Submitted:
08 January 2024
Posted:
09 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Patient Sera Samples
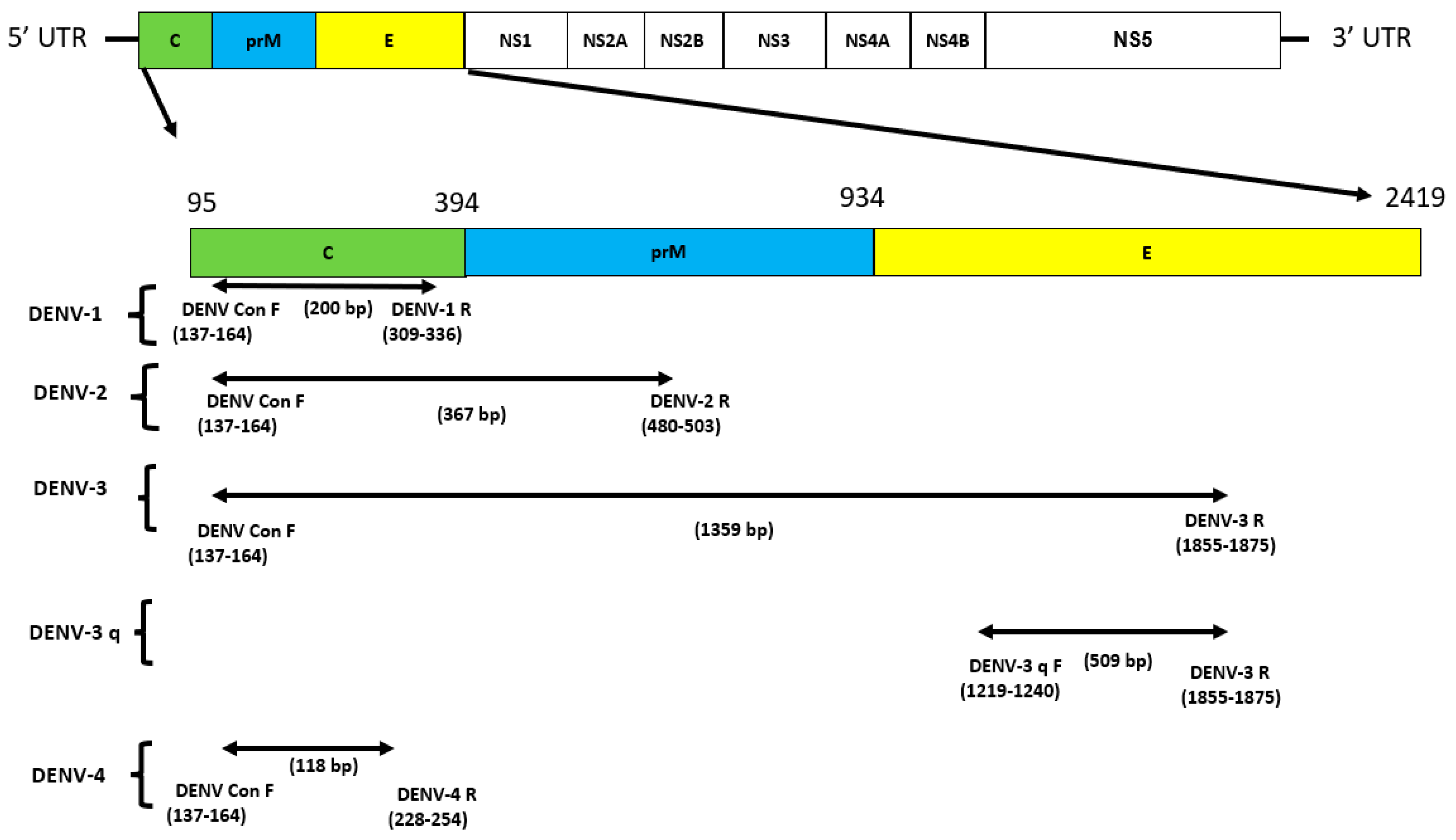
2.3. Primer Design
2.4. Viral RNA Extraction
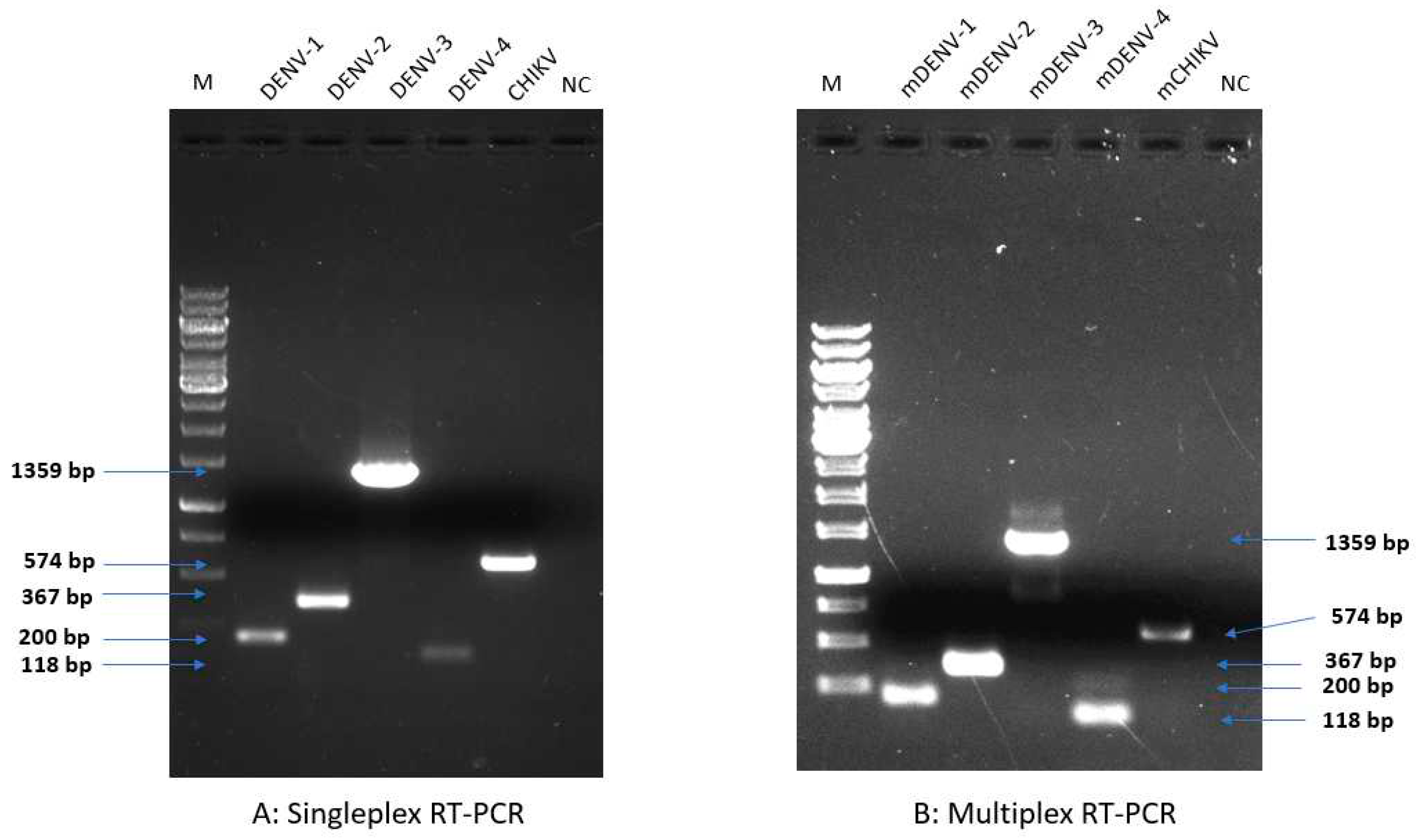
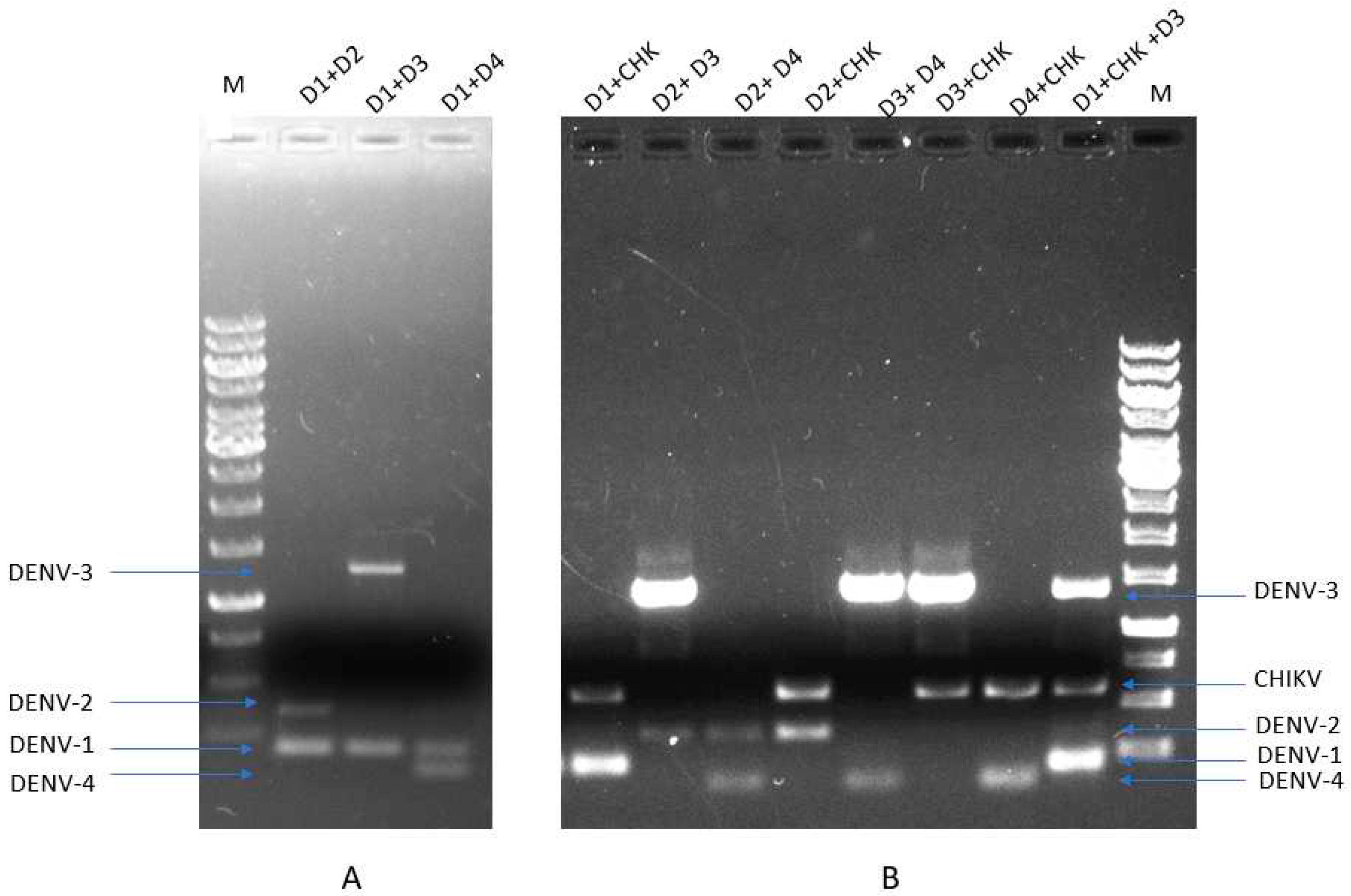
2.5. One-Step Multiplex RT-PCR Amplification
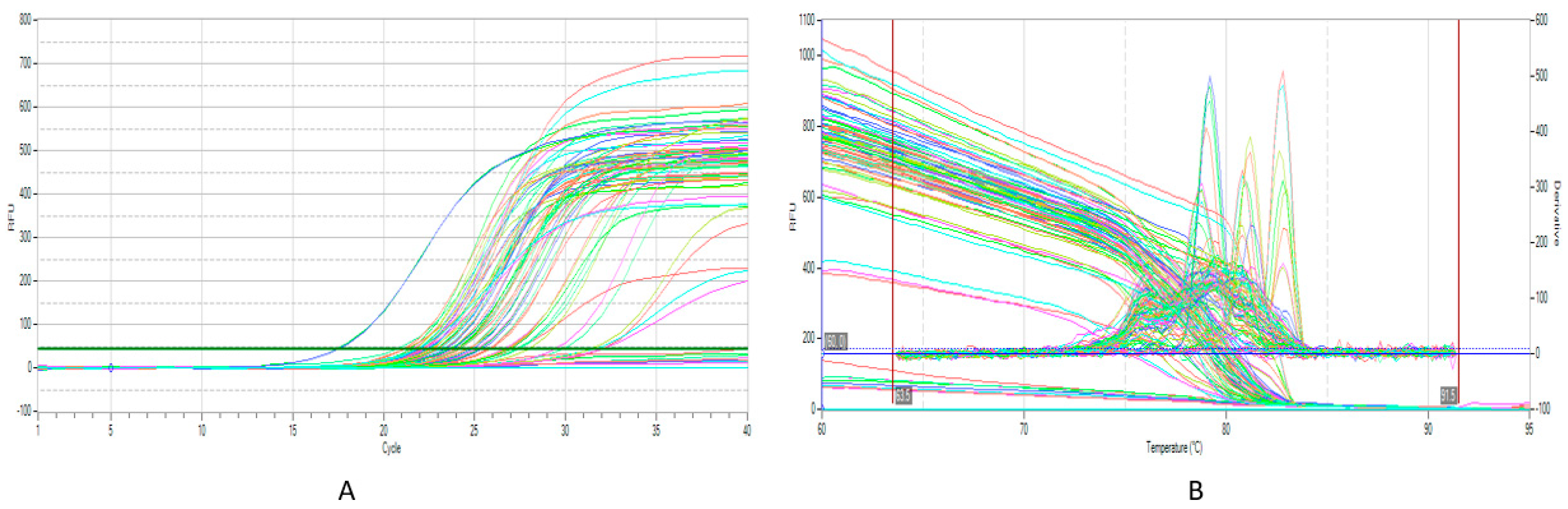
2.6. Multiplex One-Step Real-Time RT-PCR
2.7. Sequence Analysis
3. Results and Discussion
3.1. Primer Design and Their Specificity Assessment for Multiplex Detection of DENV1-4 and CHIKV:
| Family/Genus | Virus | Primers | Sequence (5′ to 3′) | Target | Genome position |
|---|---|---|---|---|---|
| Flaviviridae/Flavivirus | DENV | DENVcon F | TCAATATGCTGAAACGCGAGAGAAACCG | C | 137-164 |
| DENV-3 q F | AGGAGCTACGTGGGTTGACGTG | E | 1219-1240 | ||
| DENV-1 R | TTTGATCGCTCCATTCTTCTTGAATGAG | C | 309-336 | ||
| DENV-2 R | TTCCCTTTCTCTTGTCTACTGACG | prM | 480-503 | ||
| DENV-3 R | GTGGTGAGCATTCTAGCCCAA | E | 1855-1875 | ||
| DENV-4 R | GTGATGAATGCTAGCACCATCCGTAAG | C | 228-254 | ||
| Togaviridae/Alphavirus | CHIKV | CHIKV F | ACACGTAAACAGTGATCCCGAACAC | E1 | 9998-10022 |
| CHIKV R | CCAAACGGCGGGTAGTCCATGT | 10550-10571 |
3.2. Multiplex RT-qPCR Assessment and Sensitivity:
| Parameters | mRT-qPCR | ||||
|---|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | CHIKV | |
| Ct value | 17.28 | 23.5 | 22.06 | 21.73 | 22.75 |
| R2 | 0.93 | 0.99 | 0.99 | 0.98 | 0.98 |
| LOD (Copies/µL) |
10-2 | 10-2 | 10-2 | 10-2 | 10-2 |
| Tm | 79.17 | 80.01 | 82.7 | 78.71 | 81.84 |
3.3. Performance of mRT-PCR and mRT-qPCR Assay on Clinical Samples and Validation:
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, Y.J.S.; Higgs, S.; Vanlandingham, D.L. Biological Control Strategies for Mosquito Vectors of Arboviruses. Insects 2017, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Gao, X.; Gould, E.A. Factors Responsible for the Emergence of Arboviruses; Strategies, Challenges and Limitations for Their Control. Emerg. Microbes Infect. 2015, 4, e18. [Google Scholar] [CrossRef]
- Carrington, L.B.; Simmons, C.P. Human to Mosquito Transmission of Dengue Viruses. Front. Immunol. 2014, 5, 290. [Google Scholar] [CrossRef] [PubMed]
- Bueno, M.G.; Martinez, N.; Abdalla, L.; Duarte dos Santos, C.N.; Chame, M. Animals in the Zika Virus Life Cycle: What to Expect from Megadiverse Latin American Countries. Plos Neglect. Trop. Dis. 2016, 10, e0005073. [Google Scholar] [CrossRef] [PubMed]
- Gould, E.; Pettersson, J.; Higgs, S.; Charrel, R.; de Lamballerie, X. Emerging Arboviruses: Why Today? One Health 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Chancey, C.; Grinev, A.; Volkova, E.; Rios, M. The Global Ecology and Epidemiology of West Nile Virus. BioMed Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Glushakova, L.G.; Alto, B.W.; Kim, M.S.; Hutter, D.; Bradley, A.; Bradley, K.M.; Burkett-Cadena, N.D.; Benner, S.A. Multiplexed Kit Based on Luminex Technology and Achievements in Synthetic Biology Discriminates Zika, Chikungunya, and Dengue Viruses in Mosquitoes. BMC Infect. Dis. 2019, 19, 418. [Google Scholar] [CrossRef] [PubMed]
- Sekaran, S.D.; Artsob, H. Molecular Diagnostics for the Detection of Human Flavivirus Infections. Expert Opin. Med. Diagn. 2007, 1, 521–530. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Chikungunya. Available online: https://www.who.int/health topics/chikungunya#tab=tab_1 (accessed on 15 September 2023).
- Khongwichit, S.; Chansaenroj, J.; Chirathaworn, C.; Poovorawan, Y. Chikungunya Virus Infection: Molecular Biology, Clinical Characteristics, and Epidemiology in Asian Countries. J. Biomed. Sci. 2021, 28, 84. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; el Zowalaty, M.E.; Islam, S.; Sharif, M.; Rahman, M.R.; Amin, M.R.; Ali, M.M.; Rahman, M.T.; Morita, K.; Ashour, H.M.A. Novel Multiplex RT-PCR Assay for Simultaneous Detection of Dengue and Chikungunya Viruses. Int. J. Mol. Sci. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Murhekar, M.; Joshua, V.; Kanagasabai, K.; Shete, V.; Ravi, M.; Ramachandran, R.; Sabarinathan, R.; Kirubakaran, B.; Gupta, N.; Mehendale, S. Epidemiology of Dengue Fever in India, Based on Laboratory Surveillance Data, 2014–2017. Int. J. Infect. Dis. 2019, 84, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Jagadesh, A.; Jayaram, A.; Babu, N.; Mudgal, P.P.; Sudandiradas, R.; Sheik, S.; Shetty, U.; Verma, D.K.; Mahilkar, S.; Sunil, S.; et al. Current Status of Chikungunya in India. Front. Microbiol. 2021, 12. [Google Scholar]
- Tarnagda, Z.; Cissé, A.; Bicaba, B.W.; Diagbouga, S.; Sagna, T.; Ilboudo, A.K.; Tialla, D.; Lingani, M.; Sondo, K.A.; Yougbaré, I.; et al. Dengue Fever in Burkina Faso, 2016. Emerg. Infect. Dis. 2018, 24, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Tougma, S.A.; Yaméogo, W.N.Z.; Dahourou, D.L.; Salou Kagoné, I.A.; Compaoré, T.R.; Kaboré, A.; Kagoné, T.; Drabo, M.K.; Meda, N. Dengue Virus Infection and Pregnancy Outcomes during the 2017 Outbreak in Ouagadougou, Burkina Faso: A Retrospective Cohort Study. PLoS One 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.K.; Ridde, V.; Agnandji, S.T.; Lell, B.; Yaro, S.; Yang, J.S.; Hoinard, D.; Weaver, S.C.; Vanhomwegen, J.; Salje, H.; et al. Seroepidemiological Reconstruction of Long-Term Chikungunya Virus Circulation in Burkina Faso and Gabon. J. Infect. Dis. 2022, 227, 261–267. [Google Scholar] [CrossRef]
- Bob, N.S.; Bâ, H.; Fall, G.; Ishagh, E.; Diallo, M.Y.; Sow, A.; Sembene, P.M.; Faye, O.; el Kouri, B.; Sidi, M.L.; et al. Detection of the Northeastern African Rift Valley Fever Virus Lineage during the 2015 Outbreak in Mauritania. Open Forum Infect. Dis. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Kwagonza, L.; Masiira, B.; Kyobe-Bosa, H.; Kadobera, D.; Atuheire, E.B.; Lubwama, B.; Kagirita, A.; Katushabe, E.; Kayiwa, J.T.; Lutwama, J.J.; et al. Outbreak of Yellow Fever in Central and Southwestern Uganda, February-May 2016. BMC Infect. Dis. 2018, 18, 548. [Google Scholar] [CrossRef] [PubMed]
- Glushakova, L.G.; Bradley, A.; Bradley, K.M.; Alto, B.W.; Hoshika, S.; Hutter, D.; Sharma, N.; Yang, Z.; Kim, M.J.; Benner, S.A. High-Throughput Multiplexed XMAP Luminex Array Panel for Detection of Twenty-Two Medically Important Mosquito-Borne Arboviruses Based on Innovations in Synthetic Biology. J. Virol. Methods 2015, 214, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Prat, C.M.; Flusin, O.; Panella, A.; Tenebray, B.; Lanciotti, R.; Leparc-Goffart, I. Evaluation of Commercially Available Serologic Diagnostic Tests for Chikungunya Virus. Emerg. Infect. Dis. 2014, 20, 2129–2132. [Google Scholar] [CrossRef]
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.A.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.Y.; Vasilakis, N. Zika Virus: History, Emergence, Biology, and Prospects for Control. Antiviral Res. 2016, 130, 69–80. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and Application in Diagnostic Virology. Clin. Microbial. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, J.; Yu, N.; Yan, J.; Zhuo, Z.; Chen, M.; Su, X.; Fang, M.; He, S.; Zhang, S.; et al. Development of Multiplex Real-Time Reverse-Transcriptase Polymerase Chain Reaction Assay for Simultaneous Detection of Zika, Dengue, Yellow Fever, and Chikungunya Viruses in a Single Tube. J. Med. Virol. 2018, 90, 1681–1686. [Google Scholar] [CrossRef]
- Chen, H.; Parimelalagan, M.; Lai, Y.L.; Lee, K.S.; Koay, E.S.C.; Hapuarachchi, H.C.; Ng, L.C.; Ho, P.S.; Chu, J.J.H. Development and Evaluation of a SYBR Green-Based Real-Time Multiplex RT-PCR Assay for Simultaneous Detection and Serotyping of Dengue and Chikungunya Viruses. J. Mol. Diagn. 2015, 17, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Yaren, O.; Alto, B.W.; Bradley, K.M.; Moussatche, P.; Glushakova, L.; Benner, S.A. Multiplexed Isothermal Amplification Based Diagnostic Platform to Detect Zika, Chikungunya, and Dengue 1. J. Vis. Exp. 2018, 133, 57051. [Google Scholar] [CrossRef]
- Priye, A.; Bird, S.W.; Light, Y.K.; Ball, C.S.; Negrete, O.A.; Meagher, R.J. A Smartphone-Based Diagnostic Platform for Rapid Detection of Zika, Chikungunya, and Dengue Viruses. Sci. Rep. 2017, 7, 44778. [Google Scholar] [CrossRef] [PubMed]
- Shrinet, J.; Jain, S.; Sharma, A.; Singh, S.S.; Mathur, K.; Rana, V.; Bhatnagar, R.K.; Gupta, B.; Gaind, R.; Deb, M.; Sunil, S. Genetic Characterization of Chikungunya Virus from New Delhi Reveal Emergence of a New Molecular Signature in Indian Isolates. Virol. J. 2012, 9, 100. [Google Scholar] [CrossRef]
- Ibemgbo, S.A.; Nyodu, R.; Chaudhary, S.; Verma, D.K.; Dixit, K.; Nayak, K.; Rani, V.; Gaind, R.; Chandele, A.; Sunil, S. Short Communication: Virological and B Cell Profiles of Chikungunya and Dengue Virus Co-Infections in Delhi during 2017–2019. Virus Res. 2022, 320, 198888. [Google Scholar] [CrossRef]
- Johnson, B.W.; Goodman, C.H.; Holloway, K.; De Salazar, P.M.; Valadere, A.M.; Drebot, M.A. Evaluation of Commercially Available Chikungunya Virus Immunoglobulin M Detection Assays. Am. J. Trop. Med. Hyg. 2016, 95, 182–192. [Google Scholar] [CrossRef]
- Mat Jusoh, T.N.A.; Shueb, R.H. Performance Evaluation of Commercial Dengue Diagnostic Tests for Early Detection of Dengue in Clinical Samples. J. Trop. Med. 2017, 2017. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Calisher, C.H.; Gubler, D.J.; Chang, G.-J.; Vorndamt, A.V. Rapid Detection and Typing of Dengue Viruses from Clinical Samples by Using Reverse Transcriptase-Polymerase Chain Reaction. J. Clin. Microbiol. 1992, 30, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Yenchitsomanus, P.T.; Sricharoen, P.; Jaruthasana, I.; Pattanakitsakul, S.N.; Nitayaphan, S.; Mongkolsapaya, J.; Malasit, P. Rapid Detection and Identification of Dengue Viruses by Polymerase Chain Reaction (PCR). Southeast Asian J. Trop. Med. Public Health 1996, 27, 228–236. [Google Scholar] [PubMed]
- Yong, Y.K.; Thayan, R.; Chong, H.T.; Tan, C.T.; Sekaran, S.D. Rapid Detection and Serotyping of Dengue Virus by Multiplex RT-PCR and Real-Time SYBR Green RT-PCR. Singapore Med. J. 2007, 1, 662–668. [Google Scholar]
- Li, D.S.; Liu, W.; Guigon, A.; Mostyn, C.; Grant, R.; Aaskov, J. Rapid Displacement of Dengue Virus Type 1 by Type 4, Pacific Region, 2007-2009. Emerg. Infect. Dis. 2010, 16, 123–125. [Google Scholar] [CrossRef]
- Cummings, D.A.T.; Irizarry, R.A.; Huang, N.E.; Endy, T.P.; Nisalak, A.; Ungchusak, K.; Burke, D.S. Travelling Waves in the Occurrence of Dengue Haemorrhagic Fever in Thailand. Nature 2004, 427, 344–347. [Google Scholar] [CrossRef] [PubMed]
- van Panhuis, W.G.; Choisy, M.; Xiong, X.; Chok, N.S.; Akarasewi, P.; Iamsirithaworn, S.; Lam, S.K.; Chong, C.K.; Lam, F.C.; Phommasak, B.; et al. Region-Wide Synchrony and Traveling Waves of Dengue across Eight Countries in Southeast Asia. Proc. Natl. Acad. Sci. U S A 2015, 112, 13069–13074. [Google Scholar] [CrossRef]
| Country | Sample | Total | mRT-qPCR + N (%) | mRT-qPCR - N (%) | mRT-PCR + N (%) | mRT-PCR - N (%) |
|---|---|---|---|---|---|---|
| India | DENV positive | 16 | 16 (100.0) | 0 (0.0) | 14 (87.5) | 2 (12.5) |
| DENV negative | 16 | 0 (0.0) | 16 (100.0) | 0 (0.0) | 16 (100.0) | |
| CHIKV positive | 16 | 16 (100.0) | 0 (0.0) | 15 (93.7) | 1 (6.3) | |
| CHIKV negative | 16 | 0 (0.0) | 16 (100.0) | 0 (0.0) | 16 (100.0) | |
| Burkina Faso | DENV positive | 33 | 33 (100.0) | 0 (0.0) | 30 (90.9) | 3 (9.1) |
| DENV negative | 33 | 0 (0.0) | 33 (100.0) | 0 (0.0) | 33 (100.0) | |
| Sensitivity | 100% | 91 % | ||||
| Specificity | 100% | 100% | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





