Peptide Nanotube Scaffold. Nano Lett. 2006, 6, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Controlled patterning of aligned self-assembled peptide nanotubes. Nat. Nanotechnol. 2006, 1, 195–200. [Google Scholar] [CrossRef]
- Tiwari, O. , Gazit, E.. (2024) Characterization of amyloid-like metal-amino acid assemblies with remarkable catalytic activity.. Methods Enzymol. 697, 181-209. [CrossRef]
- Gazit, E. Self-assembled peptide nanostructures: the design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef]
- Hauser, C.A.E.; Maurer-Stroh, S.; Martins, I.C. Amyloid-based nanosensors and nanodevices. Chem. Soc. Rev. 2014, 43, 5326–5345. [Google Scholar] [CrossRef] [PubMed]
- Taheri, R.A.; Akhtari, Y.; Moghadam, T.T.; Ranjbar, B. Assembly of Gold Nanorods on HSA Amyloid Fibrils to Develop a Conductive Nanoscaffold for Potential Biomedical and Biosensing Applications. Sci. Rep. 2018, 8, 9333. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Joshi, S.; Savel’ev, S.E.; Jiang, H.; Midya, R.; Lin, P.; Hu, M.; Ge, N.; Strachan, J.P.; Li, Z.; et al. Memristors with diffusive dynamics as synaptic emulators for neuromorphic computing. Nat. Mater. 2016, 16, 101–108. [Google Scholar] [CrossRef]
- Miranda, E.; Suñé, J. Memristors for Neuromorphic Circuits and Artificial Intelligence Applications. Materials 2020, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, J.; He, X.; Ming, J.; Wang, L.; Wang, Y.; Shao, H.; Zheng, C.; Xie, L.; Ling, H. Pseudo-transistors for emerging neuromorphic electronics. Sci. Technol. Adv. Mater. 2023, 24, 2180286. [Google Scholar] [CrossRef]
- Barraj, I.; Mestiri, H.; Masmoudi, M. Overview of Memristor-Based Design for Analog Applications. Micromachines 2024, 15, 505. [Google Scholar] [CrossRef] [PubMed]
- Jariwala, D. , Sangwan, VK, Lauhon, LJ, Marks, TJ, Hersam, MC. . ( and sensing.. Chem Soc Rev. 42, 2824–2860.
- Li, C.; Mezzenga, R. The interplay between carbon nanomaterials and amyloid fibrils in bio-nanotechnology. Nanoscale 2013, 5, 6207–6218. [Google Scholar] [CrossRef]
- Terrones, H.; Terrones, M.; López–Urías, F.; Rodríguez–Manzo, J.A.; Mackay, A.L. Shape and complexity at the atomic scale: the case of layered nanomaterials. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 2004, 362, 2039–2063. [Google Scholar] [CrossRef]
- Li, C.; Mezzenga, R. The interplay between carbon nanomaterials and amyloid fibrils in bio-nanotechnology. Nanoscale 2013, 5, 6207–6218. [Google Scholar] [CrossRef]
- Mankar, S.; Anoop, A.; Sen, S.; Maji, S.K. Nanomaterials: amyloids reflect their brighter side. Nano Rev. 2011, 2. [Google Scholar] [CrossRef]
- Ramanishankar, A.; S, A.S.; Begum, R.F.; Jayasankar, N.; Nayeem, A.; Prajapati, B.G.; Nirenjen, S. Unleashing light's healing power: an overview of photobiomodulation for Alzheimer's treatment. Futur. Sci. OA 2024, 10, FSO922. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chang, Y.; Liu, L.; Wang, J. Nanomaterials for Modulating the Aggregation of β-Amyloid Peptides. Molecules 2021, 26, 4301. [Google Scholar] [CrossRef] [PubMed]
- Ender, A.M.; Kaygisiz, K.; Räder, H.-J.; Mayer, F.J.; Synatschke, C.V.; Weil, T. Cell-Instructive Surface Gradients of Photoresponsive Amyloid-like Fibrils. ACS Biomater. Sci. Eng. 2021, 7, 4798–4808. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.R.; Dobson, C.M. Chemical modification of insulin in amyloid fibrils. Protein Sci. 2003, 12, 2637–2641. [Google Scholar] [CrossRef] [PubMed]
- Morris, R. , MacPhee, C. (2013) Amyloid Protein Biomaterials. In: Roberts, G.C.K. (eds) Encyclopedia of Biophysics. Springer, Berlin, Heidelberg. [CrossRef]
- Vaziri, S.; Fazilati, M.; Arasteh, A.; Nazem, H. Amyloid Nano-biofibrils as a New Nano-Scaffold for Lipase Immobilization. Protein Pept. Lett. 2018, 25, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Roberts, S.J.; Meade, S.J.; Gerrard, J.A. Amyloid fibrils as a nanoscaffold for enzyme immobilization. Biotechnol. Prog. 2009, 26, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Wittung-Stafshede, P. Chemical catalysis by biological amyloids. Biochem. Soc. Trans. 2023, 51, 1967–1974. [Google Scholar] [CrossRef]
- Fowler, D. , Koulov, AV, Balch, WE, Kelly, JW. . ( 2007) Functional amyloid from bacteria to humans.. Trends Biochem Sci. 32, 217–224.
- Gras, S.L.; Tickler, A.K.; Squires, A.M.; Devlin, G.L.; Horton, M.A.; Dobson, C.M.; MacPhee, C.E. Functionalised amyloid fibrils for roles in cell adhesion. Biomaterials 2007, 29, 1553–1562. [Google Scholar] [CrossRef]
- Maji, S.K.; Schubert, D.; Rivier, C.; Lee, S.; Rivier, J.E.; Riek, R. Amyloid as a Depot for the Formulation of Long-Acting Drugs. PLoS Biol. 2008, 6, e17. [Google Scholar] [CrossRef]
- Pertinhez, T.A.; Conti, S.; Ferrari, E.; Magliani, W.; Spisni, A.; Polonelli, L. Reversible Self-Assembly: A Key Feature for a New Class of Autodelivering Therapeutic Peptides. Mol. Pharm. 2009, 6, 1036–1039. [Google Scholar] [CrossRef]
- Scheibel, T.; Parthasarathy, R.; Sawicki, G.; Lin, X.-M.; Jaeger, H.; Lindquist, S.L. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA 2003, 100, 4527–4532. [Google Scholar] [CrossRef] [PubMed]
- Bhak, G. , Lee, S, Park, JW, Cho, S, Paik, SR.. (2010) Amyloid hydrogel derived from curly protein fibrils of alpha-synuclein.. Biomaterials. 5986; 31. [Google Scholar]
- Herland, A.; Björk, P.; Nilsson, K.P.R.; Olsson, J.D.M.; Åsberg, P.; Konradsson, P.; Hammarström, P.; Inganäs, O. Electroactive Luminescent Self-Assembled Bio-organic Nanowires: Integration of Semiconducting Oligoelectrolytes within Amyloidogenic Proteins. Adv. Mater. 2005, 17, 1466–1471. [Google Scholar] [CrossRef]
- Tanaka, H.; Herland, A.; Lindgren, L.J.; Tsutsui, T.; Andersson, M.R.; Inganäs, O. Enhanced Current Efficiency from Bio-Organic Light-Emitting Diodes Using Decorated Amyloid Fibrils with Conjugated Polymer. Nano Lett. 2008, 8, 2858–2861. [Google Scholar] [CrossRef]
- Herland, A.; Thomsson, D.; Mirzov, O.; Scheblykin, I.G.; Inganäs, O. Decoration of amyloid fibrils with luminescent conjugated polymers. J. Mater. Chem. 2007, 18, 126–132. [Google Scholar] [CrossRef]
- Hamedi, M.; Herland, A.; Karlsson, R.H.; Inganäs, O. Electrochemical Devices Made from Conducting Nanowire Networks Self-Assembled from Amyloid Fibrils and Alkoxysulfonate PEDOT. Nano Lett. 2008, 8, 1736–1740. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M. , Kang, S, Koo, HJ, Lee, JH, Lee, YS, Paik, SR.. (2010) Nanoporous protein matrix made of amyloid fibrils of beta2-microglobulin.. Biotechnol Prog. 1759; 26. [Google Scholar]
- Preat, T.; Goguel, V. Role of Drosophila Amyloid Precursor Protein in Memory Formation. Front. Mol. Neurosci. 2016, 9, 142. [Google Scholar] [CrossRef]
- Evans, M.L.; Chapman, M.R. Curli biogenesis: Order out of disorder. Biochim. et Biophys. Acta (BBA) - Mol. Cell Res. 2014, 1843, 1551–1558. [Google Scholar] [CrossRef]
- Bhoite, S.; van Gerven, N.; Chapman, M.R.; Remaut, H. Curli Biogenesis: Bacterial Amyloid Assembly by the Type VIII Secretion Pathway. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Klauck, G. , Serra, DO, Possling, A, Hengge, R.. (2018) Spatial Organization of Different Sigma Factor Activities and C-Di-GMP Signalling within the Three-Dimensional Landscape of a Bacterial Biofilm.. Open Biol. 8. [CrossRef]
- Ulamec, S.M.; Radford, S.E. Spot the Difference: Function versus Toxicity in Amyloid Fibrils. Trends Biochem. Sci. 2020, 45, 635–636. [Google Scholar] [CrossRef] [PubMed]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Goguel, V.; Belair, A.-L.; Ayaz, D.; Lampin-Saint-Amaux, A.; Scaplehorn, N.; Hassan, B.A.; Preat, T. DrosophilaAmyloid Precursor Protein-Like Is Required for Long-Term Memory. J. Neurosci. 2011, 31, 1032–1037. [Google Scholar] [CrossRef]
- Hervás, R. , Li, L, Majumdar, A, Fernández-Ramírez Mdel, C, Unruh, JR, Slaughter, BD, Galera-Prat, A, Santana, E, Suzuki, M, Nagai, Y, Bruix, M, Casas-Tintó, S, Menéndez, M, Laurents, DV, Si, K, Carrión-Vázquez, M. . ( 2016) Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation.. PLoS Biol. 14, e1002361.
- Hervás, R.; Murzin, A.G.; Si, K. Implications of the Orb2 Amyloid Structure in Huntington’s Disease. Int. J. Mol. Sci. 2020, 21, 6910. [Google Scholar] [CrossRef]
- Garcia-Pardo, J.; Ventura, S. Cryo-EM structures of functional and pathological amyloid ribonucleoprotein assemblies. Trends Biochem. Sci. 2024, 49, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Bartolomé-Nafría, A.; García-Pardo, J.; Ventura, S. Mutations in human prion-like domains: pathogenic but not always amyloidogenic. Prion 2024, 18, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. 2001, 98, 7018–7024. [Google Scholar] [CrossRef]
- Si, K.; Kandel, E.R. The Role of Functional Prion-Like Proteins in the Persistence of Memory. Cold Spring Harb. Perspect. Biol. 2016, 8, a021774. [Google Scholar] [CrossRef]
- Kozlov, E.; Shidlovskii, Y.V.; Gilmutdinov, R.; Schedl, P.; Zhukova, M. The role of CPEB family proteins in the nervous system function in the norm and pathology. Cell Biosci. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Li, X.; Li, S.; Huang, X.; Chen, Y.; Cheng, J.; Zhan, A. Protein-mediated bioadhesion in marine organisms: A review. Mar. Environ. Res. 2021, 170, 105409. [Google Scholar] [CrossRef] [PubMed]
- Balkenende, D.W.; Winkler, S.M.; Messersmith, P.B. Marine-inspired polymers in medical adhesion. Eur. Polym. J. 2019, 116, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Ninan, L.; Monahan, J.; Stroshine, R.L.; Wilker, J.J.; Shi, R. Adhesive strength of marine mussel extracts on porcine skin. Biomaterials 2003, 24, 4091–4099. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.; Pereira, M.J.; Lee, Y.; Friehs, I.; Vasilyev, N.V.; Feins, E.N.; Ablasser, K.; O’cearbhaill, E.D.; Xu, C.; Fabozzo, A.; et al. A Blood-Resistant Surgical Glue for Minimally Invasive Repair of Vessels and Heart Defects. Sci. Transl. Med. 2014, 6, 218ra6–218ra6. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, T. , Paul, S.. (2006) Protein-misfolding diseases and chaperone-based therapeutic approaches.. FEBS J. 1331. [Google Scholar]
- Cathcart, E.S.; Shirahama, T.; Cohen, A.S. Isolation and identification of a plasma component of amyloid. Biochim. et Biophys. Acta (BBA) - Protein Struct. 1967, 147, 392–393. [Google Scholar] [CrossRef]
- Emsley, J. , White, HE, O'Hara, BP, Oliva, G, Srinivasan, N, Tickle, IJ, Blundell, TL, Pepys, MB, Wood (1994) Structure of pentameric human serum amyloid P component.. Nature.
- Pepys, M. , Booth, DR, Hutchinson, WL, Gallimore, JR, Collins, PM, Hohenester, E (1997) Amyloid P component. A critical review.. Amyloid. 4.
- Botto, M.; Hawkins, P.N.; Bickerstaff, M.C.; Herbert, J.; Bygrave, A.E.; Mcbride, A.; Hutchinson, W.L.; Tennent, G.A.; Walportz, M.J.; Pepys, M.B. Amyloid deposition is delayed in mice with targeted deletion of the serum amyloid P component gene. Nat. Med. 1997, 3, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Sproston, N.R.; Ashworth, J.J. Role of C-Reactive Protein at Sites of Inflammation and Infection. Front. Immunol. 2018, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20, 2510–2528. [Google Scholar] [CrossRef] [PubMed]
- Quittot, N.; Sebastiao, M.; Bourgault, S. Modulation of amyloid assembly by glycosaminoglycans: from mechanism to biological significance. Biochem. Cell Biol. 2017, 95, 329–337. [Google Scholar] [CrossRef]
- Sebastiao, M.; Quittot, N.; Marcotte, I.; Bourgault, S. Glycosaminoglycans Induce Amyloid Self-Assembly of a Peptide Hormone by Concerted Secondary and Quaternary Conformational Transitions. Biochemistry 2019, 58, 1214–1225. [Google Scholar] [CrossRef]
- Tsemekhman, K.; Goldschmidt, L.; Eisenberg, D.; Baker, D. Cooperative hydrogen bonding in amyloid formation. Protein Sci. 2007, 16, 761–764. [Google Scholar] [CrossRef]
- Navarro, S.; Díaz-Caballero, M.; Peccati, F.; Roldán-Martín, L.; Sodupe, M.; Ventura, S. Amyloid Fibrils Formed by Short Prion-Inspired Peptides Are Metalloenzymes. ACS Nano 2023, 17, 16968–16979. [Google Scholar] [CrossRef]
- Occhipinti, R.; Boron, W.F. Role of Carbonic Anhydrases and Inhibitors in Acid–Base Physiology: Insights from Mathematical Modeling. Int. J. Mol. Sci. 2019, 20, 3841. [Google Scholar] [CrossRef]
- Canepa, E. , Parodi-Rullan, R, Vazquez-Torres, R, Gamallo-Lana, B, Guzman-Hernandez, R, Lemon, NL, Angiulli, F, Debure, L, Ilies, MA, Østergaard, L, Wisniewski, T, Gutiérrez-Jiménez, E, Mar, AC, Fossati, S.. (2023) FDA-approved carbonic anhydrase inhibitors reduce amyloid β pathology and improve cognition, by ameliorating cerebrovascular health and glial fitness.. Alzheimers Dement. 5048; 19. [Google Scholar]
- Sapirstein, V.S.; Strocchi, P.; Gilbert, J.M. Properties and Function of Brain Carbonic Anhydrase. Ann. New York Acad. Sci. 1984, 429, 481–493. [Google Scholar] [CrossRef]
- Theparambil, S.M.; Begum, G.; Rose, C.R. pH regulating mechanisms of astrocytes: A critical component in physiology and disease of the brain. Cell Calcium 2024, 120, 102882. [Google Scholar] [CrossRef] [PubMed]
- Ruusuvuori, E. , Kaila, K.. (2014) Carbonic Anhydrases and Brain pH in the Control of Neuronal Excitability. In: Frost, S, McKenna, R. (eds) Subcellular Biochemistry. 75, Springer, Dordrecht. [CrossRef]
- Poggetti, V.; Salerno, S.; Baglini, E.; Barresi, E.; Da Settimo, F.; Taliani, S. Carbonic Anhydrase Activators for Neurodegeneration: An Overview. Molecules 2022, 27, 2544. [Google Scholar] [CrossRef]
- Wood, H.G.; Utter, M.F. The role of CO2 fixation in metabolism. . 1965, 1, 1–27. [Google Scholar] [PubMed]
- Gamble, J. , Lehninger, AL. ( NewYork, 611–622.
- Roosterman, D.; Meyerhof, W.; Cottrell, G.S. Proton Transport Chains in Glucose Metabolism: Mind the Proton. Front. Neurosci. 2018, 12, 404. [Google Scholar] [CrossRef]
- Reynolds, N.P. Amyloid-like peptide nanofibrils as scaffolds for tissue engineering: Progress and challenges (Review). Biointerphases 2019, 14, 040801. [Google Scholar] [CrossRef]
- Peña-Díaz, S.; Olsen, W.P.; Wang, H.; Otzen, D.E. Functional Amyloids: The Biomaterials of Tomorrow? Adv. Mater. 2024, 36, e2312823. [Google Scholar] [CrossRef] [PubMed]
- Aggeli, A. , Boden, N, Zhang, S (editors). . ( medicine and engineering.. Springer, Dordrecht.
- Zhang, S. (2003) More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials.. Nat Biotechnol. 1171; 21. [Google Scholar]
- Eisenberg, D.; Jucker, M. The Amyloid State of Proteins in Human Diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Daskalov, A.; Dyrka, W.; Saupe, S.J. Theme and variations: evolutionary diversification of the HET-s functional amyloid motif. Sci. Rep. 2015, 5, srep12494. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Saupe, S.J. The HET-S/s Prion Motif in the Control of Programmed Cell Death. Cold Spring Harb. Perspect. Biol. 2016, 8, a023515. [Google Scholar] [CrossRef] [PubMed]
- Lamour, G.; Nassar, R.; Chan, P.H.; Bozkurt, G.; Li, J.; Bui, J.M.; Yip, C.K.; Mayor, T.; Li, H.; Wu, H.; et al. Mapping the Broad Structural and Mechanical Properties of Amyloid Fibrils. Biophys. J. 2017, 112, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, S.; Coulary-Salin, B.; Forge, V.; Lascu, I.; Bégueret, J.; Saupe, S.J. The HET-s Prion Protein of the Filamentous Fungus Podospora anserina Aggregates in Vitro into Amyloid-like Fibrils. J. Biol. Chem. 2002, 277, 5703–5706. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, J. , Riek, R.. (2010) Biology of amyloid: structure, function and regulation. 1244; 18. [Google Scholar]
- Pham, C. , Kwan, AH, Sunde, M.. (2014) Functional amyloid: Widespread in nature, diverse in purpose. Essays Biochem. 56.
- Syed, A. , Boles, BR (2014) Fold modulating function: bacterial toxins to functional amyloids.. Frontiers in microbiology. 5.
- Barnhart, M. , Chapman, MR. . ( 2006) Curli biogenesis and function. Annu Rev Microbiol. 60, 131–147.
- Sønderby, T.V.; Zou, Y.; Wang, P.; Wang, C.; Otzen, D.E. Molecular-level insights into the surface-induced assembly of functional bacterial amyloid. Biophys. J. 2022, 121, 3422–3434. [Google Scholar] [CrossRef]
- Xuan, Q.; Wang, Y.; Chen, C.; Wang, P. Rational Biological Interface Engineering: Amyloidal Supramolecular Microstructure-Inspired Hydrogel. Front. Bioeng. Biotechnol. 2021, 9. [Google Scholar] [CrossRef]
- Morris, R. , MacPhee, C.. (2013) Amyloid Protein Biomaterials. In: Roberts, GCK (ed) Encyclopedia of Biophysics.. Springer, Berlin, Heidelberg. [CrossRef]
- Collier, J. , Messersmith, PB.. (2004) Self-assembling polymer peptide conjugates: nanostructural tailoring.. Adv Mater. 16.
- Knowles, T.P.; Fitzpatrick, A.W.; Meehan, S.; Mott, H.R.; Vendruscolo, M.; Dobson, C.M.; Welland, M.E. Role of Intermolecular Forces in Defining Material Properties of Protein Nanofibrils. Science 2007, 318, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber: spider dragline silk. Proc. Natl. Acad. Sci. 1990, 87, 7120–7124. [Google Scholar] [CrossRef]
- Yemini, M.; Reches, M.; Gazit, E.; Rishpon, J. Peptide Nanotube-Modified Electrodes for Enzyme−Biosensor Applications. Anal. Chem. 2005, 77, 5155–5159. [Google Scholar] [CrossRef]
- Scheibel, T.; Parthasarathy, R.; Sawicki, G.; Lin, X.-M.; Jaeger, H.; Lindquist, S.L. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA 2003, 100, 4527–4532. [Google Scholar] [CrossRef] [PubMed]
- Willner, I. , Katz, E, editors.. (2005) Bioelectronics: from theory to applications. Weinheim: Wiley-VCH, GmBH & Co.KGaA.
- Ligorio, C.; Mata, A. Synthetic extracellular matrices with function-encoding peptides. Nat. Rev. Bioeng. 2023, 1, 518–536. [Google Scholar] [CrossRef] [PubMed]
- Deidda, G.; Jonnalagadda, S.V.R.; Spies, J.W.; Ranella, A.; Mossou, E.; Forsyth, V.T.; Mitchell, E.P.; Bowler, M.W.; Tamamis, P.; Mitraki, A. Self-Assembled Amyloid Peptides with Arg-Gly-Asp (RGD) Motifs As Scaffolds for Tissue Engineering. ACS Biomater. Sci. Eng. 2016, 3, 1404–1416. [Google Scholar] [CrossRef] [PubMed]
- van Dalen, M.C.; Karperien, M.; Claessens, M.M.; Post, J.N. Choice of Protein, Not Its Amyloid-Fold, Determines the Success of Amyloid-Based Scaffolds for Cartilage Tissue Regeneration. ACS Omega 2023, 8, 24198–24209. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R. , Das, S, Singh, N, Patel, K, Datta, D, Sen, S, Maji, SK.. (2018) Amyloids Are Novel Cell-Adhesive Matrices.. Adv Exp Med Biol. 1112, 79-97. [CrossRef]
- Jacob, R.S.; George, E.; Singh, P.K.; Salot, S.; Anoop, A.; Jha, N.N.; Sen, S.; Maji, S.K. Cell Adhesion on Amyloid Fibrils Lacking Integrin Recognition Motif. J. Biol. Chem. 2016, 291, 5278–5298. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.S.; Ghosh, D.; Singh, P.K.; Basu, S.K.; Jha, N.N.; Das, S.; Sukul, P.K.; Patil, S.; Sathaye, S.; Kumar, A.; et al. Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation. Biomaterials 2015, 54, 97–105. [Google Scholar] [CrossRef]
- Xuan, Q.; Wang, Y.; Chen, C.; Wang, P. Rational Biological Interface Engineering: Amyloidal Supramolecular Microstructure-Inspired Hydrogel. Front. Bioeng. Biotechnol. 2021, 9. [Google Scholar] [CrossRef]
- Koo, E.H.; Park, L.; Selkoe, D.J. Amyloid beta-protein as a substrate interacts with extracellular matrix to promote neurite outgrowth. Proc. Natl. Acad. Sci. 1993, 90, 4748–4752. [Google Scholar] [CrossRef]
- Ranjan, V. , Qiu, L, Lee, JW, Chen, X, Jang, SE, Chai, C, Lim, KL, Tan, EK, Zhang, Y, Huang, WM, Zeng, L.. (2020) A microfiber scaffold-based 3D in vitro human neuronal culture model of Alzheimer's disease.. Biomater Sci. 8, 4861-4874. [CrossRef]
- Mathes, T.G.; Monirizad, M.; Ermis, M.; de Barros, N.R.; Rodriguez, M.; Kraatz, H.-B.; Jucaud, V.; Khademhosseini, A.; Falcone, N. Effects of amyloid-β-mimicking peptide hydrogel matrix on neuronal progenitor cell phenotype. Acta Biomater. 2024. [Google Scholar] [CrossRef] [PubMed]
- Barbucci, R. , editor.. (2009) Hydrogels: biological properties and applications. Milan/New York: Springer.
- Yanlian, Y.; Ulung, K.; Xiumei, W.; Horii, A.; Yokoi, H.; Shuguang, Z. Designer self-assembling peptide nanomaterials. Nano Today 2009, 4, 193–210. [Google Scholar] [CrossRef]
- Liang, Y.; Guo, P.; Pingali, S.V.; Pabit, S.; Thiyagarajan, P.; Berland, K.M.; Lynn, D.G. Light harvesting antenna on an amyloid scaffold. Chem. Commun. 2008, 6522–6524. [Google Scholar] [CrossRef]
- Channon, K.J.; Devlin, G.L.; MacPhee, C.E. Efficient Energy Transfer within Self-Assembling Peptide Fibers: A Route to Light-Harvesting Nanomaterials. J. Am. Chem. Soc. 2009, 131, 12520–12521. [Google Scholar] [CrossRef]
- Choi, Y.S.; Kim, J.; Bhak, G.; Lee, D.; Paik, S.R. Photoelectric Protein Nanofibrils of α-Synuclein with Embedded Iron and Phthalocyanine Tetrasulfonate. Angew. Chem. 2011, 123, 6194–6198. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Ebanks, K.C.; Barone, J.R. Peptide Mixtures Can Self-Assemble into Large Amyloid Fibers of Varying Size and Morphology. Biomacromolecules 2011, 12, 3770–3779. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Barone, J.R. Evolution of the Amyloid Fiber over Multiple Length Scales. ACS Nano 2012, 7, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D. , Claunch, EC, Barone, JR. (2012) The effect of processing on large, self-assembled amyloid fibers. 1029; 8. [Google Scholar]
- Diaz-Pier, S.; Carloni, P. Impact of quantum and neuromorphic computing on biomolecular simulations: Current status and perspectives. Curr. Opin. Struct. Biol. 2024, 87, 102817. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, T.; Liu, W.; Luo, B.; Li, Y.; Fan, X.; Zhang, X.; Cui, W.; Teng, Y. SemiSynBio: A new era for neuromorphic computing. Synth. Syst. Biotechnol. 2024, 9, 594–599. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, H.; Xie, D. Recent Advance in Synaptic Plasticity Modulation Techniques for Neuromorphic Applications. Nano-Micro Lett. 2024, 16, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ma, Z.; Zheng, H.; Li, T.; Chen, K.; Wang, X.; Liu, C.; Xu, L.; Wu, X.; Lin, D.; et al. The combination of brain-computer interfaces and artificial intelligence: applications and challenges. Ann. Transl. Med. 2020, 8, 712–712. [Google Scholar] [CrossRef] [PubMed]
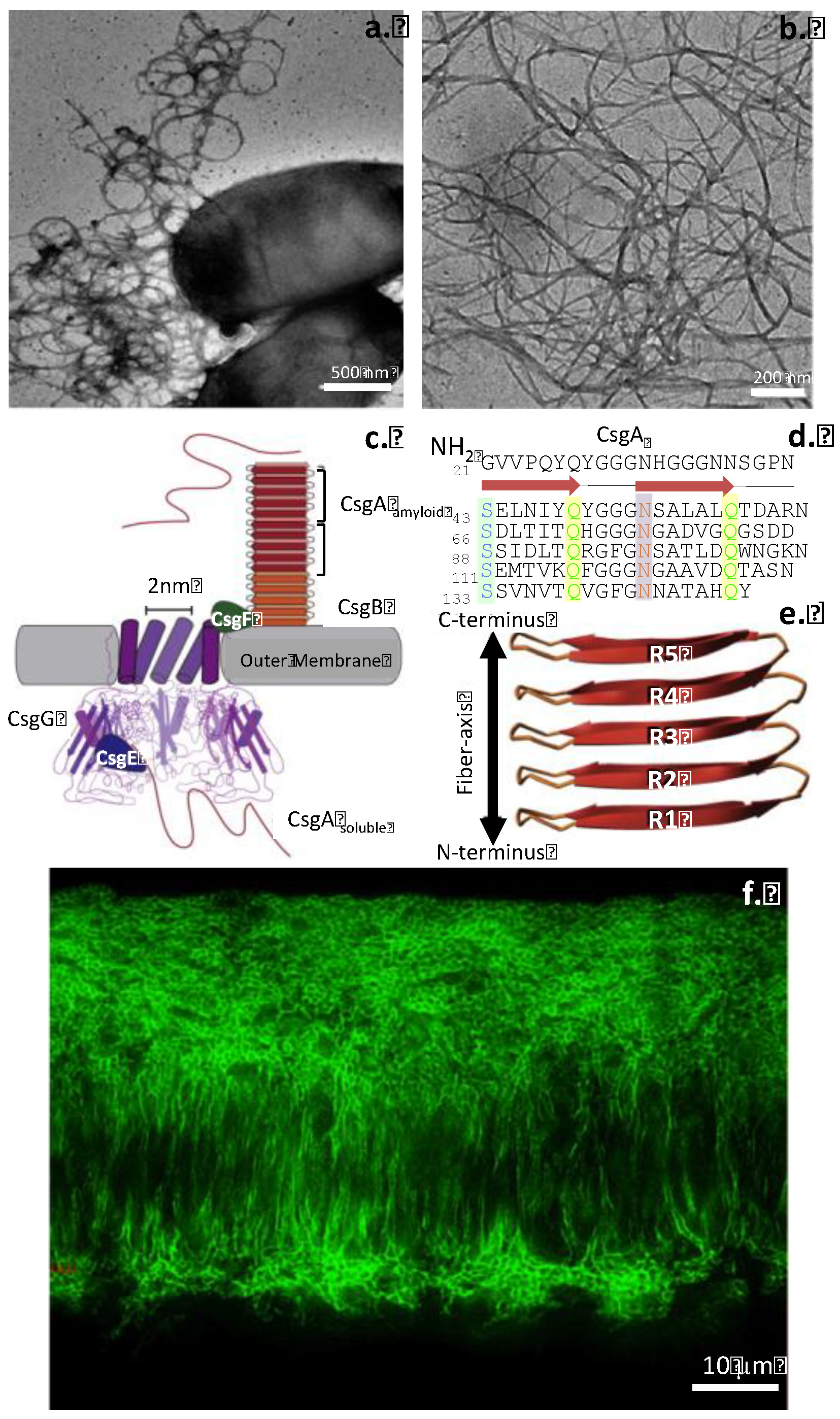
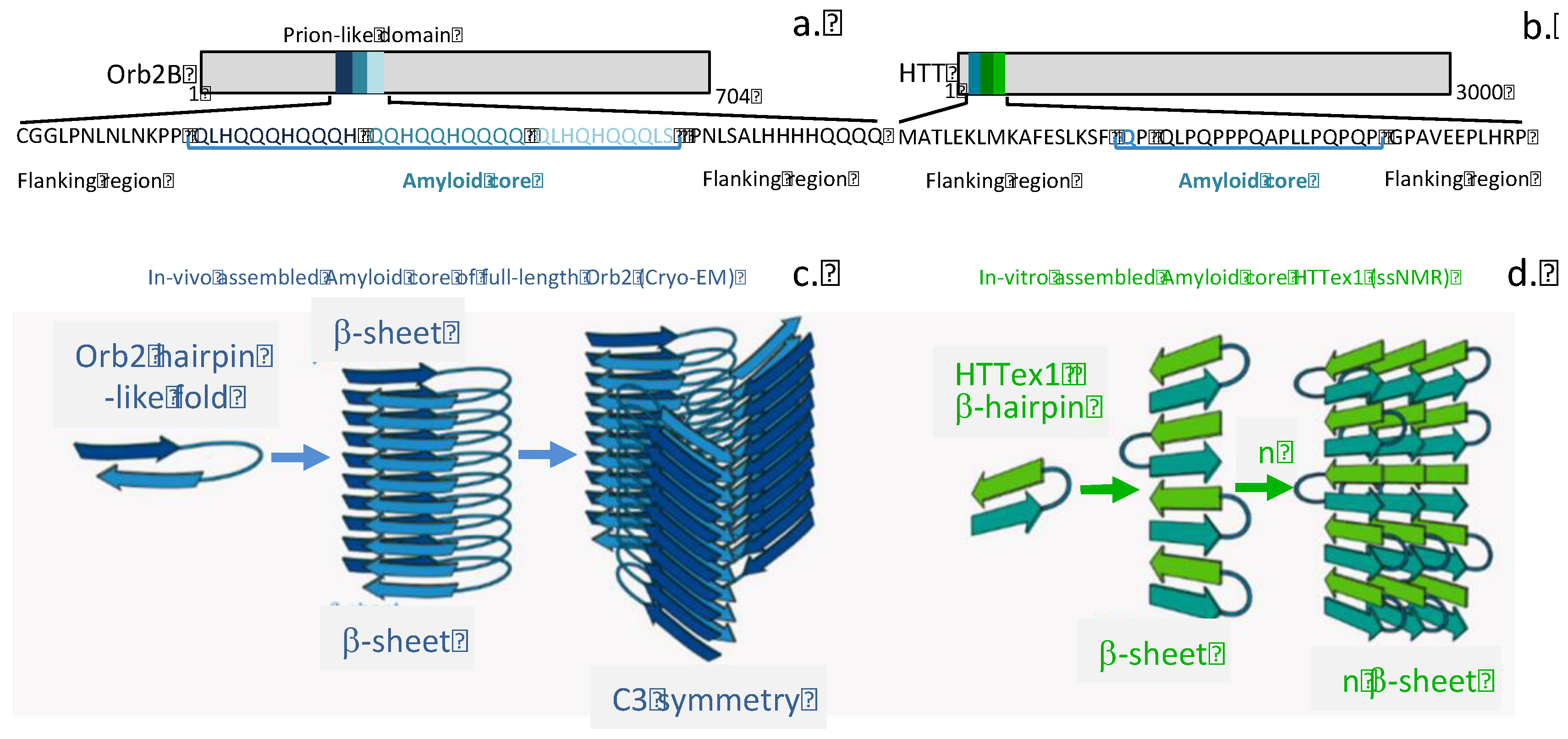
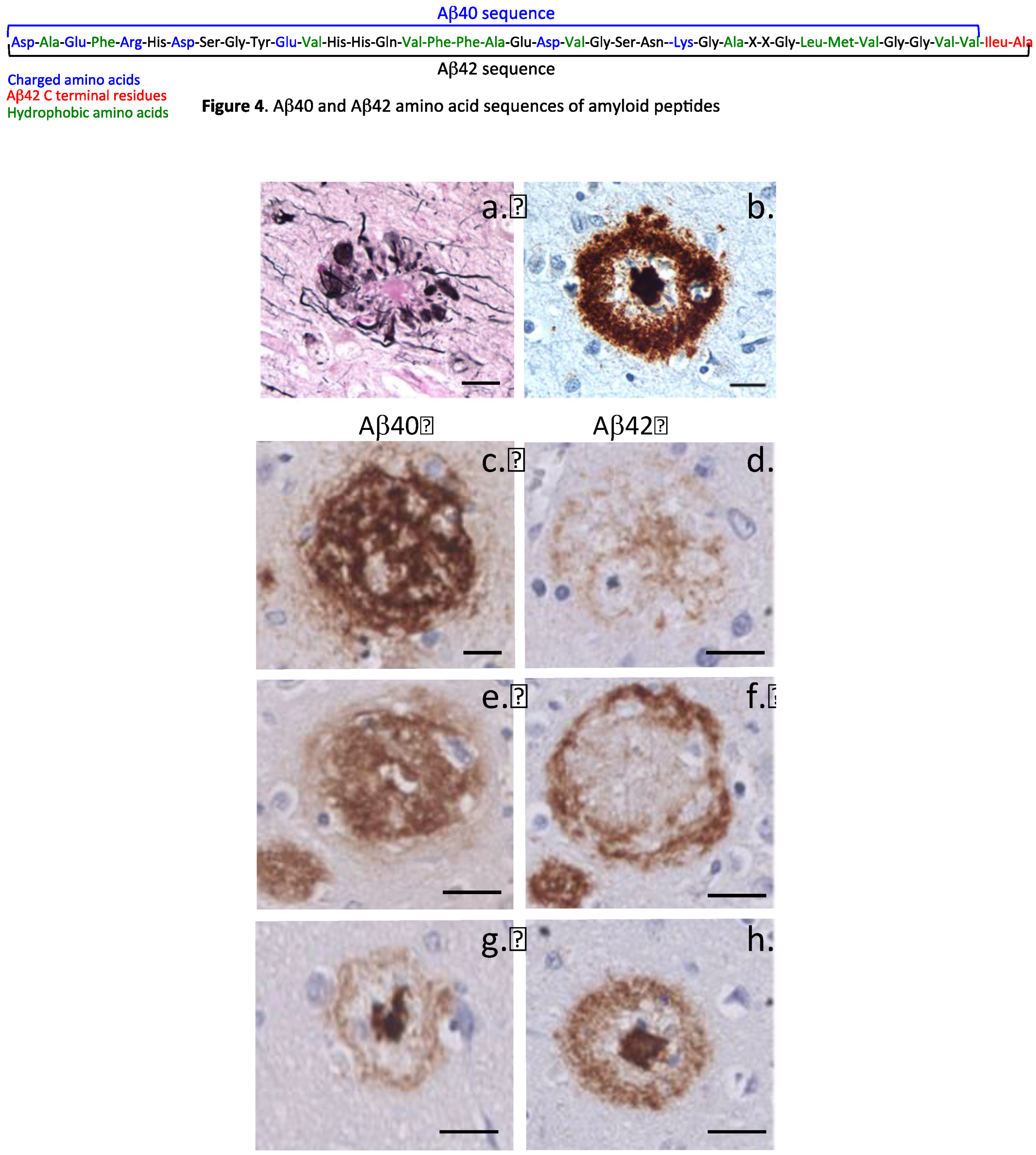
| Disease process | Amyloid precursor protein | Amyloid monomer |
|---|---|---|
| AD | APP | A |
| Atrial amyloidosis | Atrial natriuritic protein | Amyloid ATF |
| Spongiform encephalopothy | Prion protein PrPc | PrPsc |
| Primary systemic amyloidosis | Ig L and H chains | AL, AH |
| Secondary systemic amyloidosis | Apo serum amyloid A | SAA |
| Familial amyloid polyneuropathy I | Transthyretin | ATTR |
| Familial amyloid polyneuropathy II | ApoA | AApoA |
| Haemodialysis amyloidosis | 2-microglobulin | A2M |
| Hereditary systemic amyloidosis | Lysozyme | ALys |
| Diabetes type II | ProIAPP | APP/amylin |
| Insulin injection amyloidosis | Insulin | AIns |
| Cerebral amyloid angiopathy | Cystatin C | ACys |
| Finnish hereditary systemic amyloidosis | Gelsolin | AGel |
| Age associated pituitary prolactinomas | Prolactin | APro |
| Familial amyloidosis | Fibrinogen A chain | AFib |
| Amyloid | Applications | Reference |
|---|---|---|
| Transthyretin peptide (105-115) | Cell adhesive properties | [186] |
| Gonadotropin releasing hormone | Long acting peptide/protein drug depot/delivery | [187] |
| Candida albicans Killer decapeptide | Auto-delivery of therapeutic peptides | [188] |
| Yeast Sup35p NM prion domain | Nanowire development | [189] |
| -synuclein fibrils | Enzyme entrapment hydrogel | [190] |
| insulin fibril semi-conductor oligoelectrolyte |
Optoelectronic nanowire assembly | [191] |
| PPF coated Insulin fibrils | Polymer light emitting diode fibrils | [192] |
| APFO-12 coated Insulin fibrils | Optical nanowires | [193] |
| PEDOT-S coated Insulin fibrils | Conductive nanowires | [194] |
| 2-microglobulin | Nanoporous cell support matrix | [195] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





