Submitted:
26 January 2024
Posted:
29 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling Site and Physical and Chemical Parameters
2.2. Sampling
2.3. Total Cell Counts
2.4. Most Probable Number
2.5. Molecular approach for microbial enumeration
2.6. Statistical variance of the different methods
3. Results
3.1. Epilimnion
3.2. Hypolimnion
3.3. Sediment
4. Discussion
4.1. Seasonal course of bacteria and dependency on environmental parameters
4.2. Explanations on the seasonal courses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coskuner, G.; Ballinger, S.J.; Davenport, R.J.; Pickering, R.L.; Solera, R.; Head, I.M.; Curtis, T.P. Agreement between Theory and Measurement in Quantification of Ammonia-Oxidizing Bacteria. Appl. Environ. Microbiol. 2005, 71, 6325–6334. [Google Scholar] [CrossRef] [PubMed]
- Muela, A.; Gorostiza, I.; Iriberri, J.; Egea, L. Indicative Parameters of Denitrification in River Sediments. Acta Hydrochim. Et Hydrobiol. 1988, 16, 157–163. [Google Scholar] [CrossRef]
- Baldwin, B.R.; Nakatsu, C.H.; Nies, L. Detection and Enumeration of Aromatic Oxygenase Genes by Multiplex and Real-Time PCR. Appl. Environ. Microbiol. 2003, 69, 3350–3358. [Google Scholar] [CrossRef]
- Röling, W.F.M. Do Microbial Numbers Count? Quantifying the Regulation of Biogeochemical Fluxes by Population Size and Cellular Activity. FEMS Microbiol. Ecol. 2007, 62, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Trippett, C.; Dodds, W.K.; O’Brien, J.M.; Banner, E.B.K.; Head, I.M.; Smith, M.S.; Yang, R.K.; Knapp, C.W. Correlations between in Situ Denitrification Activity and Nir-Gene Abundances in Pristine and Impacted Prairie Streams. Environ. Pollut. 2010, 158, 3225–3229. [Google Scholar] [CrossRef] [PubMed]
- Christensen, P.B.; Rysgaard, S.; Sloth, N.P.; Dalsgaard, T.; Schwaerter, S. Sediment Mineralization, Nutrient Fluxes, Denitrification and Dissimilatory Nitrate Reduction to Ammonium in an Estuarine Fjord with Sea Cage Trout Farms. Aquat. Microb. Ecol. 2000, 21, 73–84. [Google Scholar] [CrossRef]
- Magalhães, C.; Bano, N.; Wiebe, W.J.; Bordalo, A.A.; Hollibaugh, J.T. Dynamics of Nitrous Oxide Reductase Genes (nosZ) in Intertidal Rocky Biofilms and Sediments of the Douro River Estuary (Portugal), and Their Relation to N-Biogeochemistry. Microb Ecol 2008, 55, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Gihring, T.M.; Canion, A.; Riggs, A.; Huettel, M.; Kostka, J.E. Denitrification in Shallow, Sublittoral Gulf of Mexico Permeable Sediments. Limnol. Oceanogr. 2010, 55, 43–54. [Google Scholar] [CrossRef]
- Chon, K.; Chang, J.-S.; Lee, E.; Lee, J.; Ryu, J.; Cho, J. Abundance of Denitrifying Genes Coding for Nitrate (narG), Nitrite (nirS), and Nitrous Oxide (nosZ) Reductases in Estuarine versus Wastewater Effluent-Fed Constructed Wetlands. Ecol. Eng. 2011, 37, 64–69. [Google Scholar] [CrossRef]
- McCrackin, M.L.; Elser, J.J. Denitrification Kinetics and Denitrifier Abundances in Sediments of Lakes Receiving Atmospheric Nitrogen Deposition (Colorado, USA). Biogeochemistry 2012, 108, 39–54. [Google Scholar] [CrossRef]
- Brankatschk, R.; Töwe, S.; Kleineidam, K.; Schloter, M.; Zeyer, J. Abundances and Potential Activities of Nitrogen Cycling Microbial Communities along a Chronosequence of a Glacier Forefield. ISME J. 2010, 5, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Abell, G.; Ross, D.; Keane, J.; Oakes, J.; Eyre, B.; Robert, S.; Volkman, J. Nitrifying and Denitrifying Microbial Communities and Their Relationship to Nutrient Fluxes and Sediment Geochemistry in the Derwent Estuary, Tasmania. Aquat. Microb. Ecol. 2013, 70, 63–75. [Google Scholar] [CrossRef]
- Bellini, M.I.; Gutiérrez, L.; Tarlera, S.; Scavino, A.F. Isolation and Functional Analysis of Denitrifiers in an Aquifer with High Potential for Denitrification. Syst. Appl. Microbiol. 2013, 36, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Veraart, A.J.; Audet, J.; Dimitrov, M.R.; Hoffmann, C.C.; Gillissen, F.; de Klein, J.J.M. Denitrification in Restored and Unrestored Danish Streams. Ecol. Eng. 2013, 66, 129–140. [Google Scholar] [CrossRef]
- Michotey, V.; Mejean, V.; Bonin, P. Comparison of Methods for Quantification of Cytochrome Cd1-Denitrifying Bacteria in Environmental Marine Samples. Appl. Environ. Microbiol. 2000, 66, 1564–1571. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.J.; Harris, D.; Dollhopf, S.L.; Gross, K.L.; Prosser, J.I.; Paul, E.A. Effects of Agronomic Treatments on Structure and Function of Ammonia-Oxidizing Communities. Appl. Environ. Microbiol. 2000, 66, 5410–5418. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Øvreås, L. Microbial Diversity and Function in Soil: From Genes to Ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Both, G.J.; Gerards, S.; Laanbroek, H.J. Enumeration of Nitrite-Oxidizing Bacteria in Grassland Soils Using a Most Probable Number Technique: Effect of Nitrite Concentration and Sampling Procedure. FEMS Microbiol. Lett. 1990, 74, 277–285. [Google Scholar] [CrossRef]
- Philippot, L. Use of Functional Genes to Quantify Denitrifiers in the Environment. Biochem. Soc. Trans. 2006, 34, 101–103. [Google Scholar] [CrossRef]
- Noble, R.T.; Weisberg, S.B.; Leecaster, M.K.; McGee, C.D.; Ritter, K.; Walker, K.O.; Vainik, P.M. Comparison of Beach Bacterial Water Quality Indicator Measurement Methods. Environ. Monit. Assess. 2003, 81, 301–312. [Google Scholar] [CrossRef]
- Belser, L.; Mays, E. Use of Nitrifier Activity Measurements to Estimate the Efficiency of Viable Nitrifier Counts in Soils and Sediments. Appl. Environ. Microbiol. 1982, 43, 945–948. [Google Scholar] [CrossRef]
- Degrange, V.; Bardin, R. Detection and Counting of Nitrobacter Populations in Soil by PCR. Appl. Environ. Microbiol. 1995, 61, 2093–2098. [Google Scholar] [CrossRef]
- Scala, D.J.; Kerkhof, L.J. Nitrous Oxide Reductase (nosZ) Gene-Specific PCR Primers for Detection of Denitrifiers and Three nosZ Genes from Marine Sediments. FEMS Microbiol. Lett. 1998, 162, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Rathsack, K.; Böllmann, J.; Martienssen, M. Comparative Study of Different Methods for Analyzing Denitrifying Bacteria in Fresh Water Ecosystems. J. Water Resour. Prot. 2014, 06, 609–617. [Google Scholar] [CrossRef]
- von Wintzingerode, F.; Göbel, U.B.; Stackebrandt, E. Determination of Microbial Diversity in Environmental Samples: Pitfalls of PCR-Based rRNA Analysis. FEMS Microbiol. Rev. 1997, 21, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Freeman, W.M.; Walker, S.J.; Vrana, K.E. Quantitative RT-PCR: Pitfalls and Potential. BioTechniques 1999, 26, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene Quantification Using Real-Time Quantitative PCR: An Emerging Technology Hits the Mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Smith, C.J.; Nedwell, D.B.; Dong, L.F.; Osborn, A.M. Evaluation of Quantitative Polymerase Chain Reaction-Based Approaches for Determining Gene Copy and Gene Transcript Numbers in Environmental Samples. Environ. Microbiol. 2006, 8, 804–815. [Google Scholar] [CrossRef]
- Schulz, S.; Peréz-de-Mora, A.; Engel, M.; Munch, J.C.; Schloter, M. A Comparative Study of Most Probable Number (MPN)-PCR vs. Real-Time-PCR for the Measurement of Abundance and Assessment of Diversity of alkB Homologous Genes in Soil. J. Microbiol. Methods 2010, 80, 295–298. [Google Scholar] [CrossRef]
- Klein, D. Quantification Using Real-Time PCR Technology: Applications and Limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Rompré, A.; Servais, P.; Baudart, J.; de-Roubin, M.-R.; Laurent, P. Detection and Enumeration of Coliforms in Drinking Water: Current Methods and Emerging Approaches. J. Microbiol. Methods 2002, 49, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Hallin, S. Finding the Missing Link between Diversity and Activity Using Denitrifying Bacteria as a Model Functional Community. Curr. Opin. Microbiol. 2005, 8, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir-Butler, K.; Jones, J.L.; Benner Jr., R. A.; Burkhardt III, W. Quantification of Total and Specific Gram-Negative Histamine-Producing Bacteria Species in Fish Using an MPN Real-Time PCR Method. Food Microbiol. 2011, 28, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Kayashima, T.; Suzuki, H.; Maeda, T.; Ogawa, H.I. Real-Time PCR for Rapidly Detecting Aniline-Degrading Bacteria in Activated Sludge. Chemosphere 2013, 91, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Picozzi, C.; Foschino, R.; Di Pilato, P. Exploiting a MPN-PCR Technique to Quantify Escherichia Coli in Minced Meat. Ann. Microbiol. 2004, 54, 343–349. [Google Scholar]
- O’Connor, B.L.; Hondzo, M.; Dobraca, D.; LaPara, T.M.; Finlay, J.C.; Brezonik, P.L. Quantity-Activity Relationship of Denitrifying Bacteria and Environmental Scaling in Streams of a Forested Watershed. J. Geophys. Res. 2006, G04014, 2005–2012. [Google Scholar] [CrossRef]
- Barer, M.R.; Harwood, C.R. Bacterial Viability and Culturability. Adv. Microb. Physiol. 1999, 41, 93–137. [Google Scholar] [PubMed]
- Oliver, J.D. Recent Findings on the Viable but Nonculturable State in Pathogenic Bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Nixdorf, B.; Rektins, A.; Mischke, U. Standards and Thresholds of the EU Water Framework Directive (WFD) — Phytoplankton and Lakes. In Standards and Thresholds for Impact Assessment; Schmidt, M., Glasson, J., Emmelin, L., Helbron, H., Eds.; Environmental Protection in the European Union; Springer Berlin Heidelberg: Berlin, Heidelberg, 2008; ISBN 978-3-540-31140-9. [Google Scholar]
- Grüneberg, B.; Rücker, J.; Nixdorf, B.; Behrendt, H. Dilemma of Non-Steady State in Lakes - Development and Predictability of in-Lake P Concentration in Dimictic Lake Scharmützelsee (Germany) after Abrupt Load Reduction. Int. Rev. Hydrobiol. 2011, 96, 599–621. [Google Scholar] [CrossRef]
- Kleeberg, A.; Grüneberg, B. Phosphorus Mobility in Sediments of Acid Mining Lakes, Lusatia, Germany. Ecol. Eng. 2005, 24, 89–100. [Google Scholar] [CrossRef]
- Hobbie, J.E.; Daley, R.J.; Jasper, S. Use of Nuclepore Filters for Counting Bacteria by Fluorescence Microscopy. Appl Env. Microbiol 1977, 33, 1225–1228. [Google Scholar] [CrossRef]
- Rathsack, K.; Böllmann, J.; Martienssen, M. Comparative Study of Different Methods for Analyzing Denitrifying Bacteria in Fresh Water Ecosystems. JWARP 2014, 06, 609–617. [Google Scholar] [CrossRef]
- Michels, J.; Stuhrmann, M.; Frey, C.; Koschitzky, H.-P. Handlungsempfehlungen Mit Methodensammlung, Natürliche Schadstoffminderung Bei Der Sanierung von Altlasten; Dechema e.V., VEGAS, Institut für Wasserbau, Universität Stuttgart: Frankfurt a.M, 2008; ISBN -13. [Google Scholar]
- Cataldo, D.A.; Maroon, M.; Schrader, L.E.; Youngs, V.L. Rapid Colorimetric Determination of Nitrate in Plant Tissue by Nitration of Salicylic Acid. Commun. Soil Sci. Plant Anal. 1975, 6, 71–80. [Google Scholar] [CrossRef]
- Bremner, J.M. Inorganic Forms of Nitrogen. In Agronomy Monographs; Norman, A.G., Ed.; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1965; pp. 1179–1237. ISBN 978-0-89118-204-7. [Google Scholar]
- Garthright, W.E.; Blodgett, R.J. FDA’s Preferred MPN Methods for Standard, Large or Unusual Tests, with a Spreadsheet. Food Microbiol. 2003, 20, 439–445. [Google Scholar] [CrossRef]
- Throbäck, I.N.; Enwall, K.; Jarvis, Ãs. ; Hallin, S. Reassessing PCR Primers Targeting nirS, nirK and nosZ Genes for Community Surveys of Denitrifying Bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Martienssen, M.; Schöps, R. Population Dynamics of Denitrifying Bacteria in a Model Biocommunity. Water Res. 1999, 33, 639–646. [Google Scholar] [CrossRef]
- Grüneberg, B.; Rücker, J.; Nixdorf, B.; Behrendt, H. Dilemma of Non-Steady State in Lakes - Development and Predictability of in-Lake P Concentration in Dimictic Lake Scharmützelsee (Germany) after Abrupt Load Reduction. Int. Rev. Hydrobiol. 2011, 96, 599–621. [Google Scholar] [CrossRef]
- Martienssen, M.; Böllmann, J.; Nixdorf, B.; Rathsack, K. Calculation of Hypolimnic Denitrification in a Dimictic Freshwater Lake during Summer Stratification. Aquat. Microb. Ecol. 2019, 83, 189–201. [Google Scholar] [CrossRef]
- Fakruddin, Md.; Mannan, K.S.B.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 1–6. [Google Scholar] [CrossRef]
- Epstein, S. The Phenomenon of Microbial Uncultivability. Curr. Opin. Microbiol. 2013, 16, 636–642. [Google Scholar] [CrossRef]
- Berg, P. Nitrifier Populations and Nitrification Rates in Agricultural Soil; Rapport - Sveriges lantbruksuniversitet, Institutionen för mikrobiologi, Ed.; Sveriges Lantbruksuniversitet: Uppsala, 1986; ISBN 91-576-2647-2. [Google Scholar]
- Bręk-Laitinen, G.; López Bellido, J.; Ojala, A. Response of a Microbial Food Web to Prolonged Seasonal Hypoxia in a Boreal Lake. Aquat. Biol. 2012, 14, 105–120. [Google Scholar] [CrossRef]
- Böllmann, J.; Rathsack, K.; Martienssen, M. The Precision of Bacterial Quantification Techniques on Different Kinds of Environmental Samples and the Effect of Ultrasonic Treatment. J. Microbiol. Methods 2016, 126, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Corinaldesi, C.; Danovaro, R.; Dell’Anno, A. Simultaneous Recovery of Extracellular and Intracellular DNA Suitable for Molecular Studies from Marine Sediments. Appl. Environ. Microbiol. 2005, 71, 46–50. [Google Scholar] [CrossRef]
- Fredslund, L.; Ekelund, F.; Jacobsen, C.S.; Johnsen, K. Development and Application of a Most-Probable-Number-PCR Assay To Quantify Flagellate Populations in Soil Samples. Appl. Environ. Microbiol. 2001, 67, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Buttner, M.P.; Cruz, P.; Stetzenbach, L.D.; Klima-Comba, A.K.; Stevens, V.L.; Cronin, T.D. Determination of the Efficacy of Two Building Decontamination Strategies by Surface Sampling with Culture and Quantitative PCR Analysis. Appl. Environ. Microbiol. 2004, 70, 4740–4747. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, F.H.; Banat, I.M.; Marchant, R. Detection and Quantification of Gene Expression in Environmental Bacteriology. Appl. Environ. Microbiol. 2004, 70, 3795–3806. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Kjelleberg, S. The Role of RNA Stability during Bacterial Stress Responses and Starvation. Minireview. Environ. Microbiol. 2000, 2, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, A.; Schmidt, I.; Saunders, A.M.; Nicolaisen, M.H. Influence of Starvation on Potential Ammonia-Oxidizing Activity and amoA mRNA Levels of Nitrosospira Briensis. Appl. Environ. Microbiol. 2005, 71, 1276–1282. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
 ), 16S rDNA gene copy number (
), 16S rDNA gene copy number ( ), b: MPN of nitrate reducing bacteria (
), b: MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), sum of nirS and nirK (
), sum of nirS and nirK ( ), nitrite concentration (grey line).
), nitrite concentration (grey line).
 ), 16S rDNA gene copy number (
), 16S rDNA gene copy number ( ), b: MPN of nitrate reducing bacteria (
), b: MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), sum of nirS and nirK (
), sum of nirS and nirK ( ), nitrite concentration (grey line).
), nitrite concentration (grey line).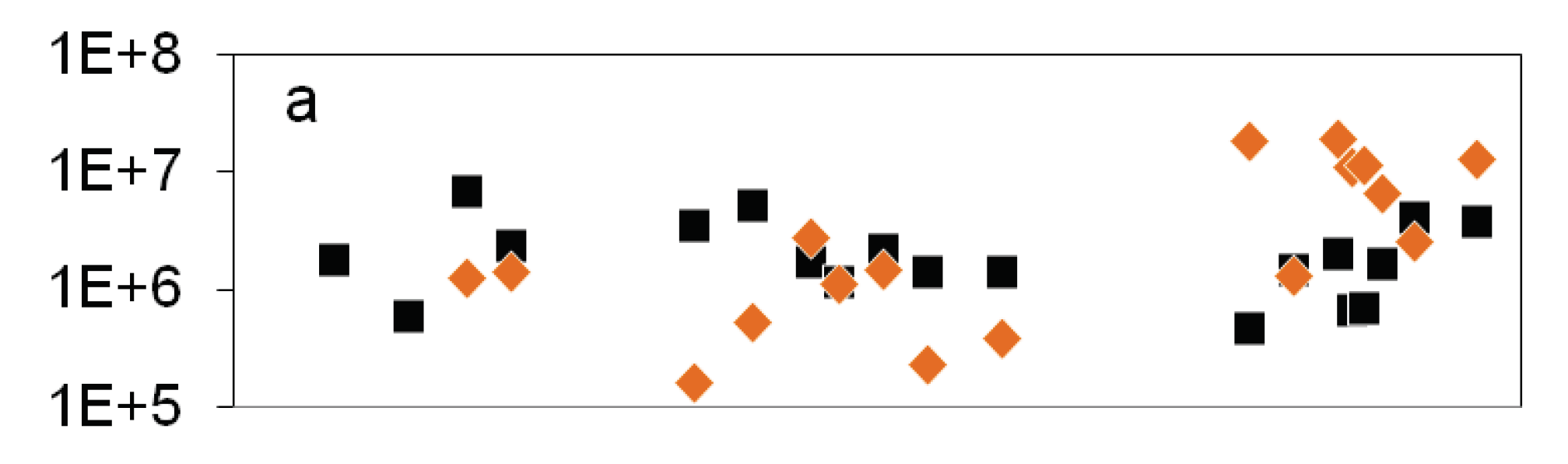
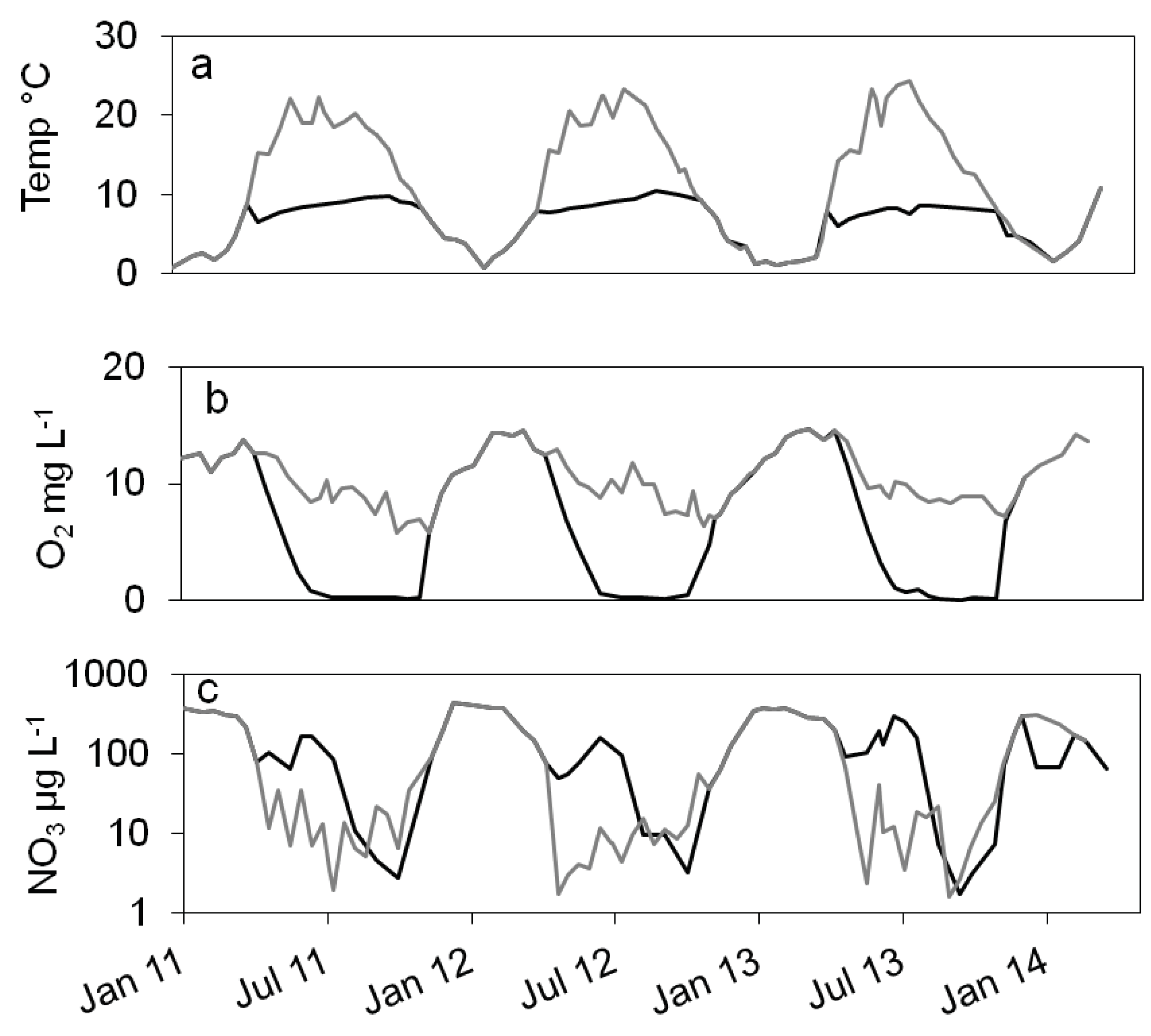
 ),16S rDNA cell number estimation (
),16S rDNA cell number estimation ( ), temperature 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line) b: numbers of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), temperature 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line) b: numbers of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrate reducing bacteria (
), MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), c: difference between log10 transformed cell numbers (nir)and MPN of denitrifying bacteria (+), nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line).
), c: difference between log10 transformed cell numbers (nir)and MPN of denitrifying bacteria (+), nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line).
 ),16S rDNA cell number estimation (
),16S rDNA cell number estimation ( ), temperature 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line) b: numbers of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), temperature 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line) b: numbers of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrate reducing bacteria (
), MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), c: difference between log10 transformed cell numbers (nir)and MPN of denitrifying bacteria (+), nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line).
), c: difference between log10 transformed cell numbers (nir)and MPN of denitrifying bacteria (+), nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line).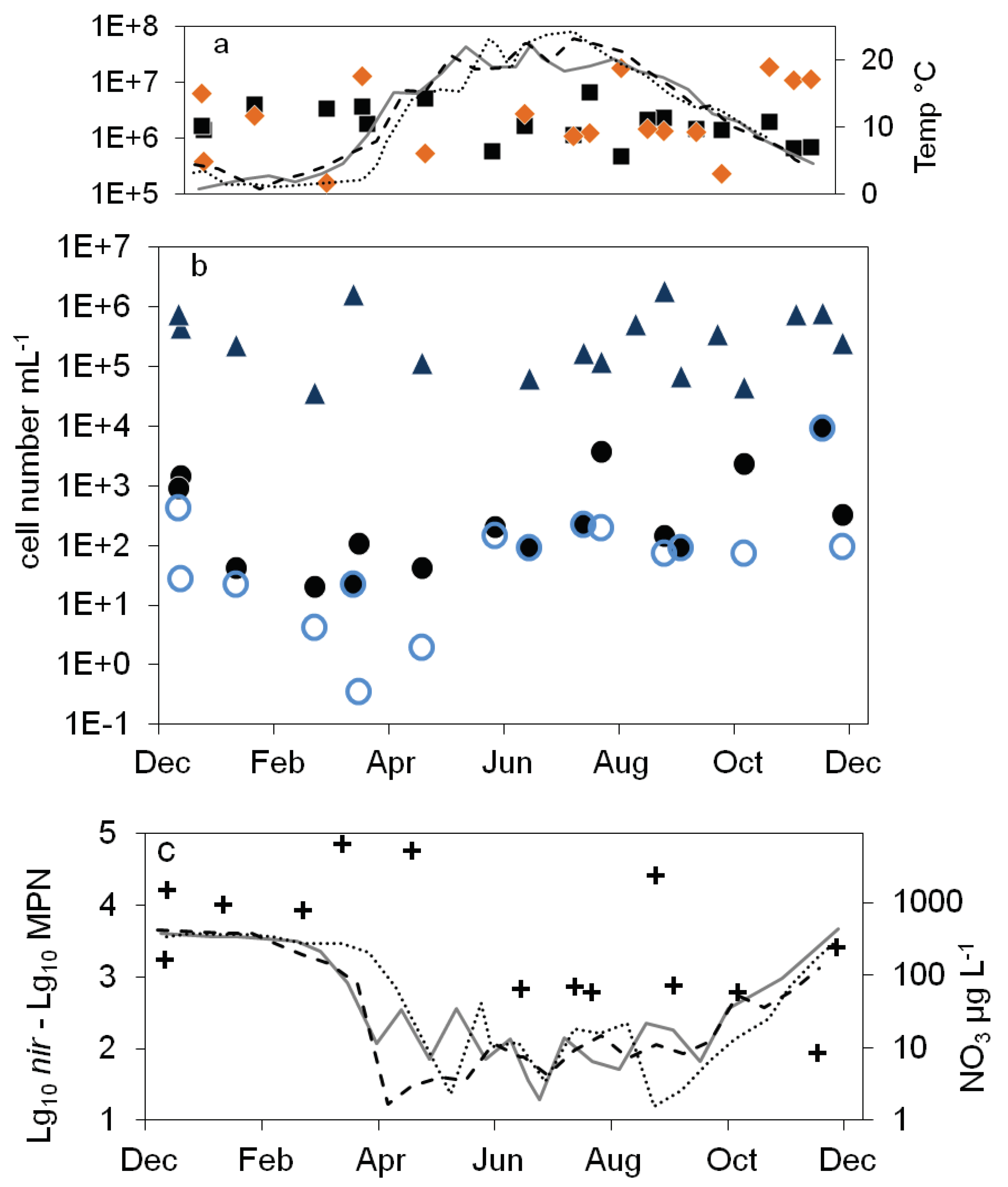
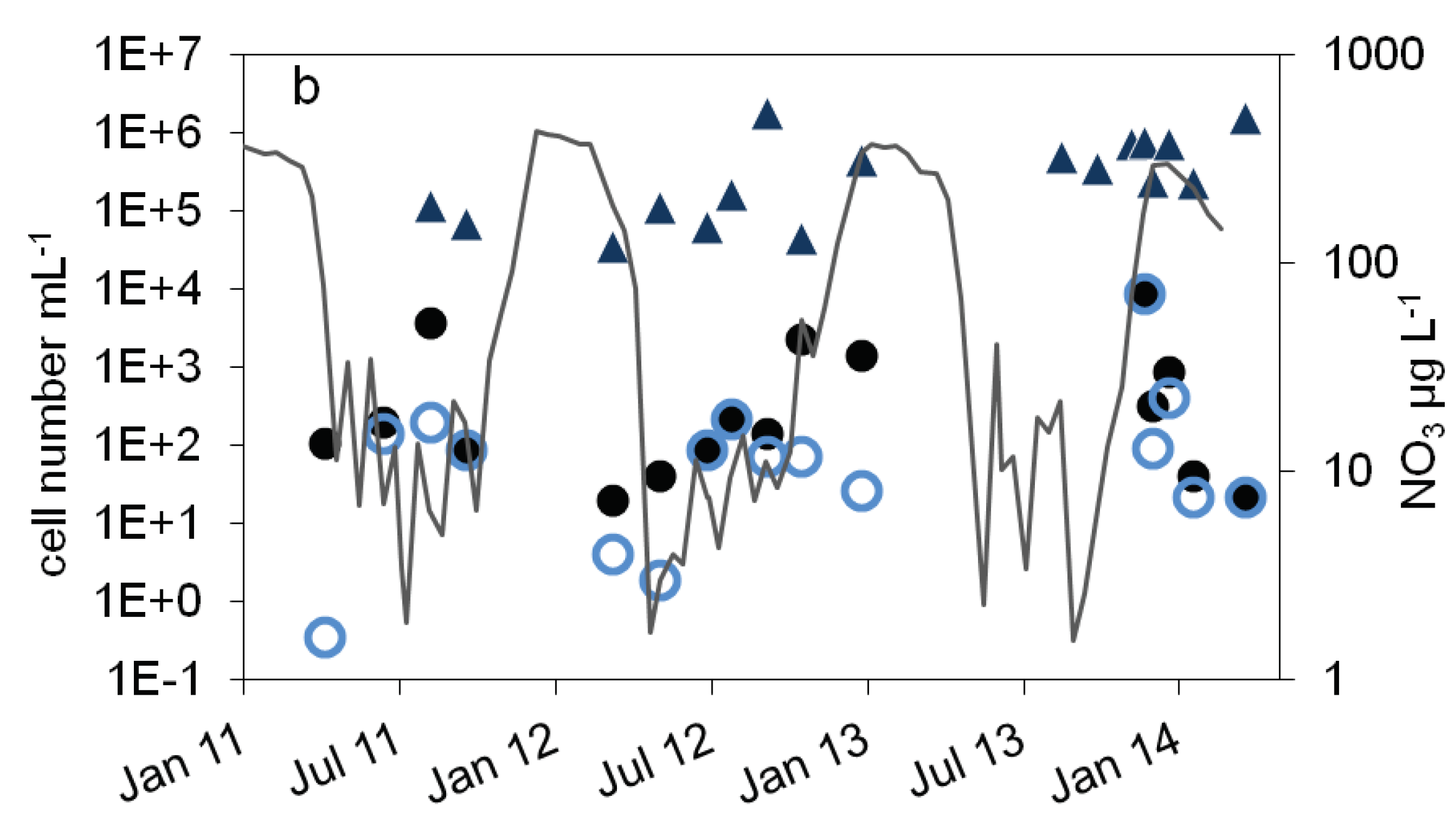
 ),16S rDNA cell number estimation (
),16S rDNA cell number estimation ( ) b: number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
) b: number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrite reducing bacteria (
), MPN of nitrite reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), nitrate concentration (black line).
), nitrate concentration (black line).
 ),16S rDNA cell number estimation (
),16S rDNA cell number estimation ( ) b: number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
) b: number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrite reducing bacteria (
), MPN of nitrite reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), nitrate concentration (black line).
), nitrate concentration (black line).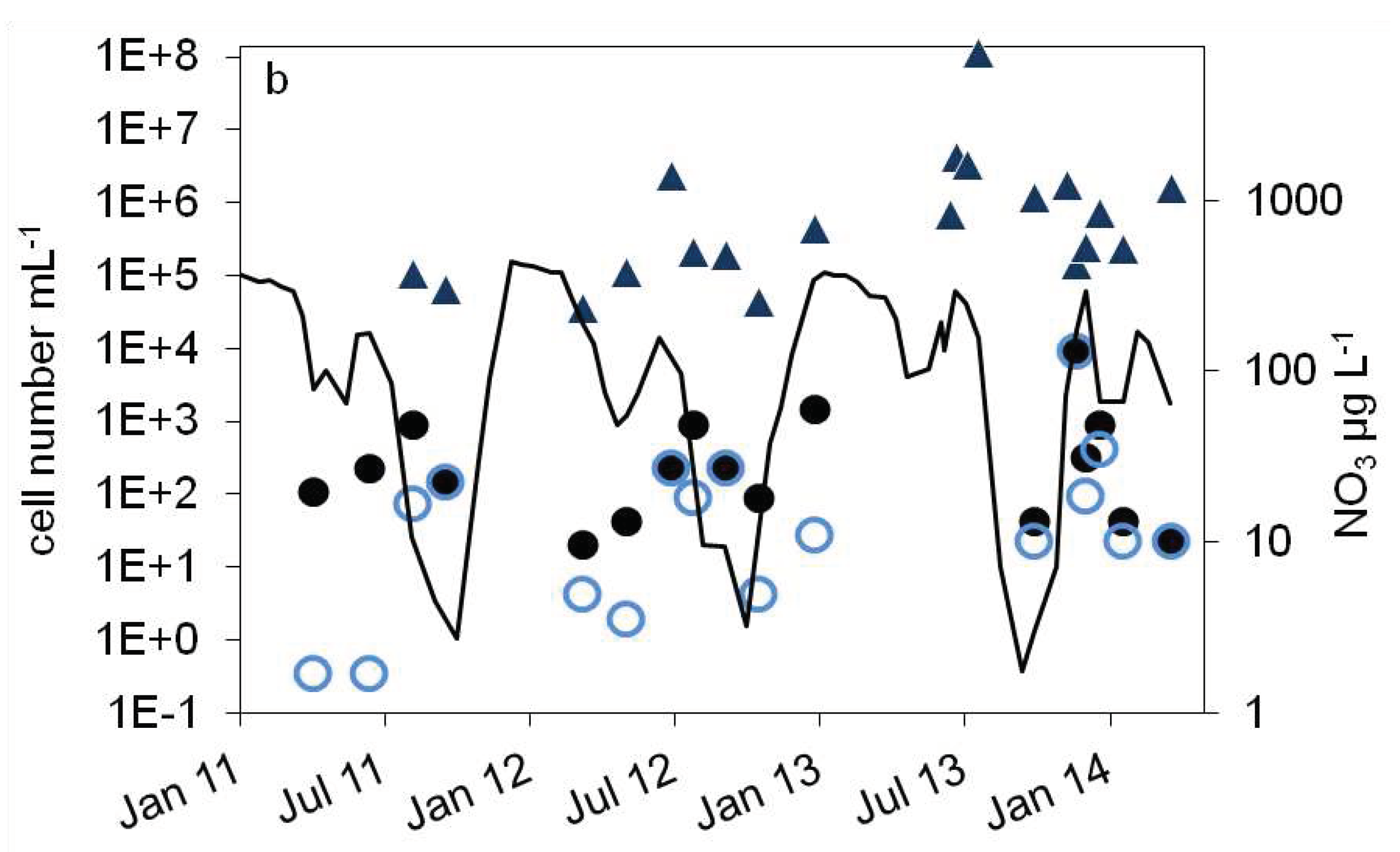
 ), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrate reducing bacteria (
), MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ),nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line), b: difference between log10 transformed cell numbers (nir) and MPN of denitrifying bacteria (+).
),nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line), b: difference between log10 transformed cell numbers (nir) and MPN of denitrifying bacteria (+).
 ), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrate reducing bacteria (
), MPN of nitrate reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ),nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line), b: difference between log10 transformed cell numbers (nir) and MPN of denitrifying bacteria (+).
),nitrate concentration 2011 (solid grey line), 2012 (dashed black line), 2013 (dotted black line), b: difference between log10 transformed cell numbers (nir) and MPN of denitrifying bacteria (+).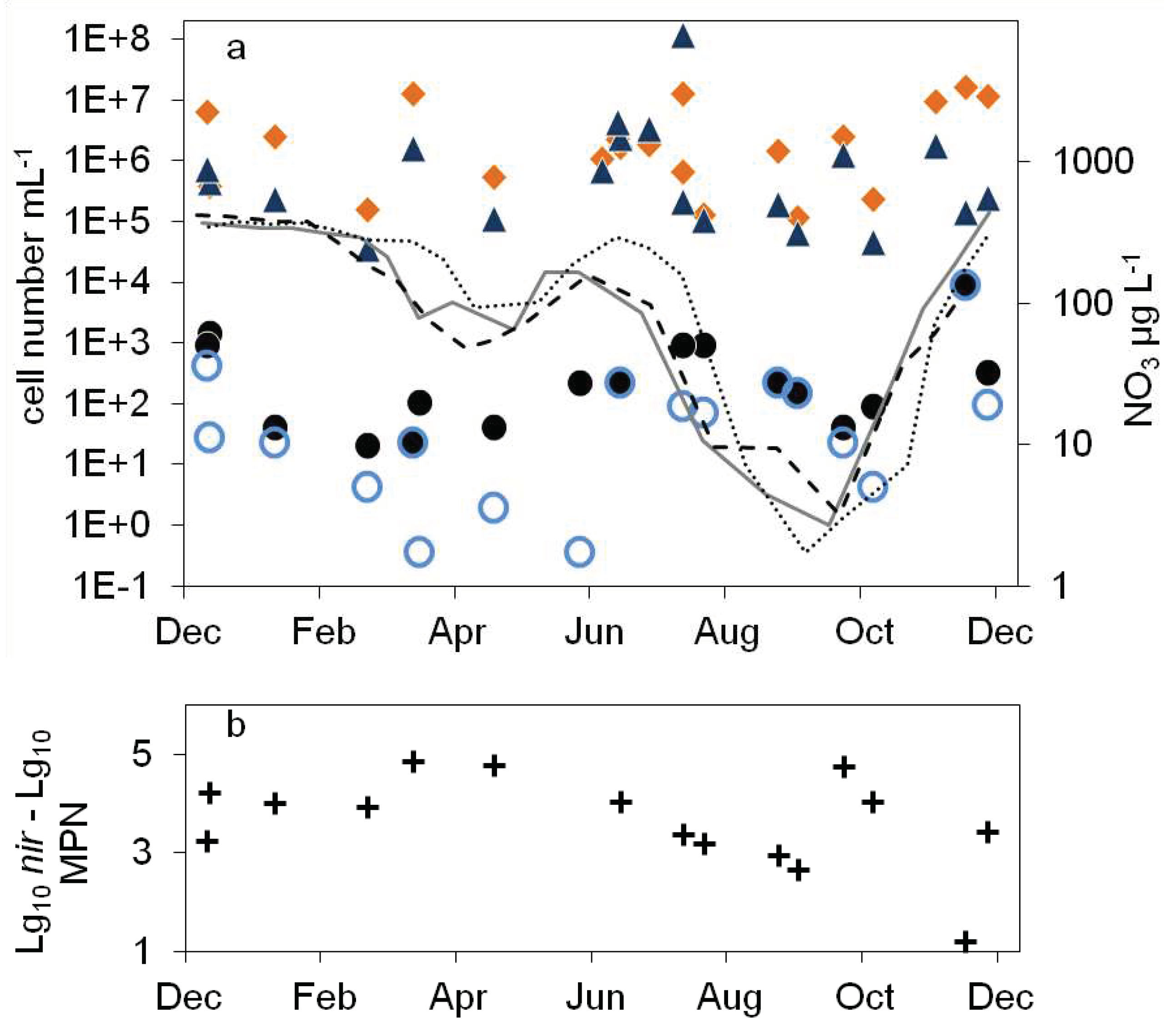
 ), 16S rDNA cell number estimation (
), 16S rDNA cell number estimation ( ), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrite reducing bacteria (
), MPN of nitrite reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), nitrate concentration (black line).
), nitrate concentration (black line).
 ), 16S rDNA cell number estimation (
), 16S rDNA cell number estimation ( ), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies (
), number of denitrifying bacteria estimated by the sum of nirS and nirK gene copies ( ), MPN of nitrite reducing bacteria (
), MPN of nitrite reducing bacteria ( ) and denitrifying bacteria (
) and denitrifying bacteria ( ), nitrate concentration (black line).
), nitrate concentration (black line).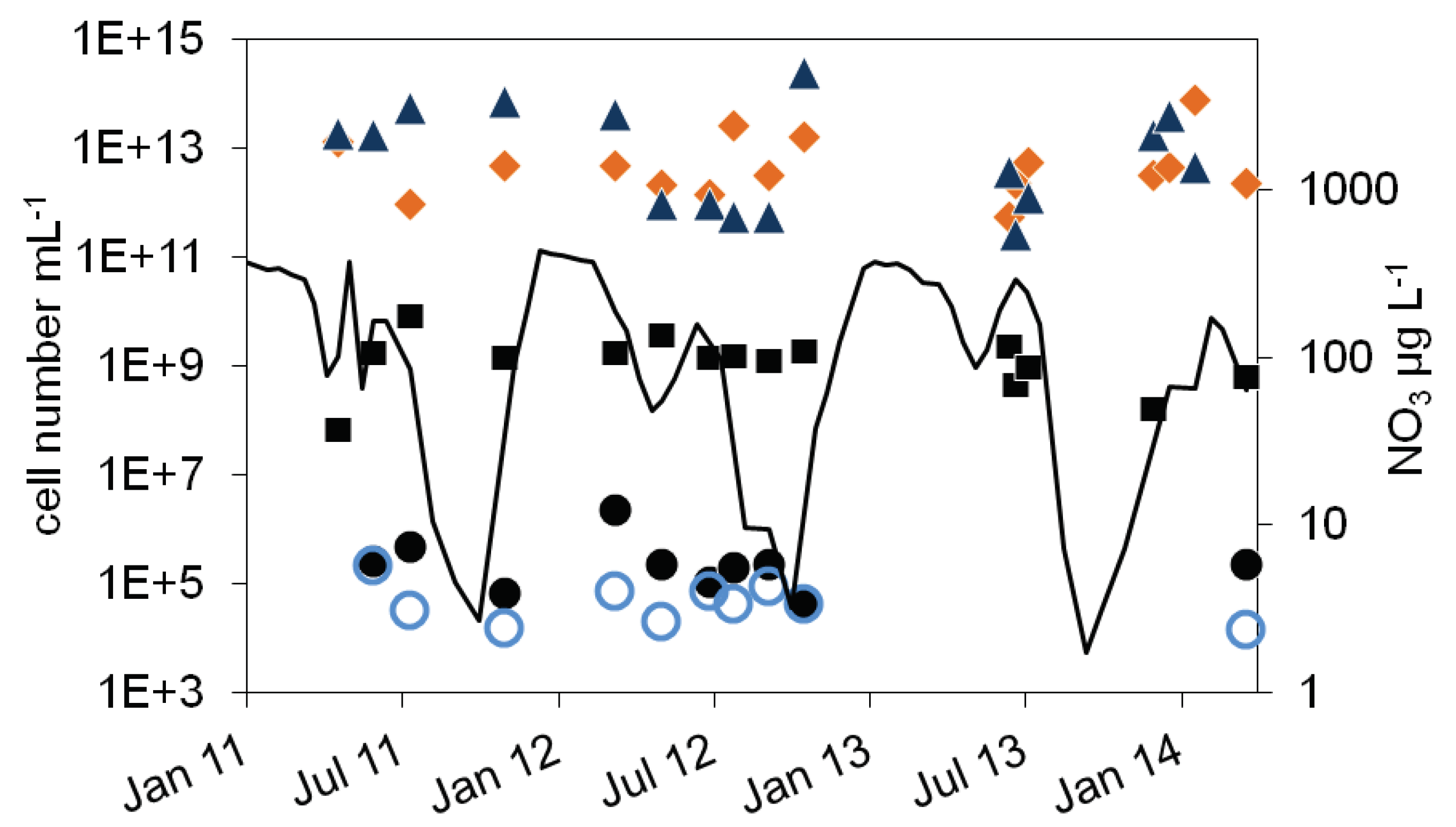
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




