1. Introduction
Time-series biosignals such as Electroencephalography (EEG), Electromyography (EMG), Electrocardiography (EKG), Electrooculography (EOG), and many more have become increasingly popular for use in commercial and research fields [
1,
2,
3,
4]. The most prevalent use for biosignals in the commercial and research domain is for the diagnosis of medical disorders. This includes research on early onset warning signs for diseases that impact physical mobility using EMG, monitoring the heart health of patients prone to cardiovascular diseases, and diagnosis of mental health disorders using neural activity [
5,
6]. Powered devices that aid mobility, such as bionic prostheses, brain-computer interfaces (BCIs), and powered orthotics, may also use EMG or EEG signals to enable more natural control of the aides [
7]. EMG and EKG have recently become a tool for use in enhancing athletic performance through optimized training results based on biosignal readings [
8]. The final large area of use for biosignals is neuroscience and behavioral research to improve our understanding of the precise functions performed by the brain to elicit emotions, thoughts, memory, and many more mental functions [
9].
Biosignals result from electrical activity emitted by the body in response to physiological processes [
10]. These include physical responses from muscle movement recorded as EMG signals, recording electrical cardiac activity from the heart as EKG signals, brain activity from EEG signals, and eye movement from EOG signals. Many biosignals are recorded as time-domain signals, in which the electrical potential variation is observed over time. Multiple applications for biosignals pair the time-domain recordings with directed tasks in order to obtain correlations between bodily function and the given task [
11]. The biosignals associated with a specific task are used as training data in a machine learning (ML) database to perform analysis or to map specific biosignal responses to outputs in mobility aides [
12].
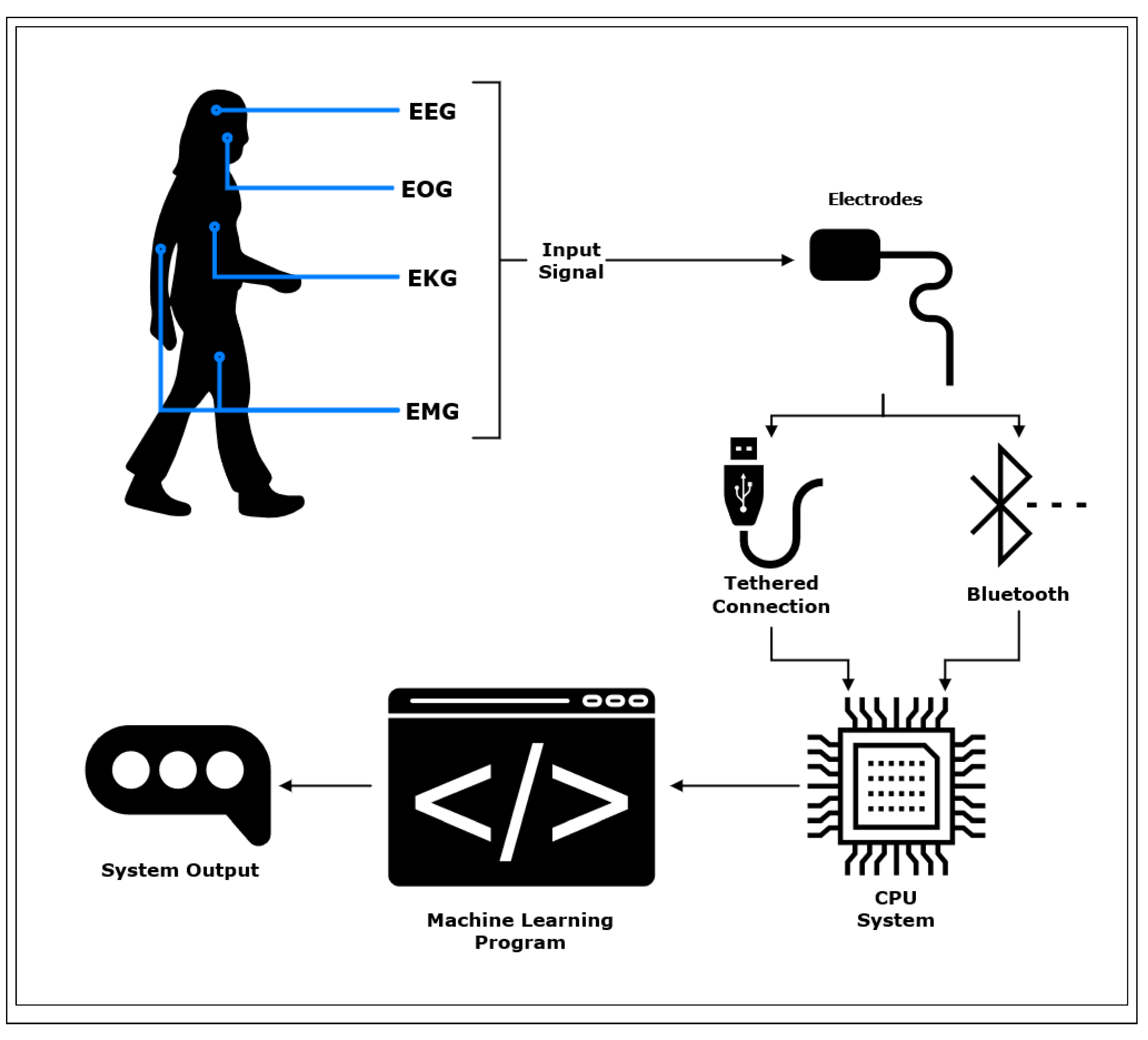
Biosignals are recorded through the use of non-invasive electrodes that are applied to the surface of the skin, or less commonly through the use of invasive needle electrodes used to collect electrical activity from underneath the surface of the skin [
13]. Electrodes may be tethered to a computational device for signal processing; however, systems are increasingly trending towards wireless models in which an electrode will transmit data over Bluetooth low-energy (BLE) to a computational unit [
14]. Electrodes used for biosignal acquisition will often employ sampling rates of 128 Hz - 2000+ Hz to construct accurate representations of physiological responses. Multiple electrodes are applied to the area(s) of interest to construct a spatio-temporal representation of physiological responses. The resolution of the recorded biosignal is dependent on the quality of the electrode, and the number of bits used to represent each sample.
As the technology and science around biosignals are developed further, the size of electrodes is decreased while the ability of the electrode to pick up higher-quality and higher-resolution data is increased [
15]. The development of more advanced electrodes, paired with current advancements in computer storage space, has allowed manufacturers of biosignal acquisition systems to introduce systems with more electrodes monitoring the areas of interest. For instance, the prevalence of increased numbers of electrodes in EEG recording devices has resulted in two new updates to the international electrode placement guidelines. The original 10-20 system was designed for 21 electrodes but was largely replaced with the 74-electrode 10-10 system since the 2000s. Modern systems often include 128-256 electrodes, which has resulted in the new 345-electrode 10-5 placement system [
16].
By increasing the number of electrodes or increasing the resolution of the signal obtained by each electrode, the amount of data that must be stored and processed by the system is greatly increased. In applications where data processing can be performed at a later point, such as performing analysis on biosignal data recorded during a clinical study, this increased processing time will result in a longer time for patient diagnosis and will result in a clinic being able to diagnose fewer patients in a given time period [
17,
18]. In real-time systems, such as a BCI or a bionic prosthesis which must react in a matter of seconds, processing large sets of matrix data produced by multi-electrode systems is not possible without dedicated hardware [
19,
20]. This makes it expensive for the user to purchase the assistive devices while also reducing the amount of research and innovation in this area due to price constraints. Many real-time systems will also reduce the number of electrodes or limit the functionality of the systems to speed up processing times [
15]. However, this often results in a lower-performance system for the end user.
Many systems that use biosignal inputs will rely on computational processing units (CPUs) or portable embedded devices such as microcontrollers (MCUs) to process the data through ML algorithms to determine the system outputs [
21]. In clinical settings, specialized machines may be developed to improve processing speed; however, due to costs, medical device regulations, and limited vending capacity, these systems are not often used outside of specialized clinics [
22]. Most CPUs and MCUs do not have the capacity to process large amounts of matrix data and are thus becoming inadequate for the speed requirements of many modern biosignal-based systems [
19]. To overcome this technological gap, many researchers have introduced Graphics Processing Units (GPUs) into biosignal-based systems to take advantage of the massively parallel matrix arithmetic capabilities of these devices [
23]. NVIDIA GPUs have been among the most readily adopted for speeding up ML processes due to the open-source Compute Unified Device Architecture (CUDA) toolkit provided by NVIDIA to run multiple popular ML algorithms at greatly accelerated rates.
The use of GPU acceleration in biosignal analysis has been practiced in numerous studies, showcasing the potential to benefit from this acceleration method in biosignal applications. In a study by Yu et al., an EMG signal is applied through a GPU-accelerated Bayesian filtering method to obtain real-time filtering [
24]. In another usage for EMG signals, a prosthetic limb used an FPGA-GPU interface in order to approach real-time muscle activity recognition [
25]. For EEG signals, GPU acceleration has been used to successfully speed up pattern recognition due to the long processing time that traditional CPUs need for large electrode array setups [
26]. GPU acceleration has also been implemented to diagnose abnormalities from remote ECG signals [
27]. These examples of GPU-accelerated biosignal implementations provide a basis on which further research may be conducted.
In this paper, the introduction of additional kernels to the CUDA toolkit for use with time-series biosignals will be investigated. By adding kernels that implement ML algorithms that are currently available for use on CPUs on a GPU device, more biosignal applications can be accelerated to produce faster outputs. This is especially critical for improving the usability of more complex real-time systems, which are currently limited by CPU or MPU processing times. Most current kernels for use on GPUs are targeted toward image processing, and as a result, researchers are limited to a small number of available ML algorithms for fast systems. By investigating current research toward introducing field-applicable and biosignal-specific ML algorithms as GPU kernels, the viability of GPU acceleration toward biosignal processing can be assessed.
In summary, the objective of this review is: (a) To provide an assessment of the benefits of GPU kernel development towards ML processes on biosignals. (b) Understand the architecture needed to design a GPU kernel. (c) Look into the speedup of GPU kernels over CPU-based algorithms. (d) Discuss the impact of GPU kernels towards the future development of biosignal systems.
2. Review Methodology
The inclusion criteria for the reviewed material are as follows: (1) Papers are published in the English language. (2) Papers are available on the Google Scholar search engine. (3) Papers are obtained from a peer-reviewed journal, conference, or university publication. (4) Articles include the keywords of biosignals, GPU kernels, kernel development, or GPU acceleration. (5) Articles are published between the years of 2014 and 2023. (6) Article pertains to the development of a GPU-based kernel for an ML algorithm. (7) GPU-accelerated algorithms must have known applications within the field of biosignal analysis.
All of the reviewed material was accessed between January 2023 and May 2023. The material of each paper was independently reviewed by the authors without the use of automation tools. Papers were reviewed for numerical speedup results where possible. In the case of non-numeric speedup results the numerical results are omitted from comparisons. Papers without numerical results indicate the final speed achieved by the GPU algorithm accompanied by confirmation that a speedup had been achieved.
For each paper that met the first 6 inclusion criteria, the GPU-accelerated algorithm was compared to recent literature in biosignal ML applications to verify field relevancy for the seventh inclusion criteria. The literature used in reference for field relevancy can be found in
Section 4.3. For the 12 papers that met all inclusion criteria, the GPU model and specifications that may impact expected acceleration performance were recorded. The speedup results were obtained from the results provided within each paper. The use of laptops, desktops, or servers was noted for each study to differentiate the results. Graphical results are displayed using graphs generated using spreadsheet software.
This systematic approach allowed for the selection of relevant studies and the extraction of critical information to evaluate the benefits of GPU kernel development in biosignal processing.
3. Architecture
In order to configure current ML algorithms into accelerated GPU kernels, the architecture of a CPU, GPU, and the CUDA Developer Toolkit must be considered. ML algorithms that are commonly used in the analysis and classification of biosignal data are traditionally designed for use with multicore CPUs that will implement the algorithm across a designated number of threads [
28]. When switching an algorithm over to the massively paralleled capabilities of a GPU kernel, the algorithm needs to be redesigned to be processed in batches of threads in order to take full advantage of the GPU design. The CUDA Developers Toolkit provided by NVIDIA provides a wide range of resources with which GPU-accelerated kernels can be coded, compiled, tested, and can interact with multiple GPU-based libraries.
3.1. Hardware Architectures
In this section, the physical architecture of both the CPU and GPU are described to showcase the attributes that differ between the two components. The difference in physical structure provides a basis on which the kernel’s structure is formed. Since more kernels are developed for use with a CPU than GPU, and given that many GPU kernels are designed to emulate the functionality of CPU-based kernels, the architecture of the CPU should be observed first.
When comparing a CPU and GPU, it should be noted that a GPU utilizes specialized clusters of arithmetic logic units (ALUs) that are able to run large matrices of numerical data through mathematical operations in parallel, whereas a CPU utilizes multi-threading to perform generalized tasks in a serial manner [
29]. While new CPU architecture may be developed to allow a greater number of threads to be available to the user, or specialized CPUs such as an AMD threadripper may be used to increase threading performance, average CPU programs do not benefit from massively increased threading [
30]. This is because not all tasks can be broken up into smaller processes that can be run massively in parallel [
31].
3.1.1. CPU Architecture
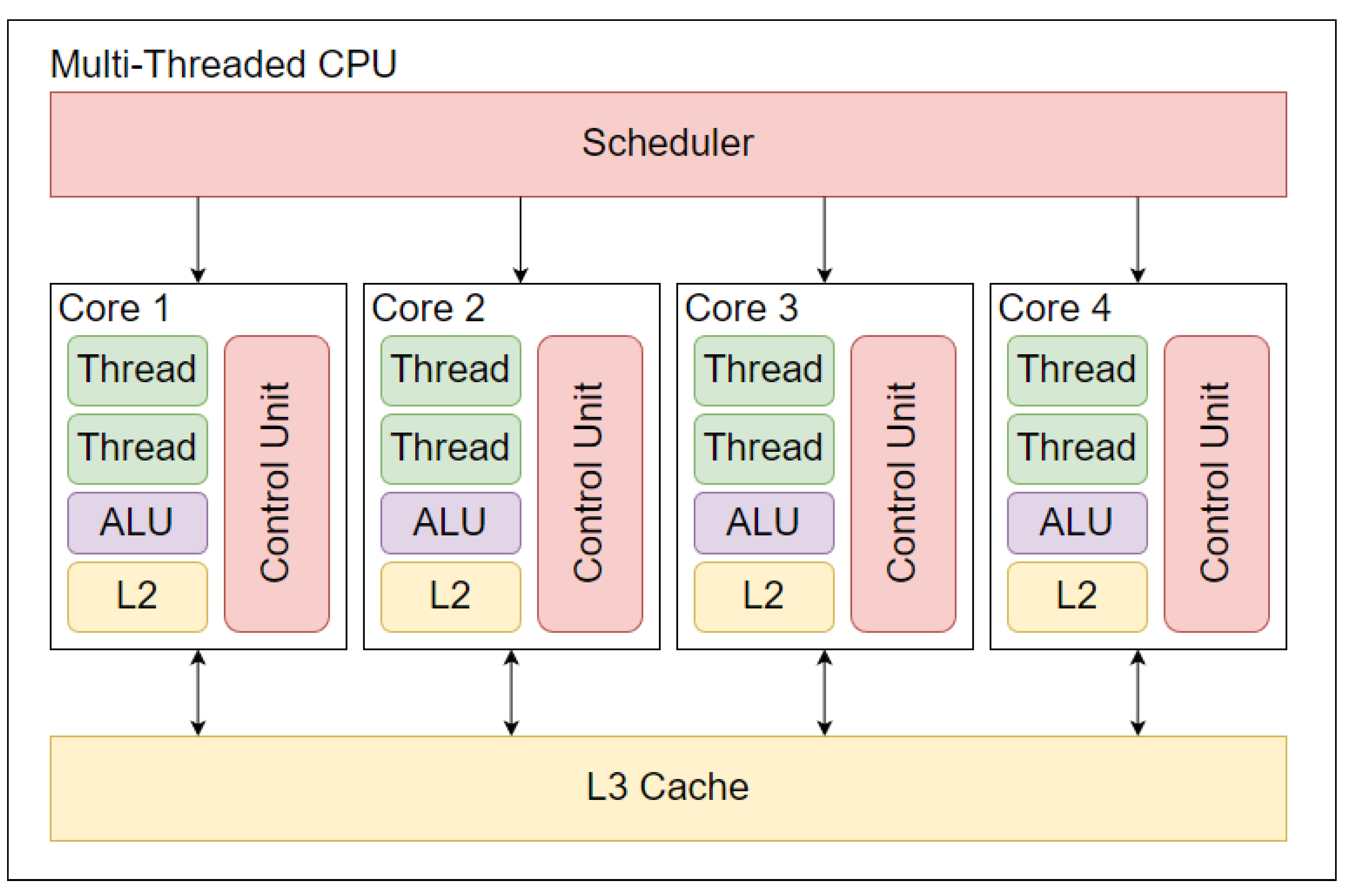
A traditional CPU architecture consists of one or more CPU cores which are controlled by a CPU scheduler. The CPU scheduler allocates tasks to the CPU by determining which processes may be executed and which tasks will be placed on hold across all available cores [
32]. The capabilities of the individual cores within the CPU vary depending on whether a single-thread or multithread unit is selected. In a single-threaded CPU, each core may execute one thread at a given time, whereas an individual multithread CPU core is associated with two logical cores that can each operate a thread in parallel [
30]. The components of a multithread CPU can be seen in
Figure 1. Due to the increased number of threads provided by a multithreaded CPU, these are most often associated with ML [
33,
34]. Many consumer-grade multithread CPU systems in the present market contain between 4-16 cores, while top market high-end CPUs such as the AMD Ryzen Threadripper offer 32 or 64 physical cores that provide 64 or 128 logical processors respectively [
35]. CPUs with 32 or more logical cores are most often observed in servers designed to perform high-volume operations in industrial settings. High-end servers may contain CPUs with up to 128 physical processors at the time of writing [
36].
A traditional CPU will contain three levels of memory caches, referred to as L3, L2, and L1 from largest to smallest capacity. Each cache is made up of high-speed Static Random Access Memory (SRAM), which provides increased data transfer speeds at reduced memory capacity [
30,
37]. The L3 cache is the largest capacity cache, storing data that is shared amongst all CPU cores. Due to the large amount of data stored in L3 relative to the L2 and L1 caches, the time needed to locate and retrieve data from the registers is the slowest of all the levels. However, a typical L3 cache will operate at double the speed of the Dynamic Random Access Memory (DRAM) that acts as the main temporary memory storage for traditional computer setups [
38]. Information that may be relevant to general processes performed by a CPU at any given time will be stored at the L3 level [
30]. Each core will have a dedicated L2 cache which has a storage capacity under half the size of the L3 cache but operates at roughly 25 times the speed of DRAM [
37]. Information pertinent to a specific core’s allocated process will be stored in the L2 cache of each core. The L1 cache size ranges significantly but has drastically less memory space available than the L2 cache. This, in turn, allows the L1 cache to operate at roughly four times the speed of the L2 cache on average [
39]. Each core will contain two L1 caches. One L1 cache stores the instructions to execute the core’s scheduled processes, while the second L1 cache contains the data used in the instructed operation [
40]. A CPU core will first search the L1 cache for the relevant data and, if not found, will go to the L2, then L3 caches, respectively, before resorting to obtaining data from the DRAM [
40]. This process allows data to be retrieved at the most efficient speed possible.
By prioritizing fast data retrieval from the available memory, a CPU is optimized to perform a wide array of general tasks at high speed. The CPU is not designed to specialize in specific data formats, and therefore being able to find relevant data is of more importance than creating highly efficient protocols for processing specific data formats [
41]. Each CPU core contains an Arithmetic Logic Unit (ALU), which is used to perform mathematical instructions which are available to perform numerical operations and may also contain other units intended for the processing of specific data formats. Having these units within the CPU cores allows for varied data to be processed by each core individually.
3.1.2. GPU Architecture
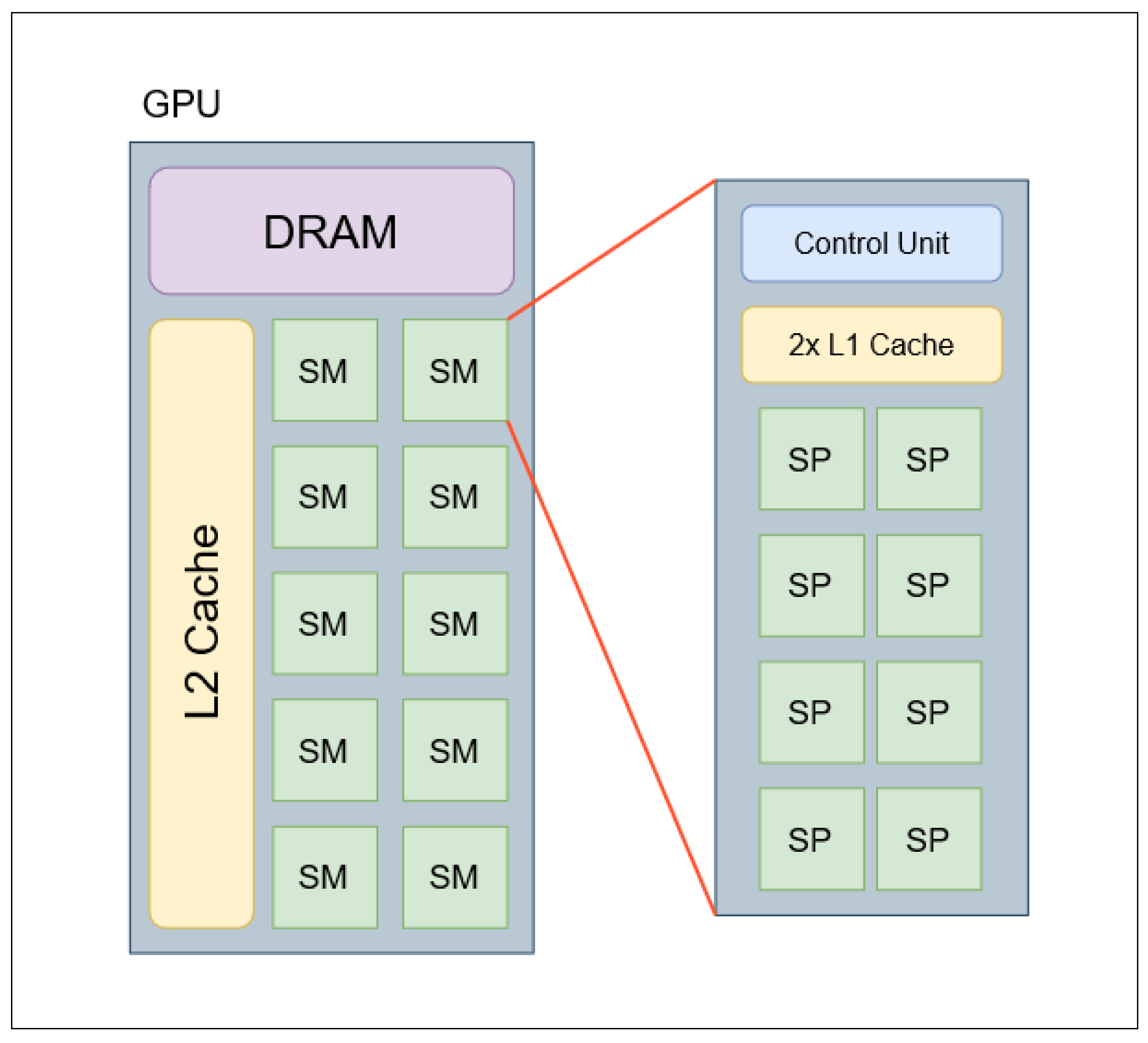
A typical GPU is comprised of multiple clusters of GPU cores arranged in units called streaming multiprocessors (SMs), a dedicated DRAM, and a large L2 cache shared amongst all SMs. The GPU contains multiple memory controllers that are linked to the SMs using network-on-chip (NoC) architecture [
43]. The NoC equips every SM and memory controller with a router containing five data input and output (I/O) ports [
44]. The routers are typically connected to each other in a grid pattern, with one I/O port transmitting data to a network interface embedded into each SM and memory controller and four I/O ports connected to a physical wire used to transmit data to neighboring routers [
45]. The NoC allows data to be retrieved from the L2 cache and sent to the appropriate SM using the memory controllers [
46]. Each memory controller is allocated to a designated portion of the L2 cache, which allows faster indexing for individual memory controllers [
47]. The GPU also employs a higher bandwidth than a CPU, allowing for larger amounts of data to be transferred at any given time [
48].
An individual SM is made up of multiple GPU cores, which are referred to as streaming processors (SPs) or CUDA cores when using NVIDIA architecture [
49]. Most modern GPUs SMs will contain multiples of 32 SPs in an SM, however, in older GPUs only 8 or 16 SPs may be present [
50]. In addition to the cluster of SPs, an SM contains two dedicated L1 memory caches - one cache contains the instructions that are distributed to the SPs in parallel, and the second L1 cache contains the data distributed to the SPs for use in calculations [
47,
57]. Each SM will contain one or more control units. The control unit has two dispatch units that receive instructions from the L1 instruction cache and allocate the threads to the appropriate SPs using a local register shared by the SPs. If all threads cannot be executed the dispatch unit will schedule additional threads to an SP in the following instruction cycle [
51]. When an SM executes 32 threads in parallel, the grouping of the 32 threads is collectively referred to as a warp. To ensure the dispatch units schedule warps correctly, the control unit contains a warp scheduler. The warp scheduler will ensure both dispatch units run in parallel by delaying the scheduled execution of threads until both dispatch units are available. The warp scheduler is also responsible for ensuring that the dispatch unit cannot send out instructions for SP units that are already busy[
52]. A simplified diagram of the base components contained in a general GPU is shown in
Figure 3.
Many variations in GPU architecture exist due to the development of new embedded technology and the variation in expected usage. Most GPUs are designed with the intent to be used near-exclusively to speed up graphics processing and therefore may contain one or more tensor cores in the SM to exchange computational accuracy for higher processing speeds [
53]. Each SP is also designated to process specific data types, most commonly for 32-bit integer (INT32), 32-bit floating point (FP32), or 64-bit floating point (FP64) data types [
54]. In addition to regular SPs, an SM may also contain a special function unit that is able to process transcendental mathematical functions while maintaining functional accuracy [
55]. The SP clusters may contain multiple Load/Store (LD/ST) units which increase the number of instructions performed per cycle by dispatching and retrieving data to and from the SP multiple times per cycle [
56]. An increasingly popular trend is the implementation of multiple 32 SP clusters, each with its own dedicated control unit and L0 instruction cache on one SM [
57].
All graphics cards in the GPUs used for testing in the reviewed studies are provided by NVIDIA. The specifications of the GPU cards used in each of the reviewed studies varies greatly. The base clock speed, DRAM size, L2 cache size, and L1 cache size of the GPU card(s) used in each study can be found in
Table 1.
3.2. CUDA Developer Toolkit
The CUDA Toolkit is a set of resources provided by NVIDIA Corporation to allow the development of GPU-accelerated computations on NVIDIA brand GPUs. All GPU-accelerated processes developed using the toolkit must use the Single Program Multiple Data (SPMD) parallelism techniques. When using SPMD parallelism a singular program is executed, and the data processed by the program is divided among multiple SPs. The resources provided by the toolkit include a GPU-syntax-modified programming language, a custom compiler, and multiple libraries that can be used to access previously developed GPU-accelerated programs.
Programs developed using the CUDA Toolkit are programmed in an extended variant of C++ named CUDA C++. The syntax of CUDA C++ follows the structural, memory, and procedural practices of C++, with the addition of GPU-specific context [
70]. The most notable additions and changes are the specifications of kernels, multi-dimensional thread hierarchy, GPU-specific memory hierarchy, and CPU-GPU interactions. All CUDA C++ programs and related files are compiled using the Nvidia CUDA Compiler (NVCC) [
71]. The NVCC also provides an array of compilation options to determine how input files are interpreted.
In the context of the CUDA Toolkit, a kernel is defined as a function that can be run in parallel over multiple SPs [
72]. Typically the kernel is developed to perform a singular process without dependencies on data resulting from the completion on other threads in different SMs [
73]. When a kernel is called by a program, the number of threads that run the kernel in parallel must also be defined. If sequential instructions must be performed on data, such as in the case of aggregating data, multiple kernels will be developed for the different instructions needed at various points in the data processing pipeline, or the same kernel will be called multiple times by the main algorithm [
73]. Due to the language associated with classic computation, an entire algorithm that calls multiple kernels may also be collectively referred to as a singular kernel.
Due to the large number of threads employed by a given CUDA application, threads can be indexed as a one, two, or three-dimensional array. The thread arrays are called thread blocks, where each block is contained to a singular SM, and the number of threads contained within the block is limited by the hardware of the GPU utilized. As of CUDA v12.1, the maximum number of threads that may be contained within a block is 1024 [
70]. Alternatively, one can implement a kernel across multiple thread blocks of the same size to utilize multiple SMs in parallel by creating a grid of thread blocks [
74].
When choosing which memory a thread will access, it must be considered which memory spaces are visible to a given thread. As each thread block is contained to a singular SM, each block is allocated shared private L1 data and instruction memory caches [
75]. All threads contained within a thread block share both read and write access to the L1 cache, and threads in other blocks will not be able to view this data. If using a graphics processing cluster (GPC), an architecture in which multiple SMs are grouped together, a Distributed Shared Memory can be created to share data from the L1 caches of different SMs [
76]. To ensure that threads may run independently of each other, the L1 cache supports atomized operations. All threads from all blocks have access to the L2 cache and will be able to publicly view any changes made to this memory space. All threads also have read-only access to the global, constant, and texture memory spaces. All three of these memory spaces remain immutable during the entire run time of a GPU-accelerated program [
77].
Unlike in traditional CPU-based programming in which only one type of processor is used, a GPU-Accelerated program is developed with two or more types of processors in use [
78]. All computing devices contain a CPU that runs both ML and general tasks. In contrast, a GPU lacks the ability to perform complex general tasks that are needed for the execution of a program [
79]. CUDA operates using heterogeneous programming in which kernels are executed on the GPU, and the CUDA C++ program is executed on a CPU [
80]. In the instance of CUDA, a GPU is referred to as a coprocessor or device, while the system CPU is referred to as a host. Because both the device and host have individual DRAM space, these are allocated as the device and host memory respectively. All global, constant, and texture data is contained within the host memory but may be transferred to the device memory by the program. Likewise, data from the device memory is transferred to the host memory by the program. In instances where data must be shared by both the device and host, a tool named Unified Memory may be used to allocate a managed memory space that is accessible to both components [
70].
4. Kernel Development
In this section, the setup and acceleration of algorithms performed in the 12 reviewed studies are analyzed. The system setup, ML algorithms that are GPU accelerated, algorithm relevance to biosignal processing, and datasets tested will be reviewed to provide a view of the factors that must be considered when analyzing the speedup results from the GPU acceleration of the ML algorithms. A summary of the system setup and GPU-accelerated kernels can be found in
Table 2.
4.1. System Setup
While all the reviewed studies use the CUDA developer toolkit to develop GPU-accelerated algorithms on NVIDIA-produced graphics cards, the system setups vary greatly from study to study. The GPU hardware used by each study can be found in
Table 1. Because GPU-accelerated programs are developed using a heterogeneous system, the GPU alone is not exclusively responsible for the expected speedup results. The CPU used by the system is also an important factor, as CUDA C++ processes performed on the CPU may delay the onset of instructions to the GPU, creating a bottleneck effect. In addition to considering the GPU and CPUs used in the reviewed studies, the operating systems (OS), and computer type, all influence the final speedup results.
Within the reviewed studies, all CPUs used to test the speedup results and act as the host for the CUDA C++ program is produced by Intel Corporation. Intel i-series processors are categorized under three labels: budget i3 microprocessors, medium-range i5 microprocessors, and high-end i7 microprocessors. None of the reviewed studies document the usage of an Intel i3 microprocessor. The Intel i5 microprocessor architecture is used by three of the reviewed studies [
62,
64,
65]. Within the architecture of the i5 microprocessor, three different CPU models are used. The Intel i7 microprocessor architecture is observed in three studies, each using a different model [
62,
63,
65]. The Intel Xeon E5-series and Quad series CPUs microprocessors are also used in multiple reviewed studies. The Intel Quad model Q6600 is used only by Da-Ming Chang & Shyan-Ming Yuan while the Intel Xeon E5 series microprocessors are used by X studies [
58,
59,
60,
61,
62,
63,
65,
66]. Within the Intel Xeon E5 microprocessor architecture, 11 different CPU models are employed.
4.2. System Setup
Two of the reviewed studies provide the operating system used. Jansson et al. has used a Windows OS over all three testing systems, using Windows 7 in two systems and Windows Server 2012 in their third testing system. Linux is explicitly used by Tsai et al. as the OS for their testing system. The chosen Linux distribution by Tsai et al. is Ubuntu 12.04.2.
Three types of computer systems are observed in the reviewed studies. Traditional desktop computers are used by all of the reviewed studies except for Tsai et al., who only use a server. A desktop provides a highly-customizable device in which many high-performance consumer-grade components may be fitted at a wide range of price points. A desktop may provide high performance for a consumer device if the right components are used; however, the results are expected to vary greatly with individual setups. A laptop is used by Gutiérrez et al. to assess GPU-accelerated performance across multiple computer types. A laptop will typically be expected to provide lower performance compared to a desktop due to the power limitations incurred by a portable device. Lastly, Gutiérrez et al., Tsai et al., and Lescano et al. all use a server as part of their test system setup. A server is typically designed using industry-grade components that provide much better performance due to their specialized design. A third-party virtual server, such as that used by Lescano et al., will assign a subsect of the available CPU and GPU cores to an individual user; thus, the specifications of the system may not be reflected in the performance obtained by the allocated processors.
4.3. ML Algorithms GPU-Accelerated
Multiple different widely-used ML algorithms have been GPU-accelerated in the reviewed studies, each with varied applications toward biosignal processing. Not all GPU kernels were developed explicitly for use in biosignal processing; rather, many are developed for use in the broader ML field while also providing increased resources for biomedical computation. The ML algorithms that have been GPU-accelerated using kernel development and the applications of the algorithms in biosignal processing are discussed in this section.
Two of the reviewed studies develop GPU kernels to accelerate ensemble-based decision tree algorithms [
61,
68]. Jansson et al. developed kernels for both the Random Forest (RF) and Extremely Randomized Trees (ERT) algorithms, which have been named gpuRF and gpuERT, respectively. Chang and Yuan developed a kernel to accelerate a Decision Tree (DT) algorithm named CUDT. All three of these algorithms are based on ensemble learning ML architecture, in which multiple models with slight variations are formed and compared to one another to determine the best classification result [
42]. Using CUDA kernel capabilities, models can be built in parallel rather than sequentially before final model aggregation is performed to determine the classification of input data. In biosignal applications, ensemble ML methods are regularly applied to classify physiological signals. This includes classifying the intended motor movements of an individual using EEG mental command detection [
81], the movement of prosthesis devices by classifying muscle activity measured by EMG [
82], or classifying arrhythmia in heartbeats using EKG signals [
83].
In addition to classical ensemble learning algorithms, many of the reviewed papers develop kernels for boosted ensemble learning algorithms. Four of the reviewed studies developed a GPU kernel to parallelize a boosted ensemble algorithm [
58,
63,
65]. A Gradient Boosted Decision Tree (GBDT) ML algorithm is accelerated by Wen et al. in a direct GPU kernel implementation. Mitchell & Frank implemented a kernel to accelerate an Extreme Gradient Boost (XGBoost) algorithm. Another popular boosted ensemble algorithm, Adaptive Boosting (AdaBoost), is accelerated by Tsai et al., while Lescano et al. uses the AdaBoost-based Viola-Jones (VJ) algorithm as the baseline for their kernel. Boosted ensemble learning maintains the same basis as classical ensemble learning; however, models are evaluated after each build instead of being collectively assessed at the end of the algorithm [
84]. This allows the creation of an ML algorithm in which the weaknesses of each individual model can be used as input to improve the construction of future models [
85]. Boosted ensemble methods are employed in biosignal classification in the same areas as classical ensemble methods. Boosted ensemble methods are increasingly popular in biosignal classification due to their model parameter-tuning capabilities that may provide more accurate results [
86].
The K-means and K-Nearest Neighbor (KNN) are two widely used distance-based algorithms that classify input data by creating clusters of locally grouped data. A K-means kernel is developed by Bhimani et al. to perform distance calculations in parallel. Gutiérrez et al. developed a Synthetic Minority Over-Sampling Technique (SMOTE) kernel, a KNN-based ML algorithm used to increase the number of available data points in one or more minority class(es) before training a classification ML model [
87]. While both kernels use distance-based algorithms, both have different applied uses in biosignal processing. KNN is used to classify biosignals into distinct categories by evaluating the degree of separation between samples or extracted variables[
4]. SMOTE is used earlier in the ML pipeline, being used to balance datasets with more samples available in some classes than others. By adding in synthetic replica samples, an ML model trained using the balanced dataset will be less prone to bias [
87]. This serves to improve databases in which motor movement or mental tasks are performed due to the abundance of resting phases present during typical data collection [
88].
The Fast Independent Component Analysis (FastICA) algorithm is accelerated in both Benko & Juhansz and Gajos-Balińska et al.’s studies. ICA algorithms operate by grouping data based on the statistical variation between data points in a system. The FastICA algorithm estimates components on a one-by-one basis compared to traditional ICA methods of performing all calculations in one sequential run [
89]. Thus, FastICA provides faster performance on traditional multithreaded CPUs and has an algorithm that is pre-dispositioned toward parallelization. FastICA is widely used in biosignal analysis due to its ability to remove white noise from recorded signals [
90]. Within the field of neurological science, FastICA is amongst the most popular methods for removing unwanted artifacts from an EEG signal [
91].
Abbasi & Rafiee and Spandana et al. both accelerated ML algorithms which rely on the procedural optimization of the provided dataset and sequential retraining of the ML models to provide associations between datapoints. Abbasi & Rafiee accelerate a Genetic Algorithm (GA) using a GPU kernel. A GA creates recombinations of data from the provided dataset to determine which data combinations provide enhanced performance and repeats this process to create further enhancements from the previous sets of best-performance data recombinations [
92]. GAs are commonly used in biosignals for the selection of an optimized feature set for use in classification ML algorithms [
93]. The Apriori Algorithm operates by locating related sets of data within the provided samples and then finding further points of correlations within the set [
94]. An Apriori algorithm allows the reconstruction of biosignals for noise removal [
95].
4.4. Datasets
Multiple datasets are used to test the performance of the GPU-accelerated algorithms. Due to the different applications and datatypes used in junctions with a given type of model, the use of a singular dataset to compare GPU acceleration results will not result in an accurate portrayal of acceleration capabilities. In addition to comparison considerations, the architecture of a CPU will produce better results when simple vectors are processed in low quantities, whereas a GPU will be expected to increase processing speed when large matrices of data are utilized. The datasets used in each of the reviewed studies can be found in
Table 3.
5. Speedup Results
A wide array of ML algorithms are GPU-accelerated in the reviewed studies. When considering the impact parallelization has on increasing the speed over a CPU-based ML algorithm, the factors impacting the speed of the CPU-based algorithms should be considered. When running a CPU-based algorithm, multithreading may be used to increase the speed at which certain processes are performed. The time that each system may take to fit, train, and evaluate a model will differ due to the system setup. As a result, the comparison of GPU time over CPU time is measured in terms of speedup within the system instead of time in milliseconds. The speedup results for GPU performance over CPU performance vary by study. The speedup is calculated using the formula:
where both CPU and GPU execution time is measured in seconds. The speedup results of the developed GPU kernels over their CPU-based algorithm counterpart are shown in
Table 4.
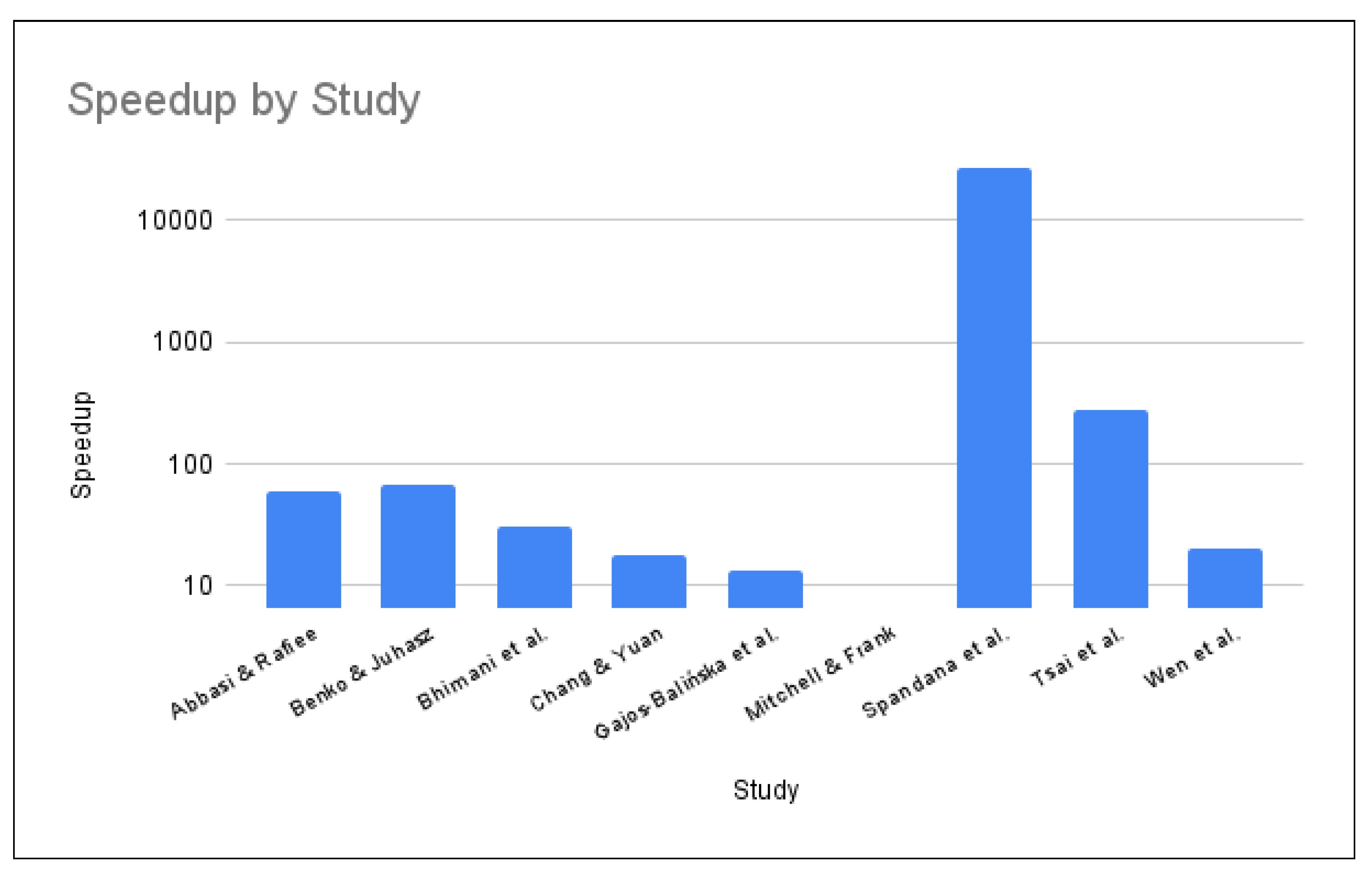
All GPU kernels provide a positive speedup over the CPU counterpart. The speedup results for the GPU kernel over the CPU algorithm are provided in nine of the reviewed papers. The greatest speedup of 27018.37 was performed by Spandana et al. by comparing their GPU Apriori kernel to a CPU-based Apriori algorithm on a 1000 transaction database. Tsai et al. obtain a best speedup of 282.62 when performing quantization feature extraction. The GPU FastICA by Benko & Juhansz provides a speedup of 67 over the 8-core CPU Matlab implementation of the FastICA algorithm when using a dataset of 128 channels being sampled at 2048 Hz for 20 seconds. Abbasi et al. obtain their best speedup of 58.35 when testing their CUDA GA kernel against a CPU-based GA algorithm using a population of 16384. Bhimani et al. obtain a speedup of 30.26 over a CPU-based K-means using K=240. Wen et al. have a best speedup of 19.88 over the xgbst-1 algorithm and a lower speedup of 1.87 over the xgbst-40 algorithms, both when using the news20 dataset. The CUDT algorithm by Chang & Yuan obtained a speedup of 18 against the SPRINT algorithm and a speedup of 6 over the Weka-j48 decision tree algorithm. Gajos-Balińska et al. obtained a speedup of 13.45 with their GPU FastICA kernel over the CPU-based FastICA algorithm. The lowest maximum speedup is achieved by Mitchell &Frank, having a speedup of 6.62 over the CPU implementation of the XGBoost algorithm.
Gutiérrez et al., Lescano et al., and Jansson et al. do not test their GPU kernels against the CPU-based counterparts for speedup. The SMOTE-GPU kernel is tested on three GPU-enabled platforms, a laptop, desktop, and server, to compare the performance across different system setups. The VJ kernel developed by Lescano et al. is tested across two different GPU setups to determine the impact of GPU architecture on the execution time as opposed to comparing GPU and CPU performance. The GPU kernel developed by Tsai et al. is compared to the VJ kernel to compare the execution times in Lescano et al.’s study. The kernel developed by Tsai et al. provides a faster execution time. Jansson et al. tests the time efficiency of the GPU kernels and CPU-based algorithms over different datasets and different forest sizes. They obtain the best time efficiency with gpuRF and gpuERT kernels across all tested datasets.
6. Discussion
All of the reviewed studies provided a positive speedup for algorithms transferred from a CPU-based algorithm toward a GPU kernel algorithm. The speedup ranges from 1.87 to 27018.37, indicating significant decreases in execution time for a given algorithm on a GPU kernel. The majority of the reviewed studies provided the largest speedups when tested with large quantities of data, scaling linearly in execution time as opposed to the exponential increase in execution time experienced by a CPU-based system under the same conditions. The algorithms with the best speedup times use massively parallel computing methods in which multiple independent analyses are performed before the results are pooled for secondary analysis. In particular, ensemble methods that perform voting or averaging are shown to greatly benefit from GPU acceleration in comparison to algorithms with a serial data processing approach. These results indicate scalability towards GPU-based kernels across a wide array of algorithms with varying expected speedups.
The improved scalability towards large data formats observed in the reviewed studies showcases the potential to improve data processing rates for biosignal data. Due to the ever-increasing number of electrodes and improved sampling rates that are integrated into biosignal acquisition systems, biosignal datasets can be expected to increase in size continuously. By being able to access systems with enhanced large-data capabilities, the bottleneck experienced by traditional CPU-system can be overcome. This is especially important for realizing real-time biosignal processing systems.
The processing time advancements offered by the GPU kernel development can be easily integrated into existing computer setups using market-available GPUs. The wide range of models and price points of GPUs on the market also allow multiple audiences with a wide array of budget and performance requirements to customize GPU acceleration into their system. This means that biosignals processing applications can be easily transferred from existing CPU-based architectures without incurring the licensing and financial restrictions associated with specialized medical devices for increased processing speeds. The transition to GPU-based systems is not only limited to desktop devices that can add a GPU to an existing system. The Nvidia Jetson series microprocessors contain integrated GPUs that can execute CUDA kernels within a self-contained miniature system. These devices provide the potential to integrate GPU-accelerated kernels into wearable biosignal processing devices.
While GPU acceleration provides multiple proposed benefits, the barriers and limitations of GPU computation should also be considered. Older computational systems may need new hardware to be able to accommodate GPU devices, including upgraded motherboards, higher-capacity power supplies, larger computer chassis, and improved computer cooling accessories. Because of the costs associated with upgrading an old system, new systems may be purchased for a better return on investment. While the inclusion of GPU units in new systems has become an increasingly popular practice, many may contain AMD or Intel brand GPUs which are not compatible with the NVIDIA CUDA architecture. When using AMD brand GPUs a user may use the AMD GPUFORT open source developers toolkit to perform GPU acceleration of ML algorithms. Despite the availability of GPUFORT, AMD GPU devices are not widely used for ML acceleration due to the relative infancy of AMD’s toolkit relative to NVIDIA’s CUDA toolkit. As the GPUFORT toolkit is improved by AMD and used by more developers the viability of this alternative may be better assessed.
7. Conclusion
The development of GPU kernels for ML algorithms with applications for biosignal processing within the 12 reviewed studies showcases a promising direction toward improved biosignal data processing capabilities. When the execution times of the GPU kernels for the selected algorithms are compared to their CPU-based algorithm counterpart, the GPUs provide positive speedups in all cases. The linear execution time with increased data dimensionality and size provides a means by which to decrease the processing time needed for multi-channel, high sampling rate biosignal data. The effectiveness of the developed GPU kernels paired with the accessibility and affordability of GPUs indicates that a future shift towards GPU-based kernels for biosignal ML algorithms may reduce the current biosignal system limitations. This is especially pertinent for real-time biosignal systems, which can be made viable by using GPU-based systems to obtain real-time reactivity.
References
- Stuart, T.; Hanna, J.; Gutruf, P. Wearable devices for continuous monitoring of biosignals: Challenges and opportunities, APL bioengineering, AIP Publishing LLC, 2022, 6, 2, 021502.
- Ancillon, L.; Elgendi, M.; Menon, C. Machine learning for anxiety detection using biosignals: a review, Diagnostics, MDPI, 2022, 12, 8, 1794.
- Sajno, E.; Bartolotta, S.; Tuena, C.; Cipresso, P.; Pedroli, E.; Riva, G. Machine learning in biosignals processing for mental health: A narrative review, Frontiers in Psychology, 2023, 13.
- Swapna, M.; Viswanadhula, U.; Aluvalu, R.; Vardharajan, V.; Kotecha, K. Bio-signals in medical applications and challenges using artificial intelligence, Journal of Sensor and Actuator Networks, MDPI, 2022, 11, 1, 17.
- Rivera, M.; Teruel, M.; Maté, A.; Trujillo, J. Diagnosis and prognosis of mental disorders by means of EEG and deep learning: a systematic mapping study, Artificial Intelligence Review, Springer, 2022, 1–43.
- Alim, A; Imtiaz, M. H. Automatic Identification of Children with ADHD from EEG Brain Waves, Signals, MDPI, 2023, 4, 1. 193–205.
- Al-Quraishi, M.; Elamvazuthi, I.; Daud, S.; Parasuraman, S.; Borboni, A. EEG-based control for upper and lower limb exoskeletons and prostheses: A systematic review, Sensors, MDPI, 2018, 18, 10, 3342.
- Catarino, A. Biosignal monitoring implemented in a swimsuit for athlete performance evaluation, Association of Universities for Textiles (AUTEX) 2011.
- Jang, E.; Park, B.; Kim, S.; Chung, M.; Park, M.; Sohn, J. Emotion classification based on bio-signals emotion recognition using machine learning algorithms, 2014 International Conference on Information Science, Electronics and Electrical Engineering, IEEE 2014, 3. 1373–1376.
- Schmidt, A. Biosignals in human-computer interaction, interactions, ACM New York, NY, USA, 2015, 23, 1, 76–79.
- Abdullah-Al-Mamun, K. Pattern identification of movement related states in biosignals, University of Southampton 2013.
- Saitis, C.; Parvez, M.; Kalimeri, K. Cognitive load assessment from EEG and peripheral biosignals for the design of visually impaired mobility aids, Wireless Communications and Mobile Computing, Hindawi Limited, 2018, 2018, 1–9.
- Ramantani, G.; Maillard, L.; Koessler, L. Correlation of invasive EEG and scalp EEG, Seizure, Elsevier, 2016, 41, 196–200.
- Lovelace, J.; Witt, T.; Beyette, F. Modular, bluetooth enabled, wireless electroencephalograph (EEG) platform, 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE 2013, 6361–6364.
- Ketola, E.; Barankovich, M.; Schuckers, S.; Ray-Dowling, A.; Hou, D.; Imtiaz, M. Channel Reduction for an EEG-Based Authentication System While Performing Motor Movements, Sensors, MDPI, 2022, 22, 23, 9156.
- Jurcak, V.; Tsuzuki, D.; Dan, I. 10/20, 10/10, and 10/5 systems revisited: their validity as relative head-surface-based positioning systems, Neuroimage, Elsevier, 2007, 34, 4, 1600–1611.
- Wade, L.; Needham, L.; McGuigan, P.; Bilzon, J. Applications and limitations of current markerless motion capture methods for clinical gait biomechanics, PeerJ, PeerJ Inc., 2022, 10, e12995.
- Wood, D.; Kafiabadi, S.; Al Busaidi, A.; Guilhem, E.; Montvila, A.; Lynch, J.; Townend, M.; Agarwal, S.; Mazumder, A.; Barker, G. Accurate brain-age models for routine clinical MRI examinations, Neuroimage, Elsevier, 2022, 249, 118871.
- Wöhrle, H.; Tabie, M.; Kim, S.K.; Kirchner, F.; Kirchner, E.A. A hybrid FPGA-based system for EEG-and EMG-based online movement prediction, Sensors, MDPI, 2017, 17, 7, 1552.
- Ketola, E.; Lloyd, C.; Shuhart, D.; Schmidt, J.; Morenz, R.; Khondker, A.; Imtiaz, M. Lessons Learned from the Initial Development of a Brain Controlled Assistive Device, 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), IEEE, 2022, 0580–0585.
- Dadebayev, D.; Goh, W.; Tan, E. EEG-based emotion recognition: Review of commercial EEG devices and machine learning techniques, Journal of King Saud University-Computer and Information Sciences, Elsevier, 2022, 34, 7, 4385–4401.
- Burnet, D.; Turner, M. Expanding EEG research into the clinic and classroom with consumer EEG Systems, Scholarworks 2017.
- Escobar, J.; Ortega, J.; González, J.; Damas, M.; Díaz, A. Parallel high-dimensional multi-objective feature selection for EEG classification with dynamic workload balancing on CPU–GPU architectures, Cluster Computing, Springer, 2017, 20, 1881–1897.
- Yu, T.; Akhmadeev, K.; Le Carpentier, E.; Aoustin, Y.; Farina, D. On-line recursive decomposition of intramuscular EMG signals using GPU-implemented Bayesian filtering, IEEE Transactions on Biomedical Engineering, IEEE, 2019, 67, 6, 1806–1818.
- Xiao, W.; Huang, H.; Sun, Y.; Yang, Q. Promise of embedded system with GPU in artificial leg control: Enabling time-frequency feature extraction from electromyography, 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE 2009, 6926–6929.
- Dohnálek, P.; Gajdoš, P.; Peterek, T.; Penhaker, M. Pattern recognition in EEG cognitive signals accelerated by
GPU, International Joint Conference CISIS‘12-ICEUTE‘12-SOCO‘12 Special Sessions, Springer Berlin Heidelberg2013, 477–485.
- Lee, D.; Lee, S.; Park, D. Efficient Signal Processing Acceleration using OpenCL-based FPGA-GPU Hybrid Cooperation for Reconfigurable ECG Diagnosis, 2021 18th International SoC Design Conference (ISOCC), IEEE 2021, 349–350.
- Steinkraus, D.; Buck, I.; Simard, P. Using GPUs for machine learning algorithms, Eighth International Conference on Document Analysis and Recognition (ICDAR’05), IEEE 2005, 1115–1120.
- Li, F.; Ye, Y.; Tian, Z.; Zhang, X. CPU versus GPU: which can perform matrix computation faster—performance comparison for basic linear algebra subprograms, Neural Computing and Applications, Springer, 2019, 31, 4353–4365.
- Hestness, J.; Keckler, S.; Wood, D. A comparative analysis of microarchitecture effects on CPU and GPU memory system behavior, 2014 IEEE International Symposium on Workload Characterization (IISWC), IEEE 2014, 150–160.
- Kumar, S. , Introduction to Parallel Programming, Cambridge University Press, 2022.
- Willhelm, T.; Dementiev, R. Intel Performance Counter Monitor - A Better Way to Measure CPU Utilization, Intel 2022.
- Lee, S. Real-time edge computing on multi-processes and multi-threading architectures for deep learning applications, Microprocessors and Microsystems, Elsevier, 2022, 92, 104554.
- Kothari, S.; Crasta, R.; Biju, A.; Lotlikar, T.; Rai, H. On-Device ML: An Efficient Approach to Classify Large Number of Images Using Multi-threading in Android Devices, Proceedings of 2nd International Conference on Artificial Intelligence: Advances and Applications: ICAIAA 2021, Springer 2022, 793–799.
- Advanced Micro Devices AMD Ryzen™ Threadripper™ Processors, amd. com 2023.
- Bizon. AMD EPYC 7003/7002 Series – Up to 128 Cores Workstation PC, https://bizon-tech.com/amd-epyc-7000-series-up-to-128-cores-workstation-pc 2023.
- Chiueh, T.; Pradhan, P. High-performance IP routing table lookup using CPU caching, IEEE INFOCOM’99. Conference on Computer Communications. Proceedings. Eighteenth Annual Joint Conference of the IEEE Computer and Communications Societies. The Future is Now (Cat. No. 99CH36320), IEEE, 1999, 3, 1421–1428. 3.
- Ahn, H.; Choi, S.; Jung, S. Evaluation of STT-MRAM L3 cache in 7nm FinFET process, 2018 International Conference on Electronics, Information, and Communication (ICEIC), IEEE 2018, 1–4.
- Komar, M. Data rate assessment on L2–L3 CPU bus and bus between CPU and RAM in modern CPUs, Automatic Control and Computer Sciences, Springer, 2017, 51, 701–708.
- Patterson, D.A. Computer Organization and Design: The Hardware/Software Interface, Morgan Kaufmann Publishers 1994.
- Lal, S.; Varma, B.; Juurlink, B. A quantitative study of locality in GPU caches for memory-divergent workloads, International Journal of Parallel Programming, Springer, 2022, 50, 2, 189–216.
- Mienye, I.; Sun, Y. A survey of ensemble learning: Concepts, algorithms, applications, and prospects, IEEE Access, IEEE, 2022, 10, 99129–99149.
- Jog, A.; Kayiran, O.; Kesten, T.; Pattnaik, A.; Bolotin, E.; Chatterjee, N.; Keckler, S.; Kandemir, M.; Das, C. Anatomy of gpu memory system for multi-application execution, Proceedings of the 2015 international symposium on memory systems, 2015, 223–234.
- Jang, H.; Kim, J.; Gratz, P.; Yum, K.; Kim, E. Bandwidth-efficient on-chip interconnect designs for GPGPUs, Proceedings of the 52nd Annual Design Automation Conference, 2015, 1–6.
- Tsai, W.; Lan, Y.; Hu, Y.; Chen, S. Networks on chips: structure and design methodologies, Journal of Electrical and Computer Engineering, Hindawi Limited London, UK, United Kingdom, 2012, 2–2.
- Zhao, X.; Jahre, M.; Eeckhout, L. Selective replication in memory-side GPU caches, 2020 53rd Annual IEEE/ACM International Symposium on Microarchitecture (MICRO), IEEE 2020, 967–980.
- Ibrahim, M.; Kayiran, O.; Eckert, Y.; Loh, G.; Jog, A. Analyzing and leveraging shared L1 caches in GPUs, Proceedings of the ACM International Conference on Parallel Architectures and Compilation Techniques, 2020, 161–173.
- Ziabari, A.; Abellan, J.; Ma, Y.; Joshi, A.; Kaeli, D. Asymmetric NoC architectures for GPU systems, Proceedings of the 9th International Symposium on Networks-on-Chip, 2015, 1–8.
- Eichstädt, J.; Peiró, J.; Moxey, D. Efficient vectorised kernels for unstructured high-order finite element fluid solvers on GPU architectures in two dimensions, Computer Physics Communications, Elsevier, 2023, 284, 108624.
- Sivalingam, K. GPU Acceleration of a Theoretical Particle Physics Application, Theses Master, The University of Edinburgh, 2010.
- Tolmachev, D. VkFFT-a performant, cross-platform and open-source GPU FFT library, IEEE Access, IEEE, 2023, 11, 12039–12058.
- Zhao, H.; Cui, W.; Chen, Q.; Zhang, Y.; Lu, Y.; Li, C.; Leng, J.; Guo, M. Tacker: Tensor-cuda core kernel fusion for improving the gpu utilization while ensuring QoS, 2022 IEEE International Symposium on High-Performance Computer Architecture (HPCA), IEEE 2022, 800–813.
- Zhuang, H.; Liu, X.; Liang, X.; Yan, Y.; He, J.; Cai, Y.; Wu, C.; Zhang, X.; Zhang, H. Tensor-CA: A high-performance cellular automata model for land use simulation based on vectorization and GPU, Transactions in GIS, Wiley Online Library, 2022, 26, 2, 755–778.
- Peddie, J. Compute Accelerators and Other GPUs, The History of the GPU-New Developments, 2023, 239–304.
- Li, A.; Song, S.; Wijtvliet, M.; Kumar, A.; Corporaal, H. SFU-driven transparent approximation acceleration on GPUs, Proceedings of the 2016 International Conference on Supercomputing, 2016, 1–14.
- Li, B.; Wei, J. REMOC: efficient request managements for on-chip memories of GPUs, Proceedings of the 19th ACM International Conference on Computing Frontiers, 2022, 1–11.
- Krashinsky, R. , Giroux, O., Jones, S., Stam, N. & Ramaswamy, S. NVIDIA ampere architecture in-depth. Nvidia Blog: Https://devblogs.nvidia.com/nvidia-ampere-architecture-in-depth(2020).
- Tsai, P.; Hsu, Y.; Chiu, C.; Chu, T. Accelerating AdaBoost algorithm using GPU for multi-object recognition, 2015 IEEE International Symposium on Circuits and Systems (ISCAS), IEEE 2015, 738–741.
- Bhimani, J.; Leeser, M.; Mi, N. Accelerating K-Means clustering with parallel implementations and GPU computing, 2015 IEEE High Performance Extreme Computing Conference (HPEC), IEEE 2015, 1–6.
- Gajos-Balińska, A.; Wójcik, G.; Stpiczynski, P. Cooperation of CUDA and Intel multi-core architecture in the independent component analysis algorithm for EEG data, Bio-Algorithms and Med-Systems, De Gruyter, 2020, 16, 3.
- Jansson, K.; Sundell, H.; Bostrp̎m, H. gpuRF and gpuERT: efficient and scalable GPU algorithms for decision tree ensembles, 2014 IEEE international parallel & distributed processing symposium workshops, IEEE 2014, 1612–1621.
- Gutiérrez, P.; Lastra, M.; Benítez, J.; Herrera, F. SMOTE-GPU: Big data preprocessing on commodity hardware for imbalanced classification, Progress in Artificial Intelligence, Springer, 2017, 6, 347–354.
- Lescano, G.; Santana-Mansilla, P.; Costaguta, R. Analysis of a GPU implementation of Viola-Jones’ algorithm for features selection, Journal of Computer Science and Technology, Universidad Nacional de La Plata, 2017, 17, 01, 68–73.
- Abbasi, M.; Rafiee, M. Efficient parallelization of a genetic algorithm solution on the traveling salesman problem with multi-core and many-core systems, International Journal of Engineering, Materials and Energy Research Center, 2020, 33, 7, 1257–1265.
- Mitchell, R.; Frank, E. Accelerating the XGBoost algorithm using GPU computing, PeerJ Computer Science, PeerJ Inc., 2017, 3, e127.
- Wen, Z.; He, B.; Kotagiri, R.; Lu, S.; Shi, J. Efficient gradient boosted decision tree training on GPUs, 2018 IEEE International Parallel and Distributed Processing Symposium (IPDPS), IEEE 2018, 234–243.
- Benko, G.; Juhasz, Z. GPU implementation of the FastICA algorithm, 2019 42nd International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), IEEE 2019, 196–199.
- Lo, W.; Chang, Y.; Sheu, R.; Chiu, C.; Yuan, S. CUDT: a CUDA based decision tree algorithm, The Scientific World Journal, Hindawi, 2014.
- Spandana, K.; Sirisha, D.; Shahida, S. Parallelizing Apriori algorithm on GPU, International Journal of Computer Applications, Foundation of Computer Science, 2016, 155, 10, 22–27.
- Nvidia; CUDA. CUDA C++ Programming Guide Release 12.1, CUDA 2023.
- Nvidia; CUDA. NVIDIA CUDA Compiler Driver Release 12.1, CUDA 2023.
- Januszewski, M.; Kostur, M. Accelerating numerical solution of stochastic differential equations with CUDA, Computer Physics Communications, Elsevier, 2010, 181, 1, 183–188.
- Gilman, G.; Walls, R. Characterizing concurrency mechanisms for NVIDIA GPUs under deep learning workloads, ACM SIGMETRICS Performance Evaluation Review, ACM New York, NY, USA, 2022, 49, 3, 32–34.
- Muller, S.; Hoffmann, J. Modeling and analyzing evaluation cost of CUDA kernels, Proceedings of the ACM on Programming Languages, ACM New York, NY, USA, 2021, 5, POPL, 1–31.
- Darabi, S.; Yousefzadeh-Asl-Miandoab, E.; Akbarzadeh, N.; Falahati, H.; Lotfi-Kamran, P.; Sadrosadati, M.; Sarbazi-Azad, H. OSM: Off-chip shared memory for GPUs, IEEE Transactions on Parallel and Distributed Systems, IEEE, 2022, 33, 12, 3415–3429.
- Yang, D.; Liu, J.; Qi, J.; Lai, J. WholeGraph: a fast graph neural network training framework with multi-GPU distributed shared memory architecture, 2022 SC22: International Conference for High Performance Computing, Networking, Storage and Analysis (SC), IEEE Computer Society 2022, 767–780.
- Huang, Y.; Zheng, X.; Zhu, Y. Optimized CPU–GPU collaborative acceleration of zero-knowledge proof for confidential transactions, Journal of Systems Architecture, Elsevier, 2023, 135, 102807.
- Rosenfeld, V. Query processing on heterogeneous CPU/GPU systems, ACM Computing Surveys (CSUR), ACM New York, NY, 2022, 55, 1, 1–38.
- Jena, B.; Nayak, G.; Saxena, S. High-Performance computing and its requirements in deep learning, High-Performance Medical Image Processing, 2022, 255–288.
- Feng, J.; Zhang, W.; Pei, Q.; Wu, J.; Lin, X. Heterogeneous computation and resource allocation for wireless powered federated edge learning systems, IEEE Transactions on Communications, IEEE, 2022, 70, 5, 3220–3233.
- Chatterjee, R.; Datta, A.; Sanyal, D. Ensemble learning approach to motor imagery EEG signal classification, Machine Learning in Bio-Signal Analysis and Diagnostic Imaging, Elsevier, 2019, 183–208.
- Subasi, A.; Qaisar, S. Surface EMG signal classification using TQWT, Bagging and Boosting for hand movement recognition, Journal of Ambient Intelligence and Humanized Computing, Springer, 2022, 13, 7, 3539–3554.
- Ecemiş, C. ; Avcu, Neslihan; Sari, Zekeriya. Classification of Imbalanced Cardiac Arrhythmia Data, Avrupa Bilim ve Teknoloji Dergisi, 2022, 34, 546–552.
- Wang, Y.; Zhang, H.; An, Y.; Ji, Z.; Ganchev, I. RG hyperparameter optimization approach for improved indirect prediction of blood glucose levels by boosting ensemble learning, Electronics, MDPI, 2021, 10, 15, 1797.
- Matthews, J.; Kim, J.; Yeo, W. Advances in Biosignal Sensing and Signal Processing Methods with Wearable Devices, Analysis & Sensing 2022, 3, 2, e202200062.
- Baydoun, M.; Safatly, L.; Ghaziri, H.; El Hajj, A. Analysis of heart sound anomalies using ensemble learning, Biomedical Signal Processing and Control, Elsevier, 2020, 62, 102019.
- Chawla, N.; Bowyer, K.; Hall, L.; Kegelmeyer, W. SMOTE: synthetic minority over-sampling technique, Journal of artificial intelligence research, 2002, 16, 321–357.
- La Fisca, L.; Vandenbulcke, V.; Wauthia, E.; Miceli, A. Biases in BCI experiments: Do we really need to balance stimulus properties across categories?, Frontiers in computational neuroscience, Frontiers, 2022, 16, 153.
- Wu, S.; Dong, J. Research of Fast-ICA Algorithm and its Improvement, IGARSS 2022-2022 IEEE International Geoscience and Remote Sensing Symposium, IEEE 2022, 3251–3254.
- Sonfack, G.; Ravier, P. Application of Single Channel Blind Source Separation Based-EEMD-PCA and Improved FastICA algorithm on Non-intrusive Appliances Load identification, Journal of Electrical and Electronic Engineering, Science Publishing Group, 2022, 10, 3, 114–120.
- Maitin, A.; Romero Muoz, J.; Garcia-Tejedor, A. Survey of machine learning techniques in the analysis of EEG signals for Parkinson’s disease: A systematic review, Applied Sciences, MDPI, 2022, 12, 14, 6967.
- Wario, F.; Avalos, O. Bio-inspired algorithms, Biosignal Processing and Classification Using Computational Learning and Intelligence, 2022, 1, 225–248.
- Saibene, A.; Gasparini, F. Genetic algorithm for feature selection of EEG heterogeneous data, Expert Systems with Applications, Elsevier, 2023, 119488.
- Prabhakar, S.; Lee, S. ENIC: Ensemble and nature inclined classification with sparse depiction based deep and transfer learning for biosignal classification, Applied Soft Computing, Elsevier, 2022, 117, 108416.
- Gao, T.; Chen, D.; Tang, Y.; Ming, Z.; Li, X. EEG Reconstruction with a Dual-scale CNN-LSTM Model for Deep Artifact Removal, IEEE Journal of Biomedical and Health Informatics, IEEE, 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).