Submitted:
25 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Treatment and Cell Viability Assays
2.2. Toluidine Blue Staining
2.3. Cell Counting Kit-8
2.4. RNA Extraction, Library Preparation, and RNA-Sequencing
2.5. Differential Gene Expression and Pathway Analysis
2.6. Real-time reverse transcription polymerase chain reaction (qRT-PCR)
2.7. Statistical Analysis
3. Results
3.1. TQ treatment for 24 hours had no significant inhibitory effect on A172 cell viability.
3.2. TQ treatment for 48 hours inhibited the viability of A172 cells in dose dependent manner.
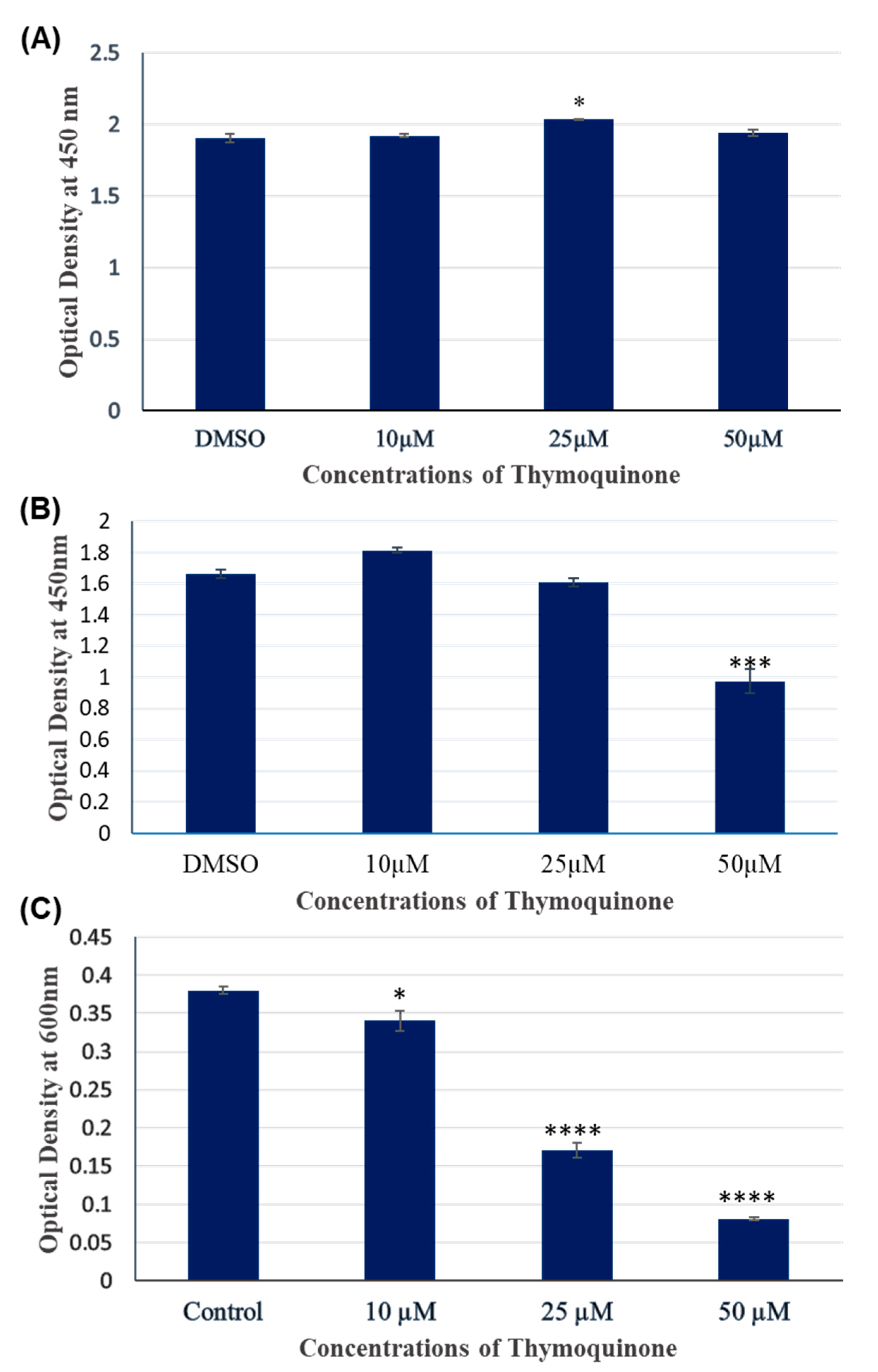
3.3. 2.5 µM and 5 µM TQ for 24 hours did not cause differential gene expression in A172 cells.
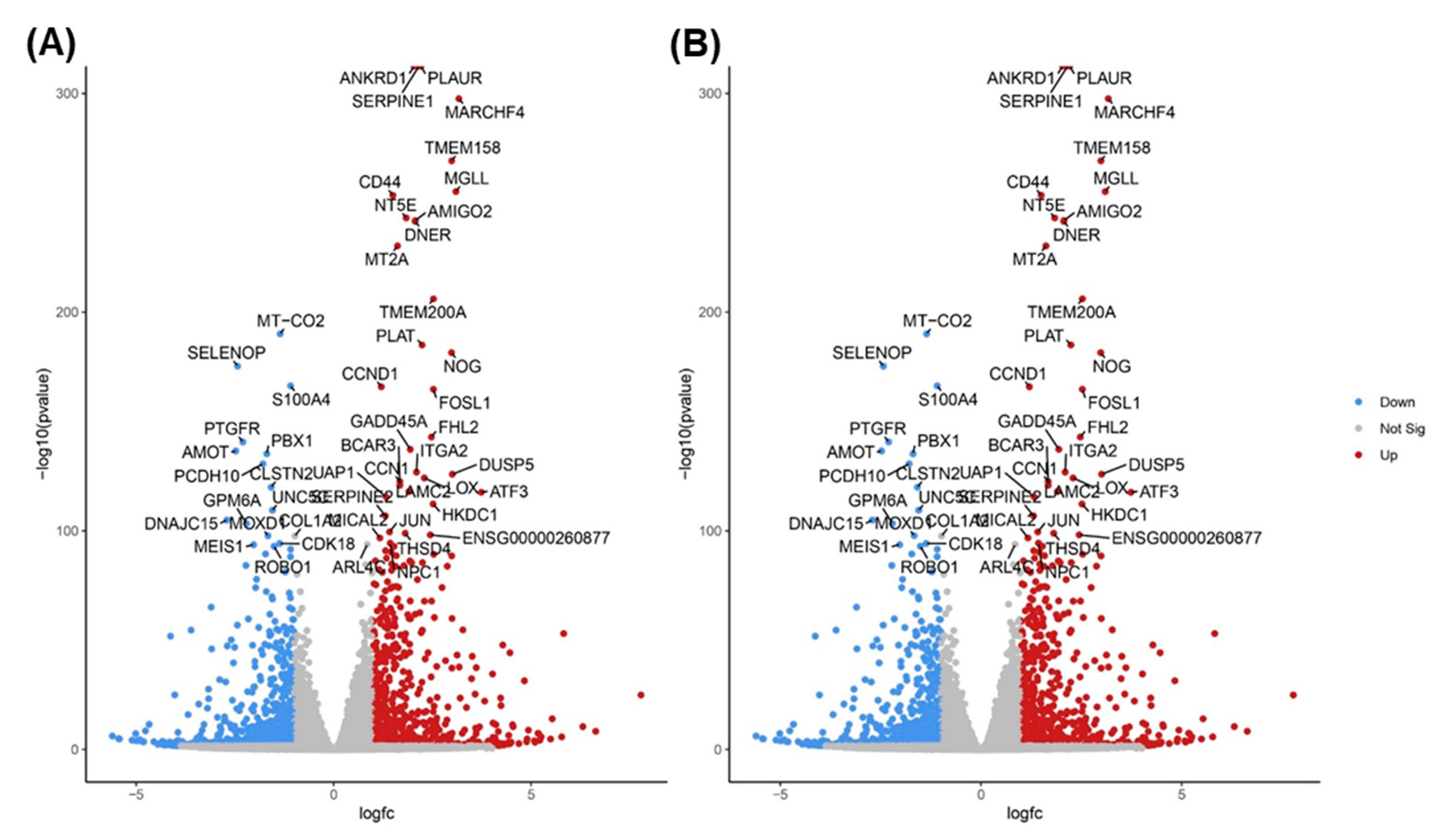
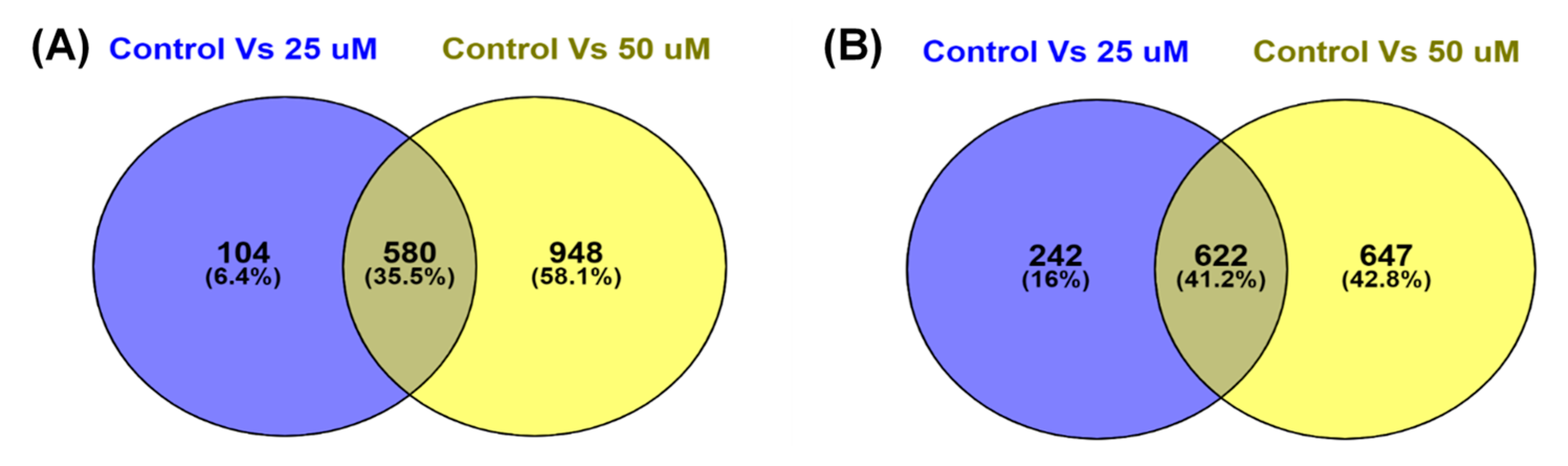
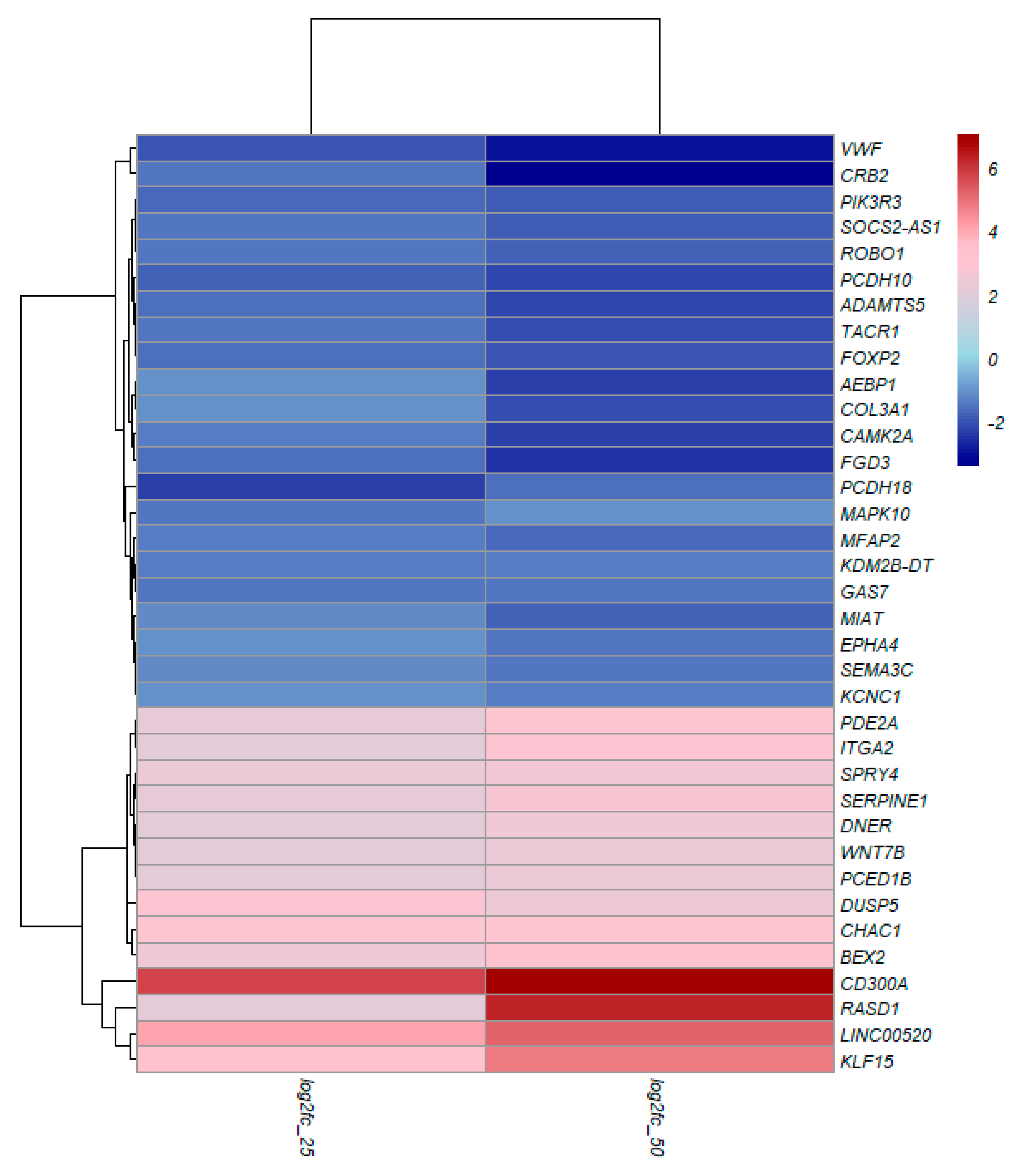
3.4. TQ treatment for 48 hours induced differentially expressed genes in A172 cells.
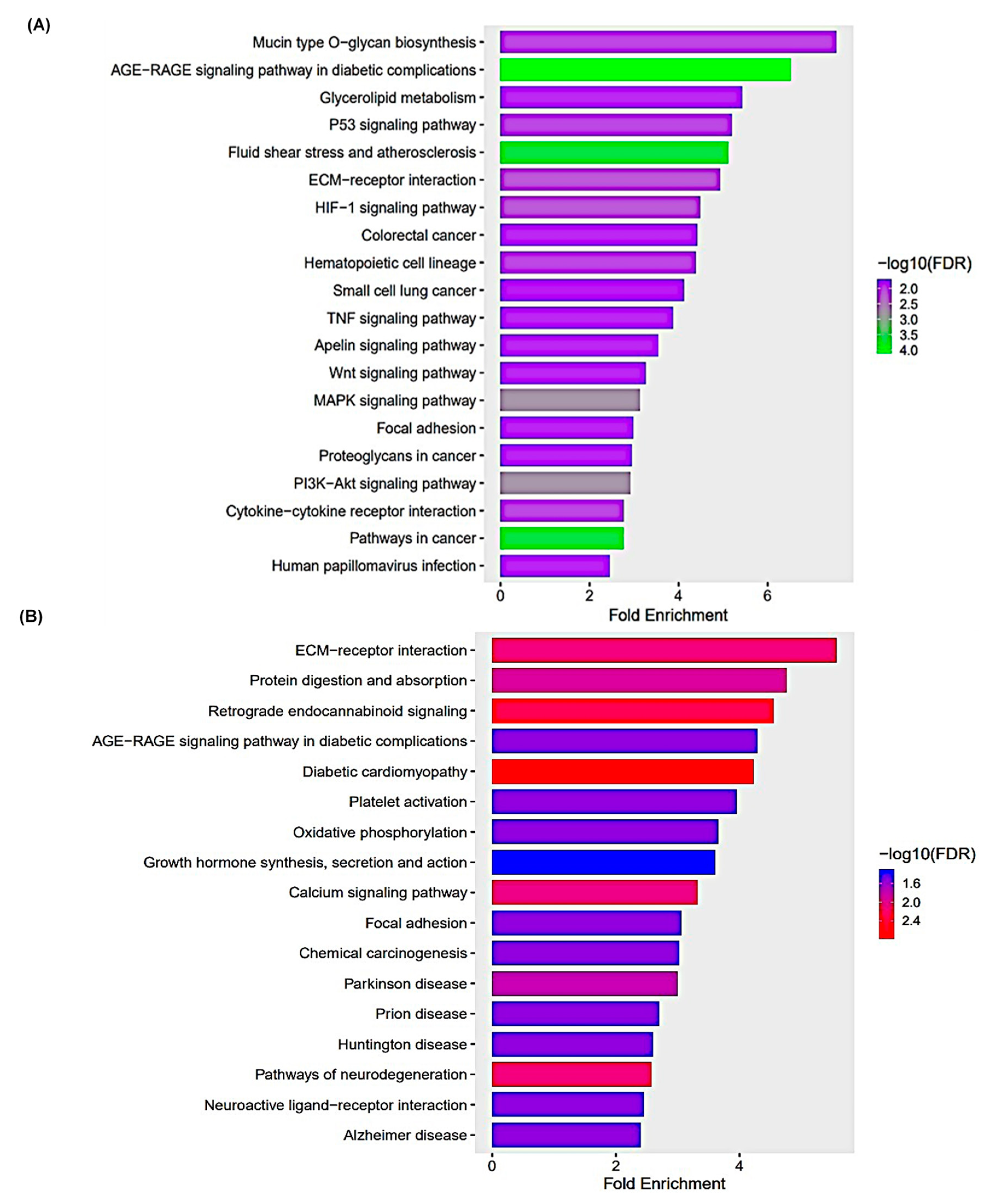
3.5. KEGG Pathway Enrichment Analysis
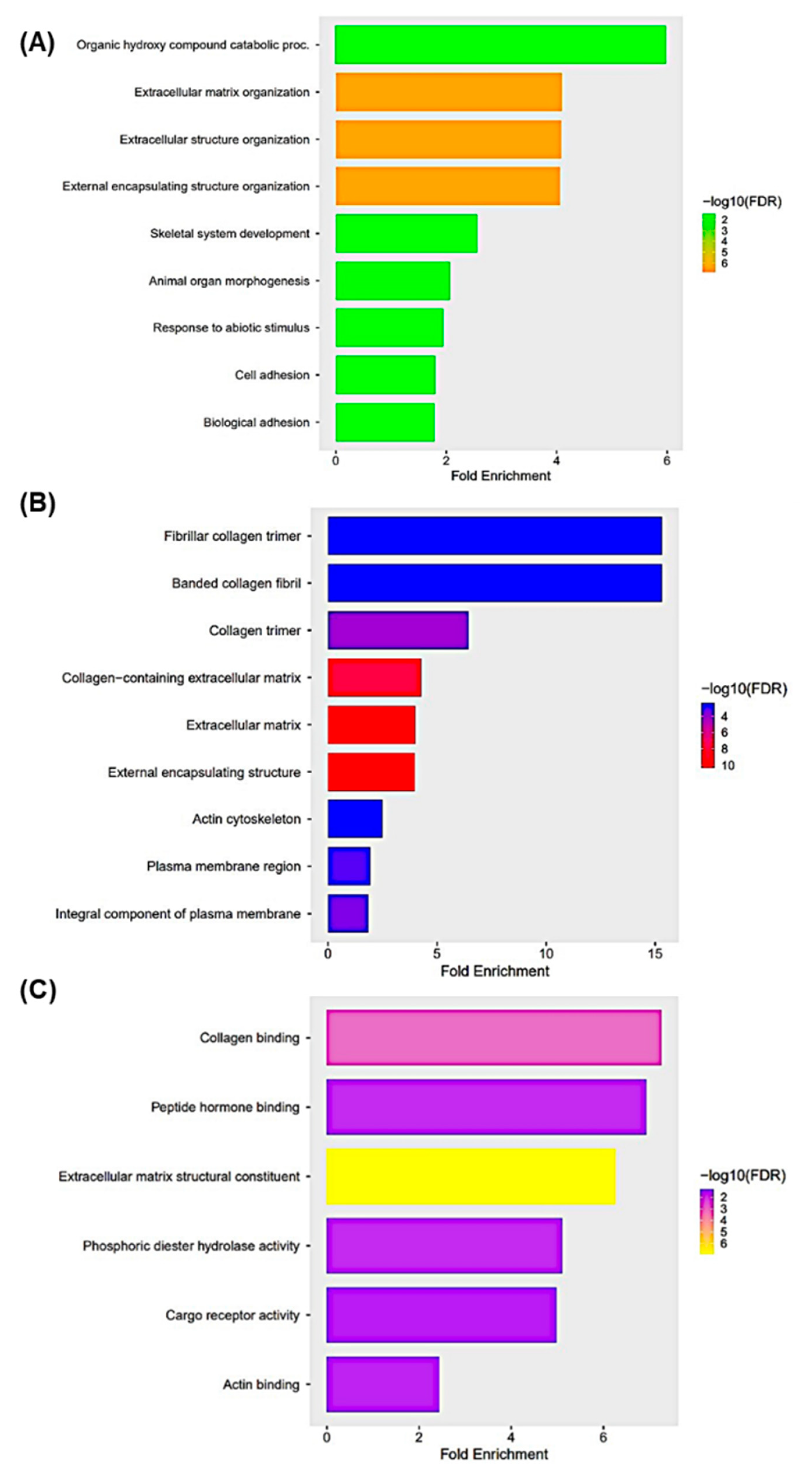
3.6. Gene Ontology Enrichment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Reardon, D.A.; Wen, P.Y. Unravelling tumour heterogeneity—implications for therapy. Nature reviews Clinical oncology 2015, 12, 69–70. [Google Scholar] [CrossRef]
- Chinot, O.L.; Wick, W.; Mason, W.; Henriksson, R.; Saran, F.; Nishikawa, R.; Carpentier, A.F.; Hoang-Xuan, K.; Kavan, P.; Cernea, D. Bevacizumab plus radiotherapy–temozolomide for newly diagnosed glioblastoma. New England Journal of Medicine 2014, 370, 709–722. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Dignam, J.J.; Armstrong, T.S.; Wefel, J.S.; Blumenthal, D.T.; Vogelbaum, M.A.; Colman, H.; Chakravarti, A.; Pugh, S.; Won, M. A randomized trial of bevacizumab for newly diagnosed glioblastoma. New England Journal of Medicine 2014, 370, 699–708. [Google Scholar] [CrossRef]
- Batash, R.; Asna, N.; Schaffer, P.; Francis, N.; Schaffer, M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Current medicinal chemistry 2017, 24, 3002–3009. [Google Scholar] [CrossRef]
- Marenco-Hillembrand, L.; Wijesekera, O.; Suarez-Meade, P.; Mampre, D.; Jackson, C.; Peterson, J.; Trifiletti, D.; Hammack, J.; Ortiz, K.; Lesser, E. Trends in glioblastoma: outcomes over time and type of intervention: a systematic evidence based analysis. Journal of neuro-oncology 2020, 147, 297–307. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro-oncology 2022, 24, v1–v95. [Google Scholar] [CrossRef]
- Davis, M.E. Glioblastoma: overview of disease and treatment. Clinical journal of oncology nursing 2016, 20, S2. [Google Scholar] [CrossRef]
- Raina, H.; Soni, G.; Jauhari, N.; Sharma, N.; Bharadvaja, N. Phytochemical importance of medicinal plants as potential sources of anticancer agents. Turkish Journal of Botany 2014, 38, 1027–1035. [Google Scholar] [CrossRef]
- Banerjee, S.; Padhye, S.; Azmi, A.; Wang, Z.; Philip, P.A.; Kucuk, O.; Sarkar, F.H.; Mohammad, R.M. Review on molecular and therapeutic potential of thymoquinone in cancer. Nutrition and cancer 2010, 62, 938–946. [Google Scholar] [CrossRef]
- Yimer, E.M.; Tuem, K.B.; Karim, A.; Ur-Rehman, N.; Anwar, F. Nigella sativa L.(black cumin): a promising natural remedy for wide range of illnesses. Evidence-Based Complementary and Alternative Medicine 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Ali, B.; Blunden, G. Pharmacological and toxicological properties of Nigella sativa. Phytotherapy Research: An international journal devoted to pharmacological and toxicological evaluation of natural product derivatives 2003, 17, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Darakhshan, S.; Pour, A.B.; Colagar, A.H.; Sisakhtnezhad, S. Thymoquinone and its therapeutic potentials. Pharmacological research 2015, 95, 138–158. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.N.; Prajapati, C.P.; Gore, P.R.; Patil, C.R.; Mahajan, U.B.; Sharma, C.; Talla, S.P.; Ojha, S.K. Therapeutic potential and pharmaceutical development of thymoquinone: a multitargeted molecule of natural origin. Frontiers in pharmacology 2017, 8, 656. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, M.A.; Alghamdi, M.S. Anticancer activity of Nigella sativa (black seed)—a review. The American journal of Chinese medicine 2011, 39, 1075–1091. [Google Scholar] [CrossRef] [PubMed]
- Shabana, A.; El-Menyar, A.; Asim, M.; Al-Azzeh, H.; Al Thani, H. Cardiovascular benefits of black cumin (Nigella sativa). Cardiovascular toxicology 2013, 13, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.C.; Kumar, A.P.; Sethi, G.; Tan, K.H.B. Thymoquinone: potential cure for inflammatory disorders and cancer. Biochemical pharmacology 2012, 83, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Tania, M.; Fu, S.; Fu, J. Thymoquinone, as an anticancer molecule: from basic research to clinical investigation. Oncotarget 2017, 8, 51907. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, H.; Parvardeh, S. Anticonvulsant effects of thymoquinone, the major constituent of Nigella sativa seeds, in mice. Phytomedicine 2004, 11, 56–64. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential analysis of count data–the DESeq2 package. Genome Biol 2014, 15, 10–1186. [Google Scholar]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bolger, A.; Giorgi, F. Trimmomatic: A flexible read trimming tool for Illumina NGS data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Ballout, F.R.; Gali-Muhtasib, H. Thymoquinone: A Potential Therapy against Cancer Stem Cells. Pharmacognosy Reviews 2020, 14. [Google Scholar] [CrossRef]
- Racoma, I.O.; Meisen, W.H.; Wang, Q.-E.; Kaur, B.; Wani, A.A. Thymoquinone inhibits autophagy and induces cathepsin-mediated, caspase-independent cell death in glioblastoma cells. PloS one 2013, 8, e72882. [Google Scholar] [CrossRef]
- Fatfat, Z.; Fatfat, M.; Gali-Muhtasib, H. Therapeutic potential of thymoquinone in combination therapy against cancer and cancer stem cells. World Journal of Clinical Oncology 2021, 12, 522. [Google Scholar] [CrossRef]
- Eid, E.E.; Alshehade, S.A.; Almaiman, A.A.; Kamran, S.; Lee, V.S.; Alshawsh, M.A. Enhancing the Anti-Leukemic Potential of Thymoquinone/Sulfobutylether-β-cyclodextrin (SBE-β-CD) Inclusion Complexes. Biomedicines 2023, 11, 1891. [Google Scholar] [CrossRef] [PubMed]
- Adilovic, A.; Mahmutović, L.; Huseinbegović, E.; Suljagic, M. Characterization of solvents and optimization of stability and solubility of bioactive compounds used in lymphoma cell culture treatments. Periodicals of Engineering and Natural Sciences 2020, 8, 2553–2563. [Google Scholar]
- Mokashi, A.A. Thymoquinone: the Evaluation of its Cytotoxic Potential, Effects on P53 Status and the Cell Cycle in Various Cancer Cell Lines. 2004.
- Lau, L.W.; Cua, R.; Keough, M.B.; Haylock-Jacobs, S.; Yong, V.W. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nature Reviews Neuroscience 2013, 14, 722–729. [Google Scholar] [CrossRef]
- Mohiuddin, E.; Wakimoto, H. Extracellular matrix in glioblastoma: Opportunities for emerging therapeutic approaches. American Journal of Cancer Research 2021, 11, 3742. [Google Scholar]
- Whatcott, C.J.; Han, H.; Posner, R.G.; Hostetter, G.; Von Hoff, D.D. Targeting the tumor microenvironment in cancer: why hyaluronidase deserves a second look. Cancer discovery 2011, 1, 291–296. [Google Scholar] [CrossRef]
- Hartmann, N.; Giese, N.A.; Giese, T.; Poschke, I.; Offringa, R.; Werner, J.; Ryschich, E. Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clinical cancer research 2014, 20, 3422–3433. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: dynamics, homeostasis and remodelling. Nature reviews Molecular cell biology 2003, 4, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, C.; Haeich, J.; Aulestia, F.J.; Kilhoffer, M.-C.; Miller, A.L.; Neant, I.; Webb, S.E.; Schaeffer, E.; Junier, M.-P.; Chneiweiss, H. Calcium signaling orchestrates glioblastoma development: Facts and conjunctures. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 2016, 1863, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Colomer, J.M.; Illario, M.; Means, A.R. The Roles of CaMKII in the Genesis of Cardiac Hypertrophy. High Blood Pressure & Cardiovascular Prevention 2007, 14, 11–19. [Google Scholar]
- Takemoto-Kimura, S.; Suzuki, K.; Horigane, S.i.; Kamijo, S.; Inoue, M.; Sakamoto, M.; Fujii, H.; Bito, H. Calmodulin kinases: essential regulators in health and disease. Journal of neurochemistry 2017, 141, 808–818. [Google Scholar] [CrossRef]
- Wang, H.; Zeng, Z.; Yi, R.; Luo, J.; Chen, J.; Lou, J. MicroRNA-3200-3p targeting CAMK2A modulates the proliferation and metastasis of glioma in vitro. Bioengineered 2022, 13, 7785–7797. [Google Scholar] [CrossRef]
- Yu, T.-J.; Liu, Y.-Y.; Li, X.-G.; Lian, B.; Lu, X.-X.; Jin, X.; Shao, Z.-M.; Hu, X.; Di, G.-H.; Jiang, Y.-Z. PDSS1-mediated activation of CAMK2A-STAT3 signaling promotes metastasis in triple-negative breast cancer. Cancer Research 2021, 81, 5491–5505. [Google Scholar] [CrossRef]
- Guan, L.; Yuan, S.; Ma, J.; Liu, H.; Huang, L.; Zhang, F. Neurokinin-1 receptor is highly expressed in cervical cancer and its antagonist induces cervical cancer cell apoptosis. European Journal of Histochemistry 2023, 67. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, X.-r.; Li, H.-y.; Zhao, Z.-j.; Liao, Y.-w.; Wang, X.-y.; Su, J.; Sang, S.-s.; Liu, Q. Anti-cancer effect of metabotropic glutamate receptor 1 inhibition in human glioma U87 cells: involvement of PI3K/Akt/mTOR pathway. Cellular Physiology and Biochemistry 2015, 35, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Yi, L.; Liu, P.; Abeysekera, I.R.; Hai, L.; Li, T.; Tao, Z.; Ma, H.; Xie, Y.; Huang, Y. Tumour cell dormancy as a contributor to the reduced survival of GBM patients who received standard therapy. Oncology Reports 2018, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Moncayo, G.; Grzmil, M.; Smirnova, T.; Zmarz, P.; Huber, R.M.; Hynx, D.; Kohler, H.; Wang, Y.; Hotz, H.-R.; Hynes, N.E. SYK inhibition blocks proliferation and migration of glioma cells and modifies the tumor microenvironment. Neuro-oncology 2018, 20, 621–631. [Google Scholar] [CrossRef]
- Pyfrom, S.C.; Quinn, C.C.; Dorando, H.K.; Luo, H.; Payton, J.E. BCALM (AC099524. 1) Is a Human B Lymphocyte–Specific Long Noncoding RNA That Modulates B Cell Receptor–Mediated Calcium Signaling. The Journal of Immunology 2020, 205, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Martin, J.; Terry, A.; Couronne, O.; Grimwood, J.; Gordon, L.A.; Scott, D.; Xie, G.; Huang, W.; Hellsten, U. The complete sequence of human chromosome 5. Nature 2004, 431. [Google Scholar] [CrossRef] [PubMed]
- Vanlandewijck, M.; Lebouvier, T.; Andaloussi Mäe, M.; Nahar, K.; Hornemann, S.; Kenkel, D.; Cunha, S.I.; Lennartsson, J.; Boss, A.; Heldin, C.-H. Functional characterization of germline mutations in PDGFB and PDGFRB in primary familial brain calcification. PloS one 2015, 10, e0143407. [Google Scholar] [CrossRef] [PubMed]
- Bazdar, D.A.; Kalinowska, M.; Panigrahi, S.; Sieg, S.F. Recycled il-7 can be delivered to neighboring t cells. The Journal of Immunology 2015, 194, 4698–4704. [Google Scholar] [CrossRef]
- Rochman, Y.; Kashyap, M.; Robinson, G.W.; Sakamoto, K.; Gomez-Rodriguez, J.; Wagner, K.-U.; Leonard, W.J. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7–induced signaling. Proceedings of the National Academy of Sciences 2010, 107, 19455–19460. [Google Scholar] [CrossRef]
- Gao, S.; Jin, L.; Liu, G.; Wang, P.; Sun, Z.; Cao, Y.; Shi, H.; Liu, X.; Shi, Q.; Zhou, X. Overexpression of RASD1 inhibits glioma cell migration/invasion and inactivates the AKT/mTOR signaling pathway. Scientific reports 2017, 7, 3202. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Słocińska, M.; Barciszewska, A.-M.; Nowak, S.; Baer-Dubowska, W. Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival. Journal of Applied Genetics 2016, 57, 189–197. [Google Scholar] [CrossRef]
- Han, J.; Goldstein, L.A.; Hou, W.; Rabinowich, H. Functional linkage between NOXA and Bim in mitochondrial apoptotic events. Journal of Biological Chemistry 2007, 282, 16223–16231. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, L.; Hwang, P.M.; Kinzler, K.W.; Vogelstein, B. PUMA induces the rapid apoptosis of colorectal cancer cells. Molecular cell 2001, 7, 673–682. [Google Scholar] [CrossRef]
- Zhang, H.; Guttikonda, S.; Roberts, L.; Uziel, T.; Semizarov, D.; Elmore, S.; Leverson, J.; Lam, L. Mcl-1 is critical for survival in a subgroup of non-small-cell lung cancer cell lines. Oncogene 2011, 30, 1963–1968. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, X.; Su, L. Parthenolide induces apoptosis via TNFRSF10B and PMAIP1 pathways in human lung cancer cells. Journal of experimental & clinical cancer research 2014, 33, 1–11. [Google Scholar]
- Salvador, J.M.; Brown-Clay, J.D.; Fornace Jr, A.J. Gadd45 in stress signaling, cell cycle control, and apoptosis. Gadd45 stress sensor genes 2013, 1–19. [Google Scholar]
- E Tamura, R.; F de Vasconcellos, J.; Sarkar, D.; A Libermann, T.; B Fisher, P.; F Zerbini, L. GADD45 proteins: central players in tumorigenesis. Current molecular medicine 2012, 12, 634–651. [Google Scholar] [CrossRef]
- Jiang, T.; Soprano, D.R.; Soprano, K.J. GADD45A is a mediator of CD437 induced apoptosis in ovarian carcinoma cells. Journal of cellular physiology 2007, 212, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Su, M.-Q.; Zhou, Y.-R.; Rao, X.; Yang, H.; Zhuang, X.H.; Ke, X.-J.; Peng, G.-Y.; Zhou, C.-L.; Shen, B.-Y.; Dou, J. Baicalein induces the apoptosis of HCT116 human colon cancer cells via the upregulation of DEPP/Gadd45a and activation of MAPKs. International journal of oncology 2018, 53, 750–760. [Google Scholar] [PubMed]
- Yin, F.; Bruemmer, D.; Blaschke, F.; Hsueh, W.A.; Law, R.E.; Van Herle, A.J. Signaling pathways involved in induction of GADD45 gene expression and apoptosis by troglitazone in human MCF-7 breast carcinoma cells. Oncogene 2004, 23, 4614–4623. [Google Scholar] [CrossRef]
- Ueda, K.; Arakawa, H.; Nakamura, Y. Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene 2003, 22, 5586–5591. [Google Scholar] [CrossRef]
- Celik-Selvi, B.E.; Stütz, A.; Mayer, C.-E.; Salhi, J.; Siegwart, G.; Sutterlüty, H. Sprouty3 and sprouty4, two members of a family known to inhibit fgf-mediated signaling, exert opposing roles on proliferation and migration of glioblastoma-derived cells. Cells 2019, 8, 808. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Sun, J.; Du, K.; Liang, N. Sprouty 4 suppresses glioblastoma invasion by inhibiting ERK phosphorylation and ETS-1-induced matrix metalloproteinase-9. Journal of neurosurgical sciences 2020. [Google Scholar] [CrossRef] [PubMed]
- Foltz, G.; Ryu, G.-Y.; Yoon, J.-G.; Nelson, T.; Fahey, J.; Frakes, A.; Lee, H.; Field, L.; Zander, K.; Sibenaller, Z. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumor suppressor genes in malignant glioma. Cancer research 2006, 66, 6665–6674. [Google Scholar] [CrossRef]
- Nie, E.; Zhang, X.; Xie, S.; Shi, Q.; Hu, J.; Meng, Q.; Zhou, X.; Yu, R. β-Catenin is involved in Bex2 down-regulation induced glioma cell invasion/migration inhibition. Biochemical and biophysical research communications 2015, 456, 494–499. [Google Scholar] [CrossRef]
- Zhang, H.; Qi, Y.; Geng, D.; Shi, Y.; Wang, X.; Yu, R.; Zhou, X. Expression profile and clinical significance of Wnt signaling in human gliomas. Oncology letters 2018, 15, 610–617. [Google Scholar] [CrossRef]
- Gonçalves, C.S.; de Castro, J.V.; Pojo, M.; Martins, E.P.; Queirós, S.; Chautard, E.; Taipa, R.; Pires, M.M.; Pinto, A.A.; Pardal, F. WNT6 is a novel oncogenic prognostic biomarker in human glioblastoma. Theranostics 2018, 8, 4805. [Google Scholar] [CrossRef]
- Liu, X.; Shan, W.; Li, T.; Gao, X.; Kong, F.; You, H.; Kong, D.; Qiao, S.; Tang, R. Cellular retinol binding protein-1 inhibits cancer stemness via upregulating WIF1 to suppress Wnt/β-catenin pathway in hepatocellular carcinoma. BMC cancer 2021, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Han, X.; Huang, H.; Yu, X.; Fang, J.; Zhao, J.; Prayson, R.A.; Bao, S.; Yu, J.S. Sema3C signaling is an alternative activator of the canonical WNT pathway in glioblastoma. nature communications 2023, 14, 2262. [Google Scholar] [CrossRef]
- Chen, P.-H.; Shen, W.-L.; Shih, C.-M.; Ho, K.-H.; Cheng, C.-H.; Lin, C.-W.; Lee, C.-C.; Liu, A.-J.; Chen, K.-C. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology 2017, 116, 300–314. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.; Li, J.; Yao, Z.; Ma, X.; Ma, J.-w. Delta and Notch-like epidermal growth factor-related receptor suppresses human glioma growth by inhibiting oncogene TOR4A. Journal of Cancer Research and Therapeutics 2022, 18, 1372–1379. [Google Scholar] [PubMed]
- Sun, P.; Xia, S.; Lal, B.; Eberhart, C.G.; Quinones-Hinojosa, A.; Maciaczyk, J.; Matsui, W.; DiMeco, F.; Piccirillo, S.M.; Vescovi, A.L. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem cells 2009, 27, 1473–1486. [Google Scholar] [CrossRef] [PubMed]
- Bountali, A.; Tonge, D.P.; Mourtada-Maarabouni, M. RNA sequencing reveals a key role for the long non-coding RNA MIAT in regulating neuroblastoma and glioblastoma cell fate. International journal of biological macromolecules 2019, 130, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wu, J.; Wang, H.; Yang, Y.; Zheng, Z.; Ni, B.; Wang, X.; Peng, Y.; Li, Y. LMO1 Plays an Oncogenic Role in Human Glioma Associated With NF-kB Pathway. Frontiers in oncology 2022, 12, 770299. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Song, L.; Chang, J.; Cao, P.; Liu, Q. AEBP1 Promotes Glioblastoma Progression and Activates the Classical NF-κB Pathway. Behavioural Neurology 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.-c.; Yang, M.; Zhu, L.; Zhou, Q.; Li, X.; Chen, Z.; Zou, C. On this page. Behavioural Neurology 2020, 2, 3. [Google Scholar]
- Verreault, M.; Segoviano Vilchis, I.; Rosenberg, S.; Lemaire, N.; Schmitt, C.; Guehennec, J.; Royer-Perron, L.; Thomas, J.l.; Lam, T.T.; Dingli, F. Identification of growth hormone receptor as a relevant target for precision medicine in low-EGFR expressing glioblastoma. Clinical and Translational Medicine 2022, 12, e939. [Google Scholar] [CrossRef]
- Echizen, K.; Nakada, M.; Hayashi, T.; Sabit, H.; Furuta, T.; Nakai, M.; Koyama-Nasu, R.; Nishimura, Y.; Taniue, K.; Morishita, Y. PCDH10 is required for the tumorigenicity of glioblastoma cells. Biochemical and biophysical research communications 2014, 444, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Fukai, J.; Yokote, H.; Yamanaka, R.; Arao, T.; Nishio, K.; Itakura, T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Molecular cancer therapeutics 2008, 7, 2768–2778. [Google Scholar] [CrossRef]
- Gao, Y.F.; Zhu, T.; Chen, J.; Liu, L.; Ouyang, R. Knockdown of collagen α-1 (III) inhibits glioma cell proliferation and migration and is regulated by miR128-3p. Oncology letters 2018, 16, 1917–1923. [Google Scholar] [CrossRef]
- Held-Feindt, J.; Paredes, E.B.; Blömer, U.; Seidenbecher, C.; Stark, A.M.; Mehdorn, H.M.; Mentlein, R. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. International journal of cancer 2006, 118, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wen-Liang, L.; Li, F.; Feng, G.; Yong-Jie, M. Slit2/Robo1 signaling in glioma migration and invasion. Neuroscience bulletin 2010, 26, 474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ohkawa, Y.; Yamamoto, S.; Momota, H.; Kato, A.; Kaneko, K.; Natsume, A.; Farhana, Y.; Ohmi, Y.; Okajima, T. St8sia1-deficiency in mice alters tumor environments of gliomas, leading to reduced disease severity. Nagoya Journal of Medical Science 2021, 83, 535. [Google Scholar] [PubMed]
- Plata-Bello, J.; Fariña-Jerónimo, H.; Betancor, I.; Salido, E. High expression of FOXP2 is associated with worse prognosis in glioblastoma. World Neurosurgery 2021, 150, e253–e278. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-Z.; Zhang, J.-H.; Li, J.-B.; Yuan, F.; Tong, L.-Q.; Wang, X.-Y.; Chen, L.-L.; Fan, X.-G.; Zhang, Y.-M.; Ren, X. MXRA5 is a novel immune-related biomarker that predicts poor prognosis in glioma. Disease Markers 2021, 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zang, J.; Zhang, D.; Sun, Z.; Qiu, B.; Wang, X. KDM2B overexpression correlates with poor prognosis and regulates glioma cell growth. OncoTargets and therapy 2018, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.a.; Yang, Q. Identification of core genes and screening of potential targets in glioblastoma multiforme by integrated bioinformatic analysis. Frontiers in Oncology 2021, 10, 615976. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Qi, P.; Xiang, W.; Yanhui, L.; Yu, L.; Qing, M. Multi-omics analysis reveals novel subtypes and driver genes in glioblastoma. Frontiers in Genetics 2020, 11, 565341. [Google Scholar] [CrossRef] [PubMed]
- Zi, J.; Wang, W.; Sun, M.; Mei, W.; Li, S.; Li, B.; Xiao, Y.; Fei, Z.; Zhang, R.; Yu, M. A high expression of MTERF3 correlates with tumor progression and predicts poor outcomes in patients with brain glioma. International Journal of Clinical and Experimental Pathology 2019, 12, 1909. [Google Scholar]
- Zhang, H.; Ma, H.; Zhang, W.; Duan, D.; Zhu, G.; Cao, W.; Liu, B. Increased expression of Sema3C indicates a poor prognosis and is regulated by miR-142-5p in glioma. Biological and Pharmaceutical Bulletin 2020, 43, 639–648. [Google Scholar] [CrossRef]
| Sample Name | No. of Raw reads | Sequence length (bp) | No. of Filtered reads | GC % | No. of Uniquely mapped reads | Mapping percentage (uniquely mapped) |
|---|---|---|---|---|---|---|
| Control_1 | 26712260 | 75 | 23196526 | 49 | 22218462 | 95.8 |
| Control_2 | 31107086 | 75 | 26775248 | 49 | 25529068 | 95.3 |
| Control_3 | 24403444 | 75 | 20604284 | 48 | 19649734 | 95.4 |
| Control_4 | 29783142 | 75 | 25852334 | 49 | 24565046 | 95.0 |
| 2.5µM_1 | 31071382 | 75 | 27008588 | 49 | 25752932 | 95.4 |
| 2.5µM_2 | 30202594 | 75 | 25898660 | 48 | 24596670 | 95.0 |
| 2.5µM_3 | 30964912 | 75 | 26766406 | 49 | 25440878 | 95.0 |
| 2.5µM_4 | 32187674 | 75 | 27893292 | 48 | 26696168 | 95.7 |
| 5µM_1 | 31265636 | 75 | 27148108 | 48 | 25957546 | 95.6 |
| 5µM_2 | 29513348 | 75 | 25496706 | 49 | 24252558 | 95.1 |
| 5µM_3 | 23419378 | 75 | 18875660 | 48 | 18043398 | 95.6 |
| 5µM_4 | 32224110 | 75 | 28173704 | 49 | 26907134 | 95.5 |
| Total | 352854966 | 303689516 | 289609594 |
| Sample Name | No. of Raw reads | Sequence length (bp) | No. of Filtered reads | GC % | No. of Uniquely mapped reads | Mapping percentage (uniquely mapped) |
|---|---|---|---|---|---|---|
| Control_1 | 25130818 | 150 | 24290286 | 49 | 23565022 | 97.0 |
| Control_2 | 25777505 | 150 | 24913016 | 49 | 24114402 | 96.8 |
| Control_3 | 33410551 | 150 | 32234977 | 50 | 31147688 | 96.6 |
| Control_4 | 32672018 | 150 | 31528490 | 51 | 30598289 | 97.0 |
| 25_1 | 26172491 | 150 | 25239490 | 51 | 24503413 | 97.1 |
| 25_2 | 22745394 | 150 | 21912988 | 51 | 21237324 | 96.9 |
| 25_3 | 27897817 | 150 | 26923593 | 51 | 26150682 | 97.1 |
| 25_4 | 27645344 | 150 | 26669748 | 51 | 25939455 | 97.3 |
| 50_1 | 34224141 | 150 | 32984120 | 51 | 31931537 | 96.8 |
| 50_2 | 26024221 | 150 | 25093850 | 51 | 24287158 | 96.8 |
| 50_3 | 25164754 | 150 | 24237707 | 51 | 23445769 | 96.7 |
| 50_4 | 26134658 | 150 | 25166726 | 49 | 24347204 | 96.7 |
| Total | 332999712 | 321194991 |
| Pathways | Fold Enrichment | No. of Genes | Pathway Genes | Genes |
|---|---|---|---|---|
| Beta-Alanine metabolism | 7.802 | 4 | 31 | ALDH3B1 UPB1 CSAD CARNS1 |
| Mineral absorption | 6.047 | 6 | 60 | ATP1A2 ATP2B3 CYBRD1 ATP7B ATP1B2 SLC34A3 |
| ECM-receptor interaction | 5.497 | 8 | 88 | COL9A3 ITGB6 COL1A2 THBS3 COL4A5 LAMA2 COL6A6 ITGA1 |
| Arrhythmogenic right ventricular cardiomyopathy | 5.497 | 7 | 77 | PKP2 ITGB6 CACNA1D SGCD LAMA2 DMD ITGA1 |
| Hypertrophic cardiomyopathy | 5.375 | 8 | 90 | TGFB2 TNNC1 ITGB6 CACNA1D SGCD LAMA2 DMD ITGA1 |
| Protein digestion and absorption | 5.284 | 9 | 103 | ATP1A2 COL11A1 COL9A3 ATP1B2 COL5A1 COL1A2 COL4A5 MME COL6A6 |
| Dilated cardiomyopathy | 5.039 | 8 | 96 | TGFB2 TNNC1 ITGB6 CACNA1D SGCD LAMA2 DMD ITGA1 |
| Cardiac muscle contraction | 4.865 | 7 | 87 | ATP1A2 TNNC1 ATP1B2 CACNA1D COX6B2 MT-CYB MT-CO1 |
| Focal adhesion | 4.233 | 14 | 200 | MYLK COL9A3 PDGFB MYL10 PDGFRB ITGB6 COL1A2 ILK THBS3 PDGFD COL4A5 LAMA2 COL6A6 ITGA1 |
| Hippo signaling pathway | 3.466 | 9 | 157 | TGFB2 WNT5A GDF5 WNT2B GDF7 GDF6 PPP2R2B TGFBR2 FZD4 |
| PI3K-Akt signaling pathway | 3.075 | 18 | 354 | LPAR2 FGF10 COL9A3 PDGFB IL2RB IL7 PDGFRB ITGB6 PPP2R2B COL1A2 THBS3 PDGFD GNG7 ERBB4 COL4A5 LAMA2 COL6A6 ITGA1 |
| Calcium signaling pathway | 3.023 | 12 | 240 | MYLK ATP2B3 FGF10 PDGFB HTR2A P2RX1 PDGFRB HRH2 TNNC1 CACNA1D PDGFD ERBB4 |
| Human papillomavirus infection | 2.740 | 15 | 331 | COL9A3 PDGFRB WNT5A ITGB6 WNT2B PPP2R2B COL1A2 THBS3 STAT2 FZD4 MAML2 COL4A5 LAMA2 COL6A6 ITGA1 |
| Pathway | Fold Enrichment | No. Genes | Pathway Genes | Genes |
|---|---|---|---|---|
| Mucin type O-glycan biosynthesis | 7.538 | 5 | 36 | ST3GAL1 GALNT12 GALNT6 GCNT3 GCNT1 |
| AGE-RAGE signaling pathway in diabetic complications | 6.513 | 12 | 100 | EDN1 SERPINE1 F3 CCND1 VEGFA IL1A EGR1 MAPK13 NOS3 PRKCE JUN THBD |
| Glycerolipid metabolism | 5.428 | 6 | 60 | DGKG MGLL LIPGGPAT3 PLPP4 PNLIPRP3 |
| P53 signaling pathway | 5.205 | 7 | 73 | CD82 BBC3 CCND1 SFN SERPINE1 PMAIP1 GADD45A |
| Fluid shear stress and atherosclerosis | 5.113 | 13 | 138 | EDN1 HMOX1 PLAT VEGFA IL1A SDC1 ARHGEF2 KLF2 MAPK13 NOS3 FOS JUN THBD |
| ECM-receptor interaction | 4.934 | 8 | 88 | CD44 LAMC2 ITGA6 SDC1 TNN COL6A3 ITGA2 LAMB3 |
| HIF-1 signaling pathway | 4.482 | 9 | 109 | EDN1 HMOX1 ANGPT4 SERPINE1 VEGFA HKDC1 HK1 NOS3 EIF4EBP1 |
| Colorectal cancer | 4.418 | 7 | 86 | BBC3 CCND1 GADD45A PMAIP1 TGFA FOS JUN |
| Hematopoietic cell lineage | 4.386 | 8 | 99 | CD44 KITLG IL4R ITGA6 IL11 IL1A ITGA2 CD55 |
| Small cell lung cancer | 4.130 | 7 | 92 | TRAF1 LAMC2 ITGA6 CCND1 GADD45A ITGA2 LAMB3 |
| TNF signaling pathway | 3.877 | 8 | 112 | TRAF1 EDN1 ATF4 JUN LIFMAPK13 CXCL3 FOS |
| Apelin signaling pathway | 3.540 | 9 | 138 | PLAT SERPINE1 PRKCE CCND1 NOS3 GNGT2 EGR1 SPHK1 KLF2 |
| Wnt signaling pathway | 3.270 | 10 | 166 | NFATC2 SFRP1 DVL1 CCND1 WNT5B LGR6 TLE3 FOSL1 JUN WNT7B |
| MAPK signaling pathway | 3.138 | 17 | 294 | KITLG ANGPT4 VEGFA DUSP4 GADD45A PGF IL1A ATF4 JUN DUSP5 FGF5 DUSP6 EPHA2 RASGRP3 MAPK13 TGFA FOS |
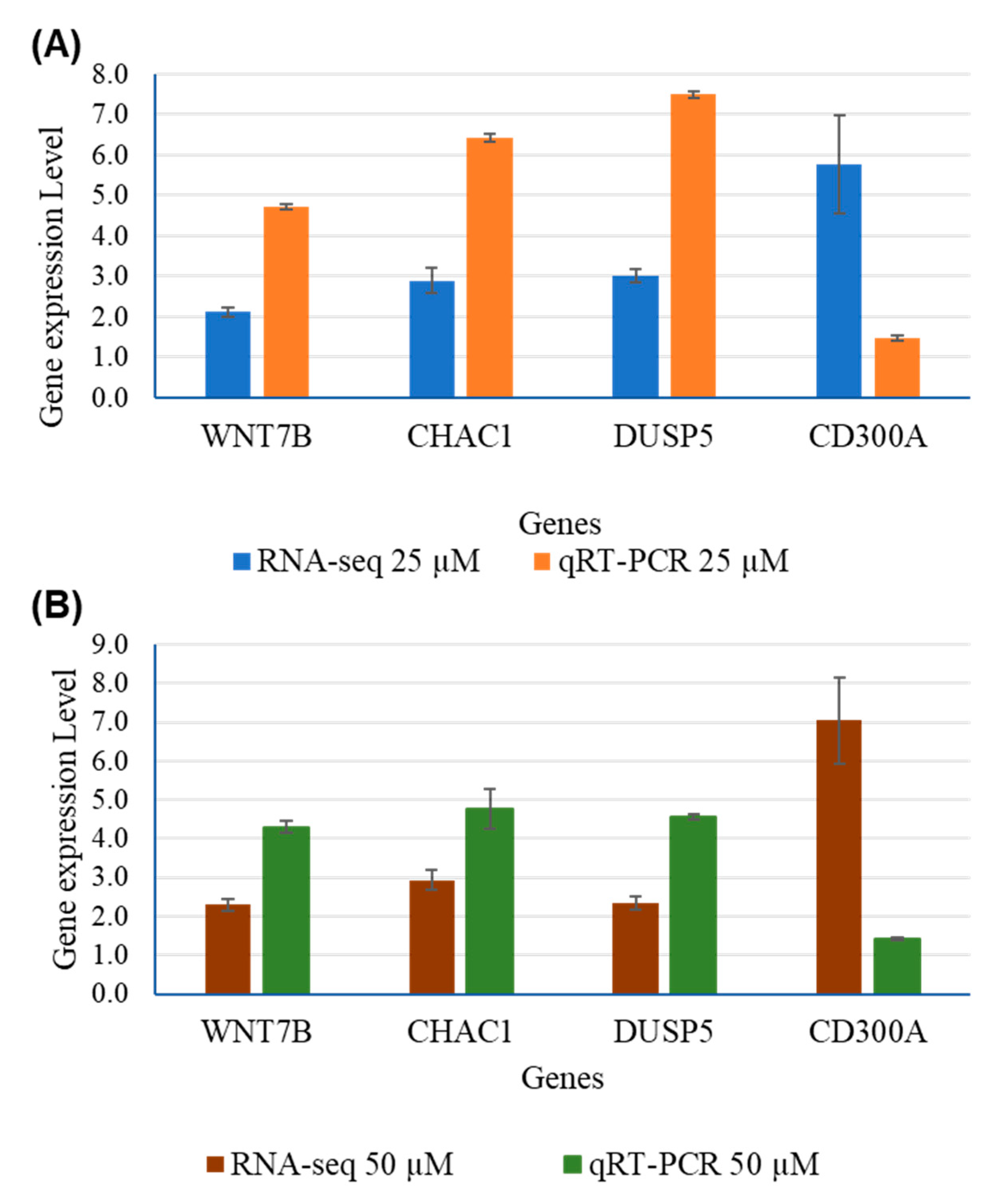
| Genes | RNA-Seq | qRT-PCR | ||||||
|---|---|---|---|---|---|---|---|---|
| 25 µM | 50 µM | 25 µM | 50 µM | |||||
| Fold Change | P-adj | Fold Change | P-adj | Fold Change | P-adj | Fold Change | P-adj | |
| WNT7B | 2.123 | 4.72E-76 | 2.2909 | 5.01E-48 | 4.7148 | 0.0003 | 4.2974 | 0.0005 |
| CHAC1 | 2.897 | 9.1E-19 | 2.9376 | 9.89E-31 | 6.4202 | 0.0264 | 4.7567 | 0.0637 |
| DUSP5 | 3.006 | 9.31E-124 | 2.3411 | 1.55E-47 | 7.4982 | 0.000006 | 4.5518 | 0.00022 |
| CD300A | 5.776 | 1,11E-05 | 7.0351 | 4.28E-09 | 1.4704 | 0.000281 | 1.4261 | 0.00049 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





