Submitted:
25 March 2024
Posted:
26 March 2024
You are already at the latest version
Abstract
Keywords:
1. Circulating Cell-Free DNA
2. CfDNA Applications in Clinical Care
2.1. Oncologic Applications
2.2. Prenatal Screening
2.3. Transplantation
3. Technical Issues for High-Quality cfDNA Analysis
3.1. The Relevance of Correct Sampling
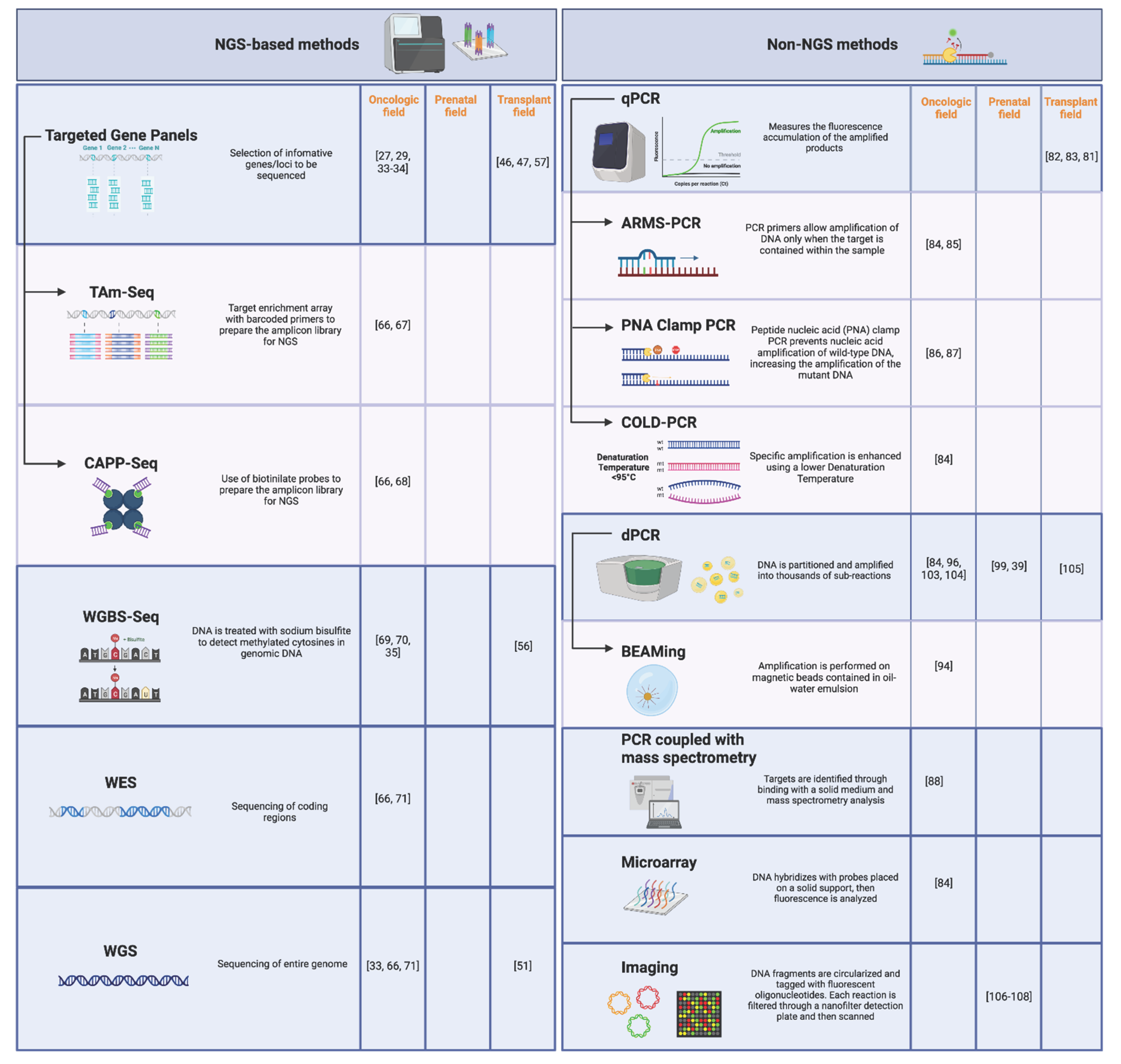
3.2. Technical Comparison of cfDNA Analysis Methods
3.2.1. NGS-Based Methods
3.2.2. Non-NGS Methods
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol OncolJ Hematol Oncol. 2022;15(1):131. [CrossRef]
- Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372(6538):eaaw3616. [CrossRef]
- Tug, S.; Helmig, S.; Deichmann, E.R. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc Immunol Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature: The diverse origins of circulating cell-free DNA. Biol Rev. 2018;93(3):1649-1683. [CrossRef]
- Grabuschnig S, Bronkhorst AJ, Holdenrieder S, et al. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int J Mol Sci. 2020;21(21):8062. [CrossRef]
- McCoubrey-Hoyer A, Okarma TB, Holman HR. Partial purification and characterization of plasma DNA and its relation to disease activity in systemic lupus erythematosus. Am J Med. 1984;77(1):23-34. [CrossRef]
- Lo YMD, Chan KCA, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. [CrossRef]
- Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20(8):1057-1067. [CrossRef]
- Rumore PM, Steinman CR. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J Clin Invest. 1990;86(1):69-74. [CrossRef]
- Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17(1):89-97. [CrossRef]
- Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347-376. [CrossRef]
- Edwards RL, Menteer J, Lestz RM, Baxter-Lowe LA. Cell-free DNA as a solid-organ transplant biomarker: technologies and approaches. Biomark Med. 2022;16(5):401-415. [CrossRef]
- Moreira VG, Prieto B, Rodríguez JSM, Alvarez FV. Usefulness of cell-free plasma DNA, procalcitonin and C-reactive protein as markers of infection in febrile patients. Ann Clin Biochem. 2010;47(Pt 3):253-258. [CrossRef]
- Burnham P, Dadhania D, Heyang M, et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun. 2018;9(1):2412. [CrossRef]
- Siljan WW, Holter JC, Nymo SH, et al. Circulating cell-free DNA is elevated in community-acquired bacterial pneumonia and predicts short-term outcome. J Infect. 2016;73(4):383-386. [CrossRef]
- Cisneros-Villanueva M, Hidalgo-Pérez L, Rios-Romero M, et al. Cell-free DNA analysis in current cancer clinical trials: a review. Br J Cancer. 2022;126(3):391-400. [CrossRef]
- O’Sullivan HM, Feber A, Popat S. Minimal Residual Disease Monitoring in Radically Treated Non-Small Cell Lung Cancer: Challenges and Future Directions. OncoTargets Ther. 2023;16:249-259. [CrossRef]
- Spigel DR, Ervin TJ, Ramlau R, et al. Final efficacy results from OAM4558g, a randomized phase II study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC. J Clin Oncol. 2011;29(15_suppl):7505-7505. [CrossRef]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735-742. [CrossRef]
- Gatzemeier U, Pluzanska A, Szczesna A, et al. Results of a phase III trial of erlotinib (OSI-774) combined with cisplatin and gemcitabine (GC) chemotherapy in advanced non-small cell lung cancer (NSCLC). J Clin Oncol. 2004;22(14_suppl):7010-7010. [CrossRef]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J Clin Oncol. 2016;34(28):3375-3382. [CrossRef]
- Paz-Ares L, Luft A, Vicente D, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med. 2018;379(21):2040-2051. [CrossRef]
- Fabrizio D, Lieber D, Malboeuf C, et al. Abstract 5706: A blood-based next-generation sequencing assay to determine tumor mutational burden (bTMB) is associated with benefit to an anti-PD-L1 inhibitor, atezolizumab. Cancer Res. 2018;78:5706-5706. [CrossRef]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33(5):843-852.e4. [CrossRef]
- Kato S, Li B, Adashek JJ, et al. Serial changes in liquid biopsy-derived variant allele frequency predict immune checkpoint inhibitor responsiveness in the pan-cancer setting. Oncoimmunology. 2022;11(1):2052410. [CrossRef]
- Zhang Q, Luo J, Wu S, et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020;10(12):1842-1853. [CrossRef]
- Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell. 2017;31(2):172-179. [CrossRef]
- Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol Off J Eur Soc Med Oncol. 2015;26(8):1715-1722. [CrossRef]
- Magbanua MJM, Swigart LB, Wu HT, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol Off J Eur Soc Med Oncol. 2021;32(2):229-239. [CrossRef]
- Cao H, Liu X, Chen Y, et al. Circulating Tumor DNA Is Capable of Monitoring the Therapeutic Response and Resistance in Advanced Colorectal Cancer Patients Undergoing Combined Target and Chemotherapy. Front Oncol. 2020;10:466. [CrossRef]
- Ma F, Zhu W, Guan Y, et al. ctDNA dynamics: a novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget. 2016;7(40):66020-66031. [CrossRef]
- Dawson SJ, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199-1209. [CrossRef]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441-1448. [CrossRef]
- Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113(13):E1826-1834. [CrossRef]
- Jamshidi A, Liu MC, Klein EA, et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. 2022;40(12):1537-1549.e12. [CrossRef]
- Rather RA, Saha SC. Reappraisal of evolving methods in non-invasive prenatal screening: Discovery, biology and clinical utility. Heliyon. 2023;9(3):e13923. [CrossRef]
- Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update. 2009;15(1):139-151. [CrossRef]
- Nectoux J. Current, Emerging, and Future Applications of Digital PCR in Non-Invasive Prenatal Diagnosis. Mol Diagn Ther. 2018;22(2):139-148. [CrossRef]
- Hui L, Bianchi DW. Fetal fraction and noninvasive prenatal testing: What clinicians need to know. Prenat Diagn. 2020;40(2):155-163. [CrossRef]
- Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2017;50(3):302-314. [CrossRef]
- De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6(241):241ra77. [CrossRef]
- De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;112(43):13336-13341. [CrossRef]
- Gielis EM, Ledeganck KJ, Dendooven A, et al. The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant Off Publ Eur Dial Transpl Assoc - Eur Ren Assoc. 2020;35(4):714-721. [CrossRef]
- Oellerich M, Budde K, Osmanodja B, et al. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front Genet. 2022;13:1031894. [CrossRef]
- Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a Clinical-Grade Assay to Measure Donor-Derived Cell-Free DNA in Solid Organ Transplant Recipients. J Mol Diagn JMD. 2016;18(6):890-902. [CrossRef]
- Sigdel TK, Archila FA, Constantin T, et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J Clin Med. 2018;8(1):19. [CrossRef]
- Sorbini M, Togliatto GM, Simonato E, et al. HLA-DRB1 mismatch-based identification of donor-derived cell free DNA (dd-cfDNA) as a marker of rejection in heart transplant recipients: A single-institution pilot study. J Heart Lung Transplant Off Publ Int Soc Heart Transplant. 2021;40(8):794-804. [CrossRef]
- Sorbini M, Togliatto G, Mioli F, et al. Validation of a Simple, Rapid, and Cost-Effective Method for Acute Rejection Monitoring in Lung Transplant Recipients. Transpl Int Off J Eur Soc Organ Transplant. 2022;35:10546. [CrossRef]
- Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108(15):6229-6234. [CrossRef]
- Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput Biol. 2017;13(8):e1005629. [CrossRef]
- Chen YB, Cutler CS. Biomarkers for acute GVHD: can we predict the unpredictable? Bone Marrow Transplant. 2013;48(6):755-760. [CrossRef]
- Waterhouse M, Pennisi S, Pfeifer D, et al. Colon and liver tissue damage detection using methylated SESN3 and PTK2B genes in circulating cell-free DNA in patients with acute graft-versus-host disease. Bone Marrow Transplant. 2021;56(2):327-333. [CrossRef]
- Moss J, Magenheim J, Neiman D, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. [CrossRef]
- Sun K, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci. 2015;112(40):E5503-E5512. [CrossRef]
- Cheng AP, Cheng MP, Loy CJ, et al. Cell-free DNA profiling informs all major complications of hematopoietic cell transplantation. Proc Natl Acad Sci. 2022;119(4):e2113476118. [CrossRef]
- Levitsky J, Kandpal M, Guo K, Kleiboeker S, Sinha R, Abecassis M. Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients. Am J Transplant Off J Am Soc Transplant Am Soc Transpl Surg. 2022;22(2):532-540. [CrossRef]
- Verhoeven JGHP, Boer K, Peeters AMA, et al. A Novel High-throughput Droplet Digital PCR-based Indel Quantification Method for the Detection of Circulating Donor-derived Cell-free DNA After Kidney Transplantation. Transplantation. 2022;106(9):1777-1786. [CrossRef]
- Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013;46(15):1561-1565. [CrossRef]
- Knüttgen F, Beck J, Dittrich M, et al. Graft-derived Cell-free DNA as a Noninvasive Biomarker of Cardiac Allograft Rejection: A Cohort Study on Clinical Validity and Confounding Factors. Transplantation. 2022;106(3):615-622. [CrossRef]
- Clausen FB, Jørgensen KMCL, Wardil LW, Nielsen LK, Krog GR. Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects. PloS One. 2023;18(2):e0282332. [CrossRef]
- Laver T, Harrison J, O’Neill PA, et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif. 2015;3:1-8. [CrossRef]
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, DePristo MA. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13(1):375. [CrossRef]
- Halloran PF, Reeve J, Madill-Thomsen KS, et al. Antibody-mediated Rejection Without Detectable Donor-specific Antibody Releases Donor-derived Cell-free DNA: Results From the Trifecta Study. Transplantation. 2023;107(3):709-719. [CrossRef]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [CrossRef]
- Li H, Jing C, Wu J, et al. Circulating tumor DNA detection: A potential tool for colorectal cancer management. Oncol Lett. 2019;17(2):1409-1416. [CrossRef]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. [CrossRef]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548-554. [CrossRef]
- Wardenaar R, Liu H, Colot V, Colomé-Tatché M, Johannes F. Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in Arabidopsis. Methods Mol Biol Clifton NJ. 2013;1067:203-224. [CrossRef]
- Hon GC, Hawkins RD, Caballero OL, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246-258. [CrossRef]
- Imperial R, Nazer M, Ahmed Z, et al. Matched Whole-Genome Sequencing (WGS) and Whole-Exome Sequencing (WES) of Tumor Tissue with Circulating Tumor DNA (ctDNA) Analysis: Complementary Modalities in Clinical Practice. Cancers. 2019;11(9):1399. [CrossRef]
- Lin C, Liu X, Zheng B, Ke R, Tzeng CM. Liquid Biopsy, ctDNA Diagnosis through NGS. Life Basel Switz. 2021;11(9):890. [CrossRef]
- Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58(4):586-597. [CrossRef]
- Levy SE, Boone BE. Next-Generation Sequencing Strategies. Cold Spring Harb Perspect Med. 2019;9(7):a025791. [CrossRef]
- Schwarz UI, Gulilat M, Kim RB. The Role of Next-Generation Sequencing in Pharmacogenetics and Pharmacogenomics. Cold Spring Harb Perspect Med. 2019;9(2):a033027. [CrossRef]
- Dengu F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant Rev. 2020;34(3):100542. [CrossRef]
- Zhao Y, Xia Q, Yin Y, Wang Z. Comparison of Droplet Digital PCR and Quantitative PCR Assays for Quantitative Detection of Xanthomonas citri Subsp. citri. PloS One. 2016;11(7):e0159004. [CrossRef]
- Feingold B, Rose-Felker K, West SC, et al. Early findings after integration of donor-derived cell-free DNA into clinical care following pediatric heart transplantation. Pediatr Transplant. 2022;26(1):e14124. [CrossRef]
- Amadio JM, Rodenas-Alesina E, Superina S, et al. Sparing the Prod: Providing an Alternative to Endomyocardial Biopsies With Noninvasive Surveillance After Heart Transplantation During COVID-19. CJC Open. 2022;4(5):479-487. [CrossRef]
- Kamath M, Shekhtman G, Grogan T, et al. Variability in Donor-Derived Cell-Free DNA Scores to Predict Mortality in Heart Transplant Recipients – A Proof-of-Concept Study. Front Immunol. 2022;13. Accessed May 24, 2023. https://www.frontiersin.org/articles/10.3389/fimmu.2022.825108.
- Dauber EM, Kollmann D, Kozakowski N, et al. Quantitative PCR of INDELs to measure donor-derived cell-free DNA-a potential method to detect acute rejection in kidney transplantation: a pilot study. Transpl Int Off J Eur Soc Organ Transplant. 2020;33(3):298-309. [CrossRef]
- García-Fernández N, Macher HC, Suárez-Artacho G, et al. Donor-Specific Cell-Free DNA qPCR Quantification as a Noninvasive Accurate Biomarker for Early Rejection Detection in Liver Transplantation. J Clin Med. 2022;12(1):36. [CrossRef]
- Fernández-Galán E, Badenas C, Fondevila C, et al. Monitoring of Donor-Derived Cell-Free DNA by Short Tandem Repeats: Concentration of Total Cell-Free DNA and Fragment Size for Acute Rejection Risk Assessment in Liver Transplantation. Liver Transplant Off Publ Am Assoc Study Liver Dis Int Liver Transplant Soc. 2022;28(2):257-268. [CrossRef]
- Galbiati S, Damin F, Burgio V, et al. Evaluation of three advanced methodologies, COLD-PCR, microarray and ddPCR, for identifying the mutational status by liquid biopsies in metastatic colorectal cancer patients. Clin Chim Acta. 2019;489:136-143. [CrossRef]
- Zhang X, Chang N, Yang G, et al. A comparison of ARMS-Plus and droplet digital PCR for detecting EGFR activating mutations in plasma. Oncotarget. 2017;8(67):112014-112023. [CrossRef]
- Simarro J, Pérez-Simó G, Mancheño N, et al. Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients. Int J Mol Sci. 2022;23(15):8526. [CrossRef]
- Watanabe K, Fukuhara T, Tsukita Y, et al. EGFR Mutation Analysis of Circulating Tumor DNA Using an Improved PNA-LNA PCR Clamp Method. Can Respir J. 2016;2016:e5297329. [CrossRef]
- Lamy PJ, van der Leest P, Lozano N, et al. Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study. Cancers. 2020;12(10):3002. [CrossRef]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. BioTechniques 1992, 13, 444–449. [Google Scholar] [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96(16):9236-9241. [CrossRef]
- Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314(5804):1464-1467. [CrossRef]
- Morrison T, Hurley J, Garcia J, et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006;34(18):e123. [CrossRef]
- Sundberg SO, Wittwer CT, Gao C, Gale BK. Spinning disk platform for microfluidic digital polymerase chain reaction. Anal Chem. 2010;82(4):1546-1550. [CrossRef]
- O’Leary B, Hrebien S, Beaney M, et al. Comparison of BEAMing and Droplet Digital PCR for Circulating Tumor DNA Analysis. Clin Chem. 2019;65(11):1405-1413. [CrossRef]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604-8610. [CrossRef]
- Dueck ME, Lin R, Zayac A, et al. Precision cancer monitoring using a novel, fully integrated, microfluidic array partitioning digital PCR platform. Sci Rep. 2019;9(1):19606. [CrossRef]
- Quan PL, Sauzade M, Brouzes E. dPCR: A Technology Review. Sensors. 2018;18(4):1271. [CrossRef]
- Verhoeven JGHP, Peeters AMA, Hesselink DA, Boer K. Pitfalls in the Detection of Donor-Derived Cell-Free DNA in Transplant Recipients. Clin Chem. 2021;67(7):1030-1032. [CrossRef]
- Sikora A, Zimmermann BG, Rusterholz C, et al. Detection of increased amounts of cell-free fetal DNA with short PCR amplicons. Clin Chem. 2010;56(1):136-138. [CrossRef]
- Verhoeven JGHP, Baan CC, Peeters AMA, Nieboer D, Hesselink DA, Boer K. A comparison of two different analytical methods for donor-derived cell-free DNA quantification. Clin Biochem. 2021;96:82-84. [CrossRef]
- Oellerich M, Christenson RH, Beck J, et al. Donor-Derived Cell-Free DNA Testing in Solid Organ Transplantation: A Value Proposition. J Appl Lab Med. 2020;5(5):993-1004. [CrossRef]
- Picard C, Frassati C, Cherouat N, et al. New methods for the quantification of mixed chimerism in transplantation. Front Immunol. 2023;14:1023116. [CrossRef]
- Ye P, Cai P, Xie J, Wei Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis. PloS One. 2021;16(3):e0248775. [CrossRef]
- Dong L, Wang S, Fu B, Wang J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci Rep. 2018;8(1):9650. [CrossRef]
- Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant Off J Am Soc Transplant Am Soc Transpl Surg. 2019;19(11):3087-3099. [CrossRef]
- Imaging single DNA molecules for high precision NIPT | Scientific Reports. Accessed June 21, 2023. https://www.nature.com/articles/s41598-018-22606-0.
- Pooh RK, Masuda C, Matsushika R, et al. Clinical Validation of Fetal cfDNA Analysis Using Rolling-Circle-Replication and Imaging Technology in Osaka (CRITO Study). Diagn Basel Switz. 2021;11(10):1837. [CrossRef]
- Saidel ML, Ananth U, Rose D, Farrell C. Non-Invasive prenatal testing with rolling circle amplification: Real-world clinical experience in a non-molecular laboratory. J Clin Lab Anal. 2023;37(6):e24870. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




