Submitted:
10 January 2024
Posted:
11 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
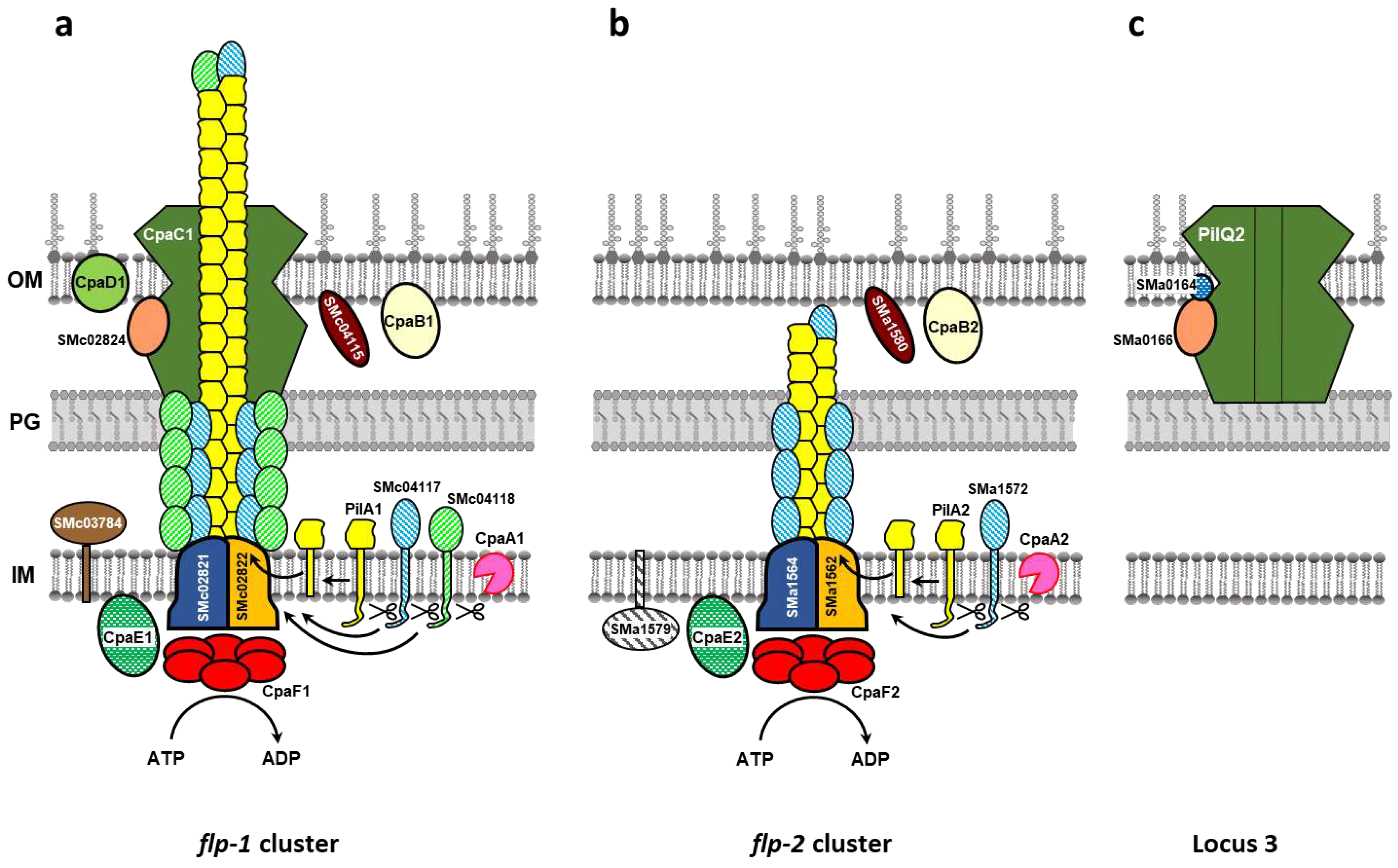
2.1. In silico analyses of Type IVc Pili (T4cP) genes of S. meliloti GR4
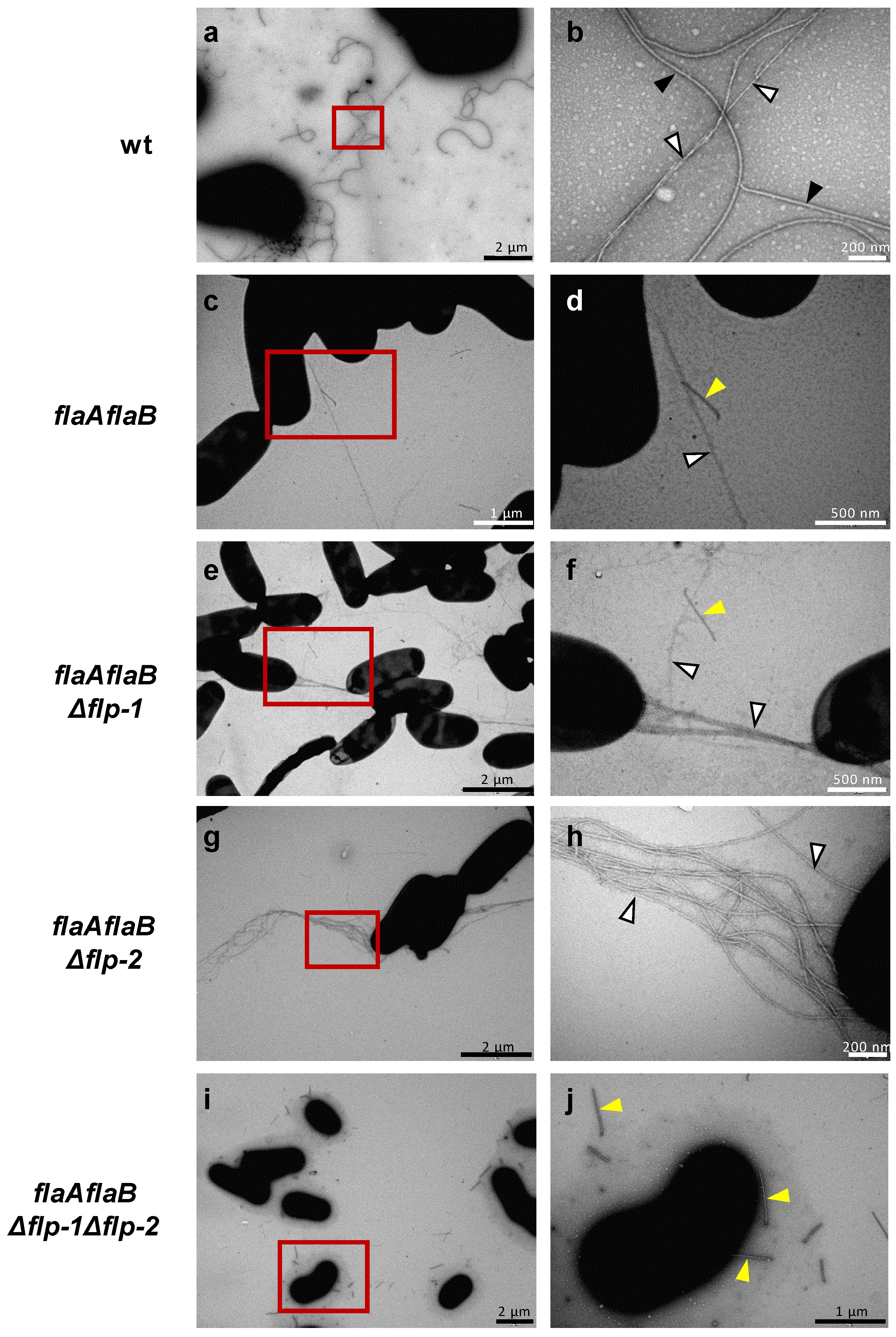
2.2. Both flp-1 and flp-2 clusters contribute to T4cP production in S. meliloti GR4
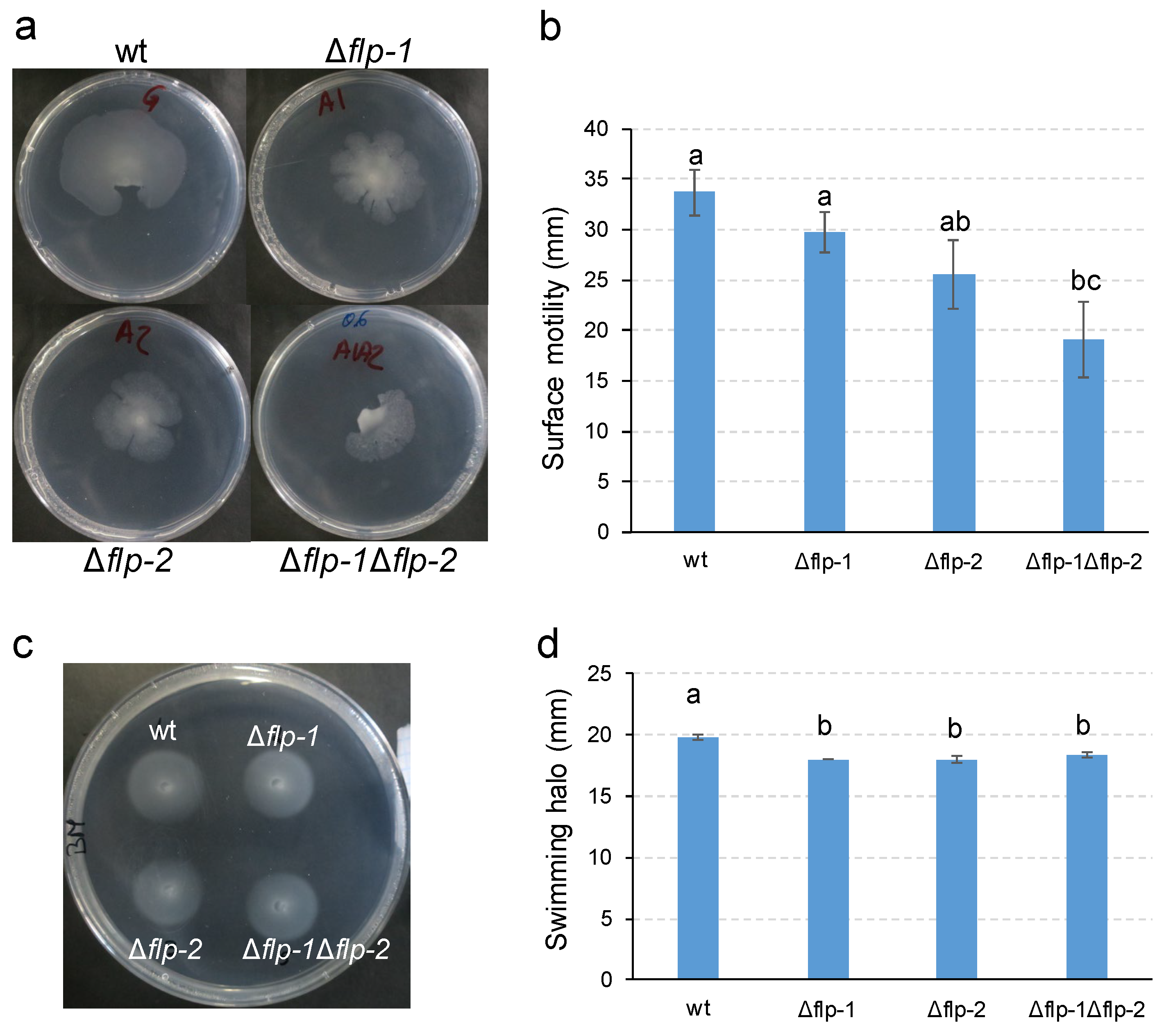
2.3. Type IVc Pili (T4cP) facilitate surface spreading of S. meliloti GR4
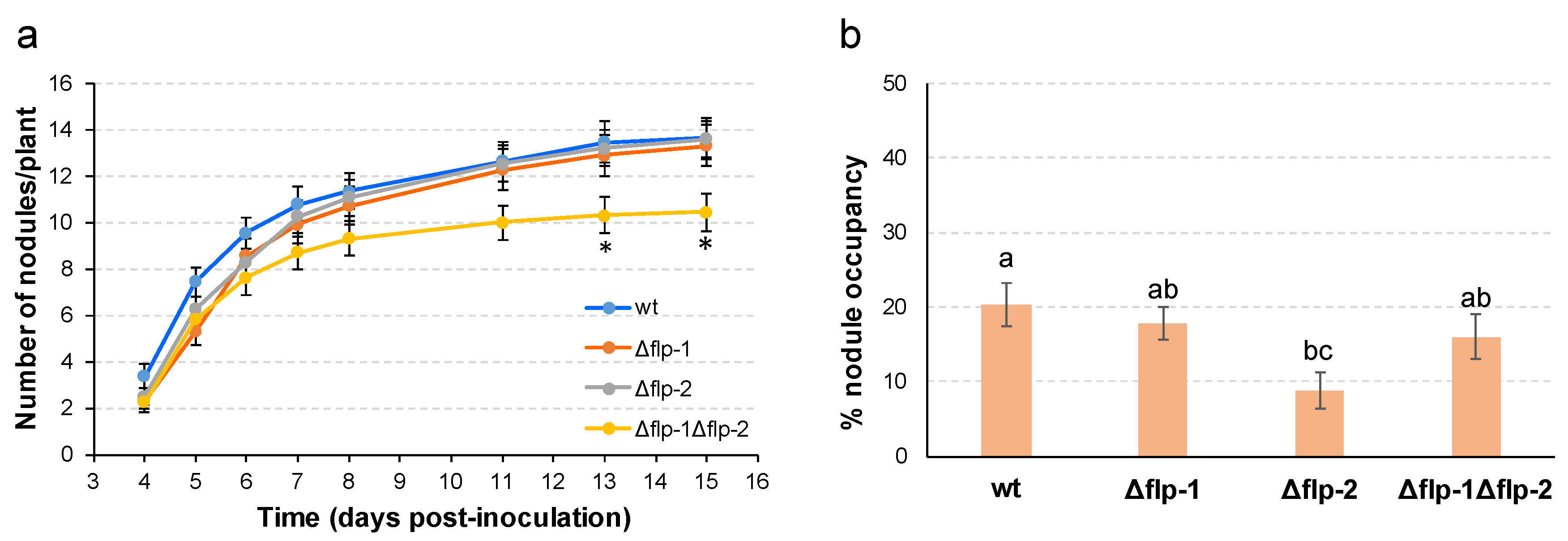
2.4. The interaction of S. meliloti GR4 with alfalfa is influenced by T4cP
3. Discussion
4. Materials and Methods
4.1. Bacterial strains, plasmids and growth conditions
4.2. In silico analyses
4.3. Construction of S. meliloti mutant strains
4.4. Transmission electron microscopy (TEM)
4.5. Motility assays
4.6. Plant assays
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 2018, 16, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Cangioli, L.; Vaccaro, F.; Fini, M.; Mengoni, A.; Fagorzi, C. Scent of a Symbiont: The Personalized Genetic Relationships of Rhizobium-Plant Interaction. Int J Mol Sci 2022, 23, 3358. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.M.; Poole, P.S. Mechanisms of bacterial attachment to roots. FEMS Microbiol Rev 2018, 42, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering bacterial mechanisms of root colonization. Environ Microbiol Rep 2021, 13, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Ames, P.; Bergman, K. Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J Bacteriol 1981, 148, 728–908. [Google Scholar] [CrossRef]
- Soby, S.; Bergman, K. Motility and Chemotaxis of Rhizobium meliloti in Soil. Appl Environ Microbiol 1983, 46, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Wrobel-Boerner, E.; Bauer, W.D. Growth and Movement of Spot Inoculated Rhizobium meliloti on the Root Surface of Alfalfa. Plant Physiol 1992, 98, 1181–1189. [Google Scholar] [CrossRef]
- Amaya-Gómez, C.V.; Hirsch, A.M.; Soto, M.J. Biofilm formation assessment in Sinorhizobium meliloti reveals interlinked control with surface motility. BMC Microbiol 2015, 15, 58. [Google Scholar] [CrossRef]
- Bernabéu-Roda, L.; Calatrava-Morales, N.; Cuéllar, V.; Soto, M. Characterization of surface motility in Sinorhizobium meliloti: regulation and role in symbiosis. Symbiosis 2015, 1–12. [Google Scholar] [CrossRef]
- Aroney, S.T.N.; Poole, P.S.; Sanchez-Canizares, C. Rhizobial Chemotaxis and Motility Systems at Work in the Soil. Front Plant Sci 2021, 12, 725338. [Google Scholar] [CrossRef]
- Soto, M.J.; Fernández-Pascual, M.; Sanjuán, J.; Olivares, J. A fadD mutant of Sinorhizobium meliloti shows multicellular swarming migration and is impaired in nodulation efficiency on alfalfa roots. Mol. Microbiol 2002, 43, 371–382. [Google Scholar] [CrossRef]
- Nogales, J.; Bernabéu-Roda, L.; Cuéllar, V.; Soto, M.J. ExpR is not required for swarming but promotes sliding in Sinorhizobium meliloti. J Bacteriol 2012, 194, 2027–2035. [Google Scholar] [CrossRef]
- Dilanji, G.E.; Teplitski, M.; Hagen, S.J. Entropy-driven motility of Sinorhizobium meliloti on a semi-solid surface. Proc Biol Sci 2014, 281, 20132575. [Google Scholar] [CrossRef]
- Ji, Y.Y.; Zhang, B.; Zhang, P.; Chen, L.C.; Si, Y.W.; Wan, X.Y.; Li, C.; Wang, R.H.; Tian, Y.; Zhang, Z.; et al. Rhizobial migration toward roots mediated by FadL-ExoFQP modulation of extracellular long-chain AHLs. ISME J 2023, 17, 417–431. [Google Scholar] [CrossRef]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat Rev Microbiol 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Mattingly, A.E.; Weaver, A.A.; Dimkovikj, A.; Shrout, J.D. Assessing Travel Conditions: Environmental and Host Influences On Bacterial Surface Motility. J Bacteriol 2018, 200, e00014–00018. [Google Scholar] [CrossRef]
- Wadhwa, N.; Berg, H.C. Bacterial motility: machinery and mechanisms. Nat Rev Microbiol 2022, 20, 161–173. [Google Scholar] [CrossRef]
- Daniels, R.; Reynaert, S.; Hoekstra, H.; Verreth, C.; Janssens, J.; Braeken, K.; Fauvart, M.; Beullens, S.; Heusdens, C.; Lambrichts, I.; et al. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci U S A 2006, 103, 14965–14970. [Google Scholar] [CrossRef]
- Tambalo, D.D.; Yost, C.K.; Hynes, M.F. Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol Lett 2010, 307, 165–174. [Google Scholar] [CrossRef]
- Covelli, J.M.; Althabegoiti, M.J.; López, M.F.; Lodeiro, A.R. Swarming motility in Bradyrhizobium japonicum. Res Microbiol 2013, 164, 136–144. [Google Scholar] [CrossRef]
- Alías-Villegas, C.; Fuentes-Romero, F.; Cuéllar, V.; Navarro-Gómez, P.; Soto, M.J.; Vinardell, J.M.; Acosta-Jurado, S. Surface motility regulation of Sinorhizobium fredii HH103 by plant flavonoids and the NodD1, TtsI, NolR, and MucR1 symbiotic bacterial regulators. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Holscher, T.; Kovacs, A.T. Sliding on the surface: bacterial spreading without an active motor. Environ Microbiol 2017, 19, 2537–2545. [Google Scholar] [CrossRef]
- Mattick, J.S. Type IV pili and twitching motility. Annu Rev Microbiol 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Maier, B.; Wong, G.C.L. How bacteria use Type IV pili machinery on surfaces. Trends Microbiol 2015, 23, 775–788. [Google Scholar] [CrossRef]
- Pelicic, V. Type IV pili: e pluribus unum? Mol Microbiol 2008, 68, 827–837. [Google Scholar] [CrossRef]
- Craig, L.; Forest, K.T.; Maier, B. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol 2019, 17, 429–440. [Google Scholar] [CrossRef]
- Denise, R.; Abby, S.S.; Rocha, E.P.C. Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol 2019, 17, e3000390. [Google Scholar] [CrossRef]
- Ellison, C.K.; Whitfield, G.B.; Brun, Y.V. Type IV Pili: dynamic bacterial nanomachines. FEMS Microbiol Rev 2022, 46. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.K.; Kan, J.; Dillard, R.S.; Kysela, D.T.; Ducret, A.; Berne, C.; Hampton, C.M.; Ke, Z.; Wright, E.R.; Biais, N.; et al. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 2017, 358, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 2012, 76, 740–772. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Planet, P.J.; Bhattacharjee, M.K.; Kollia, E.; DeSalle, R.; Fine, D.H.; Figurski, D.H. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol 2000, 182, 6169–6176. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Planet, P.J.; DeSalle, R.; Fine, D.H.; Figurski, D.H. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol 2001, 9, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Kachlany, S.C.; Planet, P.J.; Desalle, R.; Fine, D.H.; Figurski, D.H.; Kaplan, J.B. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol 2001, 40, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Tomich, M.; Planet, P.J.; Figurski, D.H. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 2007, 5, 363–375. [Google Scholar] [CrossRef]
- Roux, N.; Spagnolo, J.; de Bentzmann, S. Neglected but amazingly diverse type IVb pili. Res Microbiol 2012, 163, 659–673. [Google Scholar] [CrossRef]
- Skerker, J.M.; Shapiro, L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 2000, 19, 3223–3234. [Google Scholar] [CrossRef]
- Wang, Y.; Haitjema, C.H.; Fuqua, C. The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment. J Bacteriol 2014, 196, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Tomich, M.; Fine, D.H.; Figurski, D.H. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J Bacteriol 2006, 188, 6899–6914. [Google Scholar] [CrossRef]
- Tassinari, M.; Rudzite, M.; Filloux, A.; Low, H.H. Assembly mechanism of a Tad secretion system secretin-pilotin complex. Nat Commun 2023, 14, 5643. [Google Scholar] [CrossRef]
- Xu, Q.; Christen, B.; Chiu, H.J.; Jaroszewski, L.; Klock, H.E.; Knuth, M.W.; Miller, M.D.; Elsliger, M.A.; Deacon, A.M.; Godzik, A.; et al. Structure of the pilus assembly protein TadZ from Eubacterium rectale: implications for polar localization. Mol Microbiol 2012, 83, 712–727. [Google Scholar] [CrossRef]
- Perez-Cheeks, B.A.; Planet, P.J.; Sarkar, I.N.; Clock, S.A.; Xu, Q.; Figurski, D.H. The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol Microbiol 2012, 83, 694–711. [Google Scholar] [CrossRef]
- Clock, S.A.; Planet, P.J.; Perez, B.A.; Figurski, D.H. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J Bacteriol 2008, 190, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Seef, S.; Herrou, J.; de Boissier, P.; My, L.; Brasseur, G.; Robert, D.; Jain, R.; Mercier, R.; Cascales, E.; Habermann, B.H.; et al. A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. eLife 2021, 10. [Google Scholar] [CrossRef]
- Wairuri, C.K.; van der Waals, J.E.; van Schalkwyk, A.; Theron, J. Ralstonia solanacearum needs Flp pili for virulence on potato. Mol Plant Microbe Interact 2012, 25, 546–556. [Google Scholar] [CrossRef]
- Nykyri, J.; Mattinen, L.; Niemi, O.; Adhikari, S.; Kõiv, V.; Somervuo, P.; Fang, X.; Auvinen, P.; Mäe, A.; Palva, E.T.; et al. Role and regulation of the Flp/Tad pilus in the virulence of Pectobacterium atrosepticum SCRI1043 and Pectobacterium wasabiae SCC3193. PLOS ONE 2013, 8, e73718. [Google Scholar] [CrossRef]
- Andrade, M.; Wang, N. The Tad Pilus Apparatus of 'Candidatus Liberibacter asiaticus' and Its Regulation by VisNR. Mol Plant Microbe Interact 2019, 32, 1175–1187. [Google Scholar] [CrossRef]
- Vesper, S.J.; Bauer, W.D. Role of Pili (Fimbriae) in Attachment of Bradyrhizobium japonicum to Soybean Roots. Appl Environ Microbiol 1986, 52, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Krehenbrink, M.; Downie, J.A. Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genomics 2008, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Mongiardini, E.J.; Parisi, G.D.; Quelas, J.I.; Lodeiro, A.R. The tight-adhesion proteins TadGEF of Bradyrhizobium diazoefficiens USDA 110 are involved in cell adhesion and infectivity on soybean roots. Microbiol Res 2016, 182, 80–88. [Google Scholar] [CrossRef]
- Schmeisser, C.; Liesegang, H.; Krysciak, D.; Bakkou, N.; Le Quere, A.; Wollherr, A.; Heinemeyer, I.; Morgenstern, B.; Pommerening-Roser, A.; Flores, M.; et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol 2009, 75, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Wibberg, D.; Blom, J.; Jaenicke, S.; Kollin, F.; Rupp, O.; Scharf, B.; Schneiker-Bekel, S.; Sczcepanowski, R.; Goesmann, A.; Setubal, J.C.; et al. Complete genome sequencing of Agrobacterium sp. H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J Biotechnol 2011, 155, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Zatakia, H.M.; Nelson, C.E.; Syed, U.J.; Scharf, B.E. ExpR coordinates the expression of symbiotically important, bundle-forming Flp pili with quorum sensing in Sinorhizobium meliloti. Appl Environ Microbiol 2014, 80, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Casadesús, J.; Olivares, J. Rough and fine linkage mapping of the Rhizobium meliloti chromosome. Mol Gen Genet 1979, 174, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Bellabarba, A.; Bacci, G.; Decorosi, F.; Aun, E.; Azzarello, E.; Remm, M.; Giovannetti, L.; Viti, C.; Mengoni, A.; Pini, F. Competitiveness for Nodule Colonization in Sinorhizobium meliloti: Combined In Vitro-Tagged Strain Competition and Genome-Wide Association Analysis. mSystems 2021, 6, e0055021. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, J.P.; Reinkensmeier, J.; Barnett, M.J.; Lang, C.; Krol, E.; Giegerich, R.; Long, S.R.; Becker, A. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics 2013, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Christen, M.; Beusch, C.; Bösch, Y.; Cerletti, D.; Flores-Tinoco, C.E.; Del Medico, L.; Tschan, F.; Christen, B. Quantitative Selection Analysis of Bacteriophage φCbK Susceptibility in Caulobacter crescentus. J Mol Biol 2016, 428, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Mignolet, J.; Panis, G.; Viollier, P.H. More than a Tad: spatiotemporal control of Caulobacter pili. Curr Opin Microbiol 2018, 42, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Wu, H.C. Lipoproteins in bacteria. J Bioenerg Biomembr 1990, 22, 451–471. [Google Scholar] [CrossRef]
- Kaushik, S.; He, H.; Dalbey, R.E. Bacterial Signal Peptides- Navigating the Journey of Proteins. Front Physiol 2022, 13. [Google Scholar] [CrossRef]
- DAndrea, L.D.; Regan, L. TPR proteins: the versatile helix. Trends Biochem Sci 2003, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Sangermani, M.; Hug, I.; Sauter, N.; Pfohl, T.; Jenal, U. Tad Pili Play a Dynamic Role in Caulobacter crescentus Surface Colonization. MBio 2019, 10. [Google Scholar] [CrossRef]
- Turnbull, L.; Whitchurch, C.B. Motility Assay: Twitching Motility. In Pseudomonas Methods and Protocols, Filloux, A.; Ramos, J.-L., Eds. Springer New York: New York, NY, 2014; pp 73-86, ISBN. 978-1-4939-0473-0.
- Duong-Nu, T.M.; Jeong, K.; Hong, S.H.; Puth, S.; Kim, S.Y.; Tan, W. A stealth adhesion factor contributes to Vibrio vulnificus pathogenicity: Flp pili play roles in host invasion, survival in the blood stream and resistance to complement activation. PLoS Pathog 2019, 15, e1007767. [Google Scholar] [CrossRef] [PubMed]
- de Bentzmann, S.; Aurouze, M.; Ball, G.; Filloux, A. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J Bacteriol 2006, 188, 4851–4860. [Google Scholar] [CrossRef] [PubMed]
- Kilmury, S.L.N.; Burrows, L.L. The Pseudomonas aeruginosa PilSR Two-Component System Regulates Both Twitching and Swimming Motilities. mBio 2018, 9, e01310–01318. [Google Scholar] [CrossRef]
- Ellison, C.K.; Rusch, D.B.; Brun, Y.V. Flagellar Mutants Have Reduced Pilus Synthesis in Caulobacter crescentus. J Bacteriol 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Kühn, M.J.; Talà, L.; Inclan, Y.F.; Patino, R.; Pierrat, X.; Vos, I.; Al-Mayyah, Z.; Macmillan, H.; Negrete, J., Jr.; Engel, J.N.; et al. Mechanotaxis directs Pseudomonas aeruginosa twitching motility. Proc Natl Acad Sci U S A 2021, 118, e2101759118. [Google Scholar] [CrossRef]
- Laventie, B.J.; Jenal, U. Surface Sensing and Adaptation in Bacteria. Annu Rev Microbiol 2020, 74, 735–760. [Google Scholar] [CrossRef] [PubMed]
- Del Medico, L.; Cerletti, D.; Schachle, P.; Christen, M.; Christen, B. The type IV pilin PilA couples surface attachment and cell-cycle initiation in Caulobacter crescentus. Proc Natl Acad Sci U S A 2020, 117, 9546–9553. [Google Scholar] [CrossRef]
- Sambrook, J. , Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 1989. [Google Scholar]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Robertsen, B.K.; Aiman, P.; Darwill, A.G.; Mcneil, M.; Albersheim, P. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol 1981, 67, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016, 44, D457–462. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic-engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Calatrava-Morales, N.; Nogales, J.; Ameztoy, K.; van Steenbergen, B.; Soto, M.J. The NtrY/NtrX System of Sinorhizobium meliloti GR4 Regulates Motility, EPS I Production, and Nitrogen Metabolism but Is Dispensable for Symbiotic Nitrogen Fixation. Mol Plant Microbe Interact 2017, 30, 566–577. [Google Scholar] [CrossRef] [PubMed]
- López-Lara, I.M.; Nogales, J.; Pech-Canul, A.; Calatrava-Morales, N.; Bernabéu-Roda, L.M.; Durán, P.; Cuéllar, V.; Olivares, J.; Alvarez, L.; Palenzuela-Bretones, D.; et al. 2-Tridecanone impacts surface-associated bacterial behaviours and hinders plant-bacteria interactions. Environ Microbiol 2018, 20, 2049–2065. [Google Scholar] [CrossRef]
- Olivares, J.; Casadesús, J.; Bedmar, E.J. Method for Testing Degree of Infectivity of Rhizobium meliloti Strains. Appl Environ Microbiol 1980, 39, 967–970. [Google Scholar] [CrossRef]
- García-Rodríguez, F.M.; Toro, N. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol Plant Microbe Interact 2000, 13, 583–591. [Google Scholar] [CrossRef] [PubMed]
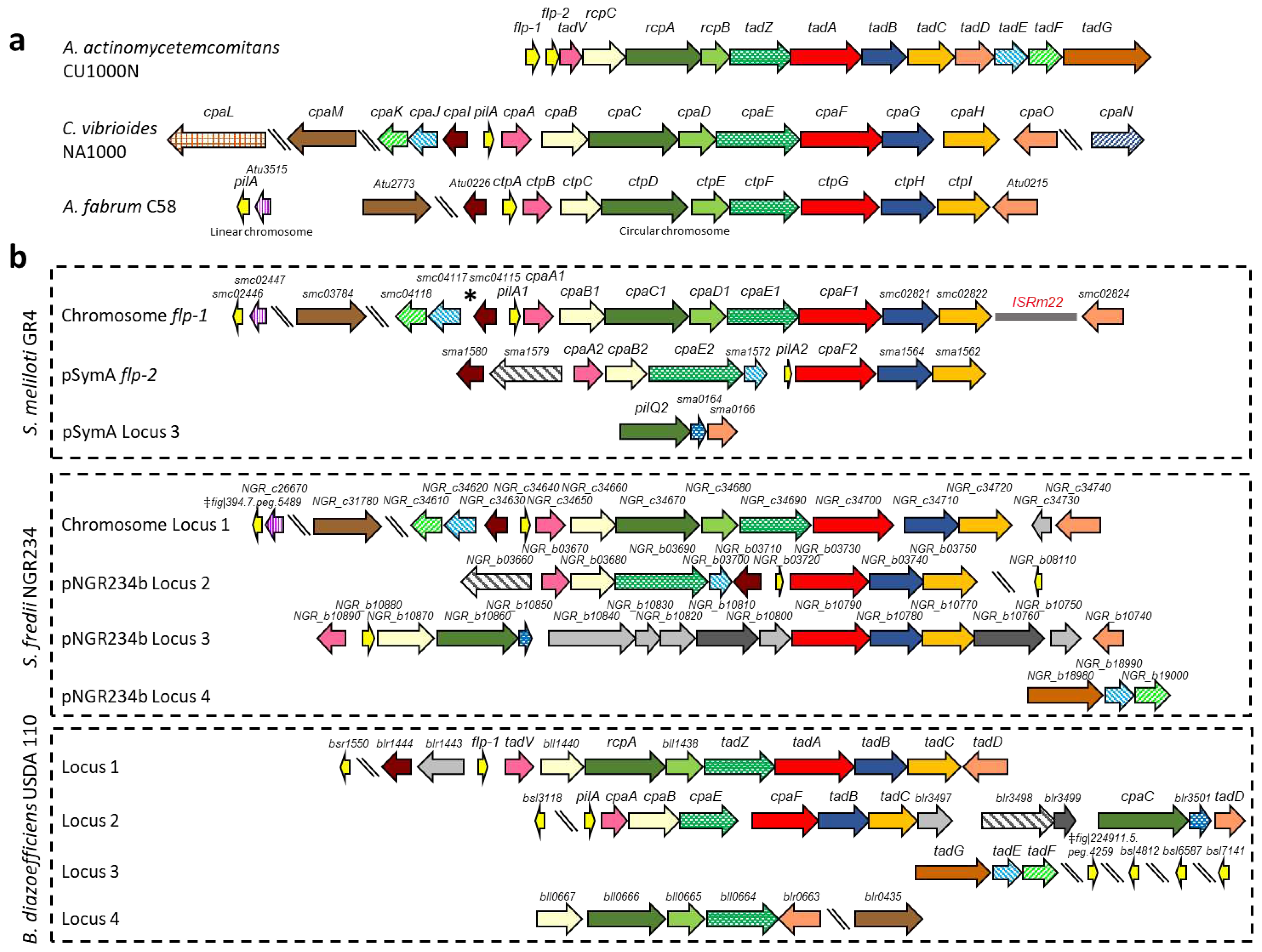

| Locus GR4/Rm1021 | Description | Aa | Cv | Af | Bd | Sf | |
|---|---|---|---|---|---|---|---|
| Chromosome flp-1 | |||||||
| C770_GR4Chr0141/smc04118 | CpaK-like pseudopilin | CpaK (25) | TadF (26) | NGR_c34610 (60) | |||
| C770_GR4Chr0142/smc04117 | CpaJ-like pseudopilin | CpaJ (32) | TadE (29) | NGR_c34620 (69) | |||
| C770_GR4Chr0143/smc04115 | CpaI-like putative pili related protein | CpaI (26) | Atu0226 (63) | Blr1444 (35) | NGR_c34630 (88) | ||
| C770_GR4Chr0144/smc04114 (pilA1) | Flp pilin subunit | Flp-2 (34) | PilA (51) | CtpA (76) | Bsr1550 (51) | NGR_c34640 (71) | |
| C770_GR4Chr0145/smc04113 (cpaA1) | Putative prepilin peptidase | TadV (31% in 65aa stretch) | CpaA (37) | CtpB (52) | TadV (43) | NGR_c34650 (71) | |
| C770_GR4Chr0146/smc04112 (cpaB1) | Component of the outer membrane secretin complex | CpaB (35) | CtpC (64) | Bll1440 (39) | NGR_c34660 (87) | ||
| C770_GR4Chr0147/smc04111* (cpaC1) | Component of the outer membrane secretin complex | RcpA (26) | CpaC (34) | CtpD (70) | RcpA (40) | NGR_c34670 (87) | |
| C770_GR4Chr0148/smc04110 (cpaD1) | Component of the outer membrane secretin complex | CpaD (39) | CtpE (58) | Bll1438 (35) | NGR_c34680 (80) | ||
| C770_GR4Chr0149/smc04109 (cpaE1) | Putative docking protein of the secretion system | TadZ (23) | CpaE (47) | CtpF (71) | TadZ (49) | NGR_c34690 (87) | |
| C770_GR4Chr0150/smc02820 (cpaF1) | Pilus assembly ATPase | TadA (50) | CpaF (72) | CtpG (81) | TadA (72) | NGR_c34700 (90) | |
| C770_GR4Chr0151/smc02821 | CpaG-like putative inner membrane platform protein | TadB (22) | CpaG (42) | CtpH (64) | TadB Locus 1 (48) | NGR_c34710 (89) | |
| C770_GR4Chr0152/smc02822 | CpaH-like putative inner membrane platform protein | TadC (21) | CpaH (48) | CtpI (69) | TadC Locus 1 (54) | NGR_c34720 (90) | |
| C770_GR4Chr0153/smc04116 (TRm22) | Insertion sequence ISRm22 | ||||||
| C770_GR4Chr0154/smc02824 | CpaO-like TPR containing protein | CpaO (37) | Atu0215 (66) | TadD Locus 1 (45) | NGR_c34740 (84) | ||
| Chromosomal orphan loci | |||||||
| C770_GR4Chr2732/smc02446 | Flp pilin subunit | Flp-2 (35) | PilA (60) | CtpA (51) | Bsl4812 (51) | ‡fig|394.7.peg.5489 (60) | |
| C770_GR4Chr2733/smc02447 | Hypothetical protein | Atu3515 (52) | NGR_c26670 (95) | ||||
| C770_GR4Chr3249/smc03784 | CpaM-like polysaccharide deacetylase | CpaM (39) | Atu2773 (59) | Blr0435 (38) | NGR_c31780 (74) | ||
| pSymA flp-2 | |||||||
| C770_GR4pC0512/sma1580* | CpaI-like putative pili related protein | CpaI (32) | Atu0226 (31) | Blr1444 (37) | NGR_b03710 (59) | ||
| C770_GR4pC0513/sma1579* | Putative pili related protein | Blr3498 (24) | NGR_b03660 (74) | ||||
| C770_GR4pC0514/sma1578* (cpaA2) | Putative prepilin peptidase | TadV (30) | CpaA (38) | CtpB (36) | TadV (43) | NGR_b03670 (71) | |
| C770_GR4pC0515/sma1576* (cpaB2) | Component of the outer membrane secretin complex | CpaB (34) | CtpC (39) | Bll1440 (40) | NGR_b03680 (78) | ||
| C770_GR4pC0516/sma1573 (cpaE2) | Putative docking protein of the secretion system | CpaE (43) | CtpF (40) | TadZ (40) | NGR_b03690 (76) | ||
| C770_GR4pC0517/sma1572 | CpaJ-like pseudopilin protein | CpaJ (41) | NGR_b03700 (75) | ||||
| C770_GR4pC0518/sma1570 (pilA2) | Flp pilin subunit | Flp-1 (34) | PilA (53) | CtpA (55) | Flp-1 (62) | NGR_b03720 (69) | |
| C770_GR4pC0519/sma1568* (cpaF2) | Pilus assembly ATPase | TadA (48) | CpaF (73) | CtpG (69) | TadA (79) | NGR_b03730 (87) | |
| C770_GR4pC0520/sma1564 | CpaG-like putative inner membrane platform protein | TadB (28) | CpaG (43) | CtpH (41) | TadB Locus 1 (46) | NGR_b03740 (71) | |
| C770_GR4pC0521/sma1562 | CpaH-like putative inner membrane platform protein | TadC (20) | CpaH (43) | CtpI (49) | TadC Locus 1 (52) | NGR_b03750 (84) | |
| pSymA Locus 3 | |||||||
| C770_GR4pC1351/sma0163 (pilQ2) | RcpA-like putative component of the outer membrane secretin complex | RcpA (28) | CpaC (31) | CtpD (31) | CpaC (40) | NGR_b10860 (40) | |
| C770_GR4pC1350/sma0164 | Hypothetical lipoprotein | Blr3501 (33) | NGR_b10850 (29) | ||||
| C770_GR4pC1349/sma0166 | CpaO/TadD-like TPR containing protein | Atu0215 (31% in 70 aa middle stretch) | TadD Locus 2 (44) | NGR_b10740 (36) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





