Submitted:
11 March 2024
Posted:
13 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Host Strains and Their Growth
2.2. Bacteriophage Isolation and Propagation
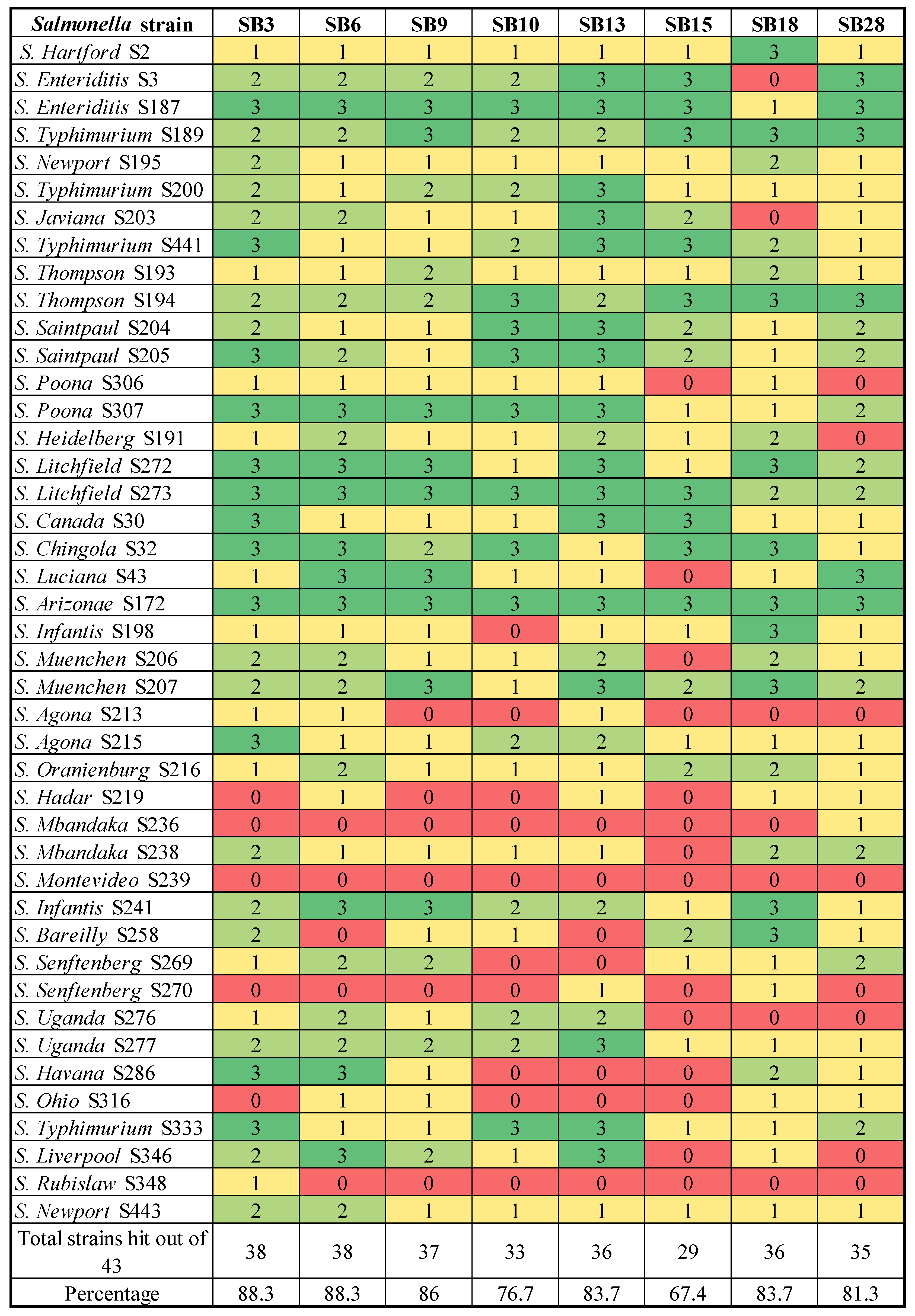
2.3. Host Range Profiles
2.4. Phage DNA Isolation, Sequencing, and Annotation
2.5. Phylogenetic and Comparative Genomic Analyses
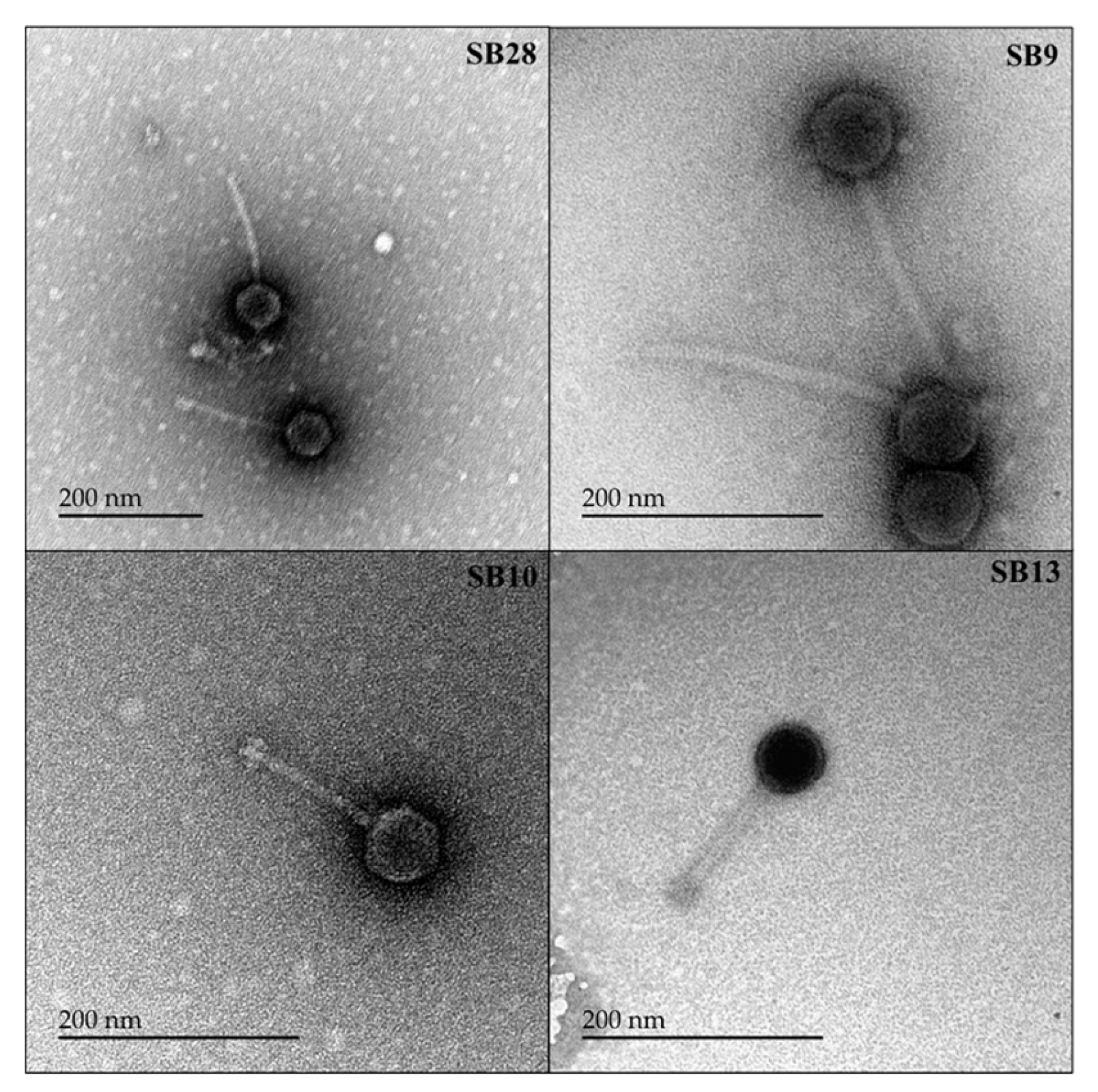
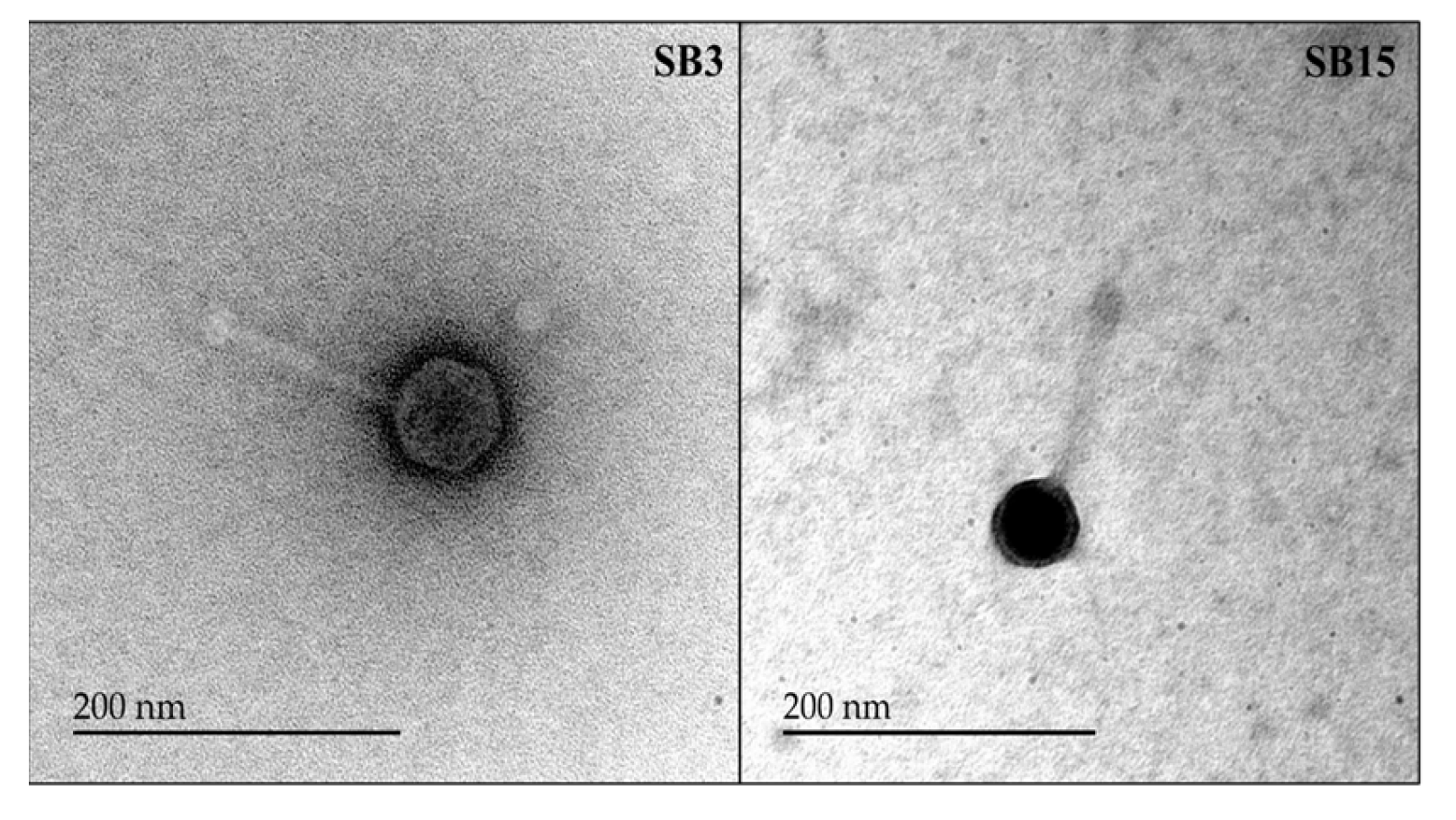
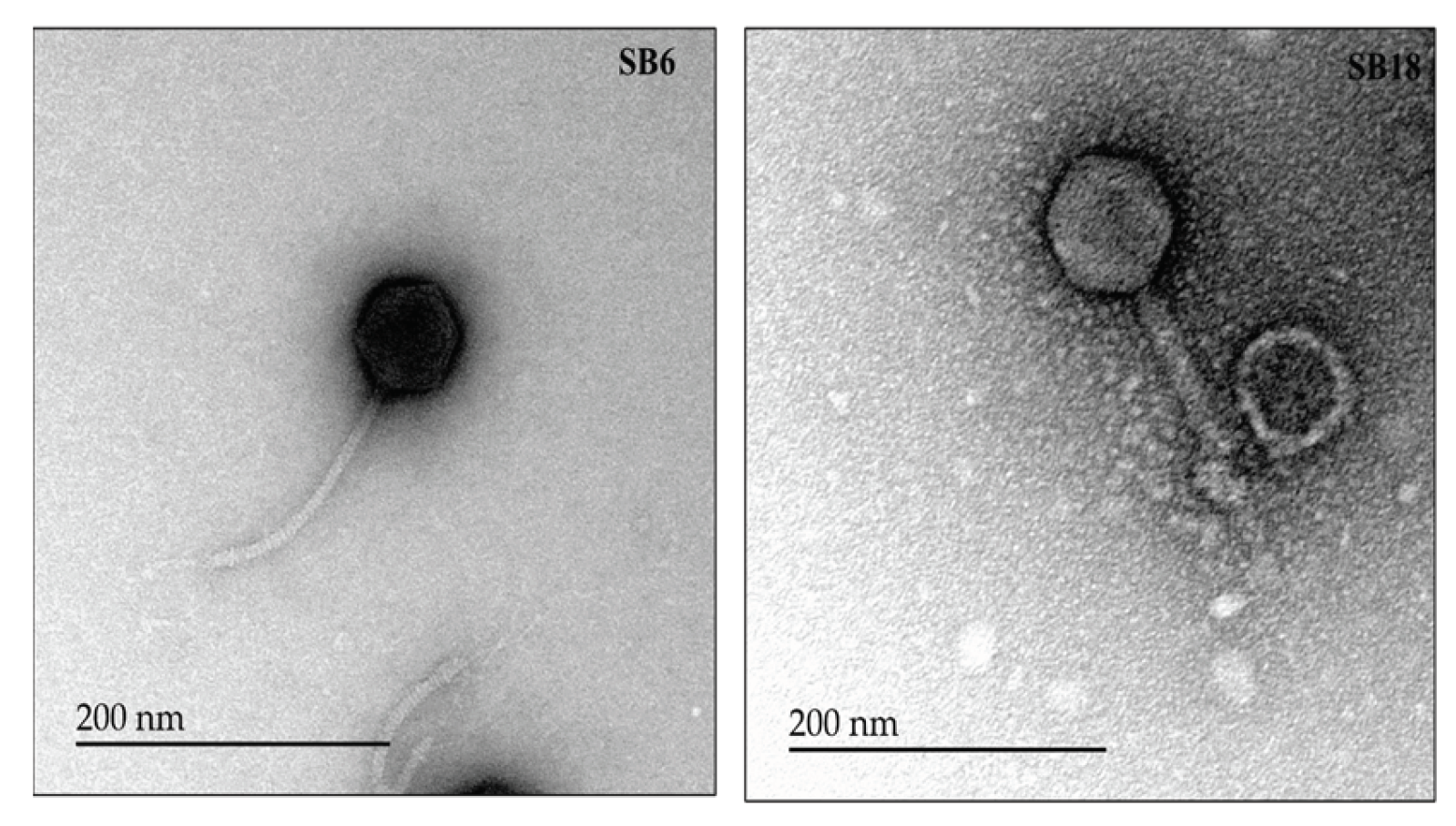
2.6. Electron Microscopic Imaging of the Phages
3. Results
3.1. Phage Isolation and Biological Characterization by Host Range Profile
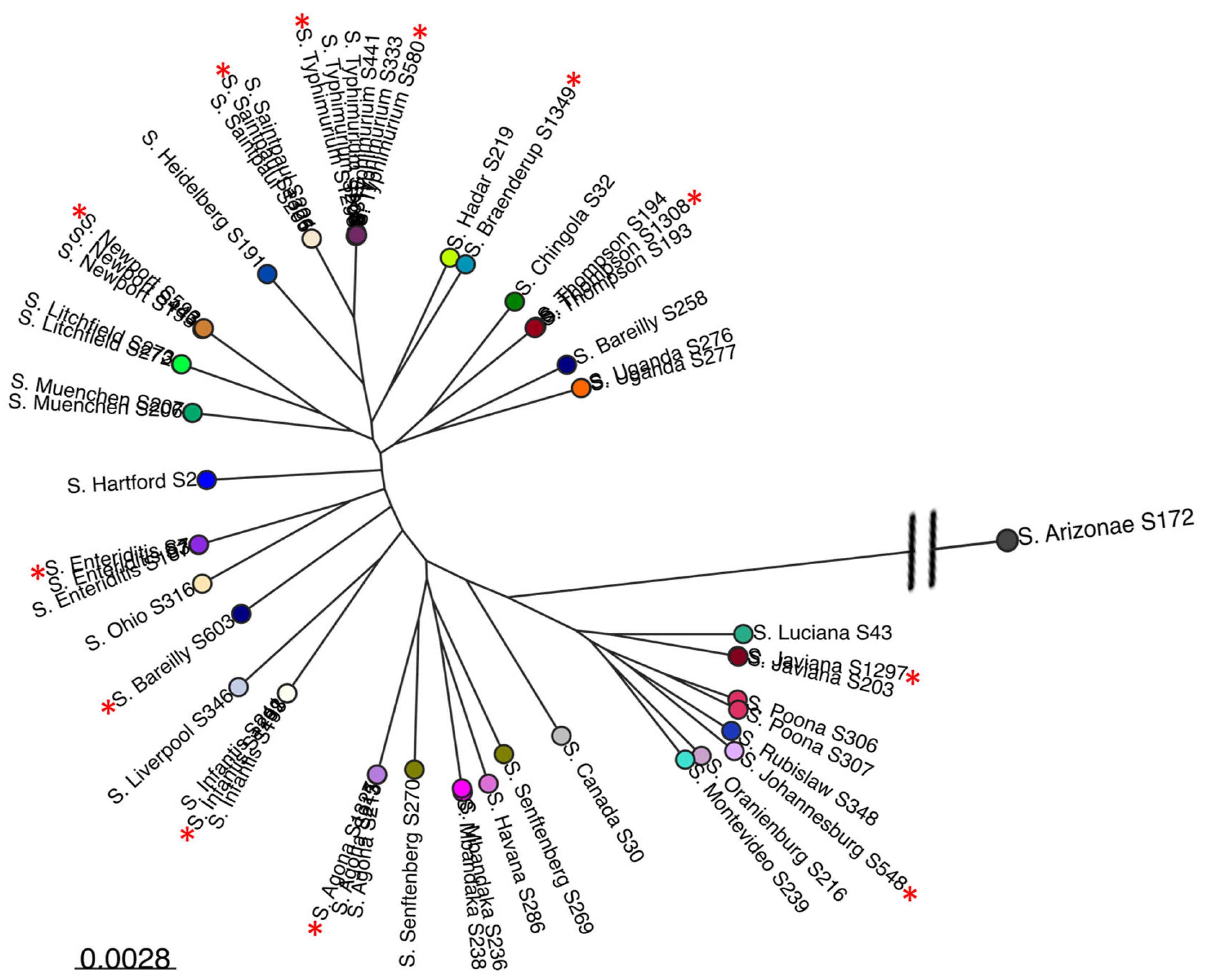
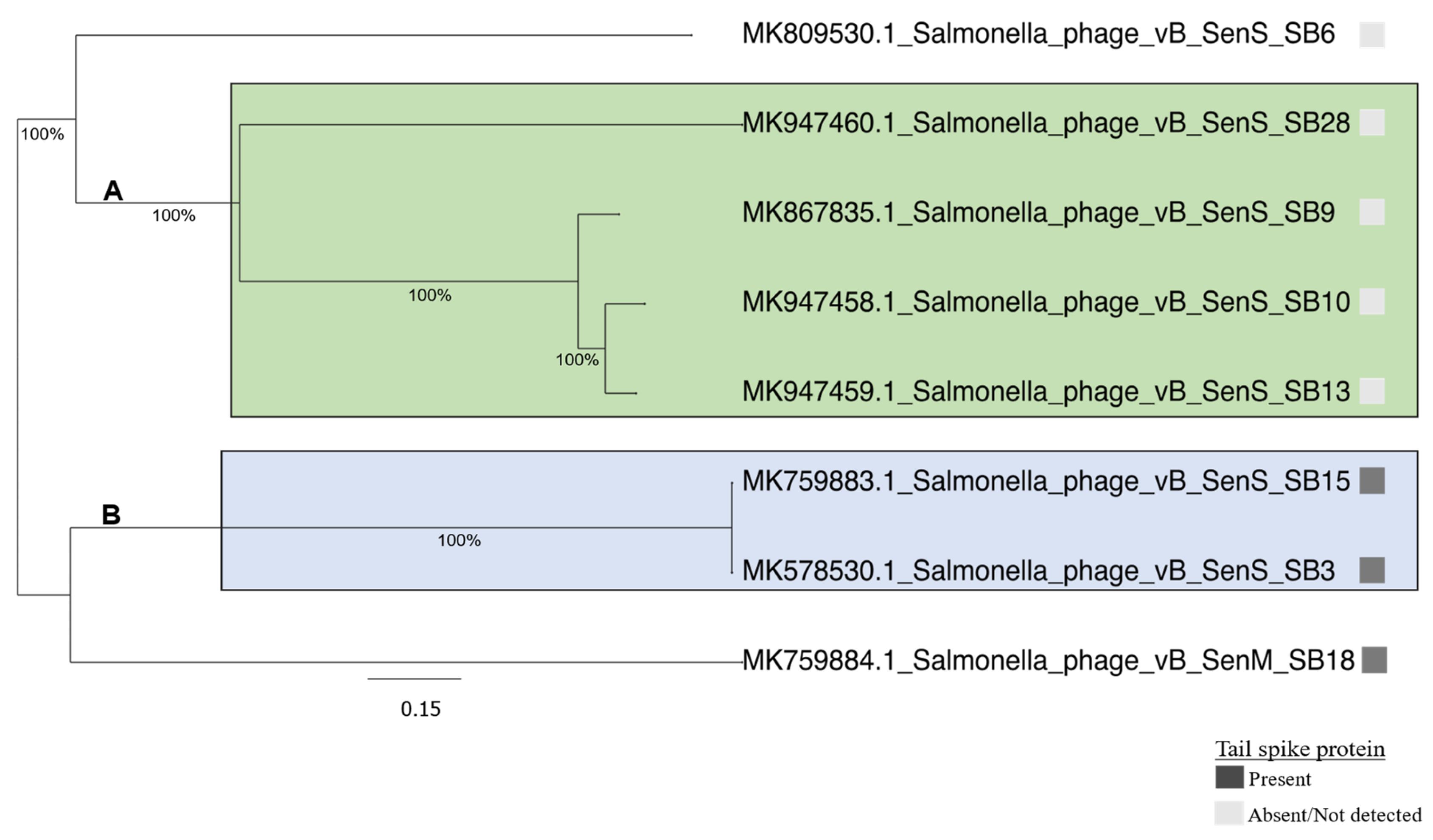
3.2. Comparative Phylogenomic Analysis of Phages Understudy
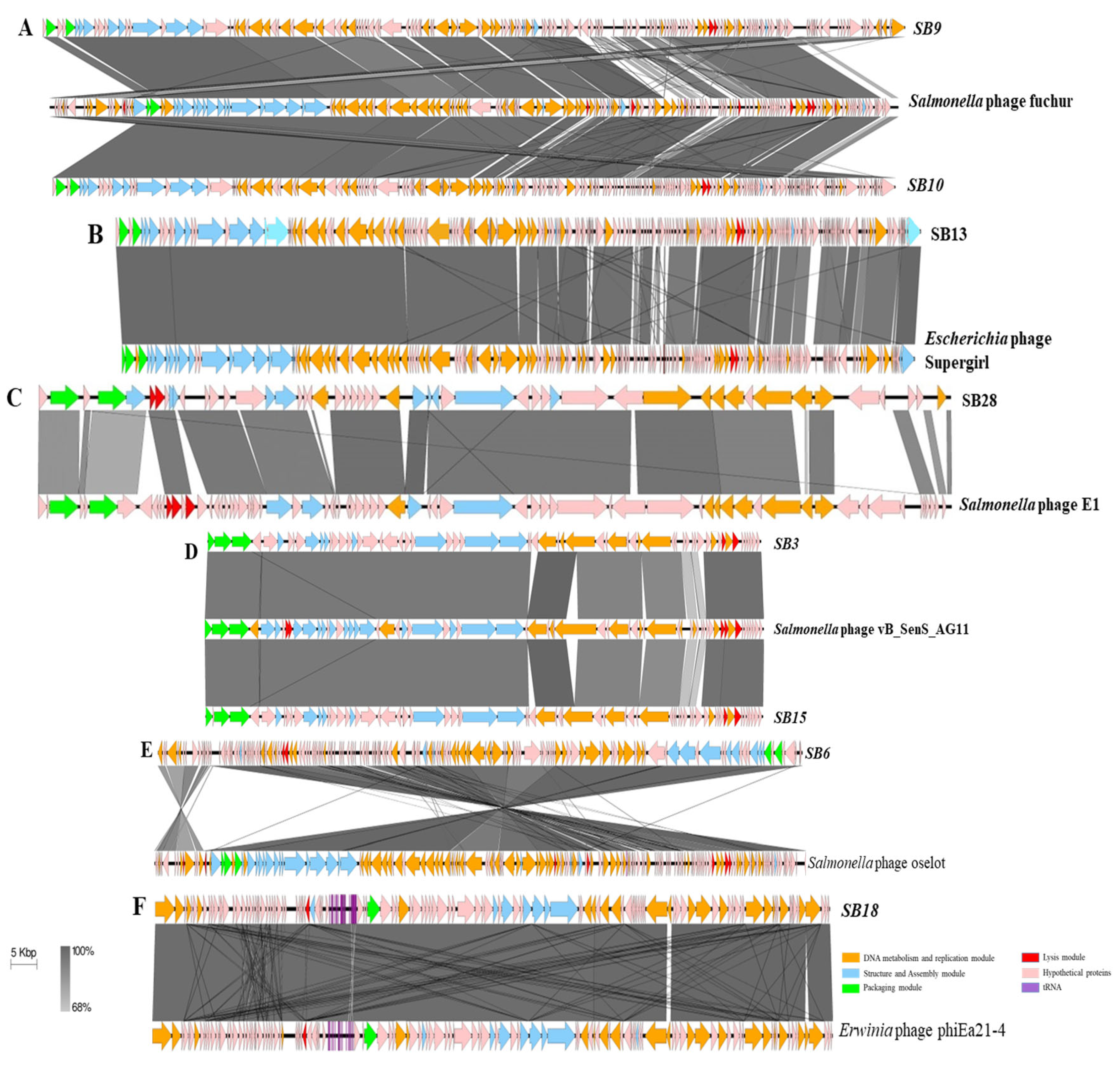
3.2.1. Comparative Gene Content Analysis of Phages under Study
3.2.2. Assessing the Genetic Relatedness of Phages under Study with Other Salmonella Phage Genomes
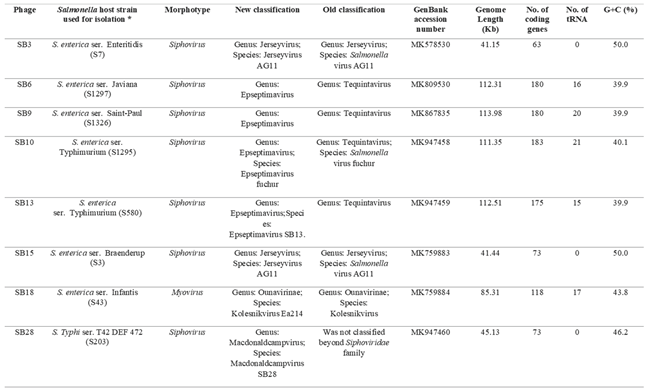
3.3. Salmonella Phages under Study Are Morphologically and Taxonomically Different
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thomas, M.K.; Murray, R.; Flockhart, L.; Pintar, K.; Fazil, A.; Nesbitt, A.; Marshall, B.; Tataryn, J.; Pollari, F. Estimates of foodborne illness-related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne pathogens and disease 2015, 12, 820-827. [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O'Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease 'Burden of Illness, S. The global burden of nontyphoidal Salmonella gastroenteritis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 2010, 50, 882-889. [CrossRef]
- Grimont, P.A.D., Weill, F-X. Antigenic formulae of the Salmonella serovars. 9th edition. http://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%20Serovars%202007%209th%20edition.pdf (Accessed on 13-2-2023) 2007.
- PHAC. Public Health Notice by Public Health Agency of Canada: Outbreak of Salmonella infections linked to peaches imported from the United States http://tinyurl.com/44wz9p9v. 2020.
- CFIA. Food safety investigation by Canadian Food Inspection Agency: Outbreak of Salmonella infections linked to red onions imported from the United States http://tinyurl.com/bddaefyn. 2020.
- PHAC. Public Health Notice by Public Health Agency of Canada: Outbreak of Salmonella infections linked to frozen whole kernel corn http://tinyurl.com/ywywz5m7. 2022.
- PHAC. Public Health Notice by Public Health Agency of Canada: Outbreak of Salmonella infections linked to Malichita and Rudy brand cantaloupes http://tinyurl.com/dm94wt82. 2024.
- Beamud, B.; Garcia-Gonzalez, N.; Gomez-Ortega, M.; Gonzalez-Candelas, F.; Domingo-Calap, P.; Sanjuan, R. Genetic determinants of host tropism in Klebsiella phages. Cell reports 2023, 42, 112048. [CrossRef]
- Bhandare, S.; Colom, J.; Baig, A.; Ritchie, J.M.; Bukhari, H.; Shah, M.A.; Sarkar, B.L.; Su, J.; Wren, B.; Barrow, P.; et al. Reviving Phage Therapy for the Treatment of Cholera. The Journal of Infectious Diseases 2018, jiy563-jiy563. [CrossRef]
- Fong, K.; LaBossiere, B.; Switt, A.I.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Danyluk, M.D.; Wang, S. Characterization of Four Novel Bacteriophages Isolated from British Columbia for Control of Non-typhoidal Salmonella in Vitro and on Sprouting Alfalfa Seeds. Frontiers in microbiology 2017, 8. [CrossRef]
- Vaz, C.S.L.; Voss-Rech, D.; Alves, L.; Coldebella, A.; Brentano, L.; Trevisol, I.M. Effect of time of therapy with wild-type lytic bacteriophages on the reduction of Salmonella Enteritidis in broiler chickens. Veterinary microbiology 2020, 240, 108527. [CrossRef]
- Wong, C.; Delaquis, P.; Goodridge, L.D.; Levesque, R.; Fong, K.; Wang, S. Inactivation of Salmonella enterica on post-harvest cantaloupe and lettuce by a lytic bacteriophage cocktail. Current Research in Food Science 2019. [CrossRef]
- Goodridge, L.; Fong, K.; Wang, S.; Delaquis, P. Bacteriophage-based weapons for the war against foodborne pathogens. Current Opinion in Food Science 2018, 20, 69-75. [CrossRef]
- Endersen, L.; Coffey, A. The use of bacteriophages for food safety. Current Opinion in Food Science 2020, 36, 1-8. [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Frontiers in microbiology 2016, 7. [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Frontiers in microbiology 2014, 5. [CrossRef]
- Switt, A.I.; den Bakker, H.C.; Vongkamjan, K.; Hoelzer, K.; Warnick, L.D.; Cummings, K.J.; Wiedmann, M. Salmonella bacteriophage diversity reflects host diversity on dairy farms. Food microbiology 2013, 36, 275-285. [CrossRef]
- Yamaki, S.; Yamazaki, K.; Kawai, Y. Broad host range bacteriophage, EscoHU1, infecting Escherichia coli O157:H7 and Salmonella enterica: Characterization, comparative genomics, and applications in food safety. International journal of food microbiology 2022, 372, 109680. [CrossRef]
- Fong, K.; Lu, Y.T.; Brenner, T.; Falardeau, J.; Wang, S. Prophage Diversity Across Salmonella and Verotoxin-Producing Escherichia coli in Agricultural Niches of British Columbia, Canada. Frontiers in microbiology 2022, 13, 853703. [CrossRef]
- Bryan, D.W.; Hudson, L.K.; Wang, J.; Denes, T.G. Characterization of a Diverse Collection of Salmonella Phages Isolated from Tennessee Wastewater. Phage (New Rochelle) 2023, 4, 90-98. [CrossRef]
- Rivera, D.; Moreno-Switt, A.I.; Denes, T.G.; Hudson, L.K.; Peters, T.L.; Samir, R.; Aziz, R.K.; Noben, J.P.; Wagemans, J.; Duenas, F. Novel Salmonella Phage, vB_Sen_STGO-35-1, Characterization and Evaluation in Chicken Meat. Microorganisms 2022, 10. [CrossRef]
- Thanki, A.M.; Brown, N.; Millard, A.D.; Clokie, M.R.J. Genomic Characterization of Jumbo Salmonella Phages That Effectively Target United Kingdom Pig-Associated Salmonella Serotypes. Frontiers in microbiology 2019, 10, 1491. [CrossRef]
- Sritha, K.S.; Bhat, S.G. Genomics of Salmonella phage PhiStp1: candidate bacteriophage for biocontrol. Virus genes 2018, 54, 311-318. [CrossRef]
- Chen, L.; Guan, G.; Liu, Q.; Yuan, S.; Yan, T.; Tian, L.; Zhou, Y.; Zhao, Y.; Ma, Y.; Wei, T.; et al. Characterization and complete genomic analysis of two Salmonella phages, SenALZ1 and SenASZ3, new members of the genus Cba120virus. Archives of virology 2019, 164, 1475-1478. [CrossRef]
- Sevilla-Navarro, S.; Catala-Gregori, P.; Marin, C. Salmonella Bacteriophage Diversity According to Most Prevalent Salmonella Serovars in Layer and Broiler Poultry Farms from Eastern Spain. Animals (Basel) 2020, 10. [CrossRef]
- Fong, K.; Tremblay, D.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Moineau, S.; Suttle, C.A.; Wang, S. Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica. Viruses 2019, 11. [CrossRef]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods in molecular biology (Clifton, N.J.) 2009, 501, 15-21. [CrossRef]
- Bhandare, S.G. Biocontrol of V. cholerae using bacteriophage. University of Nottingham, UK., Nottingham, UK, 2015.
- Phagesdb.org. Plaque Purification. Phage hunting protocols 2013.
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. Plos One 2015, 10, e0118557. [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. Methods in molecular biology (Clifton, N.J.) 2009, 501, 141-149. [CrossRef]
- Summer, E.J. Preparation of a phage DNA fragment library for whole genome shotgun sequencing. Methods in molecular biology (Clifton, N.J.) 2009, 502, 27-46. [CrossRef]
- Green, M.; Sambrook, J. Molecular cloning: A Lab Manual, 4th Edition ed.; Cold Spring Harbour Lab Press: 2012; Volume 1.
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587-589. [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Scientific reports 2015, 5, 8365. [CrossRef]
- Wattam, A.R.; Brettin, T.; Davis, J.J.; Gerdes, S.; Kenyon, R.; Machi, D.; Mao, C.; Olson, R.; Overbeek, R.; Pusch, G.D.; et al. Assembly, Annotation, and Comparative Genomics in PATRIC, the All Bacterial Bioinformatics Resource Center. Methods in molecular biology (Clifton, N.J.) 2018, 1704, 79-101. [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. Journal of molecular biology 1990, 215, 403-410. [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic acids research 2012, 40, D290-301. [CrossRef]
- Kropinski, A.M.; Borodovsky, M.; Carver, T.J.; Cerdeno-Tarraga, A.M.; Darling, A.; Lomsadze, A.; Mahadevan, P.; Stothard, P.; Seto, D.; Van Domselaar, G.; et al. In silico identification of genes in bacteriophage DNA. Methods in molecular biology (Clifton, N.J.) 2009, 502, 57-89. [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research 2002, 30, 3059-3066. [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015, 32, 268-274. [CrossRef]
- Argimon, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2016, 2, e000093. [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic acids research 2020, 48, D517-D525. [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic acids research 2016, 44, D694-697. [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic acids research 2022, 50, D912-D917. [CrossRef]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic acids research 2011, 39, D606-611. [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: a genome comparison visualizer. Bioinformatics 2011, 27, 1009-1010. [CrossRef]
- Sambrook, J.; Russell, D., (Eds.) Molecular Cloning: a Laboratory Manual. 3rd Edition ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, 2001.
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998-2008. Emerging infectious diseases 2013, 19, 1239-1244. [CrossRef]
- McCormic, Z.D.; Patel, K.; Higa, J.; Bancroft, J.; Donovan, D.; Edwards, L.; Cheng, J.; Adcock, B.; Bond, C.; Pereira, E.; et al. Bi-national outbreak of Salmonella Newport infections linked to onions: the United States experience. Epidemiol Infect 2022, 150, e199. [CrossRef]
- Centers for Disease, C.; Prevention. Outbreaks of Salmonella infections associated with eating Roma tomatoes--United States and Canada, 2004. MMWR Morb Mortal Wkly Rep 2005, 54, 325-328.
- Centers for Disease, C.; Prevention. Multistate outbreaks of Salmonella serotype Poona infections associated with eating cantaloupe from Mexico--United States and Canada, 2000-2002. MMWR Morb Mortal Wkly Rep 2002, 51, 1044-1047.
- Walsh, K.A.; Bennett, S.D.; Mahovic, M.; Gould, L.H. Outbreaks associated with cantaloupe, watermelon, and honeydew in the United States, 1973-2011. Foodborne pathogens and disease 2014, 11, 945-952. [CrossRef]
- Gencay, Y.E.; Gambino, M.; Prussing, T.F.; Brondsted, L. The genera of bacteriophages and their receptors are the major determinants of host range. Environmental microbiology 2019, 21, 2095-2111. [CrossRef]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection. PLoS biology 2021, 19, e3001424. [CrossRef]
- Pickard, D.; Thomson, N.R.; Baker, S.; Wain, J.; Pardo, M.; Goulding, D.; Hamlin, N.; Choudhary, J.; Threfall, J.; Dougan, G. Molecular characterization of the Salmonella enterica serovar Typhi Vi-typing bacteriophage E1. J Bacteriol 2008, 190, 2580-2587. [CrossRef]
- Anany, H.; Switt, A.I.; De Lappe, N.; Ackermann, H.W.; Reynolds, D.M.; Kropinski, A.M.; Wiedmann, M.; Griffiths, M.W.; Tremblay, D.; Moineau, S.; et al. A proposed new bacteriophage subfamily: "Jerseyvirinae". Archives of virology 2015, 160, 1021-1033. [CrossRef]
- Hany, A. Biocontrol of foodborne bacterial pathogens using immobilized bacteriophages. University of Guelph, 2010.
- Olsen, N.S.; Hendriksen, N.B.; Hansen, L.H.; Kot, W. A New High-throughput Screening (HiTS) Method for Phages – Enabling Crude Isolation and Fast Identification of Diverse Phages with Therapeutic Potential. bioRxiv 2020, 2020.2003.2027.011080. [CrossRef]
- Lehman, S.M.; Kropinski, A.M.; Castle, A.J.; Svircev, A.M. Complete genome of the broad-host-range Erwinia amylovora phage phiEa21-4 and its relationship to Salmonella phage felix O1. Appl Environ Microbiol 2009, 75, 2139-2147. [CrossRef]
- Gill, J.J.; Svircev, A.M.; Smith, R.; Castle, A.J. Bacteriophages of Erwinia amylovora. Appl Environ Microbiol 2003, 69, 2133-2138. [CrossRef]
- Runa, V.; Wenk, J.; Bengtsson, S.; Jones, B.V.; Lanham, A.B. Bacteriophages in Biological Wastewater Treatment Systems: Occurrence, Characterization, and Function. Frontiers in microbiology 2021, 12. [CrossRef]
- Alharbi, N.M.; Ziadi, M.M. Wastewater as a fertility source for novel bacteriophages against multi-drug resistant bacteria. Saudi Journal of Biological Sciences 2021, 28, 4358-4364. [CrossRef]
- Zhu, Y.; Shang, J.; Peng, C.; Sun, Y. Phage family classification under Caudoviricetes: A review of current tools using the latest ICTV classification framework. Frontiers in microbiology 2022, 13, 1032186. [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13. [CrossRef]
- Zerbini, F.M.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Hendrickson, R.C.; et al. Changes to virus taxonomy and the ICTV Statutes ratified by the International Committee on Taxonomy of Viruses (2023). Archives of virology 2023, 168, 175. [CrossRef]
- Colavecchio, A.; D'Souza, Y.; Tompkins, E.; Jeukens, J.; Freschi, L.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Boyle, B.; Bekal, S.; Tamber, S.; et al. Prophage Integrase Typing Is a Useful Indicator of Genomic Diversity in Salmonella enterica. Frontiers in microbiology 2017, 8, 1283. [CrossRef]
- Hayden, H.S.; Matamouros, S.; Hager, K.R.; Brittnacher, M.J.; Rohmer, L.; Radey, M.C.; Weiss, E.J.; Kim, K.B.; Jacobs, M.A.; Sims-Day, E.H.; et al. Genomic Analysis of Salmonella enterica Serovar Typhimurium Characterizes Strain Diversity for Recent U.S. Salmonellosis Cases and Identifies Mutations Linked to Loss of Fitness under Nitrosative and Oxidative Stress. mBio 2016, 7, e00154. [CrossRef]
- Lankau, E.W.; Cruz Bedon, L.; Mackie, R.I. Salmonella strains isolated from Galapagos iguanas show spatial structuring of serovar and genomic diversity. Plos One 2012, 7, e37302. [CrossRef]
- Pightling, A.W.; Pettengill, J.; Luo, Y.; Strain, E.; Rand, H. Genomic diversity of Salmonella enterica isolated from papaya samples collected during multiple outbreaks in 2017. Microbiology 2020. [CrossRef]
- Simpson, K.M.J.; Mor, S.M.; Ward, M.P.; Collins, J.; Flint, J.; Hill-Cawthorne, G.A.; Abd El Ghany, M. Genomic characterisation of Salmonella enterica serovar Wangata isolates obtained from different sources reveals low genomic diversity. Plos One 2020, 15, e0229697. [CrossRef]
- Sodagari, H.R.; Mohammed, A.B.; Wang, P.; O'Dea, M.; Abraham, S.; Robertson, I.; Habib, I. Non-typhoidal Salmonella contamination in egg shells and contents from retail in Western Australia: Serovar diversity, multilocus sequence types, and phenotypic and genomic characterizations of antimicrobial resistance. International journal of food microbiology 2019, 308, 108305. [CrossRef]
- Lamas, A.; Miranda, J.M.; Regal, P.; Vazquez, B.; Franco, C.M.; Cepeda, A. A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiological research 2018, 206, 60-73. [CrossRef]
- Majtanova, L.; Majtan, J.; Majtan, V. Trends in phage types of Salmonella enterica serovars Enteritidis and Typhimurium isolated in Slovakia from 1995 to 2009. Diagnostic microbiology and infectious disease 2011, 69, 454-456. [CrossRef]
- Bennett, S.D.; Sodha, S.V.; Ayers, T.L.; Lynch, M.F.; Gould, L.H.; Tauxe, R.V. Produce-associated foodborne disease outbreaks, USA, 1998–2013. Epidemiology & Infection 2018, 146, 1397-1406. [CrossRef]
- Aiyedun, S.O.; Onarinde, B.A.; Swainson, M.; Dixon, R.A. Foodborne outbreaks of microbial infection from fresh produce in Europe and North America: a systematic review of data from this millennium. International Journal of Food Science & Technology 2020, 56, 2215-2223. [CrossRef]
- Hanning, I.B.; Nutt, J.D.; Ricke, S.C. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne pathogens and disease 2009, 6, 635-648. [CrossRef]
- Ballesté, E.; Blanch, A.R.; Muniesa, M.; García-Aljaro, C.; Rodríguez-Rubio, L.; Martín-Díaz, J.; Pascual-Benito, M.; Jofre, J. Bacteriophages in sewage: abundance, roles, and applications. FEMS Microbes 2022, 3. [CrossRef]
- Olson, I. Retrofitting world's 3rd largest treatment plant with ozonation and improved incinerators will take 10 years. CBC News https://www.cbc.ca/news/canada/montreal/montreal-wastewater-treatment-plant-1.6711208 2023.
- GovtCanada. Census Profile, 2021 Census of Population for Montreal https://tinyurl.com/53fks2ad. 2021.
- Ramisetty, B.C.M.; Sudhakari, P.A. Bacterial ‘Grounded’ Prophages: Hotspots for Genetic Renovation and Innovation. Frontiers in genetics 2019, 10. [CrossRef]
- Colello, R.; Ruiz, M.J.; Padín, V.M.; Rogé, A.D.; Leotta, G.; Padola, N.L.; Etcheverría, A.I. Detection and Characterization of Salmonella Serotypes in the Production Chain of Two Pig Farms in Buenos Aires Province, Argentina. Frontiers in microbiology 2018, 9. [CrossRef]
- Simmonds, P.; Aiewsakun, P. Virus classification - where do you draw the line? Archives of virology 2018, 163, 2037-2046. [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9. [CrossRef]
- Shin, H.; Lee, J.H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. Plos One 2012, 7, e43392. [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Gorski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial Phage Resistance, Phage Training and Prophages in Bacterial Production Strains. Viruses 2018, 10. [CrossRef]
- Borodovich, T.; Shkoporov, A.N.; Ross, R.P.; Hill, C. Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterology Report 2022, 10. [CrossRef]
- Garvey, M. Bacteriophages and Food Production: Biocontrol and Bio-Preservation Options for Food Safety. Antibiotics (Basel) 2022, 11. [CrossRef]
- Villa, T.G.; Feijoo-Siota, L.; Rama, J.R.; Sánchez-Pérez, A.; Viñas, M. Horizontal Gene Transfer Between Bacteriophages and Bacteria: Antibiotic Resistances and Toxin Production. In Horizontal Gene Transfer: Breaking Borders Between Living Kingdoms, Villa, T.G., Viñas, M., Eds.; Springer International Publishing: Cham, 2019; pp. 97-142.
- Matsuzaki, S.; Yasuda, M.; Nishikawa, H.; Kuroda, M.; Ujihara, T.; Shuin, T.; Shen, Y.; Jin, Z.; Fujimoto, S.; Nasimuzzaman, M.D.; et al. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 2003, 187, 613-624. [CrossRef]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Current opinion in microbiology 2008, 11, 393-400. [CrossRef]
- Joseph, A.A.; Logan, G.; Honghzuan, W.; Robert, V. The E34 Phage Tailspike Protein: An in vitro characterization, Structure Prediction, Potential Interaction with <em>S. newington</em> LPS and Cytotoxicity Assessment to Animal Cell Line. bioRxiv 2021, 2021.2009.2020.461090. [CrossRef]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC microbiology 2016, 16, 207. [CrossRef]
- Miletic, S.; Simpson, D.J.; Szymanski, C.M.; Deyholos, M.K.; Menassa, R. A Plant-Produced Bacteriophage Tailspike Protein for the Control of Salmonella. Frontiers in plant science 2016, 6. [CrossRef]
- Waseh, S.; Hanifi-Moghaddam, P.; Coleman, R.; Masotti, M.; Ryan, S.; Foss, M.; MacKenzie, R.; Henry, M.; Szymanski, C.M.; Tanha, J. Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. Plos One 2010, 5, e13904. [CrossRef]
- Ibrahim, I.; Ayariga, J.A.; Xu, J.; Adebanjo, A.; Samuel-Foo, M.; Ajayi, O. CBD resistant Salmonella strains are susceptible to Epsilon 34 phage tailspike protein. 2022. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





