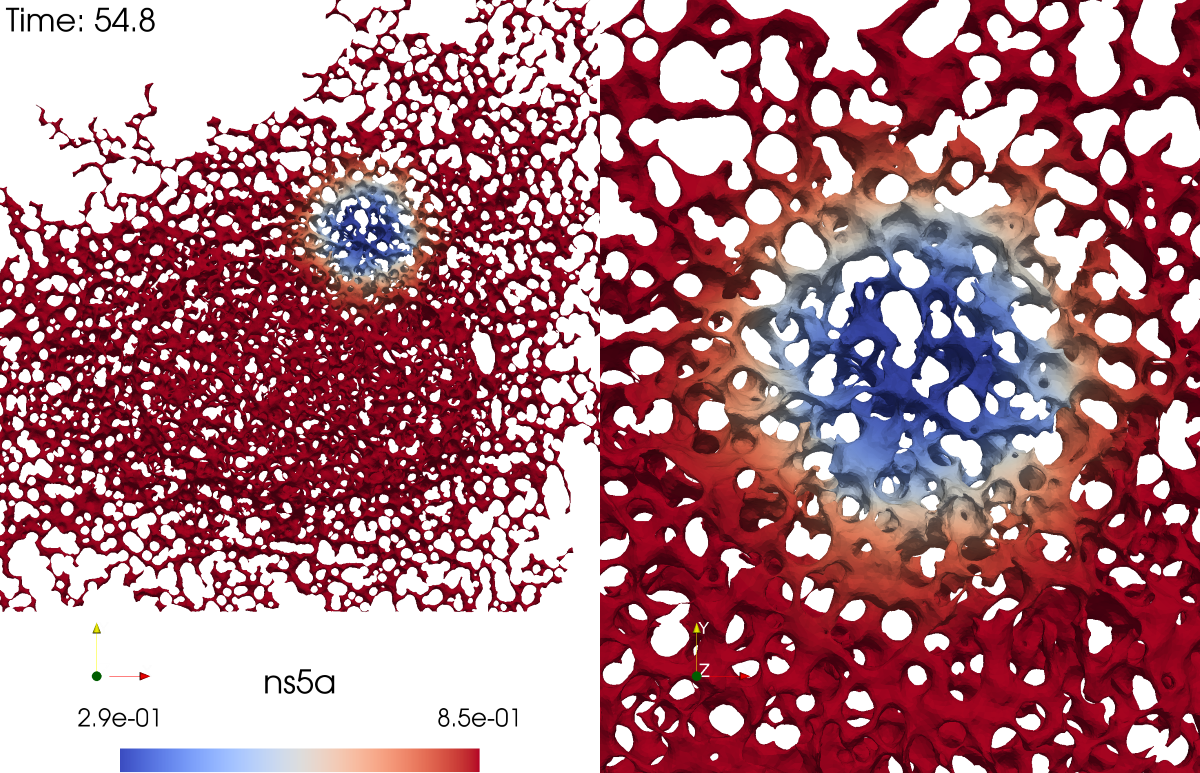
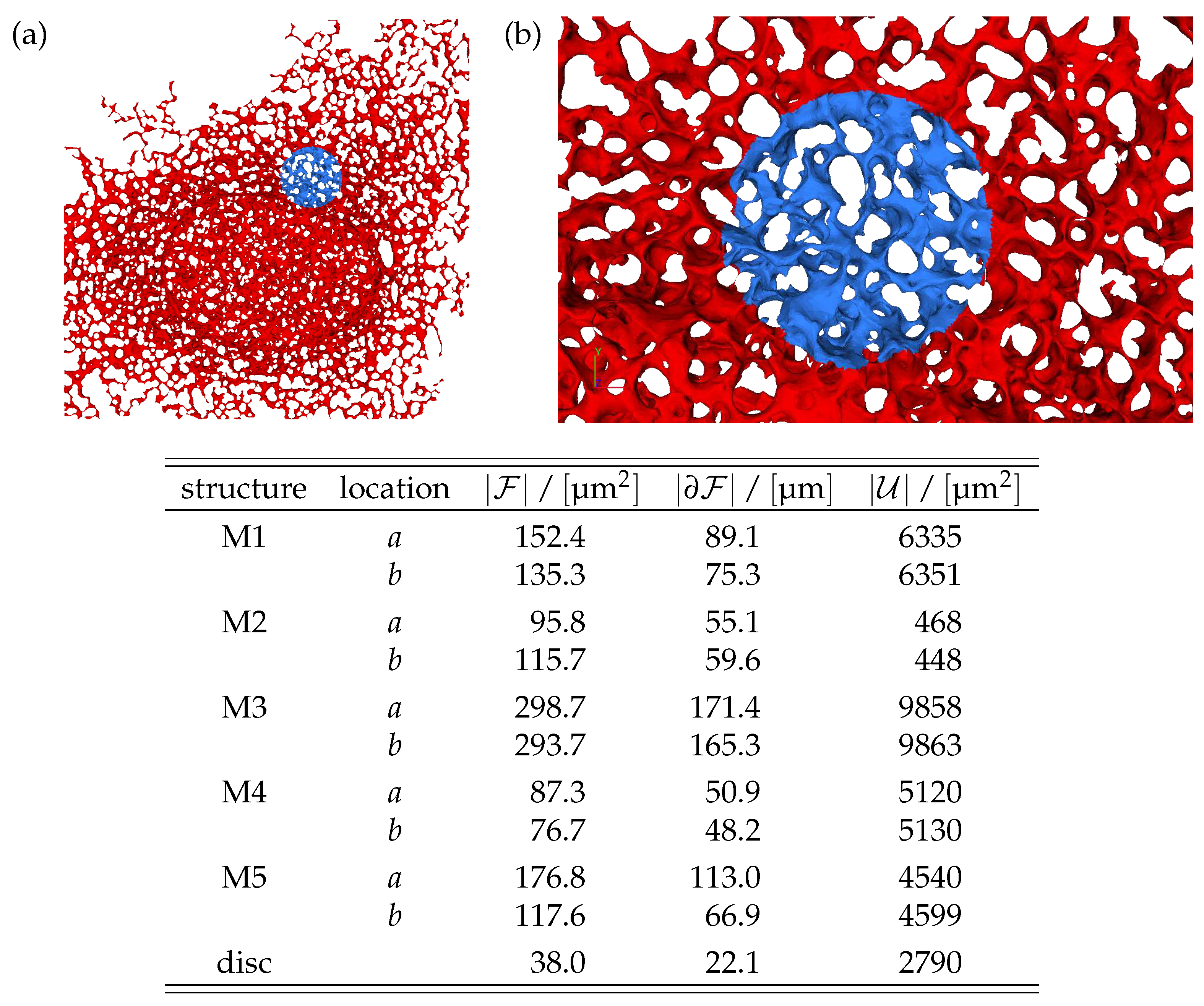
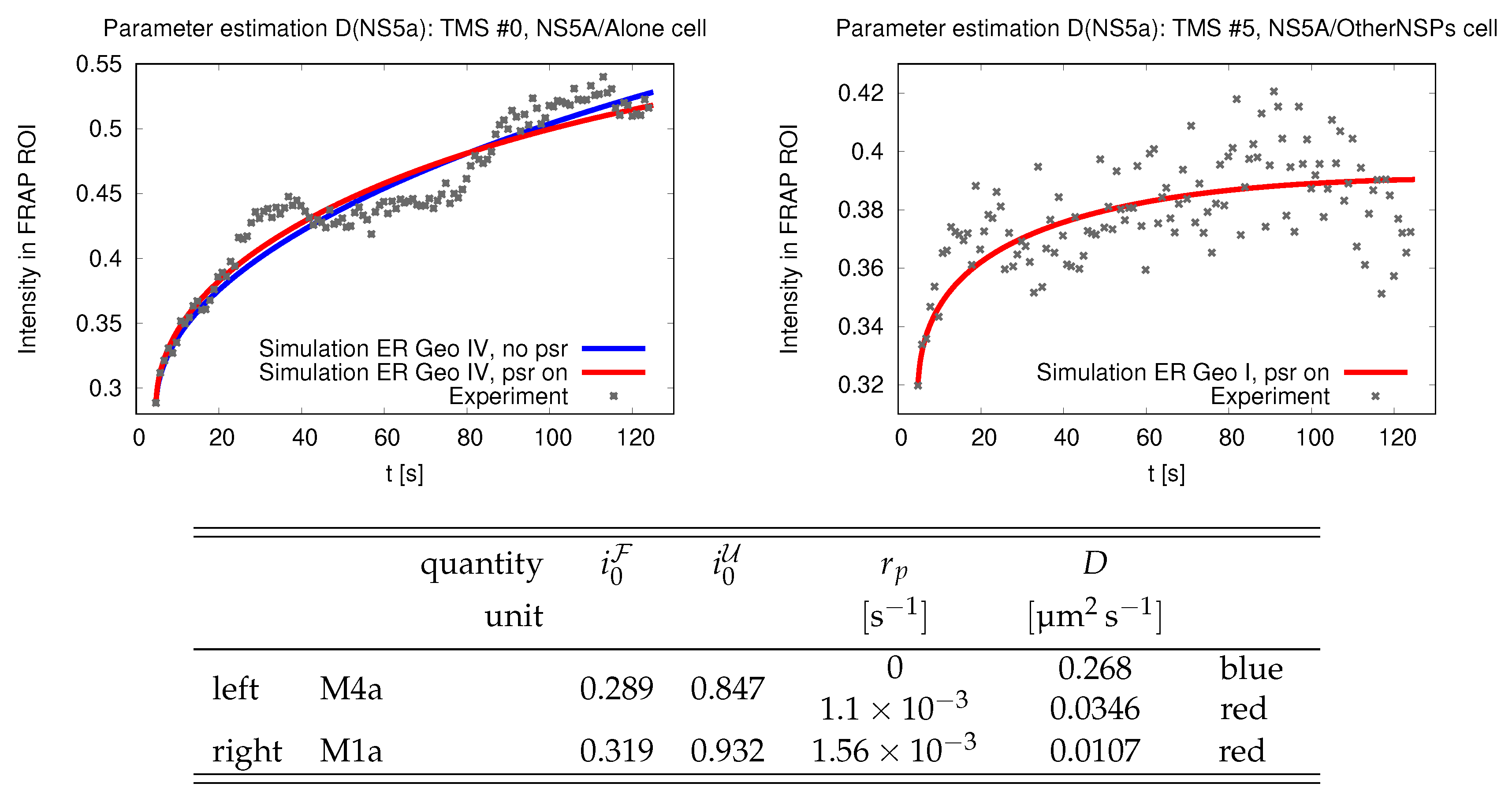
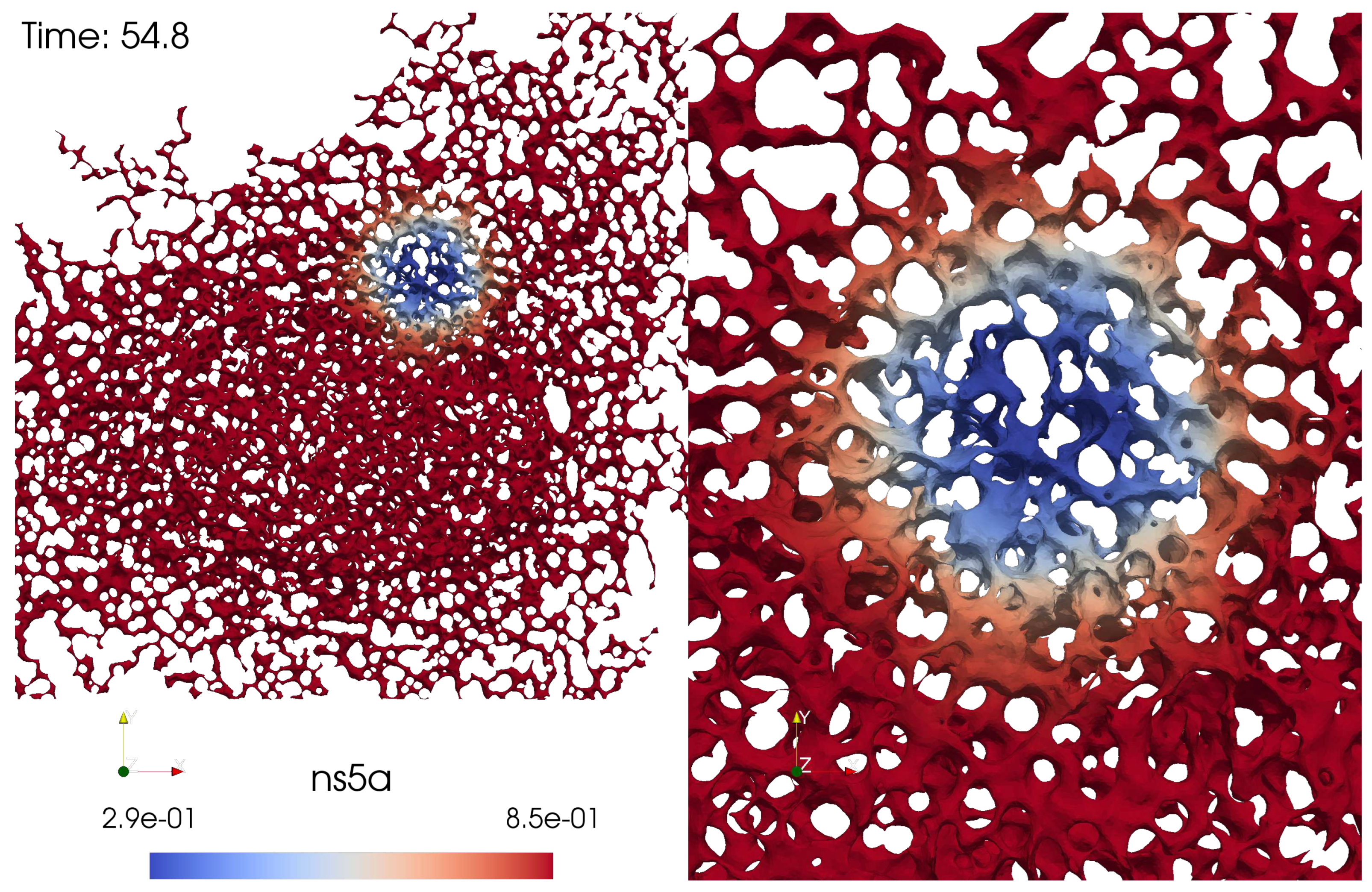
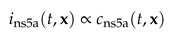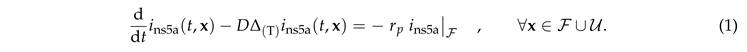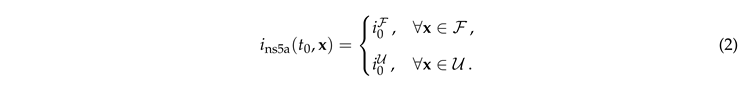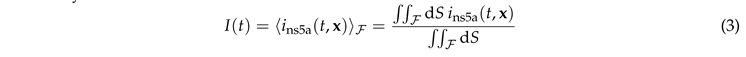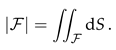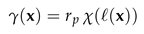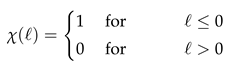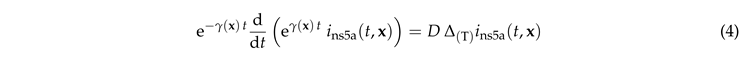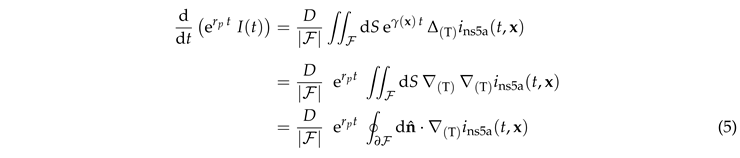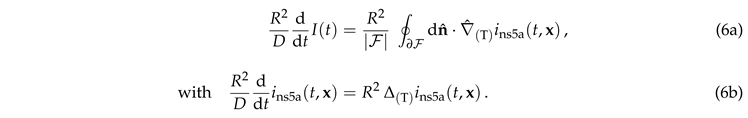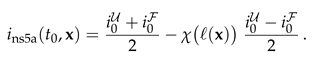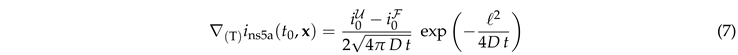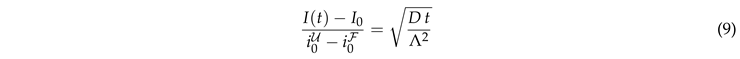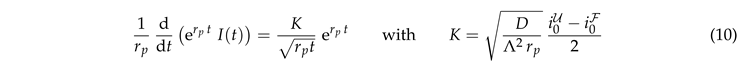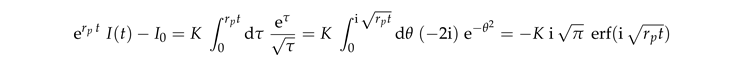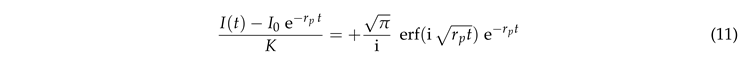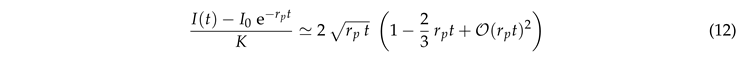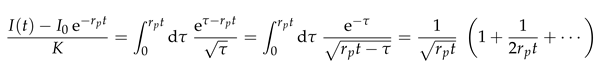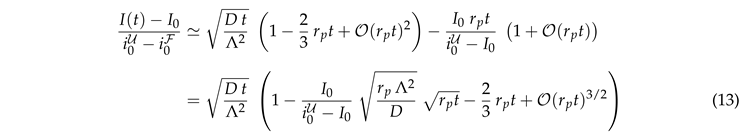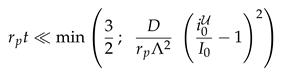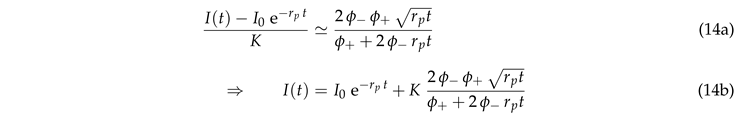In this section we discuss the non-dimensionalization of the sufPDE (1), and the scaling properties of its solutions. As a first step we establish the equations describing the intensity in .
3.2. Scaling for Negligible Photobleaching
In the absence of photobleaching,
, Equation (5) reduces to
We non-dimensionalize the equations by adopting the radius
R of the FRAP region as length scale and the dimensionless time
. Moreover, we note that the equations (
Section 3.2) are liner. As a consequence, there is a superposition principle and one can account for the different values of
and
in the initial condition by considering the normalized intensity
(see the chapter on Greens functions in Ref. [
60]).
1 As a consequence, the reduced intensity takes same evolution for every geometry of the ER, irrespective of the value of the diffusion coefficient and the initial condition.
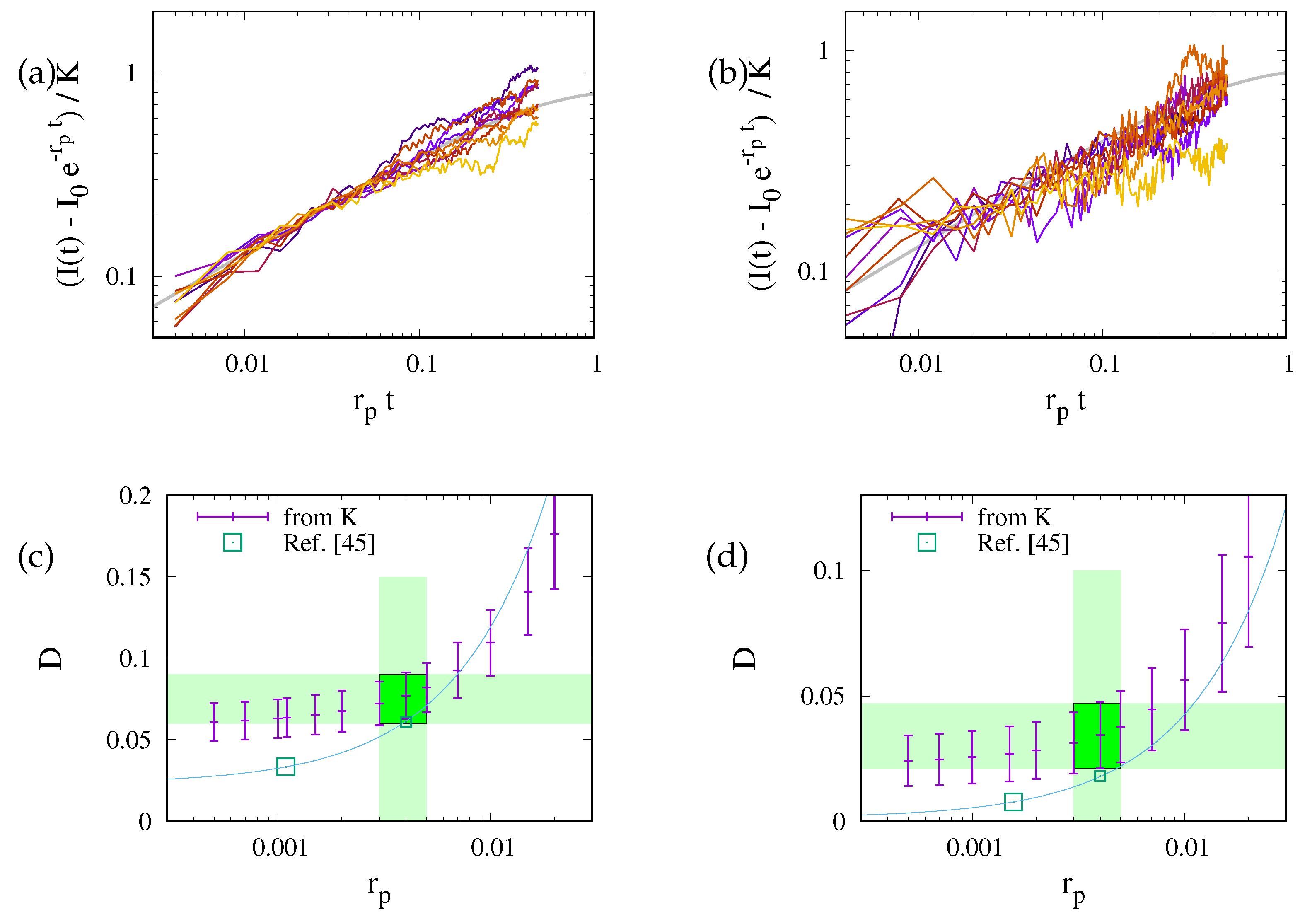
This prediction is tested in Fig. 4 where we plot data for the different geometries of the ER and for the 2D planar geometry. For each geometry the data for different diffusion coefficients collapse on a master curve. However, data for different geometries lie on distinctly different curves. The values of data for different ER geometries vary by about 20%, and those for the disk geometry (uppermost curve in the plot) take noticeably higher values. This highlights the importance of the ER geometry for the FRAP relaxation.
Figure 4.
Data collapse for the reduced density as function of the dimensionless time . Different colors refer to different values of , that take values in . The symbols refer to different geometries, as specified in the legend. (a) Simulation data with a linear scale. There are different curves for each geometry, but for any given geometry the data with different D collapse on a single line. (b) The upper panel provides a double-logarithmic plot of the data shown in (a). The lower panel shows the ratio of the data and . To a good approximation it is a horizontal line. Hence, the data lie on a power law with exponent , where the ratio provides the prefactor of the power law. For the planar geometry the prefactor takes a value slightly larger than one. For the bicontinuous structures it differs for each geometry, with values in the rage between and .
Figure 4.
Data collapse for the reduced density as function of the dimensionless time . Different colors refer to different values of , that take values in . The symbols refer to different geometries, as specified in the legend. (a) Simulation data with a linear scale. There are different curves for each geometry, but for any given geometry the data with different D collapse on a single line. (b) The upper panel provides a double-logarithmic plot of the data shown in (a). The lower panel shows the ratio of the data and . To a good approximation it is a horizontal line. Hence, the data lie on a power law with exponent , where the ratio provides the prefactor of the power law. For the planar geometry the prefactor takes a value slightly larger than one. For the bicontinuous structures it differs for each geometry, with values in the rage between and .
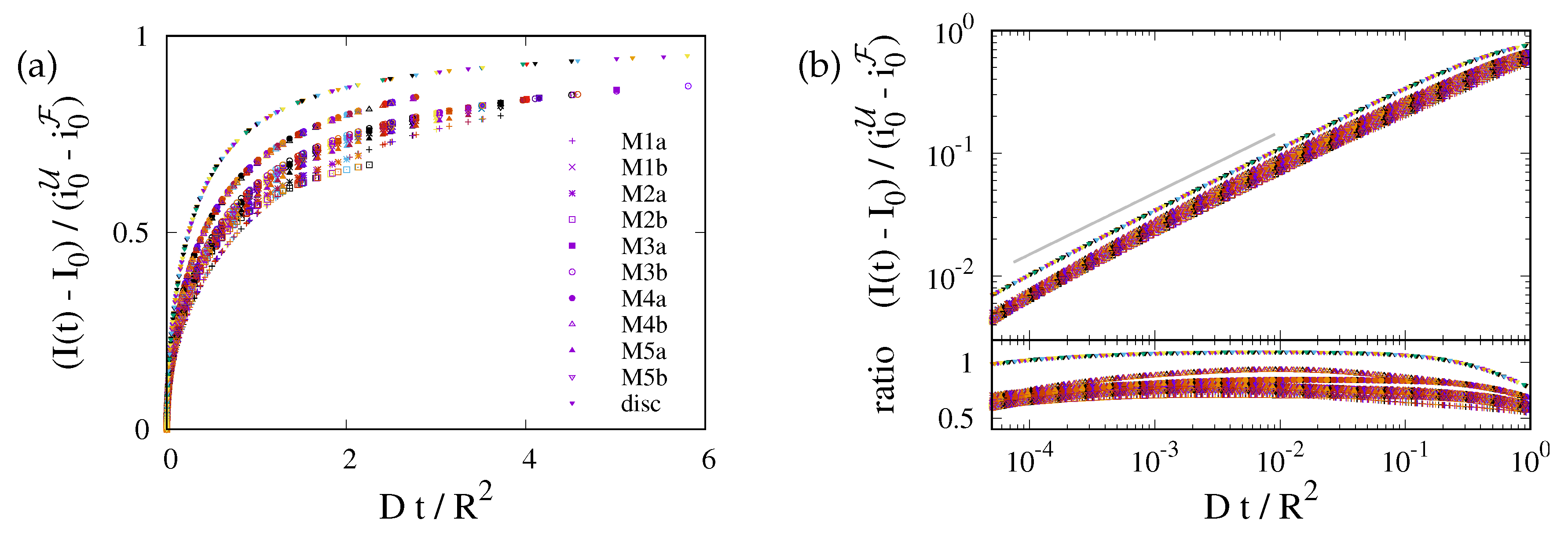
Figure 4 suggests that the relaxation of the reduced density has a time dependence proportional to the square root of the dimensionless time. We will show now that this is an immediate consequence of the form of the initial condition.
For the initial condition, Eq. (2), the intensity
takes the form of the step function,
For early times the flow is orthogonal to the surface, i.e., curvature and topology of the surface may still be neglected. The gradient of the density therefore follows the evolution of diffusive decay of a step function in one dimension
2
Right on the interface, we have
, and the exponential function takes the value of one. Hence, Eq. (6a) reduces to
Integrating time from
till
t provides
This result explains the square root dependence of the intensity observed in Fig. 4, and it provides the leading order dependence of the dependence on the surface geometry via the characteristic length scale .
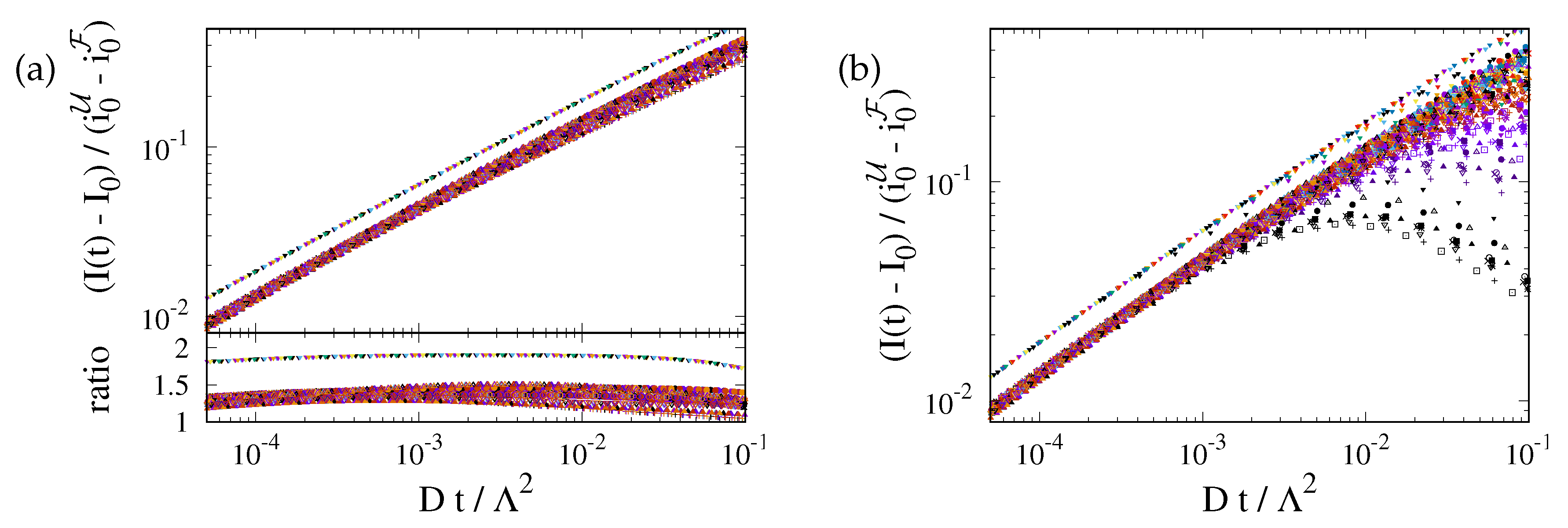
The prediction is tested in Fig. 5(a). At late time the recovery is still considerably impacted by the ER geometry. The curves for different geometries differ by up to 25%. However, at early time the transformation from a time scale
to
provides a very nice data collapse for the ER geometries. For a perfect fit the ratio of the data and
must be one. For our simulations it takes a value close to
. We attribute this difference to the impact of the nontrivial curvature of the border of
F.
3
Figure 5.
(a) Master plot, Eq. (9), for the data shown in Fig. 4(b). The values of depend on the geometry. This reduces the variance of the prefactors by roughly a factor of two. For the ER geometries they take values of about . For a 2D disk it is close to . (b) The same master plot for data with non-vanishing photobleaching, . At early times the data still follow the prediction, but now there is a cross-over to a regime where photobleaching has a severe impact.
Figure 5.
(a) Master plot, Eq. (9), for the data shown in Fig. 4(b). The values of depend on the geometry. This reduces the variance of the prefactors by roughly a factor of two. For the ER geometries they take values of about . For a 2D disk it is close to . (b) The same master plot for data with non-vanishing photobleaching, . At early times the data still follow the prediction, but now there is a cross-over to a regime where photobleaching has a severe impact.
3.3. Accounting for Photobleaching
The data collapse at early times also holds for systems with bleaching (Fig. 5(b)). However, at late times these systems cross over to a decay of the intensity that is caused by bleaching. In order to gain insight into this behavior we insert Eq. (7) into Eq. (5),
where
K is a dimensionless constant. This equation can be integrated
where erf
is the error function. Rearranging the terms provides
This prediction is tested in Fig. 6(a). Indeed we observe a data collapse. It features a cross over between different power laws for small and large values of .
For
the exponential function and the error function on the right-hand side can be expanded in a power series. This entails a square-root dependence for early times,
This is in line with Fig. 6(a) where the data for small lie close to with . Here, the correction factor accounts for the curvature effects reported in Fig. 5(a).
To find the asymptotics for
we observe that
This is in line with the data collapse in Fig. (6a) where the data for large lie close to with . Here, a correction term, , of the prefactor is expected since Eq. (7) is based on an approximation that applies only for early times.
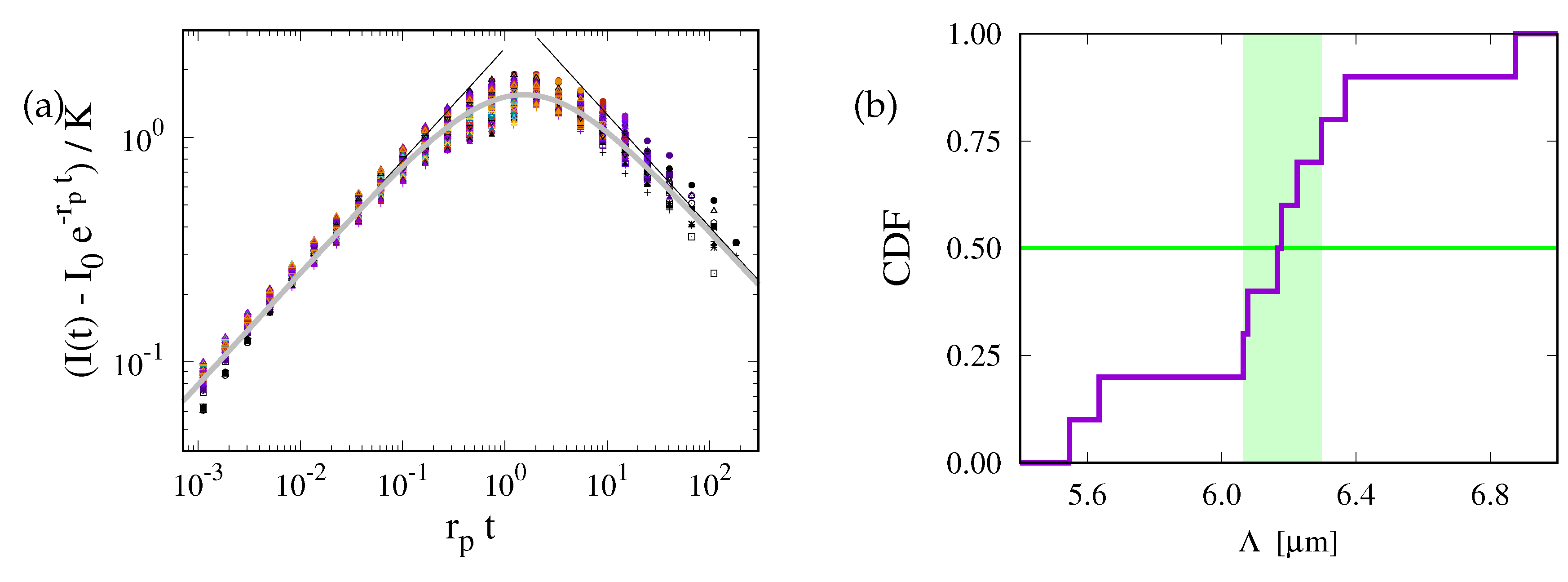
Figure 6.
(a) Data collapse of all data on a master curve predicted by Eq. (11). There is a very good collapse for small times, and the cross-over due to photobleaching is also accounted for. The two straight gray lines show the function and with and , respectively. An extrapolation that agrees very well with the observed data is provided by (thick solid gray line). (b) cumulative distribution of the values of . For with a radius of in the presently considered ER geometries the values of are sharply centered in a narrow interval with a width of (shaded region) around a median of .
Figure 6.
(a) Data collapse of all data on a master curve predicted by Eq. (11). There is a very good collapse for small times, and the cross-over due to photobleaching is also accounted for. The two straight gray lines show the function and with and , respectively. An extrapolation that agrees very well with the observed data is provided by (thick solid gray line). (b) cumulative distribution of the values of . For with a radius of in the presently considered ER geometries the values of are sharply centered in a narrow interval with a width of (shaded region) around a median of .