Submitted:
13 April 2024
Posted:
16 April 2024
You are already at the latest version
Abstract
Keywords:
MSC: 60E05; 62H30
1. Introduction
2. Distribution of Eigenvalues of Random Confusion Matrix
2.1. Distribution of trace of a random confusion matrix
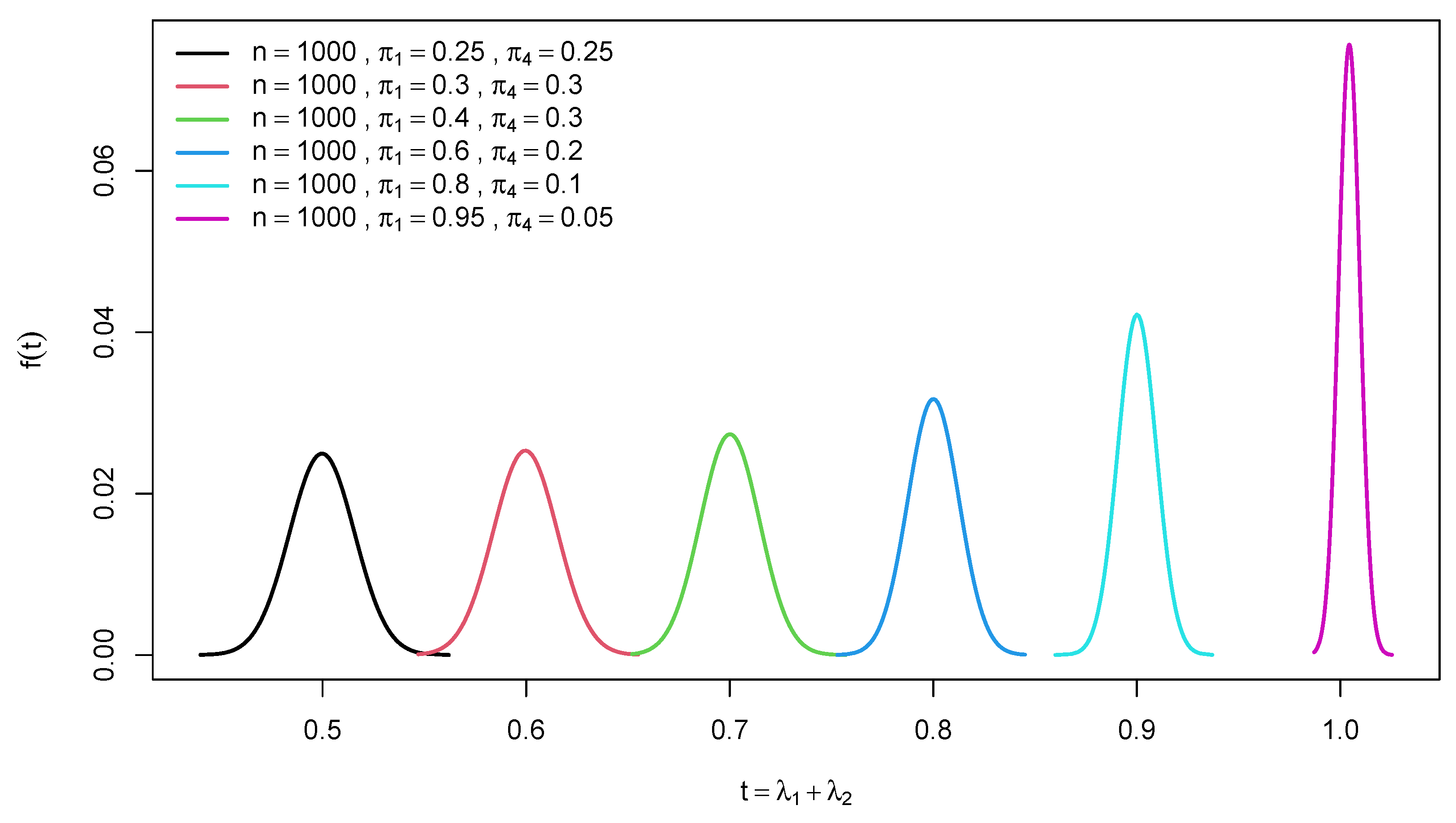
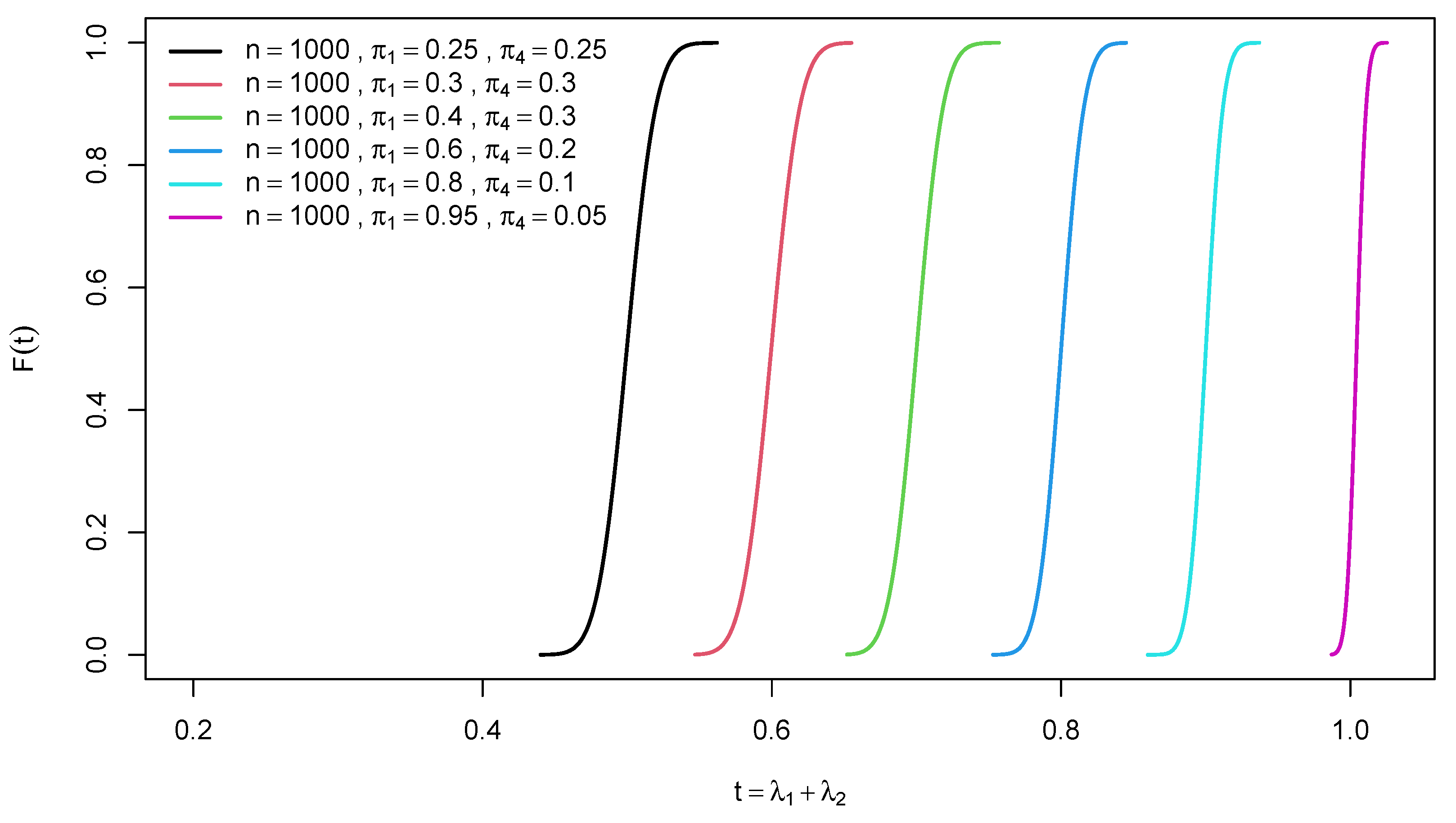
2.2. Distribution of Difference of Two Traces of Random Confusion Matrices
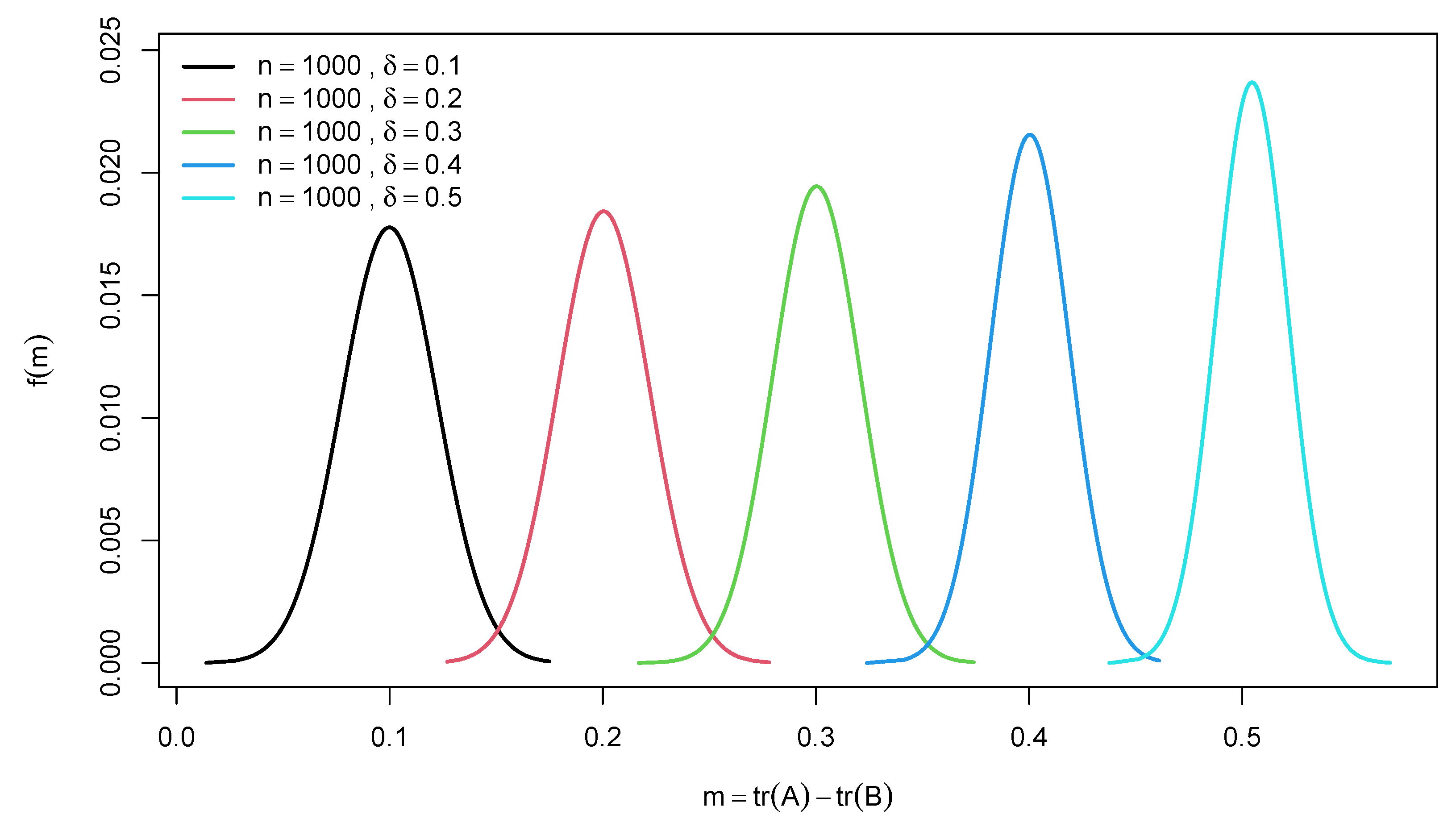
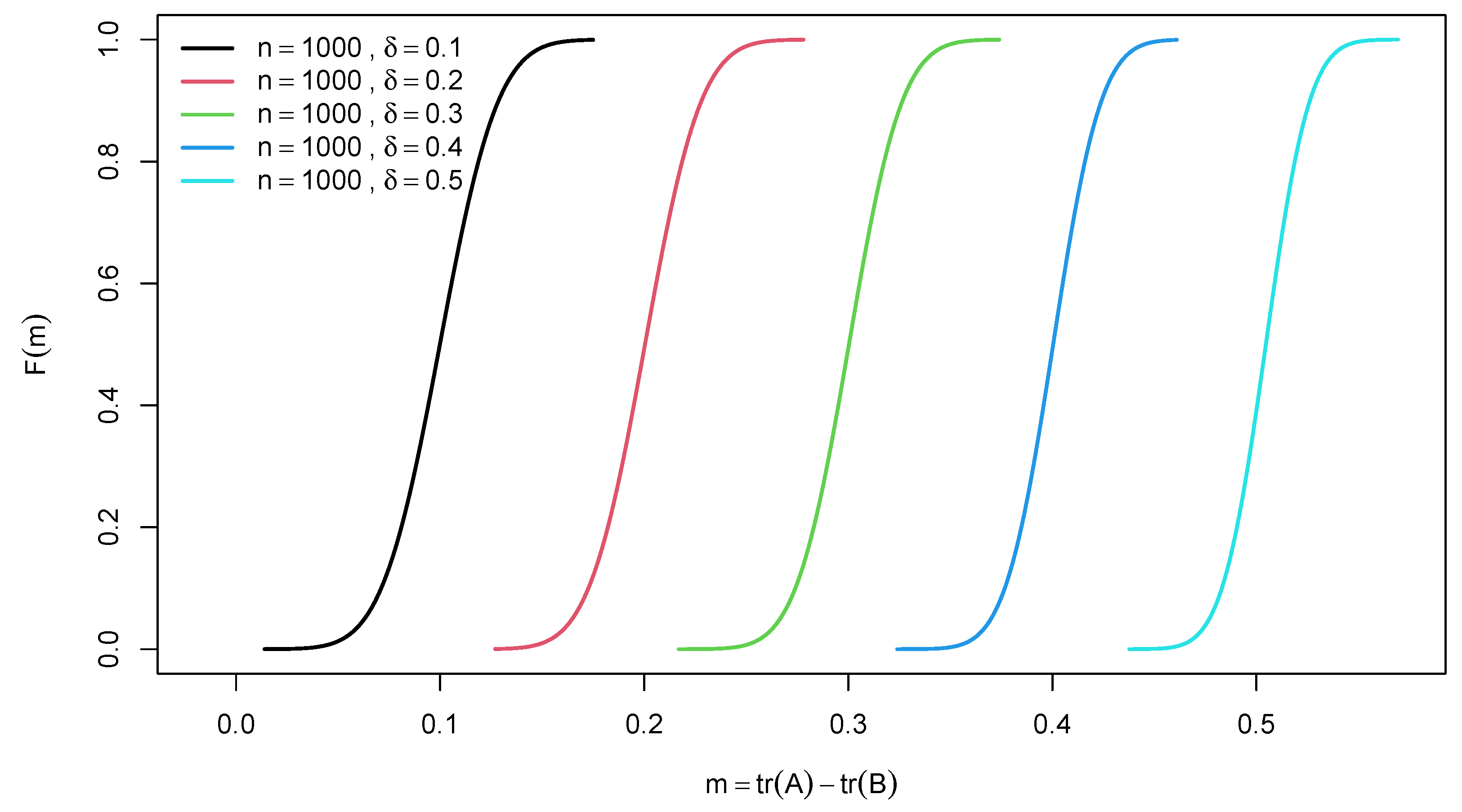
3. Example
4. Applications
- Heart disease [28]: This dataset comprises information from 303 patients with heart disease at Cleveland Hospital, including 14 features. The objective is to determine the presence or absence of heart disease.
- Breast cancer [29]: Originating from the University Medical Centre, Institute of Oncology, Ljubljana, Yugoslavia, this dataset contains data from 286 patients with breast cancer, encompassing 9 features. The goal is to predict the presence or absence of breast cancer recurrence.
- Liver disease [30]: This dataset consists of 584 patient records from the NorthEast region of Andhra Pradesh, India, across 10 features. The objective is to predict whether a patient has liver disease using various biochemical markers.
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, R.C.; Dewi, C.; Huang, S.W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. Journal of Big Data 2020, 7, 52. [Google Scholar] [CrossRef]
- Olaniran, O.R.; Abdullah, M.A.A. Bayesian weighted random forest for classification of high-dimensional genomics data. Kuwait Journal of Science 2023, 50, 477–484. [Google Scholar] [CrossRef]
- Koço, S.; Capponi, C. On multi-class classification through the minimization of the confusion matrix norm. Asian Conference on Machine Learning. PMLR, 2013, pp. 277–292.
- García-Balboa, J.L.; Alba-Fernández, M.V.; Ariza-López, F.J.; Rodríguez-Avi, J. Analysis of thematic similarity using confusion matrices. ISPRS international journal of geo-information 2018, 7, 233. [Google Scholar] [CrossRef]
- Übeyli, E.D.; Güler, İ. Features extracted by eigenvector methods for detecting variability of EEG signals. Pattern Recognition Letters 2007, 28, 592–603. [Google Scholar] [CrossRef]
- Sayyad, S.; Shaikh, M.; Pandit, A.; Sonawane, D.; Anpat, S. Confusion matrix-based supervised classification using microwave SIR-C SAR satellite dataset. Recent Trends in Image Processing and Pattern Recognition: Third International Conference, RTIP2R 2020, Aurangabad, India, January 3–4, 2020, Revised Selected Papers, Part II 3. Springer, 2021, pp. 176–187.
- Reddy, G.T.; Reddy, M.P.K.; Lakshmanna, K.; Kaluri, R.; Rajput, D.S.; Srivastava, G.; Baker, T. Analysis of dimensionality reduction techniques on big data. Ieee Access 2020, 8, 54776–54788. [Google Scholar] [CrossRef]
- Golub, G.H.; Van Loan, C.F. Matrix computations; JHU press, 2013. [Google Scholar]
- Alamsyah, A.; Fadila, T. Increased accuracy of prediction hepatitis disease using the application of principal component analysis on a support vector machine. Journal of Physics: Conference Series 2021, 1968, 012016. [Google Scholar] [CrossRef]
- Sifaou, H.; Kammoun, A.; Alouini, M.S. High-dimensional linear discriminant analysis classifier for spiked covariance model. Journal of Machine Learning Research 2020, 21, 1–24. [Google Scholar]
- Hasan, S.N.S.; Jamil, N.W. A Comparative Study of Hybrid Dimension Reduction Techniques to Enhance the Classification of High-Dimensional Microarray Data. 2023 IEEE 11th Conference on Systems, Process & Control (ICSPC). IEEE, 2023, pp. 240–245.
- Lu, J.; Lu, Y. A priori generalization error analysis of two-layer neural networks for solving high dimensional Schrödinger eigenvalue problems. Communications of the American Mathematical Society 2022, 2, 1–21. [Google Scholar] [CrossRef]
- Caelen, O. A Bayesian interpretation of the confusion matrix. Annals of Mathematics and Artificial Intelligence 2017, 81, 429–450. [Google Scholar] [CrossRef]
- Olaniran, O.R.; Alzahrani, A.R.R. On the Oracle Properties of Bayesian Random Forest for Sparse High-Dimensional Gaussian Regression. Mathematics 2023, 11, 4957. [Google Scholar] [CrossRef]
- Olaniran, O.; Abdullah, M. Subset selection in high-dimensional genomic data using hybrid variational Bayes and bootstrap priors. Journal of Physics: Conference Series. IOP Publishing, 2020, Vol. 1489, p. 012030.
- Pudjihartono, N.; Fadason, T.; Kempa-Liehr, A.W.; O’Sullivan, J.M. A review of feature selection methods for machine learning-based disease risk prediction. Frontiers in Bioinformatics 2022, 2, 927312. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, T.; Sæbø, S.; Liland, K.H. Comparison of variable selection methods in partial least squares regression. Journal of Chemometrics 2020, 34, e3226. [Google Scholar] [CrossRef]
- Chen, C.W.; Tsai, Y.H.; Chang, F.R.; Lin, W.C. Ensemble feature selection in medical datasets: Combining filter, wrapper, and embedded feature selection results. Expert Systems 2020, 37, e12553. [Google Scholar] [CrossRef]
- Wang, G.; Sarkar, A.; Carbonetto, P.; Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. Journal of the Royal Statistical Society Series B: Statistical Methodology 2020, 82, 1273–1300. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Perperoglou, A.; Schmid, M.; Abrahamowicz, M.; Becher, H.; Binder, H.; Dunkler, D.; Harrell, F.E.; Royston, P.; Heinze, G.; others. State of the art in selection of variables and functional forms in multivariable analysis—outstanding issues. Diagnostic and prognostic research 2020, 4, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.Z.I.; Turin, T.C. Variable selection strategies and its importance in clinical prediction modelling. Family medicine and community health 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Peyrache, A.; Rose, C.; Sicilia, G. Variable selection in data envelopment analysis. European Journal of Operational Research 2020, 282, 644–659. [Google Scholar] [CrossRef]
- Montoya, A.K.; Edwards, M.C. The poor fit of model fit for selecting number of factors in exploratory factor analysis for scale evaluation. Educational and psychological measurement 2021, 81, 413–440. [Google Scholar] [CrossRef]
- Greenacre, M.; Groenen, P.J.; Hastie, T.; d’Enza, A.I.; Markos, A.; Tuzhilina, E. Principal component analysis. Nature Reviews Methods Primers 2022, 2, 100. [Google Scholar] [CrossRef]
- Popoola, J.; Yahya, W.B.; Popoola, O.; Olaniran, O.R. Generalized self-similar first order autoregressive generator (gsfo-arg) for internet traffic. Statistics, Optimization & Information Computing 2020, 8, 810–821. [Google Scholar]
- Sarkar, A.; Kothiyal, M.; Kumar, S. Distribution of the ratio of two consecutive level spacings in orthogonal to unitary crossover ensembles. Physical Review E 2020, 101, 012216. [Google Scholar] [CrossRef] [PubMed]
- Grimm, U.; Römer, R.A. Gaussian orthogonal ensemble for quasiperiodic tilings without unfolding: r-value statistics. Physical Review B 2021, 104, L060201. [Google Scholar] [CrossRef]
- Janosi, Andras, S.W.P.M.; Detrano, R. Heart Disease. UCI Machine Learning Repository, 1988. [CrossRef]
- Zwitter, M.; Soklic, M. Breast Cancer. UCI Machine Learning Repository, 1988. [CrossRef]
- Ramana, B.; Venkateswarlu, N. ILPD (Indian Liver Patient Dataset). UCI Machine Learning Repository, 2012. [CrossRef]
- Ding, N.; Sadeghi, P. A submodularity-based agglomerative clustering algorithm for the privacy funnel. arXiv, arXiv:1901.06629 2019.
- Navarro, C.L.A.; Damen, J.A.; Takada, T.; Nijman, S.W.; Dhiman, P.; Ma, J.; Collins, G.S.; Bajpai, R.; Riley, R.D.; Moons, K.G.; others. Risk of bias in studies on prediction models developed using supervised machine learning techniques: systematic review. bmj 2021, 375. [Google Scholar]
- Tantithamthavorn, C.; McIntosh, S.; Hassan, A.E.; Matsumoto, K. An empirical comparison of model validation techniques for defect prediction models. IEEE Transactions on Software Engineering 2016, 43, 1–18. [Google Scholar] [CrossRef]
| Heart disease | Breast cancer | Liver disease | ||||
| Method | ||||||
| LR | 0.83 | 0.88 | 0.70 | 0.72 | 0.71 | 0.72 |
| (0.031) | (0.032) | (0.043) | (0.031) | (0.029) | (0.029) | |
| DT | 0.77 | 0.76 | 0.71 | 0.73 | 0.72 | 0.67 |
| (0.042) | (0.025) | (0.031) | (0.024) | (0.031) | (0.021) | |
| RF | 0.82 | 0.84 | 0.82 | 0.79 | 0.82 | 0.80 |
| (0.031) | (0.029) | (0.025) | (0.030) | (0.025) | (0.030) | |
| XG | 0.77 | 0.82 | 0.74 | 0.70 | 0.75 | 0.72 |
| (0.036) | (0.029) | (0.030) | (0.031) | (0.030) | (0.030) | |
| Heart disease | Breast cancer | Liver disease | ||||
| Pair | ||||||
| XG - LR | -0.05 | -0.06 | 0.04 | -0.02 | 0.03 | 0.01 |
| (0.058) | (0.408) | (0.855) | (0.473) | (0.835) | (0.509) | |
| XG - RF | -0.05 | -0.02 | -0.08 | -0.08 | -0.08 | -0.08 |
| (0.055) | (0.468) | (0.001) | (0.367) | (0.002) | (0.375) | |
| XG - DT | 0.00 | 0.06 | 0.02 | -0.03 | 0.02 | 0.05 |
| (0.481) | (0.598) | (0.728) | (0.453) | (0.719) | (0.588) | |
| LR - RF | 0.01 | 0.04 | -0.12 | -0.07 | -0.11 | -0.08 |
| (0.536) | (0.561) | (0.000) | (0.393) | (0.000) | (0.365) | |
| LR - DT | 0.06 | 0.11 | -0.02 | -0.01 | -0.01 | 0.04 |
| (0.895) | (0.685) | (0.300) | (0.481) | (0.316) | (0.579) | |
| RF - DT | 0.05 | 0.08 | 0.10 | 0.06 | 0.10 | 0.13 |
| (0.888) | (0.629) | (0.999) | (0.595) | (0.999) | (0.715) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





