1. Introduction
In cases of mixed traces found on crime scenes, sometimes DNA profiles present genotypes with more than two alleles per locus: these profiles are defined as "mixed", i.e. made up of material from more than one biological source, or rather by multiple individuals [
1,
2]. The polymorphic genetic markers of autosomal DNA (STR) and haploids of Y Chromosome (Y-STR) have been widely used in such cases to arrive at a correct interpretation and identification of the contributors [
3,
4]. Sometimes, however, there could be complex situations that can make the interpretation of the analytical data less easy or even impossible, without thus achieving a complete and correct deconvolution of the mixed profile and consequently the identification of the contributors who participated in the trace formation [
5].
In case of autosomal genetic markers, the evaluation of the electropherogram characteristics allows to establish whether it is a mixed genetic profile, and whether or not it is in a complex situation [
6]. The number of contributors that may have contributed to the genesis of a trace, in fact, is assessed through the maximum number of alleles per locus (Maximum Allele Count, MAC) [
7], and their quantitative relationship through the calculation of the Mixture Ratio (MR) and the Peak High Ratio (PHR) [
1,
8]. The degree of complexity increases with the increase in the number of alleles for each locus, and with the extent to which stochastic effects occur, mainly related to the quality and quantity of the DNA extracted from the trace [
9]. Furthermore, in the event that contributors are related, the genotypic combinations make interpretation difficult [
10].
The study of Y Chromosome’s genetic markers in mixed DNA profile, always used with autosomal data, allows instead to know the minimum number of male contributors, but not their identification [
11].
In fact, the important characteristic of the Y chromosome not to undergo recombination during meiosis, means that it is inherited over the generations in the original haploid form. The Y Chromosome’s haplotype is therefore common to all male members of a family. Thus in a mixture formed by two individuals belonging to the same paternal line, its typing is not useful for discriminatory purposes [
12].
When the mixture is made up of several contributors, male and female, related or not to each other, their discrimination can be carried out by typing the genetic markers of X Chromosome [
10,
13]. The molecular typing of the X-STRs is usually applied to population studies and parental relationship between relatives in complex cases (to complement the analysis of autosomal STRs and the Y chromosome), such as for example in investigations of deficient paternities in which female are involved [
14].
It is also applied in cases of missing persons, incest and finally in the identification of victims of attacks and mass disasters (DVI) [
14]. However their use in criminal cases, and in particular in the study of mixed traces, is still rather rare [
15].
The typing of the X chromosome on a male subject (XY), since he inherits one of the two X chromosomes from the mother (and the same Y chromosome from the father), allows to obtain directly the individual haplotype and, for a single male individual, it is represented by only one allele per typed locus. In the case of mixed profiles formed by two male contributors, the profile is represented by a maximum of two alleles for each typed locus [
16].
A female subject, on the other hand, inherits one of the two X chromosomes from the mother (or rather a combination of them), and the only X chromosome from the father: therefore the profile of the X Chromosome typed for a single female subject is represented by maximum two alleles for locus [
17]. In the case of mixed DNA profiles consisting of two individuals, one of which is male and one female, the genetic profile is represented by a maximum of three alleles for each locus analyzed, two of which belong to the female contribution and one to the male X Chromosome haplotype [
17].
Few studies have been made on forensic cases [
13,
15], in which however the use of X Chromosome markers has been decisive, such as for example mixtures from vaginal swabs of women victims of rape, in which suspects were related between them, and analysis of both autosomal and Y chromosome STRs were not conclusive [
13].
Therefore the use of X-STRs could be a very useful tool to obtain a correct interpretation of DNA mixtures. The markers of the X Chromosome, in fact, are precious tools in cases of mixtures formed by male and female cells, even if they must always be used to support the autosomal and Y Chromosome data.
In order to highlight the decisive importance of the use of X-STRs in the context of mixed genetic profiles, this work reports a judicial case related to a robbery, in which the judge's request was to understand if on the evidence (balaclava) found on the crime scene there were traces of one or more contributors and if these contributors were all male, all female or in mixed order, having the reference profile of an only one suspect available.
The first analysis of autosomal markers and Y Chromosome were not useful to clarify how many and which contributors had participated in the formation of the trace; so the application of the X Chromosome markers was used and it allowed to reach a solution.
Nowadays, however, the study of the X Chromosome profiles obtained from mixed traces or from reference samples cannot be supported by biostatistical evaluations because, since the rare use of X-STRs in criminal cases and the few population frequencies, the scientific community has not yet drawn up guidelines or created software useful to provide a statistical weight to its application in the interpretation of mixed traces [
18].
2. Materials and Methods
Sampling procedures and sample collection
On the balaclava subject of the technical assessment, two double swabs (Copan Italia spa) were made in the internal portion, at the level of the central nose-mouth and apical forehead areas.
Reference saliva sample was performed using two Whatman FTA Card.
DNA extraction and quantification
DNA extraction was carried out using innuPREP Forensic DNA Kit-IPC16 (AnalytikJena).
No modifications to the standard saliva swab protocol have been applied.
DNA quantification was performed using QuantiFluor™-ST (Promega) and QuantiFluor™ dsDNA System kit (Promega).
DNA amplification, electrophoresis and data analysis software
PCR was carried out using PowerPlex Fusion System kit (Promega) for autosomal markers, PowerPlex Y23 System kit (Promega) for Y-chromosomal markers and Investigator Argus-12X kit (Qiagen) for X-chromosomal markers on “VeritiTM Dx Thermal Cycler” (Thermo Fisher Scientific).
Electrophoresis was carried out on a ABI PRISM 310 Genetic Analyzer (Applied Biosystems), followed by GeneMapper ID v3.2 (Applied Biosystems) software.
For the interpretation of autosomal STRs and Y-STRs, the analytic threshold (AT) was set at 40 RFU (Relative Fluorescence Unit) and the stochastic threshold (ST) at 150 RFU, while the stochastic threshold for X-STRs was set at 120 RFU, as internal validation.
3. Results
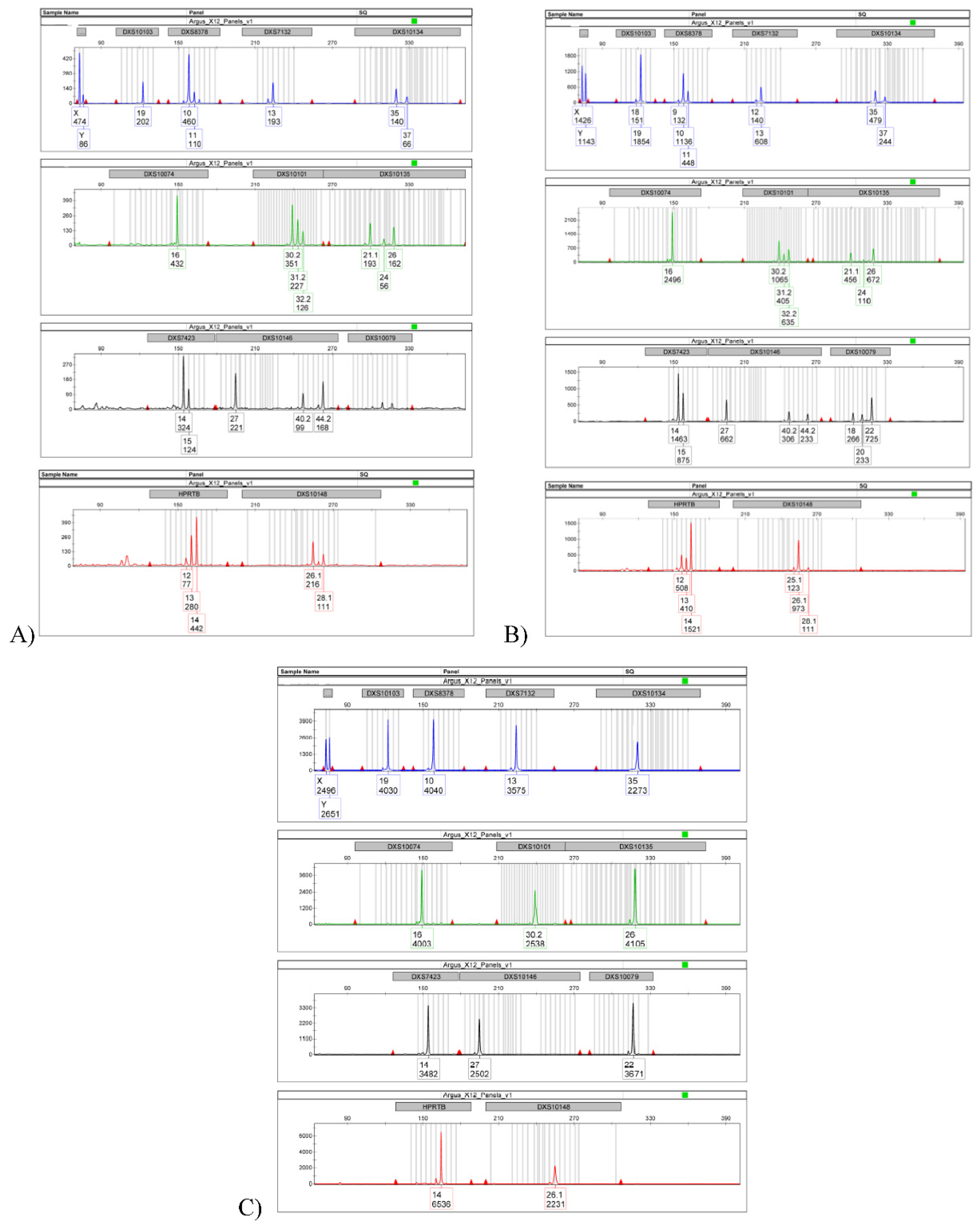
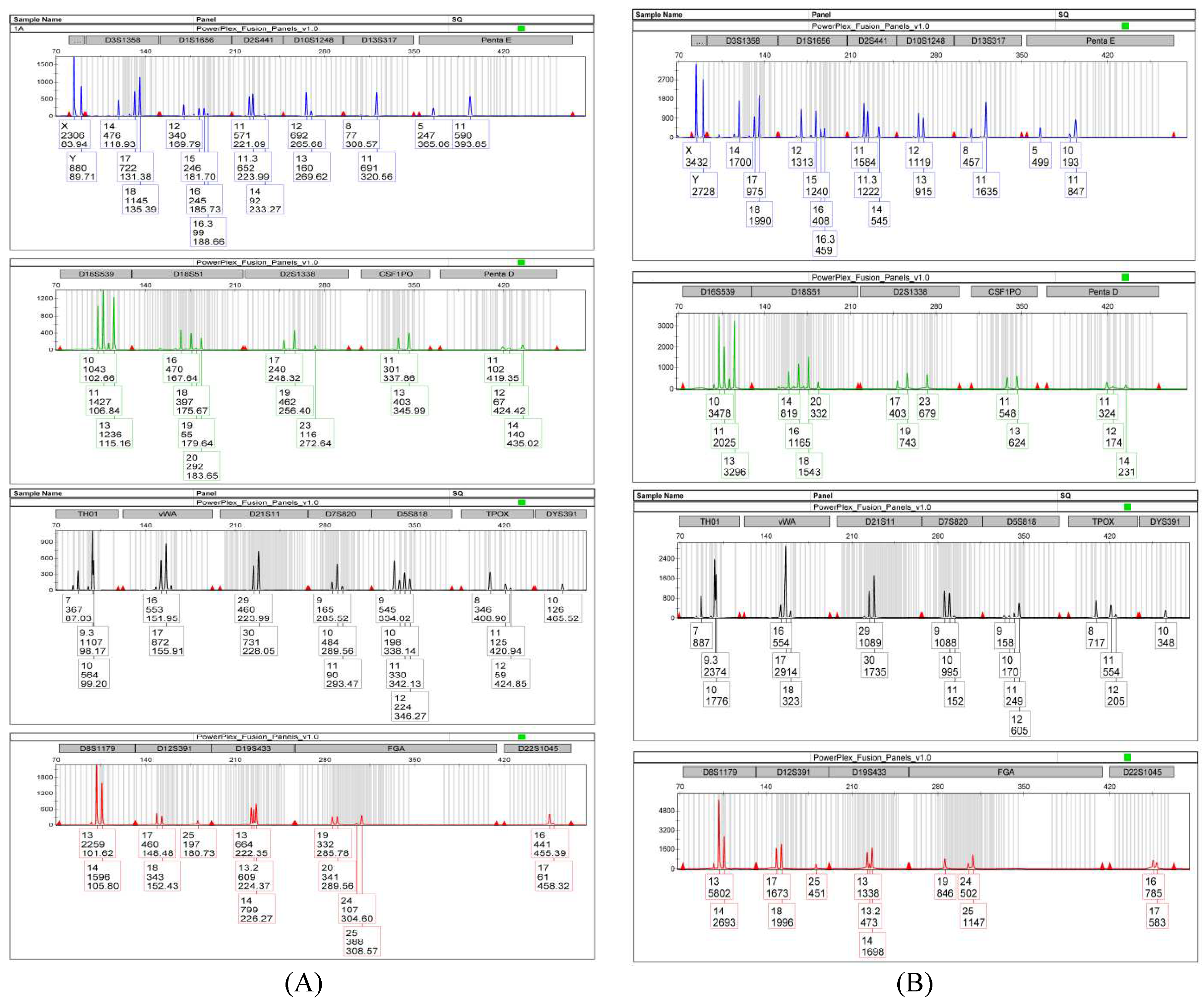
At the beginning, the autosomal and Y Chromosome STRs were analyzed on the two samples taken inside the balaclava in the nose-mouth area and in the forehead area. The results of the DNA quantification were 7,254 ng/ul and 1,237 ng/ul respectively. The profile obtained from sampling in the nose-mouth area of the balaclava highlighted the presence of an indistinguishable mixed DNA profile, of at least two contributors, of which at least one male, thanks to the presence of Y Chromosome in Amelogenin locus. An important aspect for the type of mixture was the presence of a strong allelic imbalance in Amelogenin locus, where the peak corresponding to the X Chromosome was almost three times higher than the peak of the Y Chromosome (X Chromosome: 2306 RFU; Y Chromosome: 880 RFU) (
Figure 1A and 1B).
In order to estimate the contribution of the subjects in an unbalanced mixture, the height of the peaks in loci with four alleles only was taken into account for making the proportion, assuming that the height of the peaks is proportional to the amount of DNA template. The average Mixture Ration (MR) evaluation was equal to 2,64:1. This was not sufficient to genetically discriminate the numbers of contributors within the mixture.
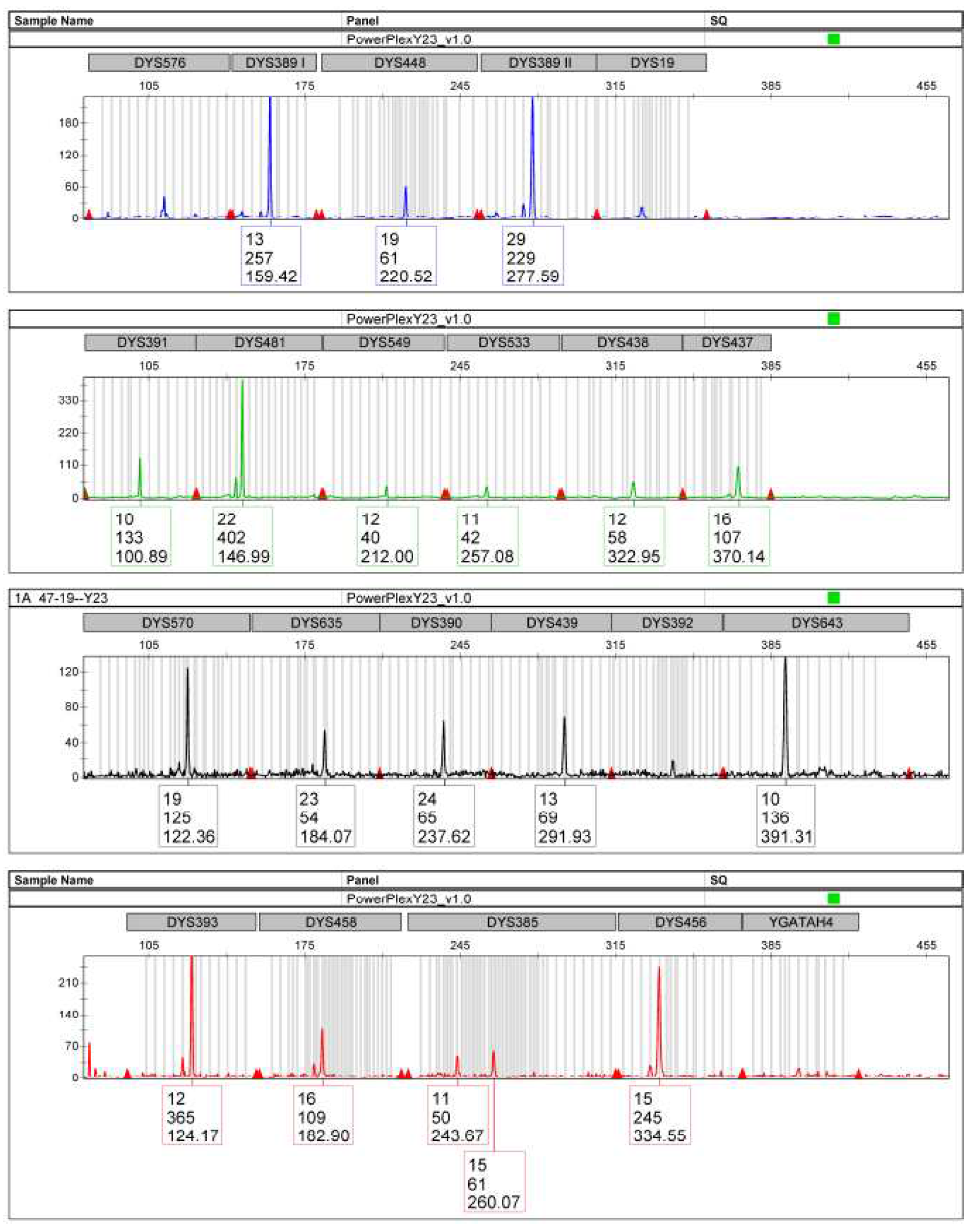
The haplotypic profile of Y Chromosome, on the other hand, provided a single haplotype: this situation, however, does not allow to understand if the male subject present in the trace is only one or if there are more male contributors related to each other through paternal line (
Figure 2).
The profile obtained from sampling in the forehead area of the balaclava, instead, highlighted the presence of the same identical indistinguishable mixed profile of the first sampling. In this second profile, however, the allelic imbalance was not present at Amelogenin locus, probably because the male subject may have contributed most to the formation of the trace.
By comparing the STR and Y-STR profiles of the traces with the reference sample of the only suspect, complete compatibility was obtained, thus being able to confirm its certain contribution to the mixtures.
This compatibility was further supported by the biostatistical evaluation of the Likelihood Ratio which for the autosomal profiles provided LR values respectively equal to 9,5888*10
12 e 1,8814*10
12 (LRMix Studio, v.2.1.5) [
19], while the haplotype profile frequency of the Y Chromosome in the reference database YHRD [
http://www.yhrd.org/, [
20] was equal to 4,7749*10
-5 ed LR was equal to 2,0943*10
4.
Given the presence of genetic mixtures, the Judge's question was also to understand how many other contributors could be present in the traces and what sex they were, in order to direct the investigations also on other fronts. The results obtained from the analysis of autosomal STRs and Y Chromosome, however, were not sufficient to completely clarify this doubt. Therefore, the STR of the X Chromosome have also been investigated, in order to better understand the distribution of the contributors in the mixtures.
From the first trace (nose-mouth area of the balaclava) a partial mixed profile was obtained. This profile again showed the allelic imbalance at the Amelogenin locus. Since a maximum of three alleles have been typed in the DXS10101, DXS10135 and DXS10146 loci, it is possible to hypothesize that the trace was formed either by two individuals, one male and one female, or by three male contributors (
Figure 3A). Comparing the mixture with the suspect’s reference sample, his haplotypic profile of Y Chromosome was found to be totally present in the mixture. Therefore, given the presence of three alleles for the loci DXS10101, DXS10135 and DXS10146, it can therefore be excluded that the second subject that contributed to the formation of the mixture is male, but it could be an unknown female subject. The allelic imbalance at the Amelogenin locus both in the autosomal profile and in the X Chromosome profile, supports this hypothesis.
From the second trace (forehead area of the balaclava), a complete mixed profile was obtained. Also in this case, because in the DXS8378, DXS10101, DXS10135, DXS10146 and DXS10079 loci a maximum of three alleles have been typed, it is possible to formulate the previous hypothesis, for which the trace was formed or by two individuals, one male and one female, or by three male contributors (
Figure 3B). Also in this trace, the haplotypic X Chromosome profile of the suspect was compatible (
Figure 3C). The only difference is the absence of the allelic imbalance at the Amelogenin locus, probably because the male subject may have contributed most to the formation of the trace. In conclusion, due to the haplotypic transmission of the X Chromosome in male and given the presence of three alleles in the DXS8378, DXS10101, DXS10135, DXS10146 and DXS10079 loci, it can therefore be excluded that the second subject that contributed to the formation of the mixture is male, but the same unknown female subject.
Figure 3.
A: X-STRs Electropherogram from first trace (balaclava mouth-nose area); B: X-STRs Electropherogram form second trace (balaclava forehead area); C) X-STRs Electropherograms from reference.
Figure 3.
A: X-STRs Electropherogram from first trace (balaclava mouth-nose area); B: X-STRs Electropherogram form second trace (balaclava forehead area); C) X-STRs Electropherograms from reference.
4. Discussion
This work reported a robbery case. On the crime scene a balaclava was found and it was the target of technical investigation. The request of the judicial authority was to investigate its genetic profile and, in the event of a mixture, determine the minimum number of contributors and their sex in a proportional relationship, having the reference sample of a single suspect available. The study of the autosomal polymorphic markers (STR), in the two sampled traces, allowed to observe mixed indistinguishable profiles, belonging to at least two contributors, one of which is certainly male and, for one of them, with the presence of strong allelic imbalance of the X Chromosome in the Amelogenin locus. The study of polymorphic markers of Y Chromosome (Y-STR), on the other hand, highlighted the presence of a single haplotypic profile. The comparison with the reference sample of the only suspect also made it possible to identify him as contributor of the traces (also thanks to the biostatistics values of LR obtained). The analysis of the autosomal STR and Y-STR, however, did not allow to establish with certainty the number of contributors present in the mixture and their relative sex.
The Judge’s needing, however, was to understand the effective gender of the contributors to the mixtures: so, the polymorphic markers of the X Chromosome (X-STRs) were analyzed. Their analysis allowed to observe always mixed DNA profiles with a maximum of three alleles per locus. This evidence can be compared to the biological status for which in a mixed genetic profile represented by maximum three alleles per locus, two would belong to a female contribution and one attributable to the X Chromosome haplotype of a male subject. Furthermore, for one of the two mixed profiles obtained, the allelic imbalance at the Amelogenin locus, both in the autosomal profile and in the X Chromosome profile, was in support of this hypothesis. Thus, thanks to the use of the X-STRs, it was possible to answer the judge's request, stating that the mixed traces found on the evidence were related to a male subject (compatible with the suspect) and an unknown female subject.
This case report should be defined as a pilot study on the use of X-STRs for the discrimination and interpretation of mixed DNA profiles, beyond its classic application on deficient parental relationships. The analysis of X-STRs markers, in fact, could lead to excellent results when neither autosomal or Y Chromosome STRs are conclusive.
Thanks to these results and biological reasoning, in the absence of further scientific work concerning the application of X-STRs in forensic cases of biological mixtures created by contributors also of different sex, this work aims to encourage the scientific forensic community to the use of X-STRs for discriminatory purposes. Their application could, in fact, be decisive for a correct deconvolution of the mixtures and consequent identification of the contributing contributors, even if of different sex [
15].
Future work, in vitro and on field, will aim to increase the case studies of the application of X-STRs in the mixtures, also trying to set up biostatistical evaluations of the likelihood ratio in order to provide scientifically and statistically relevant data.
Ethical Statement
The authors declare that they personally conducted the analyses on the evidence samples and the corresponding reference sample, authorized by the Law Court. The authors affirm that they personally collected the reference sample, which was willingly provided after giving consent for the genetic test. Furthermore, the authors consent to the anonymous use of the provided data by the Forensic Sciences Editorial Office for the purpose of publishing the article.
Author Contributions
Conceptualization, V.A. and G.M.; methodology, N.V. and S.C.; software, N.V.; validation, V.A. and G.M.; formal analysis, N.V., G.M., S.C.; investigation, N.V., G.M. and V.A.; resources, G.M.; data curation, V.A. and G.M.; writing—original draft preparation, V.A., S.C.; writing—review and editing, V.A., G.M. and S.C.; visualization, N.V.; supervision, V.A.; project administration, V.A.; All authors have read and agreed to the published version of the manuscript.”
Funding
This research received no external funding.
Conflicts of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
References
- P Gill, CH Brenner, JS Buckleton, A Carracedo, M Krawczak, WR Mayr, N Morling, M Prinz, PM Schneider, BS Weir, DNA commission of the International Society of Forensic Genetics: Recommendations on the interpretation of mixtures. Forensic Sci. Int. 160 (2006) 90-101. [CrossRef]
- PM Schneider, R Fimmers, W Keil, G Molsberger, D Patzelt, W Pflug, T Rothämel, H Schmitter, H Schneider, B Brinkmann, The German Stain Commission: recommendations for the interpretation of mixed stains, Int. J. Legal Med. 123 (2009) 1-5, (originally published in German in 2006 --Rechtsmedizin 16:401-404). [CrossRef]
- TM Clayton, JP Whitaker, R Sparkes, P Gill, Analysis and interpretation of mixed forensic stains using DNA STR profiling, Forensic Sci. Int. 91 (1998) 55–70. [CrossRef]
- J Ge, B Budowle, R Chakraborty, Interpreting Y chromosome STR haplotype mixture, Leg. Med. (Tokyo) 12 (2010) 137–143. [CrossRef]
- N Hu, B Cong, T Gao, Y Chen, J Shen, S Li, C Ma, Application of mixsep software package: performance verification of male-mixed DNA analysis, Mol. Med. Rep. 12 (2015) 2431–2442. [CrossRef]
- P Gill, H Haned, A new methodological framework to interpret complex DNA profiles using likelihood ratios, Forensic Sci. Int. Genet. 7 (2013) 251-263. [CrossRef]
- T Egeland, I Dalen, PF Mostad, Estimating the number of contributors to a DNA profile, Int. J. Legal Med. 117 (2003) 271-275. [CrossRef]
- B Leclair, CJ Frégeau, KL Bowen, RM Fourney, Systematic analysis of stutter percentages and allele peak height and peak area ratios at heterozygous STR loci for forensic casework and database samples, J. Forensic Sci. 49 (2004) 968-980. https://www.ncbi.nlm.nih.gov/pubmed/15461097.
- P Gill, L Gusmão, H Haned, WR Mayr, N Morling, W Parson, L Prieto, M Prinz, H Schneider, PM Schneider, BS Weir, DNA commission of the International Society of Forensic Genetics: Recommendations on the evaluation of STR typing results that may include drop-out and/or drop-in using probabilistic methods, Forensic Sci. Int. Genet. 6 (2012) 679-688. [CrossRef]
- G Dørum, D Kling, A Tillmar, MD Vigeland, T Egeland, Mixtures with relatives and linked markers, Int. J. Legal Med. 130 (2016) 621-634. [CrossRef]
- W Parson, H Niederstätter, A Lindinger, P Gill, ENFSI DNA Working Group, Y-STR analysis on DNA mixture samples - results of a collaborative project of the ENFSI DNA Working Group, Forensic Sci. Int. Genet. 2 (2008) 238-242. [CrossRef]
- W Parson, H Niederstätter, A Brandstätter, B Berger, Improved specificity of Y-STR typing in DNA mixture samples, Int. J. Legal Med. 117 (2003) 109-114. [CrossRef]
- M Lancia, S Severini, A Coletti, G Margiotta, M Dobosz, E Carnevali, Using X-chromosomal markers in rape investigation, Forensic Sci. Int. Genet. Suppl. Ser. 3 (2011) e55-e56. [CrossRef]
- R Szibor, M Krawczak, S Hering, J Edelmann, E Kuhlisch, D Krause, Use of X-linked markers for forensic purposes, Int. J. Legal Med. 117 (2003) 67-74. [CrossRef]
- H He, L Zha, J Cai, J Huang, The forensic value of X-linked markers in mixed-male DNA analysis, Int. J. Legal Med. 132 (2018) 1281-1285. [CrossRef]
- R Szibor, X-chromosomal markers: past present and future, Forensic Sci. Int. Genet. 1 (2007) 93-99. [CrossRef]
- D Kling, Curiosities of X chromosomal markers and haplotypes. Int. J. Legal Med. 132 (2018) 361-371. [CrossRef]
- AO Tillmar, D Kling, JM Butler, W Parson, M Prinz, PM Schneider, T Egeland, L Gusmão, DNA Commission of the International Society for Forensic Genetics (ISFG): Guidelines on the use of X-STRSs in kinship analysis, Forensic Sci. Int. Genet. 29 (2017) 269-275. [CrossRef]
- Ø Bleka, CCG Benschop, G Storvik, P Gill, A comparative study of qualitative and quantitative models used to interpret complex STR DNA profiles, Forensic Sci. Int. Genet. 25 (2016) 85-96. [CrossRef]
- L Roewer, M Krawczak, S Willuweit, M Nagy, C Alves, A Amorim, K Anslinger, C Augustin, A Betz, E Bosch, A Cagliá, A Carracedo, D Corach, AF Dekairelle, T Dobosz, BM Dupuy, S Füredi, C Gehrig, L Gusmaõ, J Henke, L Henke, M Hidding, C Hohoff, B Hoste, MA Jobling, HJ Kärgel, P de Knijff, R Lessig, E Liebeherr, M Lorente, B Martínez-Jarreta, P Nievas, M Nowak, W Parson, VL Pascali, G Penacino, R Ploski, B Rolf, A Sala, U Schmidt, C Schmitt, PM Schneider, R Szibor, J Teifel-Greding, M Kayser, Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes, Forensic Sci. Int. 118 (2001) 106-113. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).