Submitted:
22 May 2024
Posted:
05 June 2024
You are already at the latest version
Abstract
Keywords:
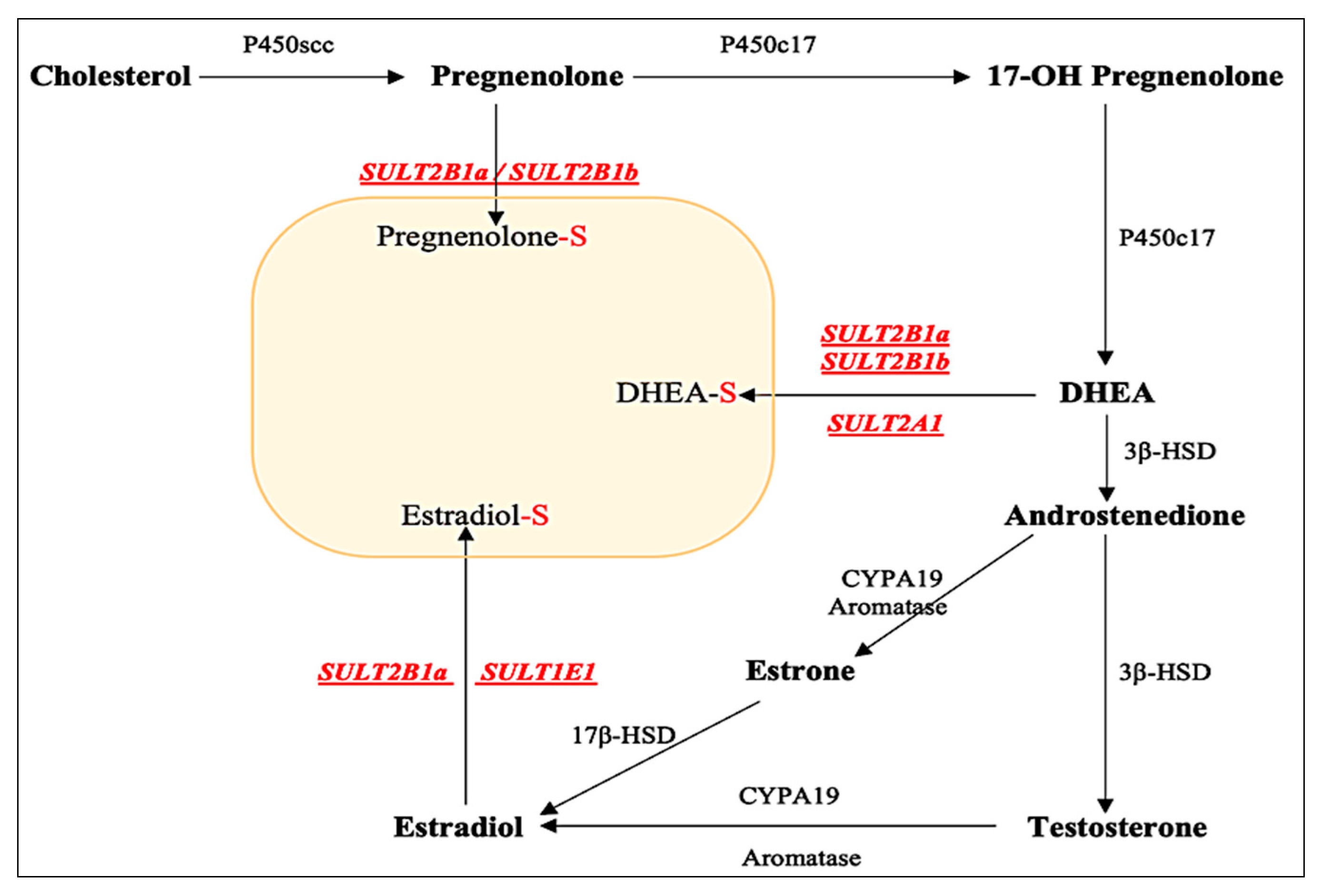
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
Enzymatic Assay for the Sulfating Activity of the Purified Recombinant SULT2B1a Allozymes
Kinetic Studies and Statistical Analysis
Data Analysis
3. Results
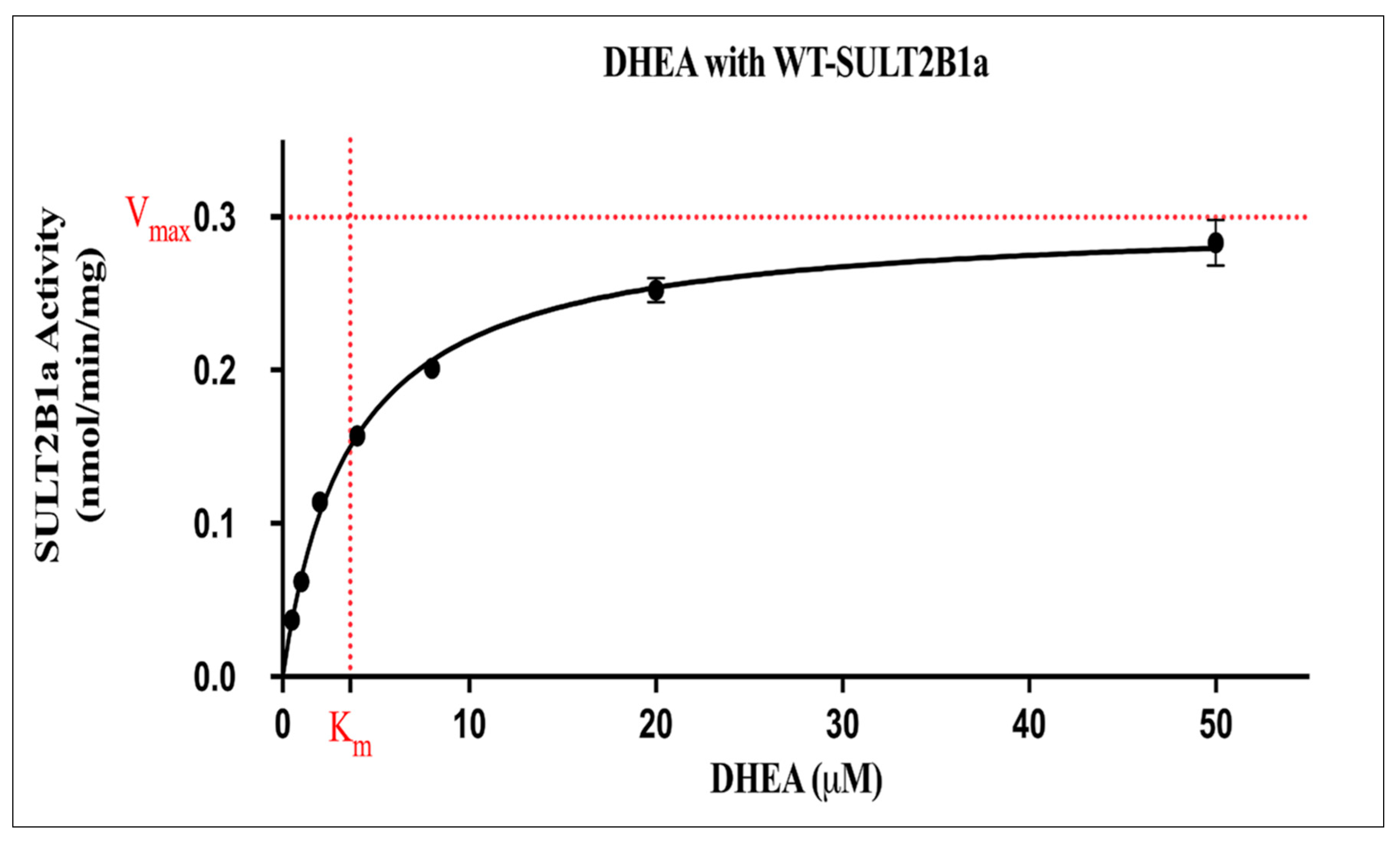
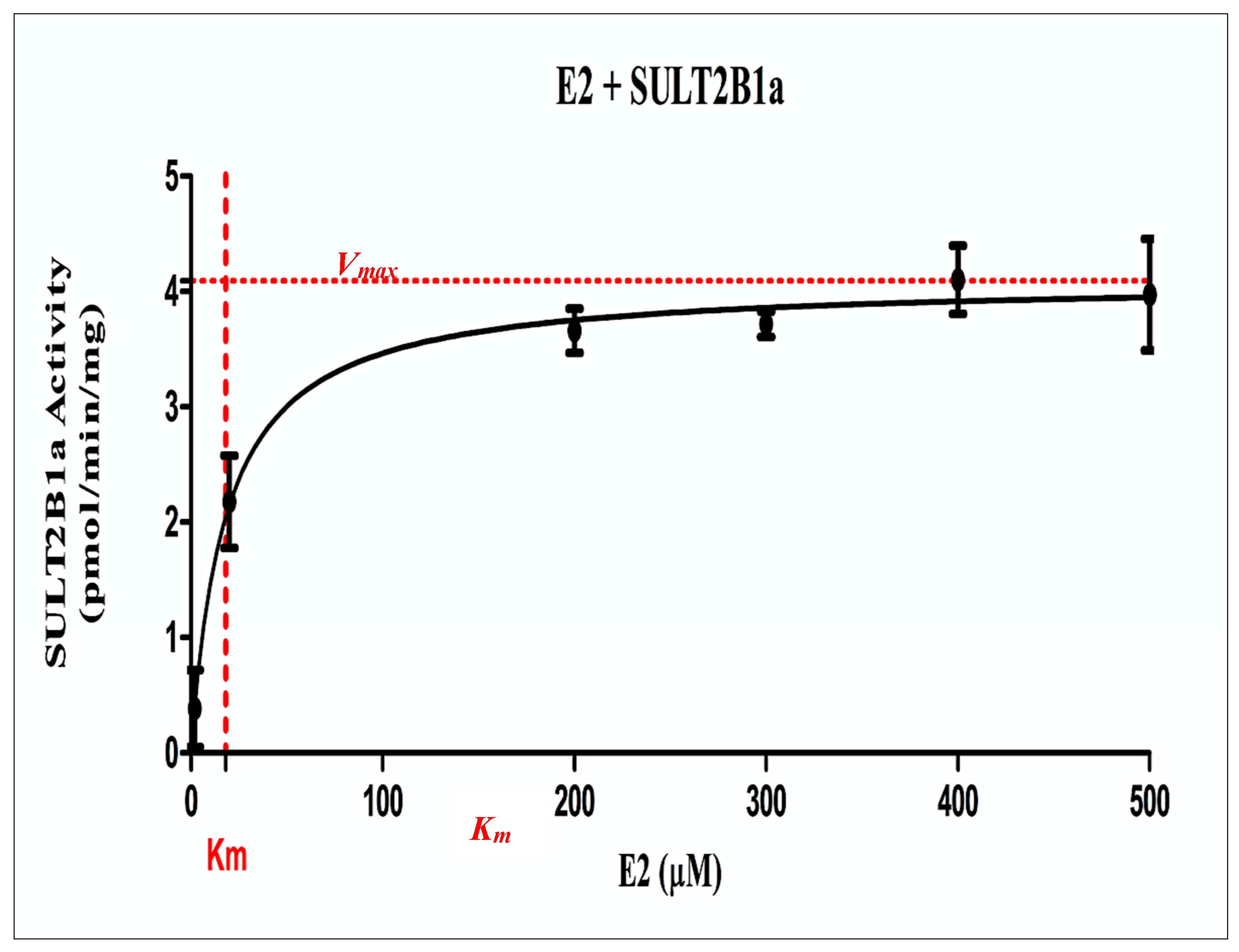
Kinetic Study on the Sulfation of DHEA and E2 by wild-Type SULT2B1a
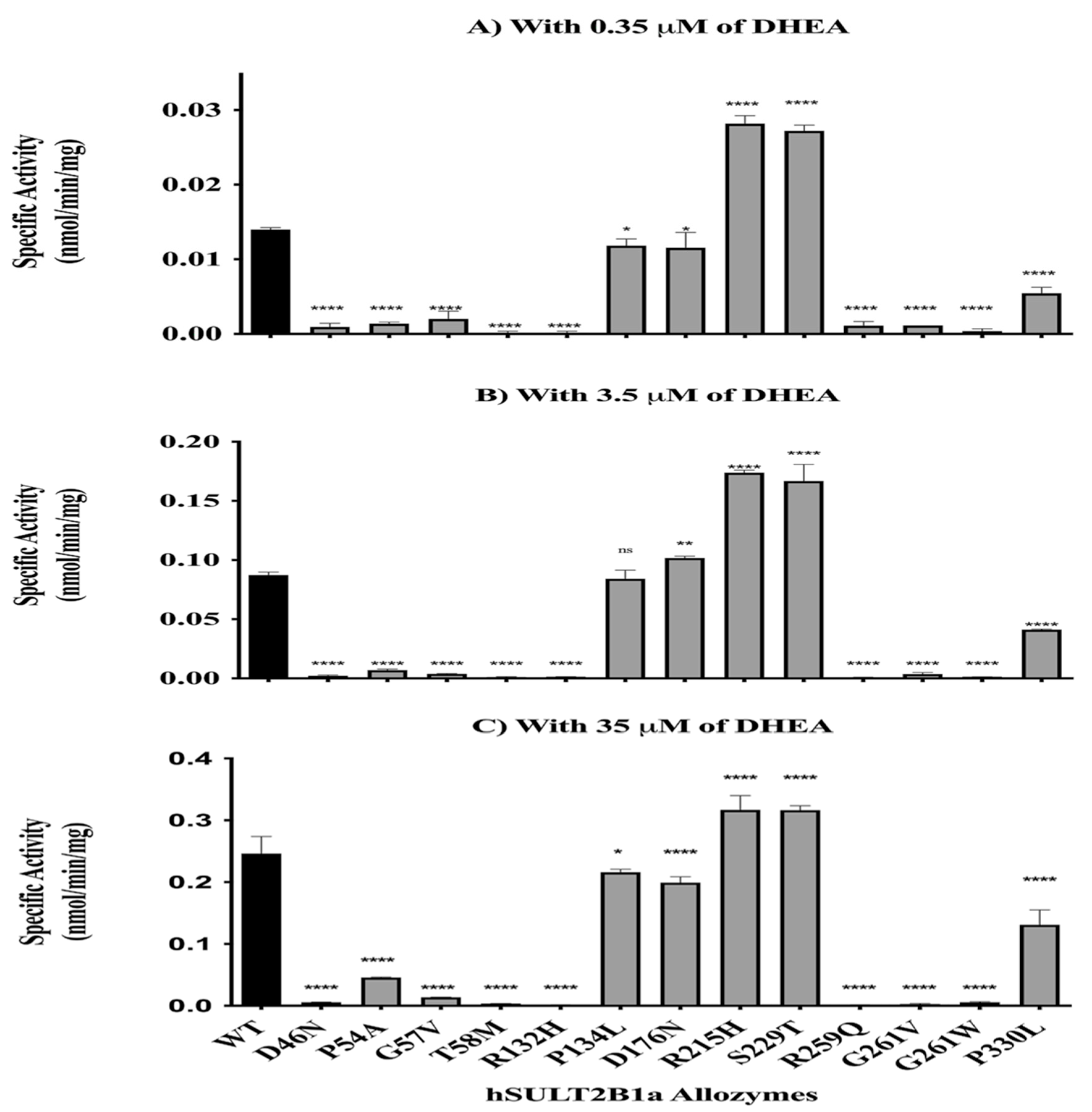
Enzymatic Characterization of SULT2B1a Allozymes Using DHEA and E2
4. Discussion
5. Conclusion
References
- Labrie, F., DHEA, important source of sex steroids in men and even more in women, in Progress in brain research. 2010, Elsevier. p. 97-148.
- Davis, S., M. Panjari, and F. Stanczyk, DHEA replacement for postmenopausal women. The Journal of Clinical Endocrinology & Metabolism, 2011. 96(6): p. 1642-1653.
- Raftogianis, R., et al., Estrogen metabolism by conjugation. JNCI Monographs, 2000. 2000(27): p. 113-124. [CrossRef]
- Ruiz-Cortés, Z.T., Gonadal sex steroids: production, action and interactions in mammals. Steroids-From Physiology to Clinical Medicine, 2012: p. 3-44.
- Schiffer, L., W. Arlt, and K.-H. Storbeck, Intracrine androgen biosynthesis, metabolism and action revisited. Molecular and Cellular Endocrinology, 2018. 465: p. 4-26.
- Stocco, C., Tissue physiology and pathology of aromatase. Steroids, 2012. 77(1-2): p. 27-35. [CrossRef]
- Falany, C.N., Enzymology of human cytosolic sulfotransferases. The FASEB Journal, 1997. 11(4): p. 206-216. [CrossRef]
- Falany, C. and J. Roth, Human Drug Metabolism; From Molecular Biology to Man. by Jeffery EH, CRC Press, Boca Raton, 1993: p. 101-115.
- Mulder, G. and W. Jakoby, Sulfation. Conjugation reactions in drug metabolism. Mulder GJ, 1990.
- Weinshilboum, R. and D. Otterness, Sulfotransferase enzymes, in Conjugation—Deconjugation Reactions in Drug Metabolism and Toxicity. 1994, Springer. p. 45-78.
- Falany, C., Properties of human cytosolic sulfotransferases involved in drug matabolism. Human Drug Metabolism: From Molecular Biology to Man, 1993: p. 101-115.
- Lipmann, F., Biological sulfate activation and transfer. Science, 1958. 128(3324): p. 575-580. [CrossRef]
- Falany, C.N. and K.J. Rohn-Glowacki, SULT2B1: unique properties and characteristics of a hydroxysteroid sulfotransferase family. Drug metabolism reviews, 2013. 45(4): p. 388-400. [CrossRef]
- Noordam, C., et al., Inactivating PAPSS2 mutations in a patient with premature pubarche. New England Journal of Medicine, 2009. 360(22): p. 2310-2318. [CrossRef]
- Vallée, M., W. Mayo, and M. Le Moal, Role of pregnenolone, dehydroepiandrosterone and their sulfate esters on learning and memory in cognitive aging. Brain Research Reviews, 2001. 37(1-3): p. 301-312. [CrossRef]
- Falany, C., et al., Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. The Journal of steroid biochemistry and molecular biology, 2006. 102(1-5): p. 214-221.
- Freimuth, R.R., et al., Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J, 2004. 4(1): p. 54-65. [CrossRef]
- HENDERSON, E., M. WEINBERG, and W.A. WRIGHT, Pregnenolone. The Journal of Clinical Endocrinology, 1950. 10(4): p. 455-474.
- Schumacher, M., et al., Pregnenolone sulfate in the brain: a controversial neurosteroid. Neurochemistry international, 2008. 52(4-5): p. 522-540. [CrossRef]
- Li, Y., et al., Structure, function and polymorphism of human cytosolic sulfotransferases. Current drug metabolism, 2008. 9(2): p. 99-105. [CrossRef]
- Eagle, K., ADHD impacted by sulfotransferase (SULT1A) inhibition from artificial food colors and plant-based foods. Physiology & behavior, 2014. 135: p. 174-179. [CrossRef]
- Eagle, K., Toxicological effects of red wine, orange juice, and other dietary SULT1A inhibitors via excess catecholamines. Food and chemical toxicology, 2012. 50(6): p. 2243-2249. [CrossRef]
- Han, D.-F., et al., Sulfotransferase 1A1 (SULT1A1) polymorphism and breast cancer risk in Chinese women. Toxicology letters, 2004. 150(2): p. 167-177. [CrossRef]
- El Daibani, A.A., et al., Impact of Human SULT1E1 Polymorphisms on the Sulfation of 17β-Estradiol, 4-Hydroxytamoxifen, and Diethylstilbestrol by SULT1E1 Allozymes. Eur J Drug Metab Pharmacokinet, 2021. 46(1): p. 105-118.
- Yanagisawa, K., et al., cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Bioscience, biotechnology, and biochemistry, 1998. 62(5): p. 1037-1040.
- Alatwi, E. and A.F. Bairam, The role of genetic polymorphisms in the sulfation of pregnenolone by human cytosolic sulfotransferase SULT2B1a. Scientific Reports, 2024. 14(1): p. 8050. [CrossRef]
- Traish, A.M., et al., Dehydroepiandrosterone (DHEA)—a precursor steroid or an active hormone in human physiology (CME). The journal of sexual medicine, 2011. 8(11): p. 2960-2982. [CrossRef]
- Glatt, H., et al., Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 2001. 482(1-2): p. 27-40.
- Freimuth, R., et al., Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. The pharmacogenomics journal, 2004. 4(1): p. 54-65. [CrossRef]
- Her, C., et al., Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics, 1998. 53(3): p. 284-295. [CrossRef]
- Falany, C.N., V. Krasnykh, and J.L. Falany, Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. The Journal of steroid biochemistry and molecular biology, 1995. 52(6): p. 529-539. [CrossRef]
- Schrag, M.L., et al., Sulfotransferase 1E1 is a low km isoform mediating the 3-O-sulfation of ethinyl estradiol. Drug metabolism and disposition, 2004. 32(11): p. 1299-1303.
- Zhang, H., et al., Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. Journal of Biological Chemistry, 1998. 273(18): p. 10888-10892. [CrossRef]
- Cui, J., Y. Shen, and R. Li, Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends in molecular medicine, 2013. 19(3): p. 197-209. [CrossRef]
- Hyland, P.L., et al., Genetic variants in sex hormone metabolic pathway genes and risk of esophageal squamous cell carcinoma. Carcinogenesis, 2013. 34(5): p. 1062-1068. [CrossRef]
- Lévesque, É., et al., Steroidogenic germline polymorphism predictors of prostate cancer progression in the estradiol pathway. Clinical Cancer Research, 2014. 20(11): p. 2971-2983. [CrossRef]
- Mostaghel, E.A., Steroid hormone synthetic pathways in prostate cancer. Translational andrology and urology, 2013. 2(3): p. 212.
- Yang, X., et al., Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo. PLoS One, 2013. 8(4): p. e60853. [CrossRef]
- Vickman, R.E., et al., Cholesterol sulfonation enzyme, SULT2B1b, modulates AR and cell growth properties in prostate cancer. Molecular Cancer Research, 2016. 14(9): p. 776-786. [CrossRef]
- Chen, W., et al., Overexpression of SULT2B1b promotes angiogenesis in human gastric cancer. Cellular Physiology and Biochemistry, 2016. 38(3): p. 1040-1054. [CrossRef]
- Hu, L., et al., Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Laboratory investigation, 2015. 95(9): p. 1005-1018. [CrossRef]
- Heinz, L., et al., Mutations in SULT2B1 cause autosomal-recessive congenital ichthyosis in humans. The American Journal of Human Genetics, 2017. 100(6): p. 926-939. [CrossRef]
- Lee, K.A., et al., Crystal Structure of Human Cholesterol Sulfotransferase (SULT2B1b) in the Presence of Pregnenolone and 3′-Phosphoadenosine 5′-Phosphate RATIONALE FOR SPECIFICITY DIFFERENCES BETWEEN PROTOTYPICAL SULT2A1 AND THE SULT2B1 ISOFORMS. Journal of Biological Chemistry, 2003. 278(45): p. 44593-44599.
- Ji, Y., et al., Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. Journal of Pharmacology and Experimental Therapeutics, 2007. 322(2): p. 529-540. [CrossRef]
- Fuda, H., et al., Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. Journal of Biological Chemistry, 2002. 277(39): p. 36161-36166. [CrossRef]
- Betts, M.J. and R.B. Russell, Amino acid properties and consequences of substitutions. Bioinformatics for geneticists, 2003. 317: p. 289.
| Substrate | Final Concentrations (µM) |
|---|---|
| Dehydroepiandrosterone$(DHEA) | 0.5, 1, 2, 4, 8, 20, 50 |
| 17-β estradiol$(E2) | 2, 20, 200, 300, 400, 500 |
| Substrate | Vmax | Km (µM) |
|---|---|---|
| Dehydroepiandrosterone (DHEA) | 0.299 ± 0.004 (nmol/min/mg). | 3.616 ± 0.18 |
| 17-β estradiol$(E2) | 4.092 ± 0.081 (pmol/min/mg). | 18.12 ± 2.46 |
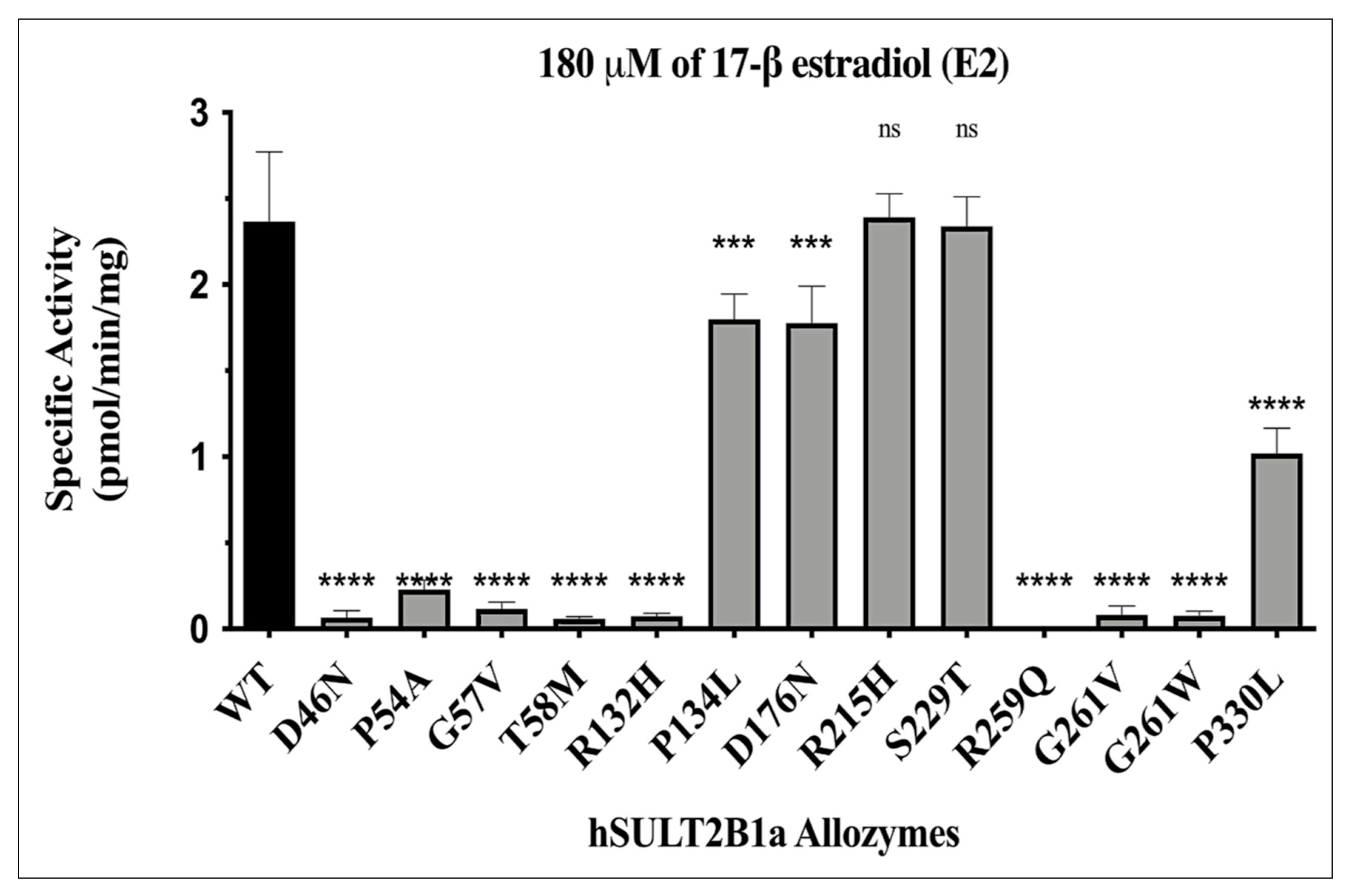
| Allozyme | Relative activity (% of wild-type SULT2B1a) |
|---|---|
| SULT2B1a-D46N | ≈2-3 |
| SULT2B1a-P54A | ≈10-20 |
| SULT2B1a-G57V | ≈5 |
| SULT2B1a-T58M | ≈3-2 |
| SULT2B1a-R132H | ≈1-3 |
| SULT2B1a-P134L | ≈75-85 |
| SULT2B1a-D176N | ≈75-80 |
| SULT2B1a-R215H | ≈100-130 |
| SULT2B1a-S229T | ≈100-130 |
| SULT2B1a-R259Q | ND |
| SULT2B1a-G261V | ≈1-3 |
| SULT2B1a-G261W | ≈2-3 |
| SULT2B1a-P330L | ≈45-50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




