Submitted:
25 April 2024
Posted:
26 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
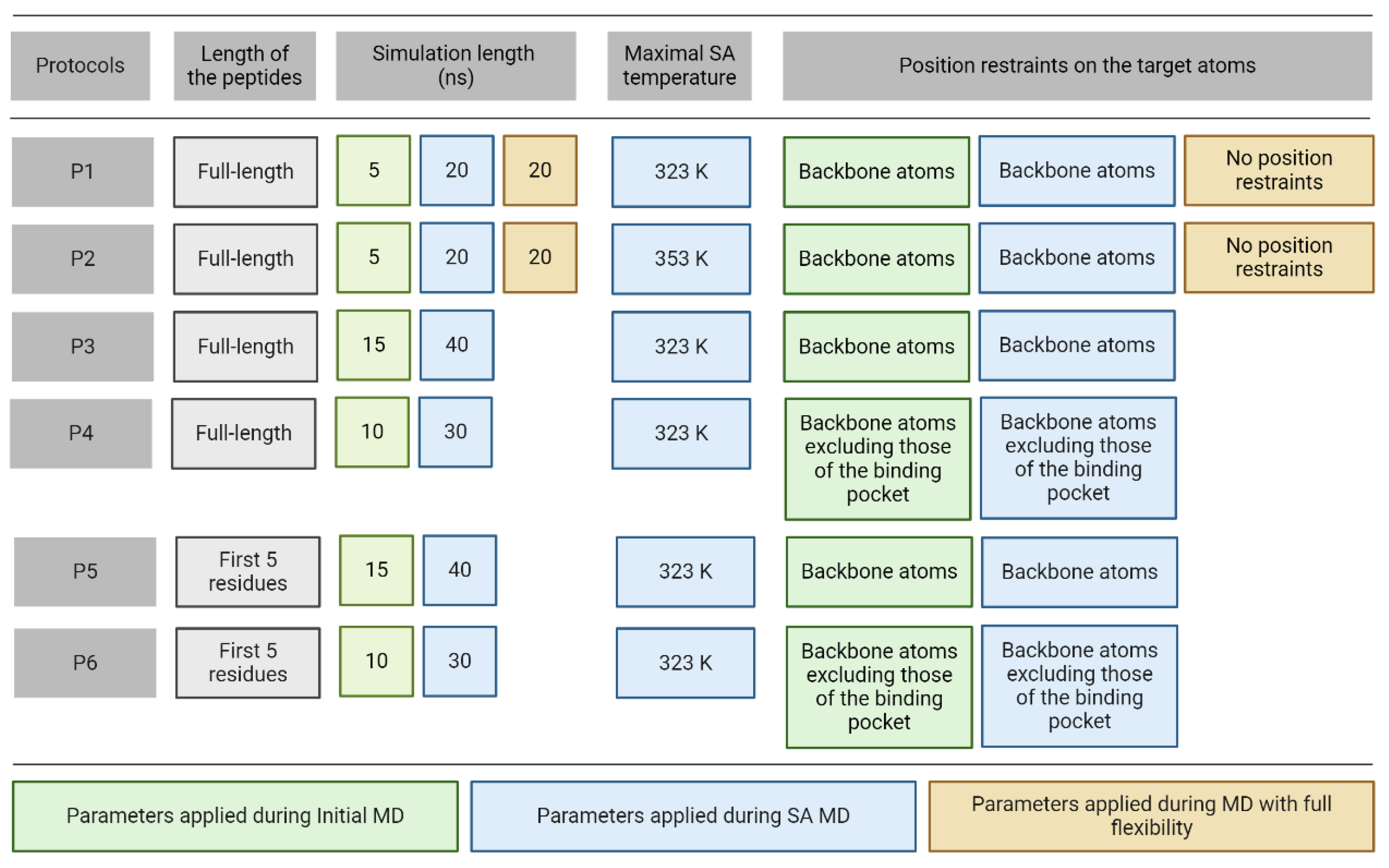
2.1. Systems and MD Protocols
2.2. Structural Performance
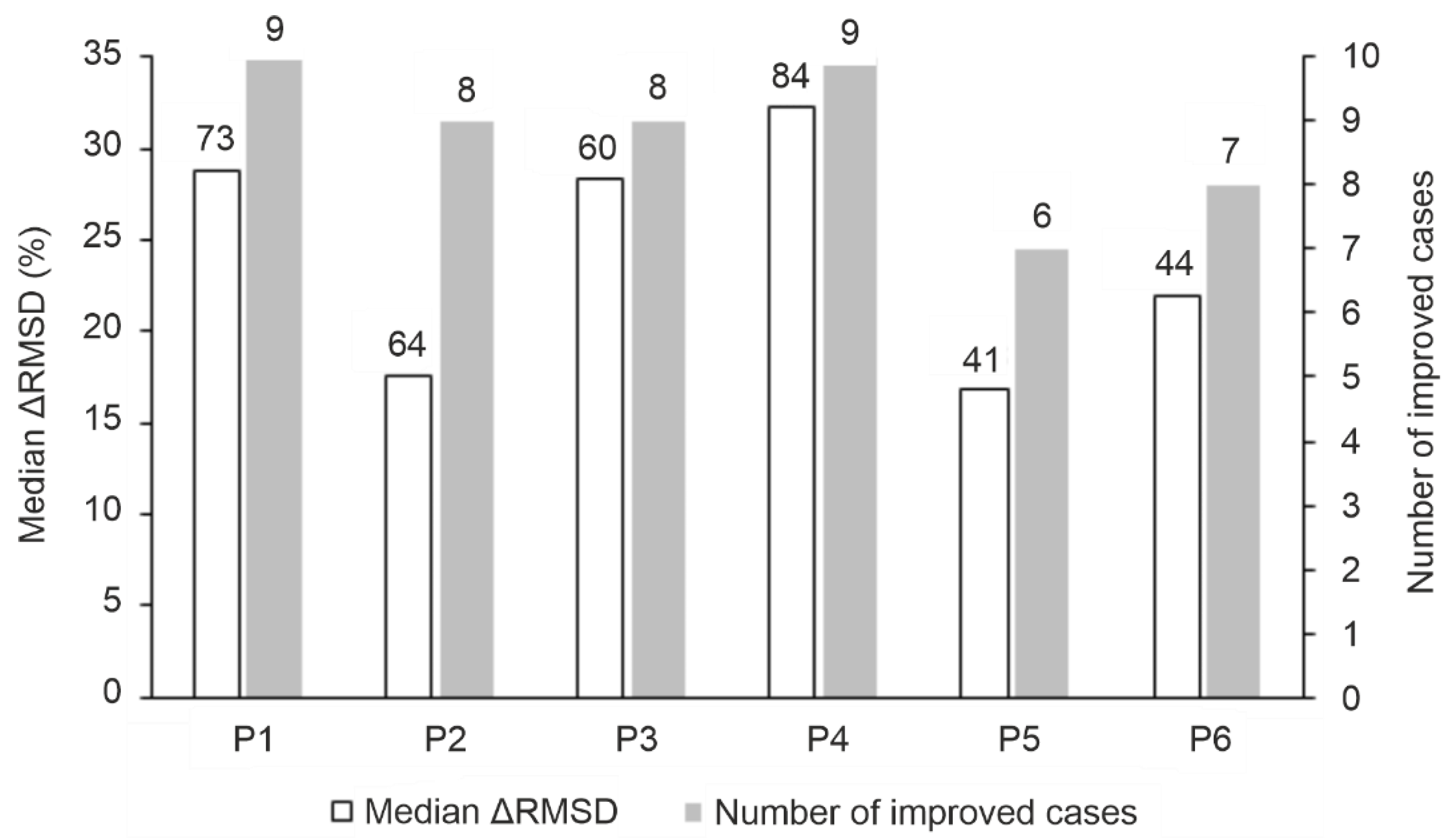
2.2.1. The Overall Performance of the MD Protocols
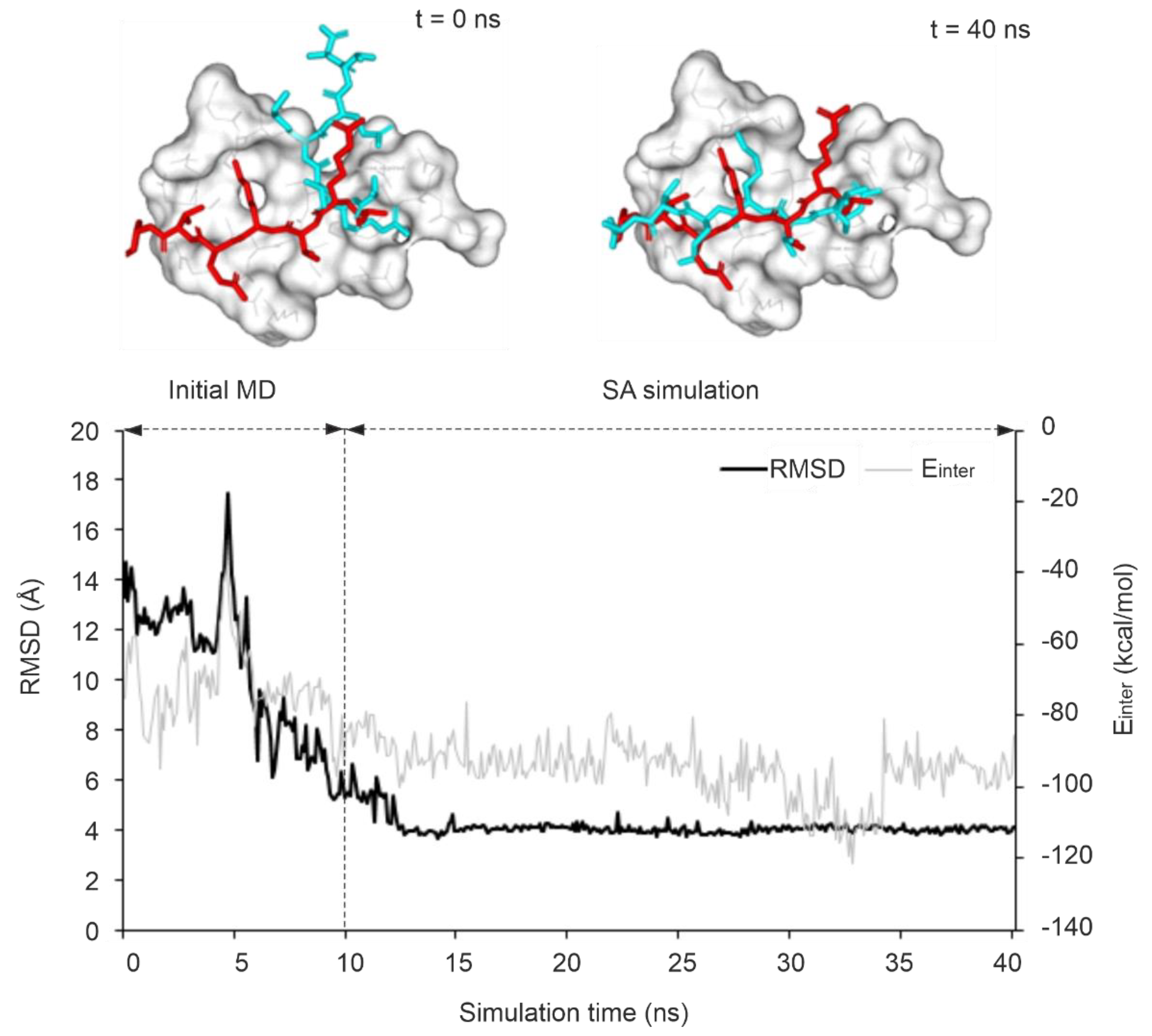
2.2.2. The Kinetic Stability of the MD-Refined Complex Structure
2.2.3. Comparison with the Results of Other Post-Docking Refinement Studies
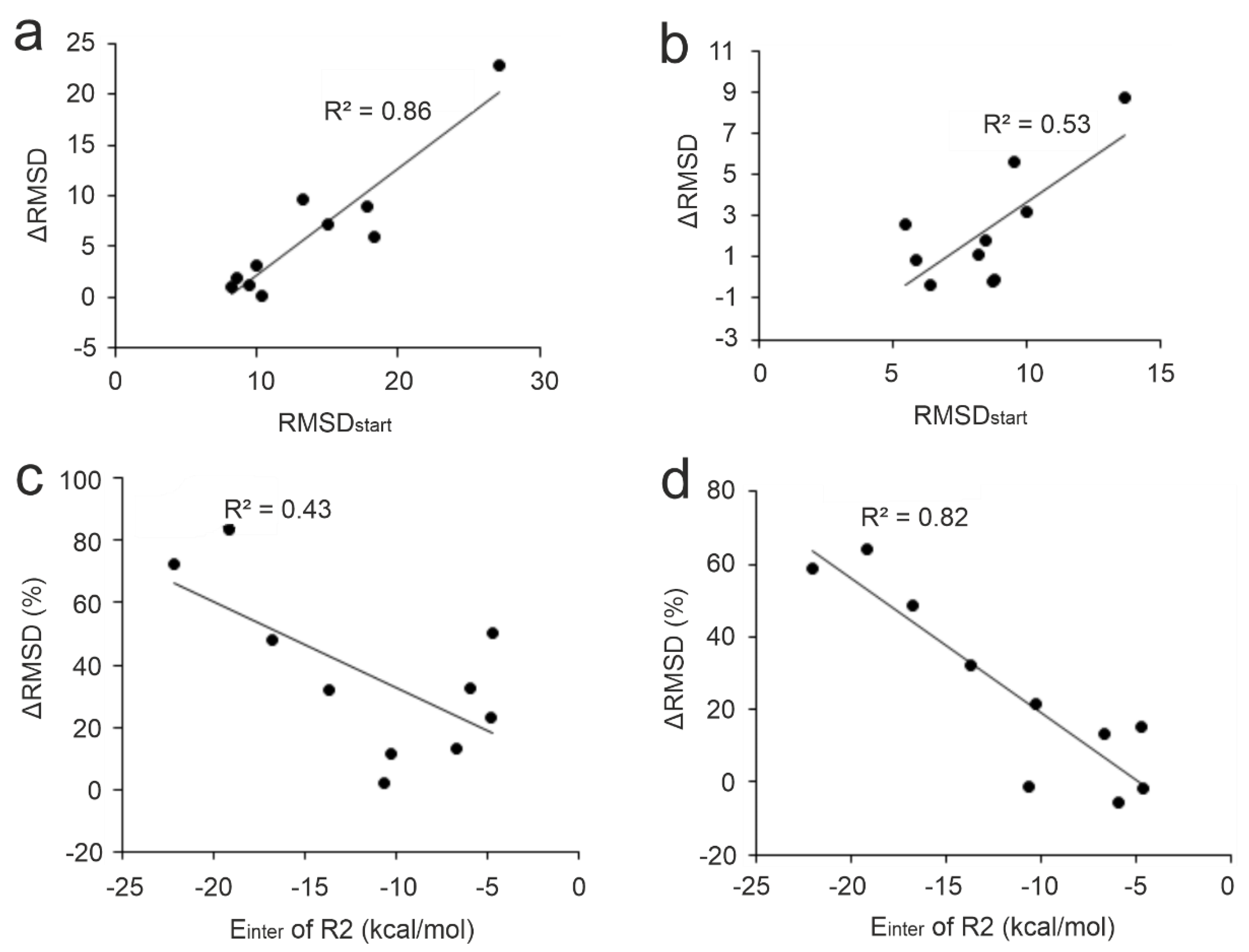
2.3. Factors Influencing MD-Refinement
2.3.1. Target Conformation
2.3.2. Initial Ligand Binding Mode
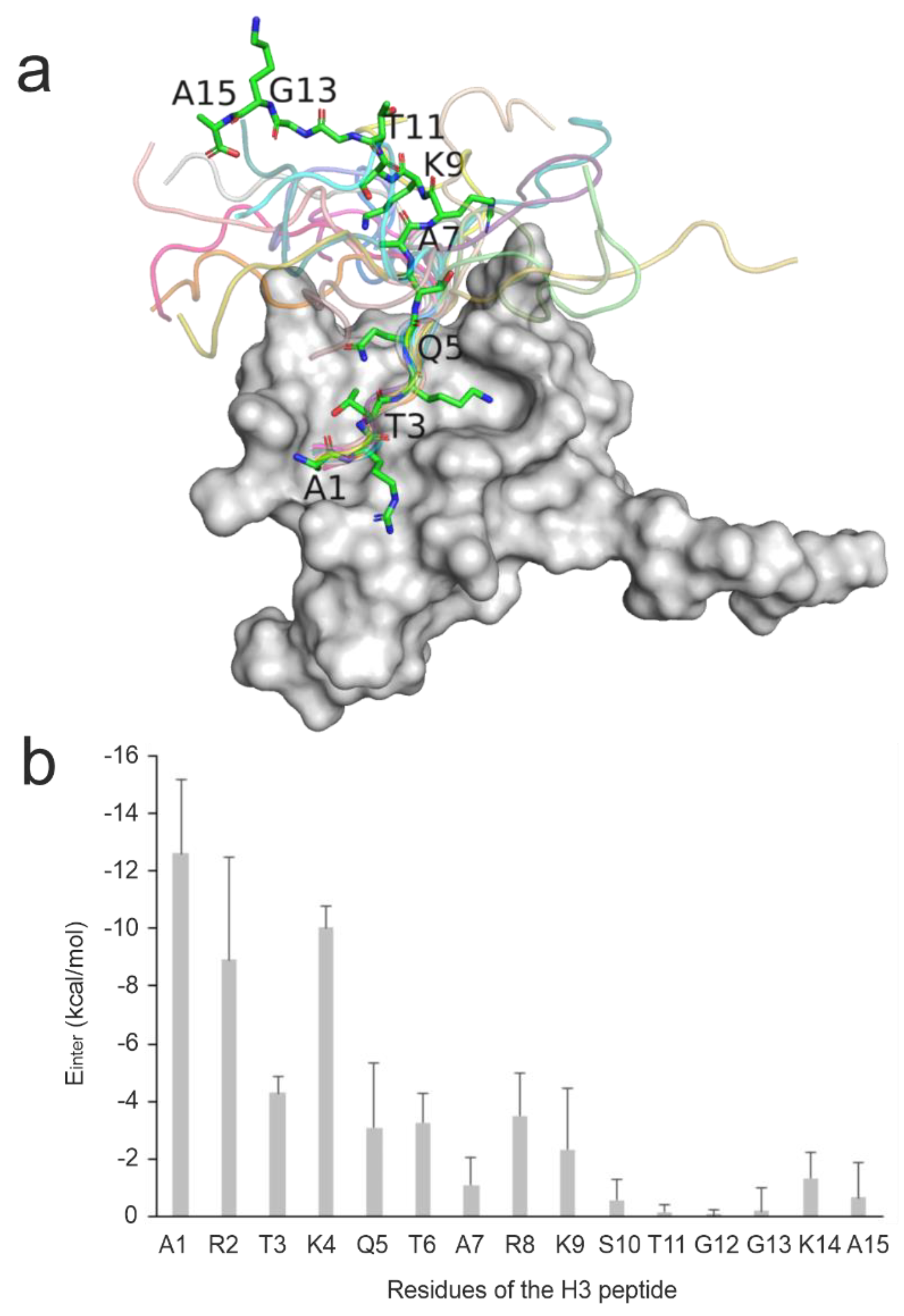
2.3.3. Anchoring Residues
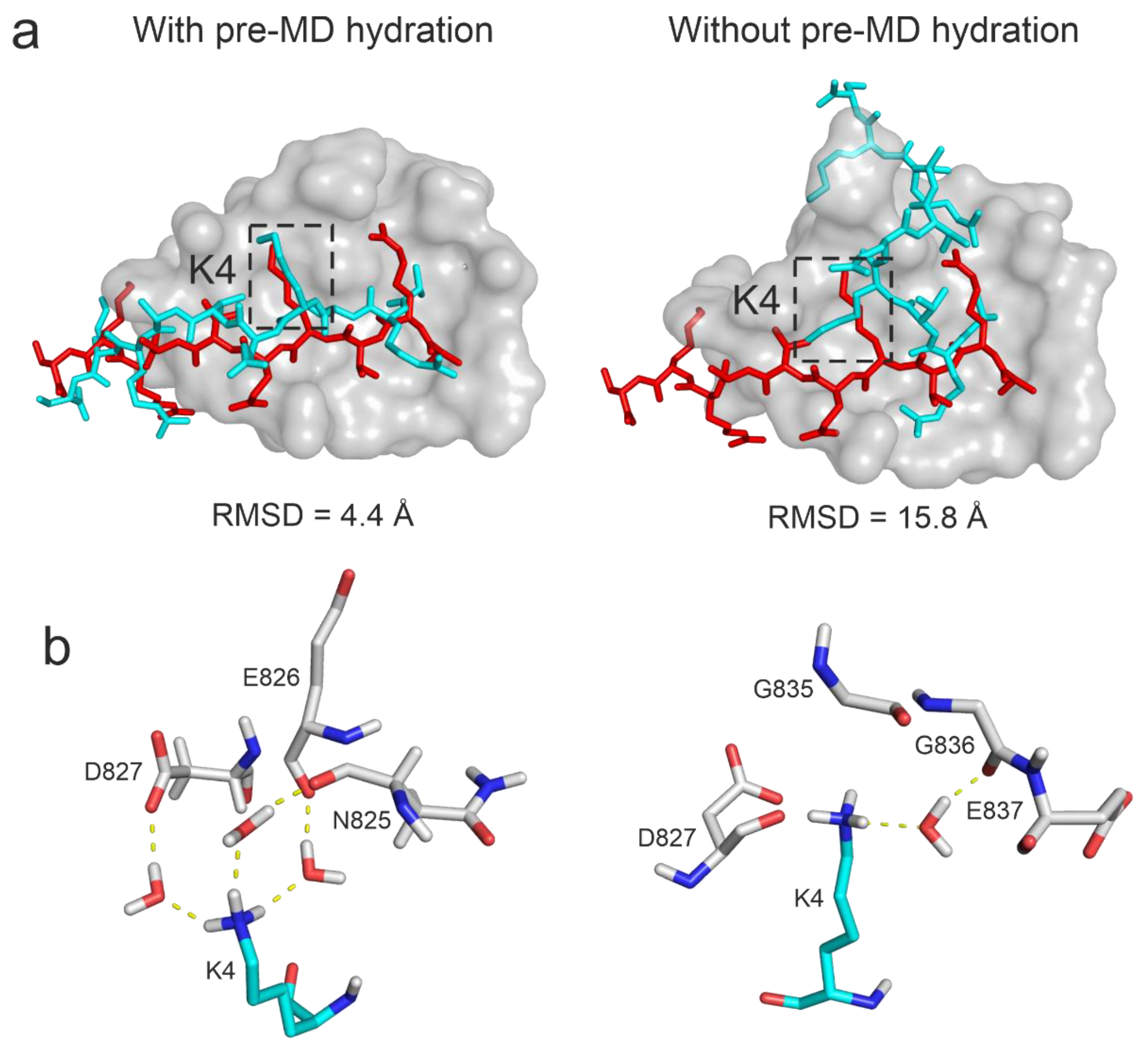
2.3.4. Interfacial Water Network
3. Materials and Methods
3.1. Refinement Protocols
3.2. Evaluation Metrics
3.3. Per-Residue Interaction Energy Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drwal, M.N.; Griffith, R. Combination of Ligand- and Structure-Based Methods in Virtual Screening. Drug Discov Today Technol 2013, 10, e395–e401. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and Scoring in Virtual Screening for Drug Discovery: Methods and Applications. Nat Rev Drug Discov 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Dias, R.; de Azevedo, W.F. Molecular Docking Algorithms. Curr Drug Targets 2008, 9, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr Comput Aided Drug Des 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.; Hamilton, A.D. Targeting Protein-Protein Interactions by Rational Design: Mimicry of Protein Surfaces. J R Soc Interface 2006, 3, 215–233. [Google Scholar] [CrossRef]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva-Jr, F.P. Key Topics in Molecular Docking for Drug Design. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- de Ruyck, J.; Brysbaert, G.; Blossey, R.; Lensink, M.F. Molecular Docking as a Popular Tool in Drug Design, an in Silico Travel. Adv Appl Bioinform Chem 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Li, C.; Sun, J.; Li, L.-W.; Wu, X.; Palade, V. An Effective Swarm Intelligence Optimization Algorithm for Flexible Ligand Docking. IEEE/ACM Trans Comput Biol Bioinform 2022, 19, 2672–2684. [Google Scholar] [CrossRef]
- Pawson, T.; Nash, P. Assembly of Cell Regulatory Systems through Protein Interaction Domains. Science 2003, 300, 445–452. [Google Scholar] [CrossRef]
- Petsalaki, E.; Russell, R.B. Peptide-Mediated Interactions in Biological Systems: New Discoveries and Applications. Curr Opin Biotechnol 2008, 19, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically Unstructured Proteins and Their Functions. Nat Rev Mol Cell Biol 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Das, A.A.; Sharma, O.P.; Kumar, M.S.; Krishna, R.; Mathur, P.P. PepBind: A Comprehensive Database and Computational Tool for Analysis of Protein-Peptide Interactions. Genomics Proteomics Bioinformatics 2013, 11, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Fatoki, T.H.; Chukwuejim, S.; Udenigwe, C.C.; Aluko, R.E. In Silico Exploration of Metabolically Active Peptides as Potential Therapeutic Agents against Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences 2023, 24, 5828. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Fleishman, S.J. Computational Design of Protein-Protein Interactions. Curr. Opin. Struct. Biol. 2013, 23, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, S.; Fernández-Recio, J. Protein-Protein Docking and Hot-Spot Prediction for Drug Discovery. Curr. Pharm. Des. 2012, 18, 4607–4618. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.C.; Burgoyne, N.J.; Jackson, R.M. Predicting Druggable Binding Sites at the Protein-Protein Interface. Drug Discov. Today 2009, 14, 155–161. [Google Scholar] [CrossRef]
- Kann, M.G. Protein Interactions and Disease: Computational Approaches to Uncover the Etiology of Diseases. Brief. Bioinformatics 2007, 8, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Bienstock, R.J. Computational Drug Design Targeting Protein-Protein Interactions. Curr. Pharm. Des. 2012, 18, 1240–1254. [Google Scholar] [CrossRef]
- Silva, M.; Philadelpho, B.; Santos, J.; Souza, V.; Souza, C.; Santiago, V.; Silva, J.; Souza, C.; Azeredo, F.; Castilho, M.; et al. IAF, QGF, and QDF Peptides Exhibit Cholesterol-Lowering Activity through a Statin-like HMG-CoA Reductase Regulation Mechanism: In Silico and In Vitro Approach. Int J Mol Sci 2021, 22, 11067. [Google Scholar] [CrossRef]
- Ahrens, V.M.; Bellmann-Sickert, K.; Beck-Sickinger, A.G. Peptides and Peptide Conjugates: Therapeutics on the Upward Path. Future Med Chem 2012, 4, 1567–1586. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kahler, U.; Fuchs, J.E.; Goettig, P.; Liedl, K.R. An Unexpected Switch in Peptide Binding Mode: From Simulation to Substrate Specificity. J Biomol Struct Dyn 2018, 36, 4072–4084. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int J Mol Sci 2019, 20, 2383. [Google Scholar] [CrossRef]
- Koehler, A.N. A Complex Task? Direct Modulation of Transcription Factors with Small Molecules. Curr Opin Chem Biol 2010, 14, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Hong, S.-Y.; Kim, S.-G.; Park, C.-M. Competitive Inhibition of Transcription Factors by Small Interfering Peptides. Trends Plant Sci 2011, 16, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Oleinikov, P.D.; Fedulova, A.S.; Armeev, G.A.; Motorin, N.A.; Singh-Palchevskaia, L.; Sivkina, A.L.; Feskin, P.G.; Glukhov, G.S.; Afonin, D.A.; Komarova, G.A.; et al. Interactions of Nucleosomes with Acidic Patch-Binding Peptides: A Combined Structural Bioinformatics, Molecular Modeling, Fluorescence Polarization, and Single-Molecule FRET Study. International Journal of Molecular Sciences 2023, 24, 15194. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-Y. Search Strategies and Evaluation in Protein-Protein Docking: Principles, Advances and Challenges. Drug Discov. Today 2014, 19, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Ciemny, M.; Kurcinski, M.; Kamel, K.; Kolinski, A.; Alam, N.; Schueler-Furman, O.; Kmiecik, S. Protein-Peptide Docking: Opportunities and Challenges. Drug Discov. Today 2018, 23, 1530–1537. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, C.; Ren, Y.; Yang, C.; Tian, F. Computational Peptidology: A New and Promising Approach to Therapeutic Peptide Design. Curr. Med. Chem. 2013, 20, 1985–1996. [Google Scholar] [CrossRef]
- Wodak, S.J.; Méndez, R. Prediction of Protein-Protein Interactions: The CAPRI Experiment, Its Evaluation and Implications. Curr. Opin. Struct. Biol. 2004, 14, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Zsidó, B.Z.; Hetényi, C. The Role of Water in Ligand Binding. Current Opinion in Structural Biology 2021, 67, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Swails, J.M.; Manas, E.S. SPAM: A Simple Approach for Profiling Bound Water Molecules. J Chem Theory Comput 2013, 9, 5539–5549. [Google Scholar] [CrossRef] [PubMed]
- Bodnarchuk, M.S.; Viner, R.; Michel, J.; Essex, J.W. Strategies to Calculate Water Binding Free Energies in Protein–Ligand Complexes. J. Chem. Inf. Model. 2014, 54, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, J.; Leriche, C.; Chamiot-Clerc, P.; Feutrill, J.; Halley, F.; Papin, D.; Derimay, N.; Mugler, C.; Grépin, C.; Schio, L. Preparation and Optimization of Pyrazolo[1,5-a]Pyrimidines as New Potent PDE4 Inhibitors. Bioorg Med Chem Lett 2016, 26, 454–459. [Google Scholar] [CrossRef]
- Bortolato, A.; Tehan, B.G.; Bodnarchuk, M.S.; Essex, J.W.; Mason, J.S. Water Network Perturbation in Ligand Binding: Adenosine A(2A) Antagonists as a Case Study. J Chem Inf Model 2013, 53, 1700–1713. [Google Scholar] [CrossRef]
- Chrencik, J.E.; Patny, A.; Leung, I.K.; Korniski, B.; Emmons, T.L.; Hall, T.; Weinberg, R.A.; Gormley, J.A.; Williams, J.M.; Day, J.E.; et al. Structural and Thermodynamic Characterization of the TYK2 and JAK3 Kinase Domains in Complex with CP-690550 and CMP-6. J Mol Biol 2010, 400, 413–433. [Google Scholar] [CrossRef]
- Mason, J.S.; Bortolato, A.; Congreve, M.; Marshall, F.H. New Insights from Structural Biology into the Druggability of G Protein-Coupled Receptors. Trends Pharmacol Sci 2012, 33, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Tshilande, N.; Mammino, L.; Bilonda, M.K. The Study of Molecules and Processes in Solution: An Overview of Questions, Approaches and Applications. Computation 2024, 12, 78. [Google Scholar] [CrossRef]
- Wei, L.; Chen, Y.; Liu, J.; Rao, L.; Ren, Y.; Xu, X.; Wan, J. Cov_DOX: A Method for Structure Prediction of Covalent Protein–Ligand Bindings. J. Med. Chem. 2022, 65, 5528–5538. [Google Scholar] [CrossRef]
- Zsidó, B.Z.; Börzsei, R.; Szél, V.; Hetényi, C. Determination of Ligand Binding Modes in Hydrated Viral Ion Channels to Foster Drug Design and Repositioning. J Chem Inf Model 2021, 61, 4011–4022. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Lill, M.A. Protein Pharmacophore Selection Using Hydration-Site Analysis. J Chem Inf Model 2012, 52, 1046–1060. [Google Scholar] [CrossRef]
- Murphy, R.B.; Repasky, M.P.; Greenwood, J.R.; Tubert-Brohman, I.; Jerome, S.; Annabhimoju, R.; Boyles, N.A.; Schmitz, C.D.; Abel, R.; Farid, R.; et al. WScore: A Flexible and Accurate Treatment of Explicit Water Molecules in Ligand-Receptor Docking. J. Med. Chem. 2016, 59, 4364–4384. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, C.; Bernacki, K.; Jacobson, M.P. Virtual Screening against Highly Charged Active Sites: Identifying Substrates of Alpha−Beta Barrel Enzymes. Biochemistry 2005, 44, 2059–2071. [Google Scholar] [CrossRef] [PubMed]
- Anighoro, A.; Rastelli, G. Enrichment Factor Analyses on G-Protein Coupled Receptors with Known Crystal Structure. J Chem Inf Model 2013, 53, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Jeszenői, N.; Bálint, M.; Horváth, I.; van der Spoel, D.; Hetényi, C. Exploration of Interfacial Hydration Networks of Target-Ligand Complexes. J Chem Inf Model 2016, 56, 148–158. [Google Scholar] [CrossRef]
- Kapla, J.; Rodríguez-Espigares, I.; Ballante, F.; Selent, J.; Carlsson, J. Can Molecular Dynamics Simulations Improve the Structural Accuracy and Virtual Screening Performance of GPCR Models? PLoS Comput Biol 2021, 17, e1008936. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved Prediction of Protein-Protein Interactions Using AlphaFold2. Nat Commun 2022, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ko, J.; Lee, J. Can AlphaFold2 Predict Protein-Peptide Complex Structures Accurately? 07.27.45 3972.
- Johansson-Åkhe, I.; Wallner, B. Improving Peptide-Protein Docking with AlphaFold-Multimer Using Forced Sampling. Front Bioinform 2022, 2, 959160. [Google Scholar] [CrossRef] [PubMed]
- de Brevern, A.G. An Agnostic Analysis of the Human AlphaFold2 Proteome Using Local Protein Conformations. Biochimie 2023, 207, 11–19. [Google Scholar] [CrossRef]
- Al-Masri, C.; Trozzi, F.; Lin, S.-H.; Tran, O.; Sahni, N.; Patek, M.; Cichonska, A.; Ravikumar, B.; Rahman, R. Investigating the Conformational Landscape of AlphaFold2-Predicted Protein Kinase Structures. Bioinform Adv 2023, 3, vbad129. [Google Scholar] [CrossRef]
- Tong, A.B.; Burch, J.D.; McKay, D.; Bustamante, C.; Crackower, M.A.; Wu, H. Could AlphaFold Revolutionize Chemical Therapeutics? Nat Struct Mol Biol 2021, 28, 771–772. [Google Scholar] [CrossRef]
- Skolnick, J.; Gao, M.; Zhou, H.; Singh, S. AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function. J Chem Inf Model 2021, 61, 4827–4831. [Google Scholar] [CrossRef] [PubMed]
- Buttenschoen, M. ; M. Morris, G.; M. Deane, C. PoseBusters: AI-Based Docking Methods Fail to Generate Physically Valid Poses or Generalise to Novel Sequences. Chemical Science 2024, 15, 3130–3139. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Janson, G.; Feig, M. Physics-Based Protein Structure Refinement in the Era of Artificial Intelligence. Proteins 2021, 89, 1870–1887. [Google Scholar] [CrossRef]
- Arantes, P.R.; Polêto, M.D.; Pedebos, C.; Ligabue-Braun, R. Making It Rain: Cloud-Based Molecular Simulations for Everyone. J. Chem. Inf. Model. 2021, 61, 4852–4856. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.; Wu, P.; Wu, S.; Lee, T.-Y.; Bai, C. Application of Computational Biology and Artificial Intelligence in Drug Design. International Journal of Molecular Sciences 2022, 23, 13568. [Google Scholar] [CrossRef]
- Adiyaman, R.; Edmunds, N.S.; Genc, A.G.; Alharbi, S.M.A.; McGuffin, L.J. Improvement of Protein Tertiary and Quaternary Structure Predictions Using the ReFOLD Refinement Method and the AlphaFold2 Recycling Process. Bioinform Adv 2023, 3, vbad078. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat Struct Biol 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Norberg, J.; Nilsson, L. Advances in Biomolecular Simulations: Methodology and Recent Applications. Q Rev Biophys 2003, 36, 257–306. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat Protoc 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X Public Web Server for Protein-Protein Docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J Am Chem Soc 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, G.; Pinzi, L. Refinement and Rescoring of Virtual Screening Results. Front Chem 2019, 7, 498. [Google Scholar] [CrossRef] [PubMed]
- Guterres, H.; Im, W. Improving Protein-Ligand Docking Results with High-Throughput Molecular Dynamics Simulations. J Chem Inf Model 2020, 60, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Jo, S.; Lim, H.-S.; Im, W. Application of Binding Free Energy Calculations to Prediction of Binding Modes and Affinities of MDM2 and MDMX Inhibitors. J Chem Inf Model 2012, 52, 1821–1832. [Google Scholar] [CrossRef]
- Radom, F.; Plückthun, A.; Paci, E. Assessment of Ab Initio Models of Protein Complexes by Molecular Dynamics. PLoS Comput Biol 2018, 14, e1006182. [Google Scholar] [CrossRef]
- Wang, J.; Alekseenko, A.; Kozakov, D.; Miao, Y. Improved Modeling of Peptide-Protein Binding Through Global Docking and Accelerated Molecular Dynamics Simulations. Front Mol Biosci 2019, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Wang, J.; Cieplak, P.; Kollman, P.A.; Kuntz, I.D. Molecular Dynamics and Free Energy Analyses of Cathepsin D-Inhibitor Interactions: Insight into Structure-Based Ligand Design. J Med Chem 2002, 45, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de Novo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Raveh, B.; London, N.; Schueler-Furman, O. Sub-Angstrom Modeling of Complexes between Flexible Peptides and Globular Proteins. Proteins 2010, 78, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2. 2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Branches, A.D.S.; Da Silva, J.N.; De Oliveira, M.D.L.; Bezerra, D.P.; Soares, M.B.P.; Costa, E.V.; Oliveira, K.M.T. DFT Calculations, Molecular Docking, Binding Free Energy Analysis and Cytotoxicity Assay of 7,7-Dimethylaporphine Alkaloids with Methylenedioxy Ring in Positions 1 and 2. Computational and Theoretical Chemistry 2024, 1233, 114483. [Google Scholar] [CrossRef]
- Cavalli, A.; Bottegoni, G.; Raco, C.; De Vivo, M.; Recanatini, M. A Computational Study of the Binding of Propidium to the Peripheral Anionic Site of Human Acetylcholinesterase. J Med Chem 2004, 47, 3991–3999. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Yeom, M.S.; Lee, S. Loop Flexibility and Solvent Dynamics as Determinants for the Selective Inhibition of Cyclin-Dependent Kinase 4: Comparative Molecular Dynamics Simulation Studies of CDK2 and CDK4. Chembiochem 2004, 5, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Ogrizek, M.; Turk, S.; Lešnik, S.; Sosič, I.; Hodošček, M.; Mirković, B.; Kos, J.; Janežič, D.; Gobec, S.; Konc, J. Molecular Dynamics to Enhance Structure-Based Virtual Screening on Cathepsin B. J Comput Aided Mol Des 2015, 29, 707–712. [Google Scholar] [CrossRef]
- Bálint, M.; Jeszenői, N.; Horváth, I.; van der Spoel, D.; Hetényi, C. Systematic Exploration of Multiple Drug Binding Sites. Journal of Cheminformatics 2017, 9, 65. [Google Scholar] [CrossRef]
- Fu, I.; Geacintov, N.E.; Broyde, S. Molecular Dynamics Simulations Reveal How H3K56 Acetylation Impacts Nucleosome Structure to Promote DNA Exposure for Lesion Sensing. DNA Repair (Amst) 2021, 107, 103201. [Google Scholar] [CrossRef]
- Mosammaparast, N.; Shi, Y. Reversal of Histone Methylation: Biochemical and Molecular Mechanisms of Histone Demethylases. Annu. Rev. Biochem. 2010, 79, 155–179. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Zhang, Y. The Diverse Functions of Histone Lysine Methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Zsidó, B.Z.; Hetényi, C. Molecular Structure, Binding Affinity, and Biological Activity in the Epigenome. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Bortoluzzi, A.; Amato, A.; Lucas, X.; Blank, M.; Ciulli, A. Structural Basis of Molecular Recognition of Helical Histone H3 Tail by PHD Finger Domains. Biochem J 2017, 474, 1633–1651. [Google Scholar] [CrossRef] [PubMed]
- Ruthenburg, A.J.; Wang, W.; Graybosch, D.M.; Li, H.; Allis, C.D.; Patel, D.J.; Verdine, G.L. Histone H3 Recognition and Presentation by the WDR5 Module of the MLL1 Complex. Nat. Struct. Mol. Biol. 2006, 13, 704–712. [Google Scholar] [CrossRef]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to de Novo Methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef]
- Iwase, S.; Xiang, B.; Ghosh, S.; Ren, T.; Lewis, P.W.; Cochrane, J.C.; Allis, C.D.; Picketts, D.J.; Patel, D.J.; Li, H.; et al. ATRX ADD Domain Links an Atypical Histone Methylation Recognition Mechanism to Human Mental-Retardation Syndrome. Nat. Struct. Mol. Biol. 2011, 18, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Rajakumara, E.; Wang, Z.; Ma, H.; Hu, L.; Chen, H.; Lin, Y.; Guo, R.; Wu, F.; Li, H.; Lan, F.; et al. PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression. Mol. Cell 2011, 43, 275–284. [Google Scholar] [CrossRef]
- Tsai, W.-W.; Wang, Z.; Yiu, T.T.; Akdemir, K.C.; Xia, W.; Winter, S.; Tsai, C.-Y.; Shi, X.; Schwarzer, D.; Plunkett, W.; et al. TRIM24 Links a Non-Canonical Histone Signature to Breast Cancer. Nature 2010, 468, 927–932. [Google Scholar] [CrossRef]
- Chignola, F.; Gaetani, M.; Rebane, A.; Org, T.; Mollica, L.; Zucchelli, C.; Spitaleri, A.; Mannella, V.; Peterson, P.; Musco, G. The Solution Structure of the First PHD Finger of Autoimmune Regulator in Complex with Non-Modified Histone H3 Tail Reveals the Antagonistic Role of H3R2 Methylation. Nucleic Acids Res. 2009, 37, 2951–2961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Guo, X.; Rong, N.; Song, Y.; Xu, Y.; Lan, W.; Zhang, X.; Liu, M.; Xu, Y.; et al. The PHD1 Finger of KDM5B Recognizes Unmodified H3K4 during the Demethylation of Histone H3K4me2/3 by KDM5B. Protein Cell 2014, 5, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, H.; Huang, S.-Y. Dynamics and Mechanisms in the Recruitment and Transference of Histone Chaperone CIA/ASF1. International Journal of Molecular Sciences 2019, 20, 3325. [Google Scholar] [CrossRef] [PubMed]
- Zsidó, B.Z.; Bayarsaikhan, B.; Börzsei, R.; Hetényi, C. Construction of Histone–Protein Complex Structures by Peptide Growing. International Journal of Molecular Sciences 2023, 24, 13831. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.A.; Devaurs, D.; Kavraki, L.E. Understanding the Challenges of Protein Flexibility in Drug Design. Expert Opin Drug Discov 2015, 10, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int J Mol Sci 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Windshügel, B. LEADS-PEP: A Benchmark Data Set for Assessment of Peptide Docking Performance. J Chem Inf Model 2016, 56, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qin, S.; Chen, T.-Y.; Lei, M.; Dhar, S.S.; Ho, J.C.; Dong, A.; Loppnau, P.; Li, Y.; Lee, M.G.; et al. Structural Insights into Trans-Histone Regulation of H3K4 Methylation by Unique Histone H4 Binding of MLL3/4. Nat Commun 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, R.; Renard, B.Y. Docking Small Peptides Remains a Great Challenge: An Assessment Using AutoDock Vina. Brief. Bioinformatics 2015, 16, 1045–1056. [Google Scholar] [CrossRef]
- Peterson, L.X.; Roy, A.; Christoffer, C.; Terashi, G.; Kihara, D. Modeling Disordered Protein Interactions from Biophysical Principles. PLoS Comput Biol 2017, 13, e1005485. [Google Scholar] [CrossRef]
- Alam, N.; Goldstein, O.; Xia, B.; Porter, K.A.; Kozakov, D.; Schueler-Furman, O. High-Resolution Global Peptide-Protein Docking Using Fragments-Based PIPER-FlexPepDock. PLoS Comput. Biol. 2017, 13, e1005905. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-Dock Web Server for the Flexible Docking of Peptides to Proteins without Prior Knowledge of the Binding Site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A Web Server for Blind Peptide-Protein Docking Based on a Hierarchical Algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A Web Server for Protein-Protein and Protein-DNA/RNA Docking Based on a Hybrid Strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Brenke, R.; Comeau, S.R.; Vajda, S. PIPER: An FFT-Based Protein Docking Program with Pairwise Potentials. Proteins 2006, 65, 392–406. [Google Scholar] [CrossRef]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef] [PubMed]
- Bálint, M.; Jeszenői, N.; Horváth, I.; Ábrahám, I.M.; Hetényi, C. Dynamic Changes in Binding Interaction Networks of Sex Steroids Establish Their Non-Classical Effects. Sci Rep 2017, 7, 14847. [Google Scholar] [CrossRef]
- Trellet, M.; Melquiond, A.S.J.; Bonvin, A.M.J.J. A Unified Conformational Selection and Induced Fit Approach to Protein-Peptide Docking. PLoS One 2013, 8, e58769. [Google Scholar] [CrossRef]
- de Vries, S.J.; Rey, J.; Schindler, C.E.M.; Zacharias, M.; Tuffery, P. The pepATTRACT Web Server for Blind, Large-Scale Peptide-Protein Docking. Nucleic Acids Res 2017, 45, W361–W364. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.E.M.; de Vries, S.J.; Zacharias, M. Fully Blind Peptide-Protein Docking with pepATTRACT. Structure 2015, 23, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Lamothe, G.; Malliavin, T.E. Re-TAMD: Exploring Interactions between H3 Peptide and YEATS Domain Using Enhanced Sampling. BMC Struct Biol 2018, 18, 4. [Google Scholar] [CrossRef]
- Cuendet, M.A.; Zoete, V.; Michielin, O. How T Cell Receptors Interact with Peptide-MHCs: A Multiple Steered Molecular Dynamics Study. Proteins 2011, 79, 3007–3024. [Google Scholar] [CrossRef] [PubMed]
- Sheik Amamuddy, O.; Veldman, W.; Manyumwa, C.; Khairallah, A.; Agajanian, S.; Oluyemi, O.; Verkhivker, G.M.; Tastan Bishop, Ö. Integrated Computational Approaches and Tools for Allosteric Drug Discovery. Int J Mol Sci 2020, 21, 847. [Google Scholar] [CrossRef]
- Porter, K.A.; Xia, B.; Beglov, D.; Bohnuud, T.; Alam, N.; Schueler-Furman, O.; Kozakov, D. ClusPro PeptiDock: Efficient Global Docking of Peptide Recognition Motifs Using FFT. Bioinformatics 2017, 33, 3299–3301. [Google Scholar] [CrossRef] [PubMed]
- Keskin, O. Binding Induced Conformational Changes of Proteins Correlate with Their Intrinsic Fluctuations: A Case Study of Antibodies. BMC Struct Biol 2007, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Conformational Transitions upon Ligand Binding: Holo-Structure Prediction from Apo Conformations. PLoS Comput Biol 2010, 6, e1000634. [Google Scholar] [CrossRef] [PubMed]
- Birch, L.; Murray, C.W.; Hartshorn, M.J.; Tickle, I.J.; Verdonk, M.L. Sensitivity of Molecular Docking to Induced Fit Effects in Influenza Virus Neuraminidase. J Comput Aided Mol Des 2002, 16, 855–869. [Google Scholar] [CrossRef]
- Erickson, J.A.; Jalaie, M.; Robertson, D.H.; Lewis, R.A.; Vieth, M. Lessons in Molecular Recognition: The Effects of Ligand and Protein Flexibility on Molecular Docking Accuracy. J Med Chem 2004, 47, 45–55. [Google Scholar] [CrossRef]
- Gervasoni, S.; Guccione, C.; Fanti, V.; Bosin, A.; Cappellini, G.; Golosio, B.; Ruggerone, P.; Malloci, G. Molecular Simulations of SSTR2 Dynamics and Interaction with Ligands. Sci Rep 2023, 13, 4768. [Google Scholar] [CrossRef]
- Neduva, V.; Linding, R.; Su-Angrand, I.; Stark, A.; de Masi, F.; Gibson, T.J.; Lewis, J.; Serrano, L.; Russell, R.B. Systematic Discovery of New Recognition Peptides Mediating Protein Interaction Networks. PLoS Biol 2005, 3, e405. [Google Scholar] [CrossRef] [PubMed]
- Puntervoll, P.; Linding, R.; Gemünd, C.; Chabanis-Davidson, S.; Mattingsdal, M.; Cameron, S.; Martin, D.M.A.; Ausiello, G.; Brannetti, B.; Costantini, A.; et al. ELM Server: A New Resource for Investigating Short Functional Sites in Modular Eukaryotic Proteins. Nucleic Acids Res 2003, 31, 3625–3630. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Pache, R.A.; Bernadó, P.; Pons, M.; Aloy, P. Dynamic Interactions of Proteins in Complex Networks: A More Structured View. FEBS J 2009, 276, 5390–5405. [Google Scholar] [CrossRef] [PubMed]
- Scaramozzino, D.; Khade, P.M.; Jernigan, R.L. Protein Fluctuations in Response to Random External Forces. Applied Sciences 2022, 12, 2344. [Google Scholar] [CrossRef]
- Dutkiewicz, Z.; Mikstacka, R. Hydration and Structural Adaptations of the Human CYP1A1, CYP1A2, and CYP1B1 Active Sites by Molecular Dynamics Simulations. International Journal of Molecular Sciences 2023, 24, 11481. [Google Scholar] [CrossRef] [PubMed]
- Jeszenői, N.; Horváth, I.; Bálint, M.; van der Spoel, D.; Hetényi, C. Mobility-Based Prediction of Hydration Structures of Protein Surfaces. Bioinformatics 2015, 31, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Dahlström, K.M.; Salminen, T.A. Apprehensions and Emerging Solutions in ML-Based Protein Structure Prediction. Curr Opin Struct Biol 2024, 86, 102819. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Maffucci, I.; Contini, A. Advances in the Treatment of Explicit Water Molecules in Docking and Binding Free Energy Calculations. Curr Med Chem 2019, 26, 7598–7622. [Google Scholar] [CrossRef]
- Park, H.; Lee, H.; Seok, C. High-Resolution Protein-Protein Docking by Global Optimization: Recent Advances and Future Challenges. Curr Opin Struct Biol 2015, 35, 24–31. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. The Journal of Chemical Physics 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J Chem Phys 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. The Journal of Chemical Physics 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant Pressure Molecular Dynamics for Molecular Systems. Molecular Physics 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. Journal of Applied Physics 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J Chem Theory Comput 2008, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. Journal of Computational Chemistry 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC.: New York, NY, USA, 2021. [Google Scholar]
- Gasteiger, J.; Marsili, M. Iterative Partial Equalization of Orbital Electronegativity—a Rapid Access to Atomic Charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. Journal of Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cieplak, P.; Li, J.; Cai, Q.; Hsieh, M.-J.; Luo, R.; Duan, Y. Development of Polarizable Models for Molecular Mechanical Calculations. 4. van Der Waals Parametrization. J Phys Chem B 2012, 116, 7088–7101. [Google Scholar] [CrossRef] [PubMed]
- Mehler, E.L.; Solmajer, T. Electrostatic Effects in Proteins: Comparison of Dielectric and Charge Models. Protein Engineering, Design and Selection 1991, 4, 903–910. [Google Scholar] [CrossRef] [PubMed]
| PDB ID (Apo) | Res (Å) |
PDB ID (Holo) | Res (Å) |
Target | Histone H3 peptide sequence1 | Kd (µM) |
RMSDstart (Å) |
|---|---|---|---|---|---|---|---|
| 1xwh | NMR2 | 2ke1 | NMR2 | AIRE PHD finger | ARTKQTARKS | 6.5 | 8.56 |
| 2fui | NMR2 | 2fuu | NMR2 | BPTF PHD finger | ARTKQTARKSTGGKA | 2.7 | 17.75 |
| 2gnq | 1.8 | 2co0 | 2.25 | WDR5 | ARTKQTARKSTGGKA | 3.3 | 8.21 |
| 2mny | NMR2 | 2mnz | NMR2 | KDM5B PHD1 finger | ARTKQTARKS | 6.4 | 18.33 |
| 2pv0 | 3.3 | 2pvc | 3.69 | DNMT3L | ARTKQTA | 2.1 | 9.51 |
| 3o33 | 2.0 | 3o37 | 2.0 | TRIM24 PHD-Bromo complex | ARTKQTARKS | 8.6 | 27.18 |
| 3qln | 1.90 | 3qlc | 2.5 | ARTX ADD | ARTKQTARKSTGGKA | 3.7 | 13.28 |
| 3sox | 2.65 | 3sou | 1.8 | UHRF1 PHD finger | ARTKQTARK | 2.1 | 10.32 |
| 4ljn | 3.0 | 4lk9 | 1.6 | MOZ double PHD finger | ARTKQTARKSTGGKAPRKQLA | - | 15.02 |
| 4qf2 | 1.7 | 4q6f | 1.91 | BAZ2A PHD Zinc finger | ARTKQ | 2.51 | 9.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




