Submitted:
28 March 2024
Posted:
29 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
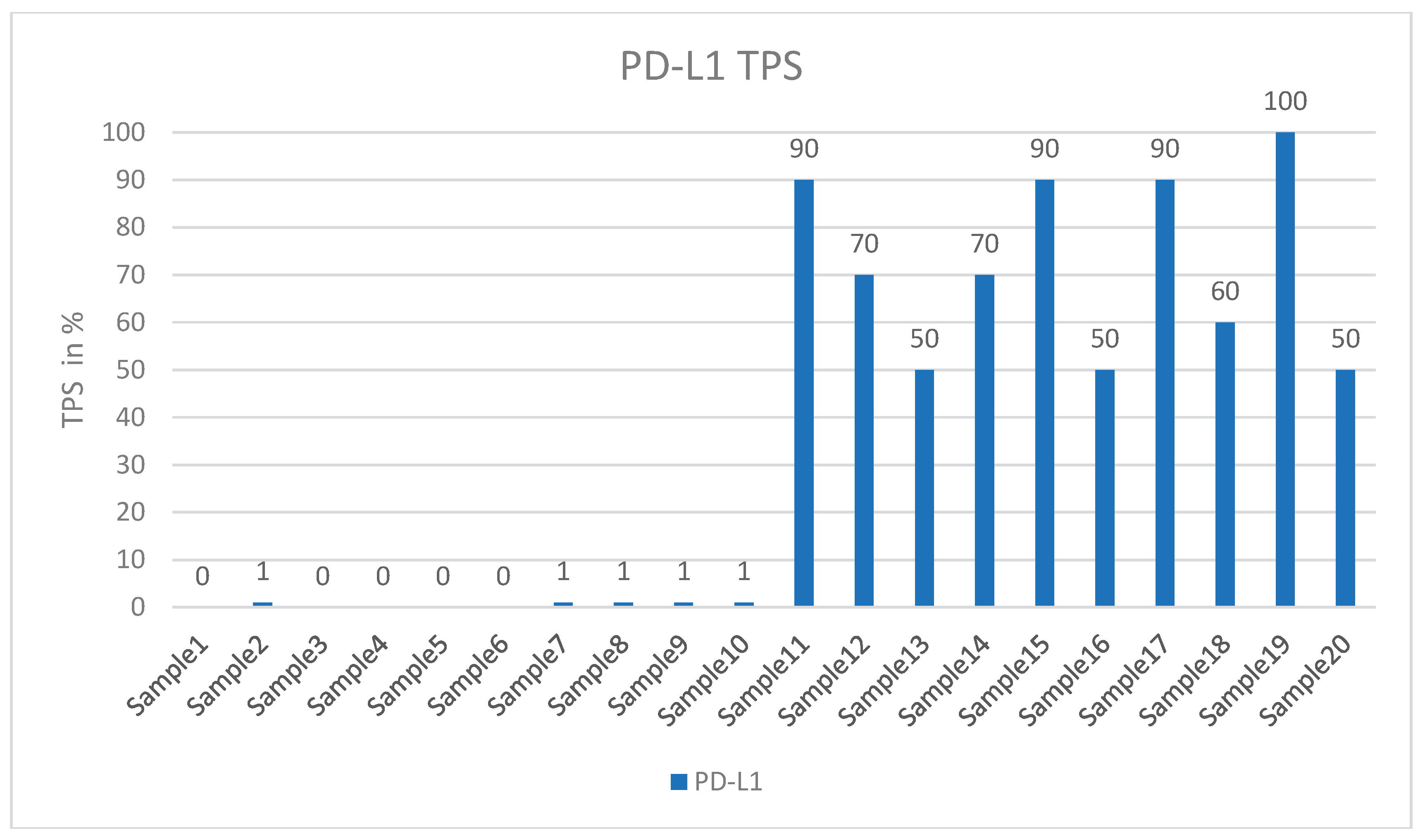
2.1. Tissue Collection and Immunohistochemical Analysis
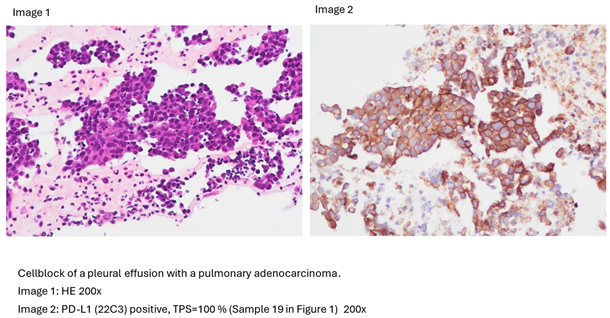
2.2. Molecular and Genetic Analysis
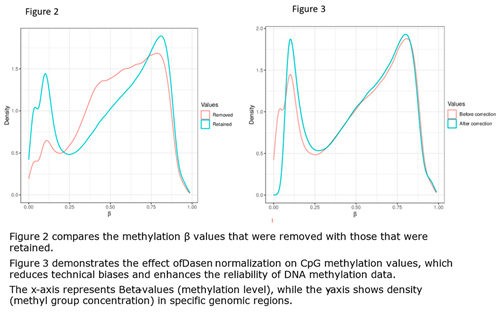
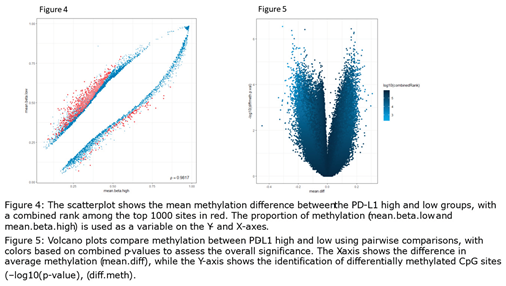
2.3. Computational Data Analysis
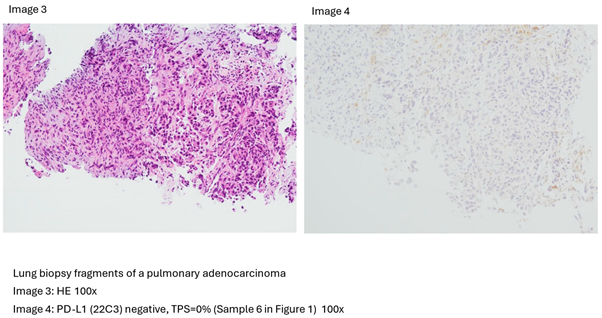
3. Results
- Hypermethylated genes and promotors in PD-L1 high-expressing cases:
- Hypomethylated genes and promotors in PD-L1 high-expressing cases:
- Hypermethylated genes and promotors in PD-L1 low-expressing cases:
- Hypomethylated genes and promotors in PD-L1 low-expressing cases:
4. Discussion
Pathobiological Mechanisms of Hypermethylated Genes and Promotors in the PD-L1 High-Expressing Tumors
Pathobiological Mechanisms of Hypomethylated Genes and Promotors in the PD-L1 High-Expression Group
Pathobiological Mechanisms of Hypermethylated Genes and Promotors in PD-L1 Low-Expressing Cases
Pathobiological Mechanisms of Hypomethylated Genes and Promotors in the PD-L1 Low-Expressing Group
Hypermethylated Genes and Promotors with the Same Methylation Status Simultaneously in PD-L1 High and Low-Expressing Carcinomas
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chi, S.A.; Yu, H.; Choi, Y.L.; Park, S.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Ahn, M.J.; Choi, D.H.; Kim, K.; et al. Trends in Survival Rates of Non-Small Cell Lung Cancer With Use of Molecular Testing and Targeted Therapy in Korea, 2010-2020. JAMA Netw Open 2023, 6, e232002. [CrossRef]
- Paci, E.; Puliti, D.; Lopes Pegna, A.; Carrozzi, L.; Picozzi, G.; Falaschi, F.; Pistelli, F.; Aquilini, F.; Ocello, C.; Zappa, M.; et al. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax 2017, 72, 825-831. [CrossRef]
- Hardtstock, F.; Myers, D.; Li, T.; Cizova, D.; Maywald, U.; Wilke, T.; Griesinger, F. Real-world treatment and survival of patients with advanced non-small cell lung Cancer: a German retrospective data analysis. BMC Cancer 2020, 20, 260. [CrossRef]
- Godoy, L.A.; Chen, J.; Ma, W.; Lally, J.; Toomey, K.A.; Rajappa, P.; Sheridan, R.; Mahajan, S.; Stollenwerk, N.; Phan, C.T.; et al. Emerging precision neoadjuvant systemic therapy for patients with resectable non-small cell lung cancer: current status and perspectives. Biomark Res 2023, 11, 7. [CrossRef]
- Tang, Q.; Chen, Y.; Li, X.; Long, S.; Shi, Y.; Yu, Y.; Wu, W.; Han, L.; Wang, S. The role of PD-1/PD-L1 and application of immune-checkpoint inhibitors in human cancers. Front Immunol 2022, 13, 964442. [CrossRef]
- Wojtukiewicz, M.Z.; Rek, M.M.; Karpowicz, K.; Gorska, M.; Politynska, B.; Wojtukiewicz, A.M.; Moniuszko, M.; Radziwon, P.; Tucker, S.C.; Honn, K.V. Inhibitors of immune checkpoints-PD-1, PD-L1, CTLA-4-new opportunities for cancer patients and a new challenge for internists and general practitioners. Cancer Metastasis Rev 2021, 40, 949-982. [CrossRef]
- Bruno, R.; Fontanini, G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics (Basel) 2020, 10. [CrossRef]
- Czajka-Francuz, P.; Prendes, M.J.; Mankan, A.; Quintana, A.; Pabla, S.; Ramkissoon, S.; Jensen, T.J.; Peiro, S.; Severson, E.A.; Achyut, B.R.; et al. Mechanisms of immune modulation in the tumor microenvironment and implications for targeted therapy. Front Oncol 2023, 13, 1200646. [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124-128. [CrossRef]
- Ruschoff, J.; Schildhaus, H.U.; Ruschoff, J.H.; Johrens, K.; Bocker-Edmonston, T.; Dietmaier, W.; Blaker, H.; Baretton, G.; Horst, D.; Dietel, M.; et al. [Not Available]. Pathologie (Heidelb) 2023. [CrossRef]
- Schildhaus, H.U. [Predictive value of PD-L1 diagnostics]. Pathologe 2018, 39, 498-519. [CrossRef]
- Twomey, J.D.; Zhang, B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J 2021, 23, 39. [CrossRef]
- Turchan, W.T.; Pitroda, S.P.; Weichselbaum, R.R. Radiotherapy and Immunotherapy Combinations in the Treatment of Patients with Metastatic Disease: Current Status and Future Focus. Clin Cancer Res 2021, 27, 5188-5194. [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J Clin 2019, 69, 7-34. [CrossRef]
- Romero-Garcia, S.; Prado-Garcia, H.; Carlos-Reyes, A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front Oncol 2020, 10, 1152. [CrossRef]
- De Marchi, P.; Leal, L.F.; Duval da Silva, V.; da Silva, E.C.A.; Cordeiro de Lima, V.C.; Reis, R.M. PD-L1 expression by Tumor Proportion Score (TPS) and Combined Positive Score (CPS) are similar in non-small cell lung cancer (NSCLC). J Clin Pathol 2021, 74, 735-740. [CrossRef]
- Kaur, D.; Lee, S.M.; Goldberg, D.; Spix, N.J.; Hinoue, T.; Li, H.-T.; Dwaraka, V.B.; Smith, R.; Shen, H.; Liang, G.; et al. Comprehensive evaluation of the Infinium human MethylationEPIC v2 BeadChip. Epigenetics Communications 2023, 3, 6. [CrossRef]
- Bock, C. Analysing and interpreting DNA methylation data. Nat Rev Genet 2012, 13, 705-719. [CrossRef]
- Wang, Y.; Gorrie-Stone, T.J.; Grant, O.A.; Andrayas, A.D.; Zhai, X.; McDonald-Maier, K.D.; Schalkwyk, L.C. InterpolatedXY: a two-step strategy to normalize DNA methylation microarray data avoiding sex bias. Bioinformatics 2022, 38, 3950-3957. [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J Mol Biol 1987, 196, 261-282. [CrossRef]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Springer New York: New York, NY, 2005; pp. 397-420.
- Flanagan, J.M. Epigenome-wide association studies (EWAS): past, present, and future. Methods Mol Biol 2015, 1238, 51-63. [CrossRef]
- Campagna, M.P.; Xavier, A.; Lechner-Scott, J.; Maltby, V.; Scott, R.J.; Butzkueven, H.; Jokubaitis, V.G.; Lea, R.A. Epigenome-wide association studies: current knowledge, strategies and recommendations. Clin Epigenetics 2021, 13, 214. [CrossRef]
- Bhootra, S.; Jill, N.; Shanmugam, G.; Rakshit, S.; Sarkar, K. DNA methylation and cancer: transcriptional regulation, prognostic, and therapeutic perspective. Med Oncol 2023, 40, 71. [CrossRef]
- Lee, C.J.; Ahn, H.; Jeong, D.; Pak, M.; Moon, J.H.; Kim, S. Impact of mutations in DNA methylation modification genes on genome-wide methylation landscapes and downstream gene activations in pan-cancer. BMC Med Genomics 2020, 13, 27. [CrossRef]
- Frost, H.R. Analyzing cancer gene expression data through the lens of normal tissue-specificity. PLoS Comput Biol 2021, 17, e1009085. [CrossRef]
- Chen, Y.; Breeze, C.E.; Zhen, S.; Beck, S.; Teschendorff, A.E. Tissue-independent and tissue-specific patterns of DNA methylation alteration in cancer. Epigenetics Chromatin 2016, 9, 10. [CrossRef]
- Stahl, M.; Kohrman, N.; Gore, S.D.; Kim, T.K.; Zeidan, A.M.; Prebet, T. Epigenetics in Cancer: A Hematological Perspective. PLoS Genet 2016, 12, e1006193. [CrossRef]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Zhang, S.; Gong, Z.; Li, X.; Cao, K.; Deng, H.; He, Y.; et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res 2020, 39, 204. [CrossRef]
- Surace, A.E.A.; Hedrich, C.M. The Role of Epigenetics in Autoimmune/Inflammatory Disease. Front Immunol 2019, 10, 1525. [CrossRef]
- de Mendoza, A.; Nguyen, T.V.; Ford, E.; Poppe, D.; Buckberry, S.; Pflueger, J.; Grimmer, M.R.; Stolzenburg, S.; Bogdanovic, O.; Oshlack, A.; et al. Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability. Genome Biol 2022, 23, 163. [CrossRef]
- Mourksi, N.E.; Morin, C.; Fenouil, T.; Diaz, J.J.; Marcel, V. snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management. Cells 2020, 9. [CrossRef]
- Han, P.; Liu, Q.; Xiang, J. Monitoring methylation-driven genes as prognostic biomarkers in patients with lung squamous cell cancer. Oncol Lett 2020, 19, 707-716. [CrossRef]
- Yeung, Y.T.; Fan, S.; Lu, B.; Yin, S.; Yang, S.; Nie, W.; Wang, M.; Zhou, L.; Li, T.; Li, X.; et al. CELF2 suppresses non-small cell lung carcinoma growth by inhibiting the PREX2-PTEN interaction. Carcinogenesis 2020, 41, 377-389. [CrossRef]
- Cuttano, R.; Colangelo, T.; Guarize, J.; Dama, E.; Cocomazzi, M.P.; Mazzarelli, F.; Melocchi, V.; Palumbo, O.; Marino, E.; Belloni, E.; et al. miRNome profiling of lung cancer metastases revealed a key role for miRNA-PD-L1 axis in the modulation of chemotherapy response. J Hematol Oncol 2022, 15, 178. [CrossRef]
- Guz, M.; Rivero-Muller, A.; Okon, E.; Stenzel-Bembenek, A.; Polberg, K.; Slomka, M.; Stepulak, A. MicroRNAs-role in lung cancer. Dis Markers 2014, 2014, 218169. [CrossRef]
- Jiang, J.; Lu, Y.; Zhang, F.; Huang, J.; Ren, X.L.; Zhang, R. The Emerging Roles of Long Noncoding RNAs as Hallmarks of Lung Cancer. Front Oncol 2021, 11, 761582. [CrossRef]
- Gong, S.; Xu, M.; Zhang, Y.; Shan, Y.; Zhang, H. The Prognostic Signature and Potential Target Genes of Six Long Non-coding RNA in Laryngeal Squamous Cell Carcinoma. Front Genet 2020, 11, 413. [CrossRef]
- Liu, X.L.; Zuo, R.; Ou, W.B. The hippo pathway provides novel insights into lung cancer and mesothelioma treatment. J Cancer Res Clin Oncol 2018, 144, 2097-2106. [CrossRef]
- Padilla, L.; Dakhel, S.; Adan, J.; Masa, M.; Martinez, J.M.; Roque, L.; Coll, T.; Hervas, R.; Calvis, C.; Llinas, L.; et al. S100A7: from mechanism to cancer therapy. Oncogene 2017, 36, 6749-6761. [CrossRef]
- Zhang, Y.; Lu, X.; Zhang, Y.; Zhao, D.; Gong, H.; Du, Y.; Sun, H. The Effect of Extracellular Superoxide Dismutase (SOD3) Gene in Lung Cancer. Front Oncol 2022, 12, 722646. [CrossRef]
- Xiao-Jie, L.; Ai-Mei, G.; Li-Juan, J.; Jiang, X. Pseudogene in cancer: real functions and promising signature. J Med Genet 2015, 52, 17-24. [CrossRef]
- Pang, B.; Wu, N.; Guan, R.; Pang, L.; Li, X.; Li, S.; Tang, L.; Guo, Y.; Chen, J.; Sun, D.; et al. Overexpression of RCC2 Enhances Cell Motility and Promotes Tumor Metastasis in Lung Adenocarcinoma by Inducing Epithelial-Mesenchymal Transition. Clin Cancer Res 2017, 23, 5598-5610. [CrossRef]
- Herath, S.; Sadeghi Rad, H.; Radfar, P.; Ladwa, R.; Warkiani, M.; O’Byrne, K.; Kulasinghe, A. The Role of Circulating Biomarkers in Lung Cancer. Front Oncol 2021, 11, 801269. [CrossRef]
- Sheng, X.; Bowen, N.; Wang, Z. GLI pathogenesis-related 1 functions as a tumor-suppressor in lung cancer. Mol Cancer 2016, 15, 25. [CrossRef]
- Zhang, D.; Wang, H.; He, H.; Niu, H.; Li, Y. Interferon induced transmembrane protein 3 regulates the growth and invasion of human lung adenocarcinoma. Thorac Cancer 2017, 8, 337-343. [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Watari, H. Tumor-Intrinsic PD-L1 Signaling in Cancer Initiation, Development and Treatment: Beyond Immune Evasion. Front Oncol 2018, 8, 386. [CrossRef]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, A.J. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci 2019, 20. [CrossRef]
- Zhu, Q.; Zhang, Y.; Li, M.; Zhang, Y.; Zhang, H.; Chen, J.; Liu, Z.; Yuan, P.; Yang, Z.; Wang, X. MiR-124-3p impedes the metastasis of non-small cell lung cancer via extracellular exosome transport and intracellular PI3K/AKT signaling. Biomark Res 2023, 11, 1. [CrossRef]
- Loedige, I.; Gaidatzis, D.; Sack, R.; Meister, G.; Filipowicz, W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res 2013, 41, 518-532. [CrossRef]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiss, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol Cell 2017, 66, 270-284 e213. [CrossRef]
- Jin, J.; Lu, Z.; Wang, X.; Liu, Y.; Han, T.; Wang, Y.; Wang, T.; Gan, M.; Xie, C.; Wang, J.; Yu, B. E3 ubiquitin ligase TRIM7 negatively regulates NF-kappa B signaling pathway by degrading p65 in lung cancer. Cell Signal 2020, 69, 109543. [CrossRef]
- Schuck, S. Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 2020, 133. [CrossRef]
- Kravic, B.; Behrends, C.; Meyer, H. Regulation of lysosome integrity and lysophagy by the ubiquitin-conjugating enzyme UBE2QL1. Autophagy 2020, 16, 179-180. [CrossRef]
- Garcia-Heredia, J.M.; Verdugo Sivianes, E.M.; Lucena-Cacace, A.; Molina-Pinelo, S.; Carnero, A. Numb-like (NumbL) downregulation increases tumorigenicity, cancer stem cell-like properties and resistance to chemotherapy. Oncotarget 2016, 7, 63611-63628. [CrossRef]
- Haider, T.; Pandey, V.; Banjare, N.; Gupta, P.N.; Soni, V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol Rep 2020, 72, 1125-1151. [CrossRef]
- Shen, X.; He, Z.; Li, H.; Yao, C.; Zhang, Y.; He, L.; Li, S.; Huang, J.; Guo, Z. Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS One 2012, 7, e44822. [CrossRef]
| PD-L1 Expression | Methylation Status | Gene | Methylation | P value | Chr |
| PD-L1 high | Highest methylation in gene | SNORD114-14 (, C/D Box 114-14). Small Nucleolar RNAs (SnoRNAs) | 0,9 | 0,04 | 14 |
| Highest methylation in promotor | DCAF4L2 (DDB1 Associated Factor 4 Like 2 ) | 0,82 | 0,02 | 8 | |
| CELF2-AS1 (CELF2 Antisense RNA 1) | 0,8 | 0.02 | 10 | ||
| LINCMD1 (Long Intergenic Non-Protein Coding RNA, Muscle Differentiation1, MIR133BHG) | 0,8 | 0,01 | 6 | ||
| Long Intergenic Non-Protein Coding RNA 528 (LINC00528) | 0,73 | 0,001 | 22 | ||
| Lowest methylation in gene | GLIPR1L2 (GLIPR1 Like 2) | 0,21 | 0,007 | 12 | |
| Lowest methylation in promotor | CAPS2 (Calcyphosine 2). | 0,14 | 0,005 | 12 | |
| GLIPR1L2 (GLI Pathogenesis Related 1 Like 2) | 0,19 | 0,008 | 12 | ||
| IFITM3 (Interferon Induced Transmembrane Protein 3 ) | 0,21 | 0,03 | 11 | ||
| PD-L1 low | Highest methylation in gene | SNORD114-14, (C/D Box 114-14) Small Nucleolar RNAs (SnoRNAs) |
0,9 | 0,04 | 14 |
| Highest methylation in promotor | SNORD114-14, (C/D Box 114-14) Small Nucleolar RNAs (SnoRNAs) |
0,75 | 0,04 | 14 | |
| Lowest methylation in gene | MIR124-3 (MicroRNA124-3) | 0,21 | 0,01 | 20 | |
| Lowest methylation in promotor | TRIM71 (Tripartite Motif Containing 71, LIN41) | 0,15 | 0,02 | 3 | |
| CAPS2 ( Calcyphosine 2). | 0,26 | 0;005 | 12 | ||
| UBE2QL1 (Ubiquitin Conjugating Enzyme E2 Q Family Like 1) | 0,3 | 0,007 | 5 | ||
| GLIPR1L2 (GLIPR1 Like 2) | 0,3 | 0,008 | 12 | ||
| PDL1 high/low | Difference* in gene methylation | S100A7L2 gene, also known as S100 Calcium Binding Protein A7 Like 2 | 0,74/0,57 Delta 17% |
0,002 | 1 |
| Difference* in promotor methylation | SOD1P3 (Superoxide Dismutase 1 Pseudogene 3), | 0,68/0,54 Delta 14% |
0,02 | 3 | |
| PDL1 low/high | Difference* in gene methylation | Long Intergenic Non-Protein Coding RNA 528 (LINC00528) | 0,54/0,73 Delta 19 % |
0,001 | 22 |
| Difference* in promotor methylation | NUMB gene, (NUMB Endocytic Adaptor Protein) | 0,31/0,51 Delta 20 % |
0,0005 | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





