1. Introduction
Candida is a yeast like fungi present as normal flora of skin, mucosa and various internal organs. They commonly reside in oral cavity, throat, gut and vagina without causing any clinical problems. But they are responsible for variety of opportunistic infections when the host’s defense mechanism is compromised. Opportunistic candidiasis is ranging from skin and mucosal infection to invasive and allergic manifestations in humans. Currently, candidiasis is one of the globally found common infections that of growing medical concern. According to a large study conducted in 2013 with more than 1800 clinical fungal isolates from 31 countries, it was reported that 82% of which was caused by
Candida spp. [
1]. Globally invasive candidiasis is one of the commonest opportunistic infection with high rate of morbidity and mortality. It accounts for more than 250,000 cases and a mortality of more than 50,000 people worldwide. According to a multicentre study in five tertiary care teaching hospitals in Italy and Spain during 2008 to 2010, 995 cases have been reported. Here the overall incidence was 1.55 cases in 1000 admissions [
2]. In India, the occurrence is 6-18%, among which
Candida tropicalis is the predominant causative agent of invasive candidiasis [
3]. In most of the regions, incidence rates is either stable or increasing. But in some regions, the disease is controlled by proper patient management, hygienic practices and nutritional improvement among infants [
4].
In most of the cases, invasive candidiasis is associated with prolonged ICU stay, granulocytopenia, abdominal surgery, necrotizing pancreatitis, haematological malignancies, tumors, solid organ transplantation, preterm and malnourished infants, haemodialysis, cancer chemotherapy and other prolonged antimicrobial therapy, extremes of age, diabetes, total parenteral nutrition and presence of central vascular catheters [
5]. Invasive candidiasis includes both candidemia and deep-seated tissue candidiasis. The most common type is candidemia. Candidemia in humans is caused by more than 15 different
Candida species. The most common among them are
Candida albicans, Candida tropicalis, Candida parapsilosis, Candida glabrata, Candida krusei and
Candida auris. Haematogenous dissemination or direct inoculation of
Candida species to a sterile region, such as the peritoneal cavity, causes deep-seated candidiasis [
6].
Distribution of
C. albicans and other non-albicans
Candida (NAC) species varies greatly according to the geographical area, underlying medical conditions of patients, antifungal agents received and other conditions in hospital and patient’s immune status.
C. albicans was found to be the most frequently isolated pathogen in Northern and Central Europe and the United States, while NAC species were found to predominate in Asia, Southern Europe, and South America, according to a recent systematic review that evaluated the geographical distribution of
Candida species in blood samples from inpatients in various parts of the world [
5,
7]. Shift of
Candida species from commensal flora to pathogenic agent is a result from interplay between host’s immune system and fungal virulence. Clinically invasive candidiasis ranging from asymptomatic to deep-seated candidiasis with more than 70% mortality rate. Deep seated candidiasis includes intra-abdominal abscess, peritonitis, osteomyelitis etc. [
8].
Candida species can resist host’s defense mechanism by expressing several virulence factors including adhesins and invasins, hydrolytic enzymes, toxins and pseudohyphal formation. Expression of these virulence factors may vary according to different species, geographical origin, type of infection and susceptibility to the host. The resistant mechanisms are different such as expression of adhesins and invasins, thigmotropism, biofilm formation, secretion of hydrolytic enzymes such as hemolysins, aspartyl protease and serine proteases and phenotypic switching mechanism. In addition to that adaptation to fluctuations in environmental pH and temperature, metabolic flexibility, powerful nutrient acquisition system, and stress response mechanism are also added to their pathogenesis [
9].
Even though candidiasis is a global infection, currently the treatment options are limited to only four antifungal drug families, which are azoles, polyenes, echinocandins and pyrimidine analogues [
5]. Candidiasis is a curable condition with accurate diagnosis and management of patients. The clinical practice guidelines for management of candidiasis is released by the Infectious Disease Society of America (IDSA) in 2016. Generally, for oral and mucocutaneous candidiasis topical application of azole creams like cotrimoxazole, miconazole, ketoconazole and econazole are recommended. In addition, itraconazole solution for topical application may be used. While for systemic infections, echinocandins or intravenous administration of amphotericin B are used. Some azole drugs like ketoconazole, fluconazole and itraconazole can be given orally [
10].
Resistance to various azole drugs to different species of
Candida are emerging nowadays. Hence, antifungal susceptibility testing (AFST) is crucial in providing targeted therapy. Antifungal resistance can be either clinical or microbiological. Clinical resistance refers to the persistence of a fungal infection even after the complete course of treatment and microbiological resistance is the in vitro resistance towards some particular drugs compared to the other isolates of the same species. Microbiological resistance can be investigated and can be determined. Furthermore, microbiological resistance may be either intrinsic or acquired. Intrinsic resistance occur naturally without any prior exposure to drugs. In contrast, acquired resistance develops either after repeated exposure to an antimicrobial drug or due to altered gene expressions due to mutation [
11]. Among the four groups of anticandidal drugs resistance to azole group of drugs are more common. Resistance to polyene group of drugs like amphotericin B is less commonly reported [
12].
Azoles generally inhibit enzymes like lanosterol 14α-demethylase, which is involved in final stage of ergosterol synthesis resulting in accumulation of sterol precursors that leads to cell membrane instability and impaired fungal growth. Azole resistance in
Candida species is evolved due to the combination of various mechanisms [
13]. It includes decrease in the intracellular concentration of the drug due to expression of ATP-binding cassette (ABC) or major facilitator superfamily (MFS) transporters, mutation in drug target like mutation in ERG 11 gene and increased production of lanosterol 14α-demethylase due to multiple factors like gene duplication, mutation in promoter gene or mutation in gene encoding for the target enzyme [
14]. CDR1 and CDR2 are two important transporters responsible for azole resistance in
Candida spp. [
15]. Deletion of both alleles in CDR1, over expression of either CDR1 or CDR2 or both, fungal homologues of CDR1 and CDR2 such as CgCDR1, CgCDR2, CgSNQ2 in
C. glabrata, CdCDR1 and CdCDR2 in
C. dubliniensis and ABC1 in
C. krusei are some of the mechanisms described for resistance [
14]. MFS transporters are involved in the transport of molecules by means of a proton gradient mechanism in the plasma membrane [
16]. Ninety-five MFS transporters has been identified in
C. albicans among which only the product of multidrug resistance 1 gene (MDR1) is associated with clinical azole resistance [
15]. Expression of transcription factor multidrug-resistance regulator 1 (MRR1), inactivation of MRR1 in clinical
C. albicans strains blocked MDR1 expression, gain-of-function mutations in MRR1 causing constitutive up-regulation of MDR1 are some of the mechanism involved in MDR1 expression [
14].
Increased production of ergosterol by over expression of ERG 11 gene have also found to be one of the mechanism in azole resistance predominantly in
C. albicans. ERG 11 overexpression is linked to mainly two mechanisms in
C. albicans: (i) an increase in the expression of the transcription factor Upc2, which controls the expression of the majority of genes involved in biosynthesis of ergosterol and (ii) production of an isochromosome with two copies of left arm of chromosome 5, which contains the ERG 11 gene, or the chromosome is duplicated in its entirety [
13]. In
C. dubliniensis, C. glabrata, C. krusei, C. parapsilosis, and
C. tropicalis, there has also been a link between enhanced ERG 11 expression and azole resistance [
17].
Changes in target site of enzyme resulted by mutations are the frequently encountered mechanism in azole resistance. Therefore, most of the study were conducted to determine mutation in ERG 11 gene and in
C. albicans isolates. Here ergosterol synthesis of the yeasts get impaired due to mutation in ERG 11 gene by changing the structure of lanosterol 14α- demethylase which result in decreased affinity of this molecule to azoles [
14,
15]. There are more than 140 amino acid substitutions reported in
C. albicans. Some of those substitutions resulting only in azole resistance are K143R, S405F, G464S, I471T and R467K [
18]. Other than
C. albicans, mutation in ERG 11 gene have also been reported in
C. dubliniensis, C. tropicalis, C. krusei, C. kefyr and
C. parapsilosis isolates [
14]. Currently, many clinical isolates with reduced fluconazole susceptibilities carry mutations in ERG 11 gene. In a study conducted by Stephanie A et al, among the 63 isolates that were determined to be resistant to fluconazole, 55 carried a mutation in ERG 11 that led to at least one amino acid substitution [
19]. Fluconazole resistant
Candida species (FRCS) are responsible for 44,800 infections and 2,200 deaths in the United States, according to a 2019 report by the Centres for Disease Control and Prevention (CDC) [
20].
C. albicans was the most common causative agent in the study.
There is a challenge in successful treatment if the infection is caused by the azole resistant isolates. Therefore, determining the local epidemiology of the resistant isolates and understanding the mechanism of resistance will help in better treatment and management of patients. There are limited studies available in India for detection of mutation in ERG 11 gene in fluconazole resistant Candida albicans (FRCA) isolates. Therefore, this study was aimed to determine the profile of FRCS isolates and to detect the mutation in ERG 11 gene in FRCA isolates obtained from patients admitted in a tertiary care hospital, South India.
2. Materials and Methods
2.1. Inclusion criteria
A cross-sectional study was conducted from April to December 2021 in the Mycology section of the Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education & Research (JIPMER), Puducherry, India. All non-duplicate 150 FRCS isolates which were stocked and available in the Mycology section, which were collected from the patients admitted in JIPMER from September 2019 to December 2021 were included in the study.
2.2. Ethics
The study was conducted after getting approval from Institute Ethics Committee (IEC) - Project no. JIP/IEC/2021/068 with waiver of consent.
2.3. Clinicodemographic details
Clinicodemographic details including age, gender, ward, sample, diabetic status and HIV status of the patient from whom FRCS was isolated, were collected from the laboratory register or hospital information system (HIS) and recorded.
2.4. Identification of Candida species by MALDI-TOF MS
All the 150 FRCS isolates were revived by subculturing onto sabouraud dextrose agar (SDA). Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry - MALDI-TOF MS (VITEK ® MS, Biomerieux, France) was used for the identification of FRCS isolates revived on SDA as per the manufacturer’s instruction.
2.5. Detection of fluconazole susceptibility by VITEK 2 system
Fluconazole susceptibility of the all FRCS isolates were confirmed by VITEK 2 (VITEK® 2, BIOMERIEUX, USA) automated system by using YS08 (VITEK® 2 AST- YS08, 2881896403) as per the manufacturer’s instruction.
2.6. DNA Extraction and amplification of the FRCA isolates
DNA extraction of the FRCA isolates were performed by QIAamp DNA Mini kit (Qiagen - 51304, Hilden, Germany). Following which PCR was performed for each FRCA isolates as per the method described by Xu et al [
21], and optimized in our laboratory. There are three primer sets which amplify whole length of ERG 11 gene. However, we could optimize amplification from one primer set - ERGSec3A (5’ -AGGTGGTG ATTTGAATGATTTGACTT-3’) and ERGSec3B (5’ -GAACTATA ATCAGGGTCAGGCA CTTT-3’) targeting for an expected PCR product extending from 1067 to 1576 bp of the ERG 11 gene. There was a limited financial support, which restricts us for amplifying PCR products from the other two primer sets. Optimization was done with thirty- three cycles of amplification were done with a total reaction volume of 25 µl. Total duration of PCR was 2 hours and 26 minutes.
2.7. Quality control
A positive clinical FRCA strain was used as positive control and nuclease free water was used as negative control.
2.8. Gel electrophoresis and documentation
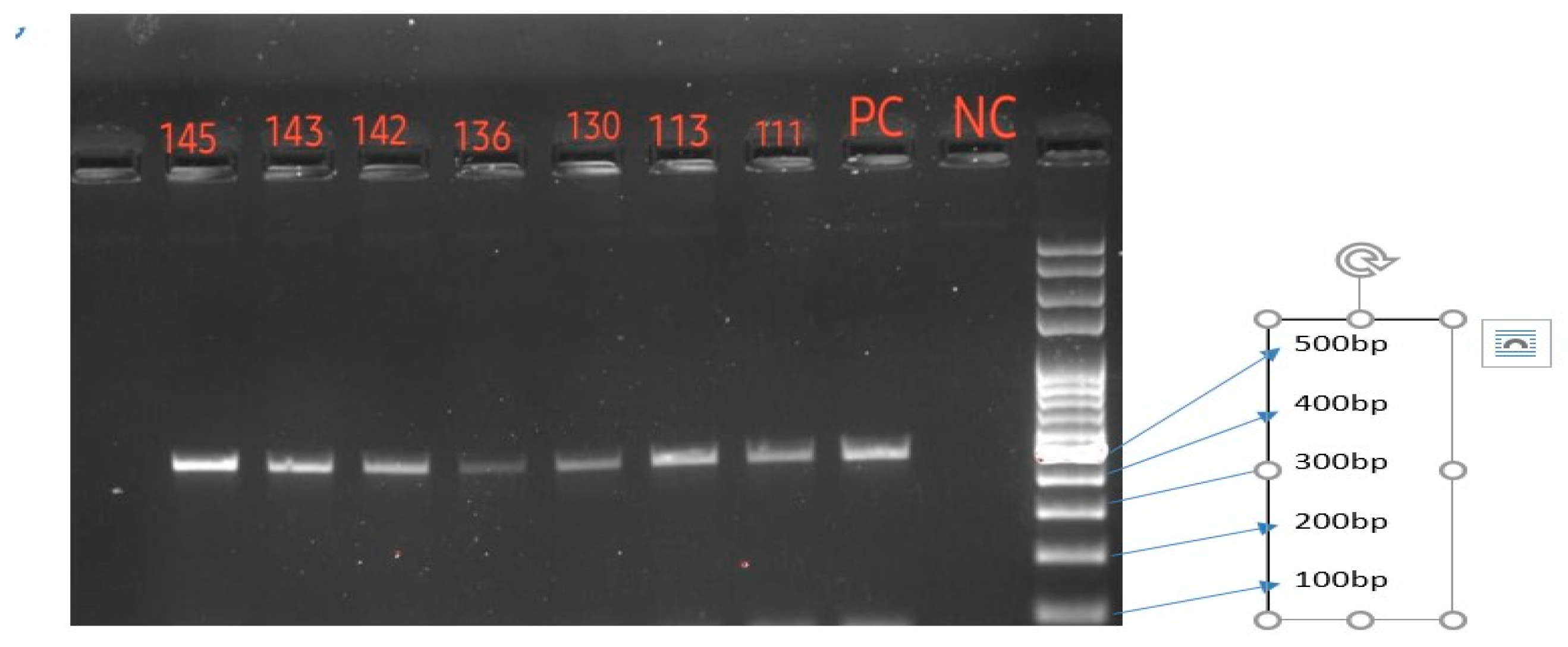
Gel electrophoresis was performed using gel electrophoretic equipment -BIO-RAD, PowerPacTM –HV, USA. Amplification products of ERG 11 gene were observed using gel documentation system - BIO-RAD, Gel Doc XR System, USA. Amplicon product sizes were recorded and compared with 100 bp DNA ladder. FRCA isolates which showed 510 bp band in gel electrophoresis were considered successful amplification of the part of ERG 11 gene.
2.9. Detection of mutation in ERG 11 gene
The ERG 11 gene products were purified and subjected for sanger sequencing with an ABI PRISM DNA analyzer (Applied Biosystems) and mutation were detected by using Mega (version 7) software by comparing with the published GenBank sequence AF153844.1 of Candida albicans strain ATCC 28516.
2.10. Phylogenetic tree
Phylogenetic tree was constructed by using published GenBank sequences from five different continents. Forty nucleic acid sequences from Asia including twelve sequences from this study, 10 sequences each from Africa, Australia and Europe, seven sequences from South America and one sequence of
Candida albicans ATCC 28516 were used for the phylogenetic tree construction. The evolutionary history was inferred using the Neighbor-Joining method [
22]. The bootstrap consensus tree inferred from 1000 replicates were taken to represent the evolutionary history of the taxa analyzed [
23]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed.
2.11. Statistics
Categorical variables like gender, ward, sample, diabetic status, Human Immunodeficiency virus (HIV) status, presence of ERG 11 gene, and mutation in ERG 11 gene were expressed in percentage. Continuous variable like age was expressed as mean with standard deviation.
4. Discussion
Candidiasis is a growing medical problem. Candidemia is the most serious clinical presentation among candidiasis. A limited study is available which describes incidence or prevalence of candidemia involving multiple hospitals and especially in India. In South Asia, prevalence of candidemia were observed ranging from 0.16 to 4.53 cases per 1000 hospital discharges and 11.7 cases per 1000 ICU discharges [
24]. Invasive candidiasis includes candidemia as well as subcutaneous and deep-seated tissue candidiasis. The important causative agents are
Candida albicans, Candida tropicalis, Candida parapsilosis, Candida glabrata, Candida krusei and
Candida auris. Fluconazole is the most common antifungal drug used in the treatment of candidiasis. Indiscriminate use of fluconazole results in increase in the FRCS isolates. CDC observed 7%
Candida spp. isolates were resistant to fluconazole during 2012 to 2016 and
C. albicans was the most common causative agent [
20]. Mutation in the ERG 11 gene of
C. albicans is considered for the fluconazole resistance and therefore mutation in the ERG 11 gene of
C. albicans isolates were studied most. Molecular mechanism were not studied much in India. Therefore, here the study was conducted on FRCS isolates and the mutation in a part of ERG 11 gene of FRCA isolates. A total of 150 FRCS isolates were available in the Mycology section of Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India from September 2019 to December 2021 and were included in the study. Isolates were received from both male and female patients almost in an equal proportion. It indicates that FRCS causes infection equally to both genders. FRCS isolates were recovered from patients from newborn infants to elderly up to 80 years with a predominant age group between 21 years to 30 years. Very few studies were encountered on FRCS. A similar study conducted in Iran by Sardari et al observed 23% FRCA isolates were isolated from the patients with age group of 31 to 40 followed by 20% from 21 to 30 [
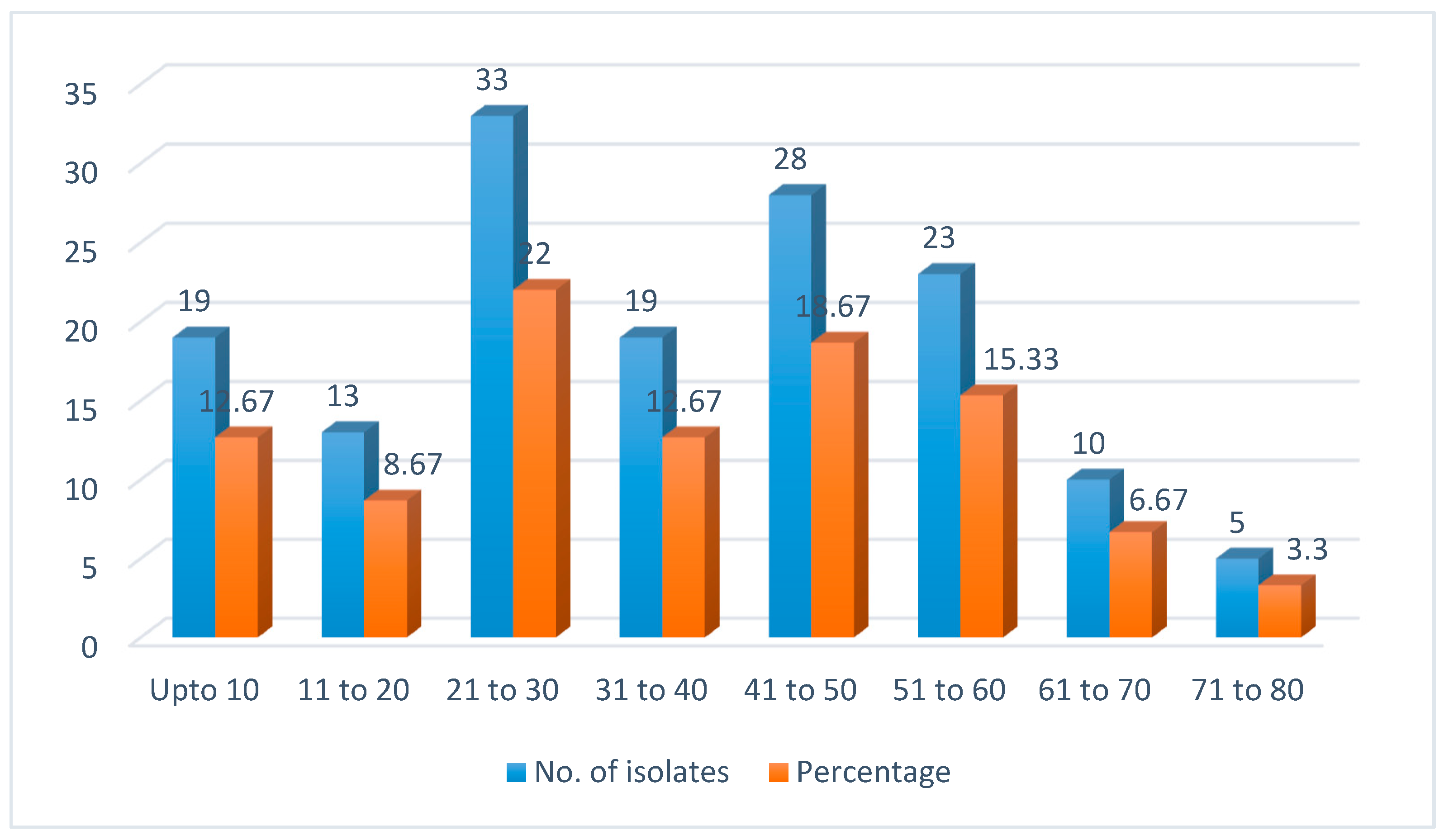
25]. Both the studies showed that candidiasis caused by fluconazole resistant isolates commonly affect adult and earning population. It needs further evaluation to prevent it.
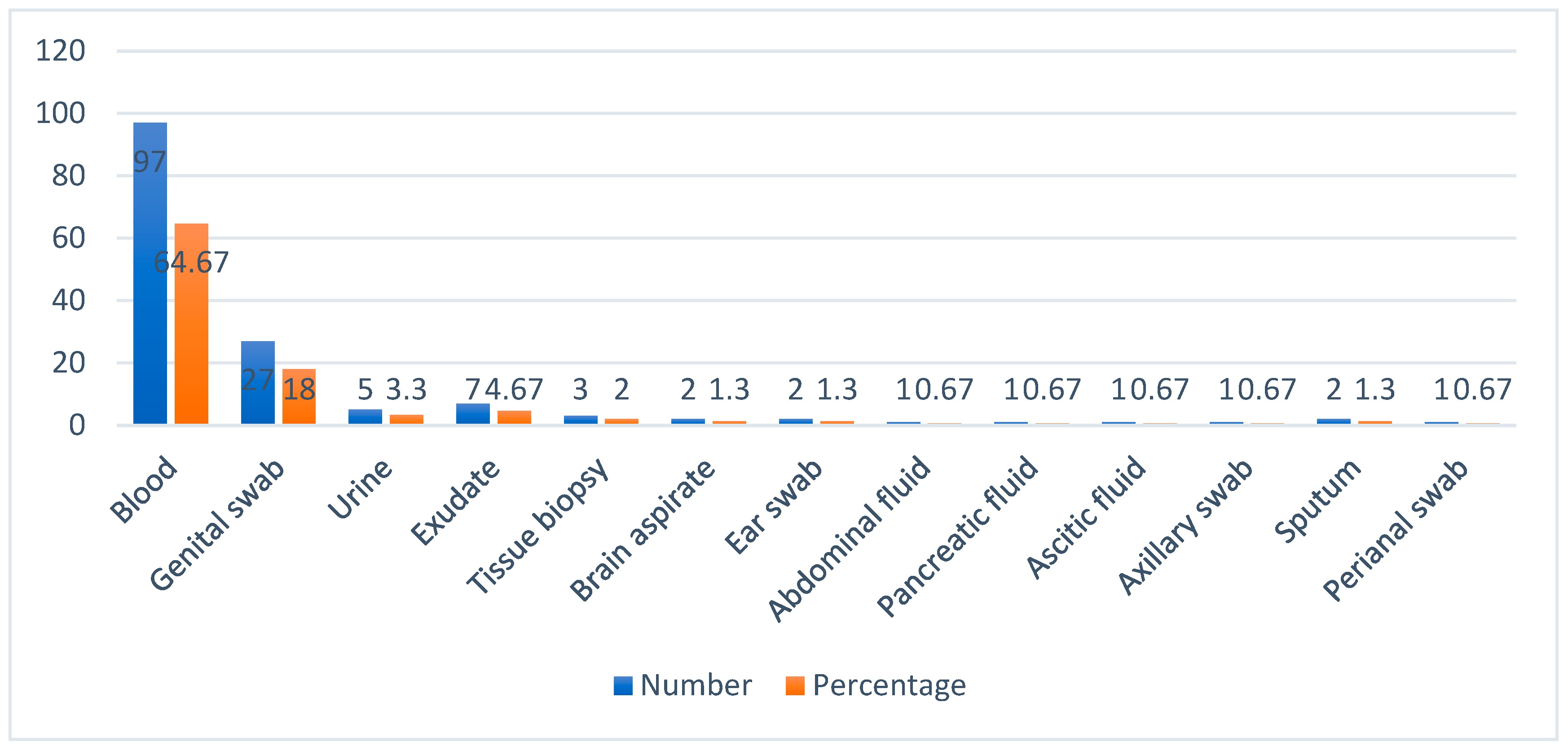
Candidemia was the predominant form of candidiasis (64.7%) followed by vaginal candidiasis (18%) in this study. Similar finding was observed by Chakrabarti et al in 2014. He reviewed candidemia as the most prevalent clinical condition in hospitalized patients with a higher mortality rate. He also identified abdominal candidiasis as one of the common form of candidiasis in South India whereas we identified only 2% gastro intestinal candidiasis in our study [
24]. Earlier study from our Institute observed 27.3 % cases of vulvovaginal candidiasis during 2017 [
26]. Vijaya et al identified 17.7% cases of vaginal candidiasis in 2014 [
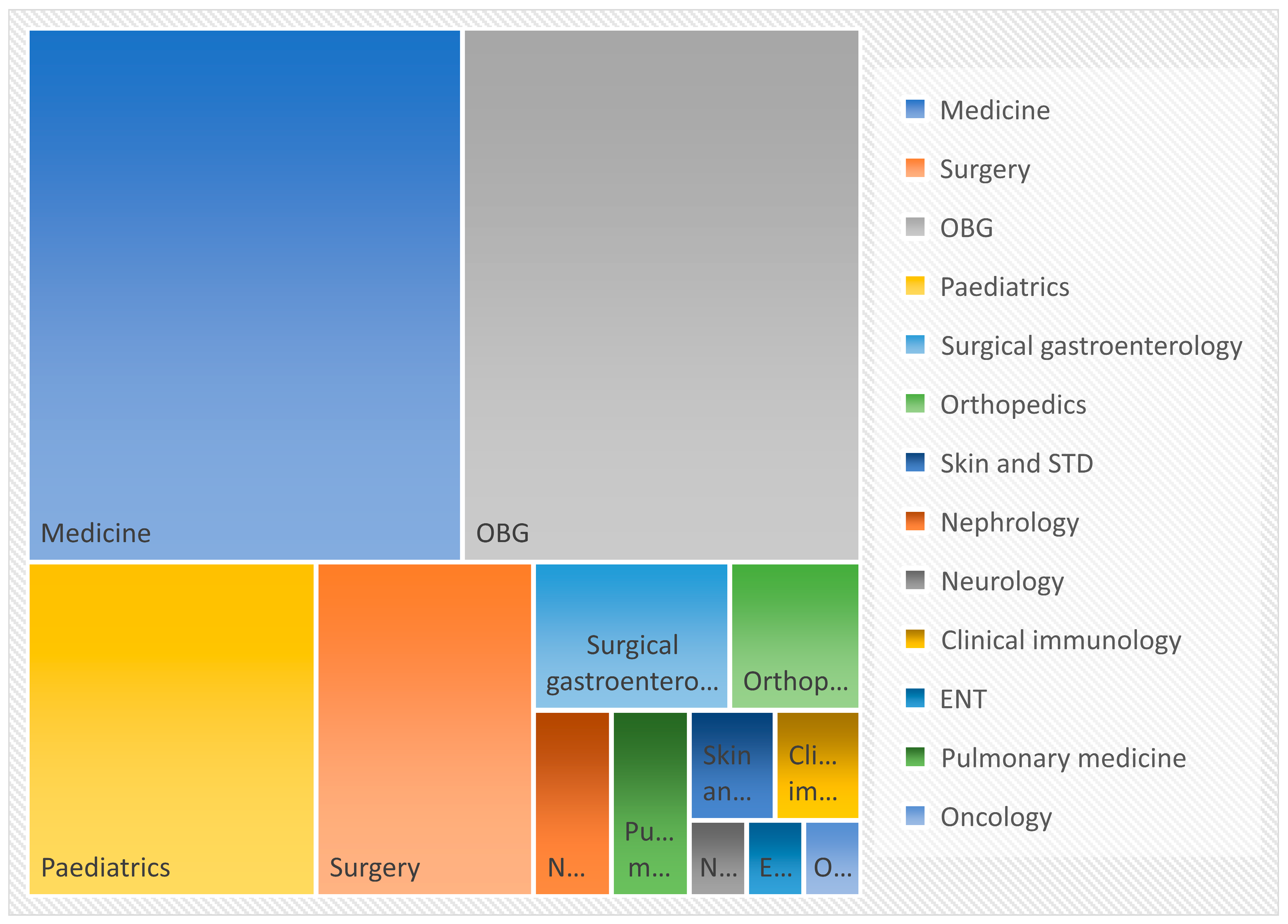
27]. Most of the isolates were recovered from samples received from Medicine department i.e., 48 out of 150 (32%) followed by Obstetrics and Gynaecology, Paediatrics, Surgery, Surgical gastroenterology, Orthopaedics, Nephrology, Pulmonary Medicine, Skin and STD, Clinical Immunology department, Neurology, ENT and Oncology departments. According to a prospective multicentric study conducted in Spain between 2011 and 2016, 204 (55.3%) were from medical wards, while 165 (44.7%) of the 369 cases admitted in a hospital were from surgical wards [
28]. The most common presentation of candidiasis is candidemia who are admitted in medical speciality and the same was observed in this study.
Candidiasis is linked to a number of comorbid illnesses, including diabetes, HIV/AIDS, indwelling catheters, and antibiotic overuse. Here, 92 (61.3%) patients were diabetic and was the most common predisposing factor. India has now become a diabetic hub and therefore diabetes is the commonest risk factor for the development of candidal infection. Goswami et al. found a statistically significant difference in the isolation rate of various
Candida spp. in diabetic patients (46%) compared to control subjects (23%) [
29]. Even though candidiasis is a prevalent opportunistic illness in HIV patients, none of the isolates in our investigation was from HIV patients. Khan et al. did a prospective study on AIDS patients and found that, out of a total 165 HIV-positive individuals, 80 were diagnosed with candidiasis [
30]. It may be due to active antiretroviral therapy provided by the government hospitals to HIV infected persons which prevents candidal infection in our region.
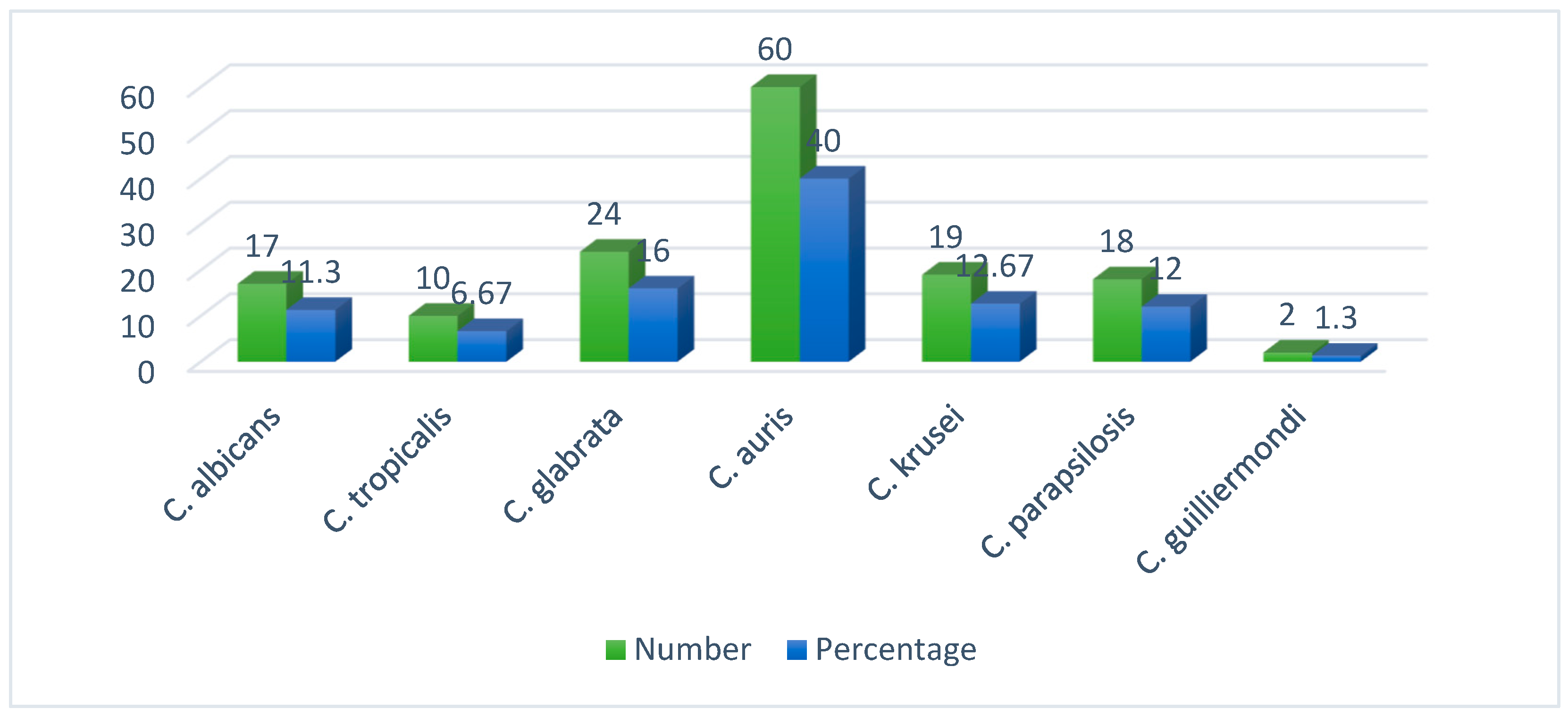
All 150 FRCS isolates were identified by MALDI-TOF MS.
C. auris was the predominant species isolated in this study. Forty percentage (60/150) isolates were of
C. auris followed by 16% (24/150)
C. glabrata, 12.7% (19/150)
C. krusei, 12% (18/150)
C. parapsilosis, 11.3% (17/150)
C. albicans, 6.7% (10/150)
C. tropicalis and 1.3% (2/150)
C. guilliermondii. According to a retrospective study of six years from China, the most common
Candida species identified was
C. albicans (46.3%), followed by
C. parapsilosis (19.5%),
C. glabrata (15.9%), and
C. tropicalis (14.6%). Few isolates were found resistant to fluconazole and maximum number were observed in
C. parapsilosis, followed by
C. tropicalis and
C. glabrata. None of the
C. albicans isolates were resistant to fluconazole [31}. Whereas, there is a shift towards non-albicans candidemia in India and here
C. tropicalis is the most common causative agent of candidemia. Tak et al in his study identified
C. tropicalis (39%) as the commonest isolate followed by
C. parapsilosis (18-20%),
C. albicans (12-14%) and
C. glabrata (11-12%) [
32]. It is found that fluconazole resistance in
C. albicans is rare in the United States, affecting just 0.5–2% of cases. Conversely
, C. parapsilosis,
C. tropicalis, and
C. glabrata had greater rates, ranging from 4-9%, 2-6%, and 11-13%, respectively [
33,
34]. In contrast to these resistance patterns the newly emerging
Candida species
, C. auris have shown a higher resistance rate of 93% [
35].
C. krusei have shown intrinsic resistance to fluconazole [
36]. Similarly in another recent USA study describing that the least resistant species to fluconazole is
C. tropicalis (2.3%), followed by
C. parapsilosis (3.4%),
C. albicans (3.5%) and
C. glabrata (7.8%) [
37]. Rise of antimicrobial resistance is considered a global public health problem. Therefore, Indian Council of Medical Research (ICMR) is monitoring antimicrobial resistance in India through a network of diagnostic laboratories. According to the ICMR annual report 2022 January to 2022 December, antifungal susceptibility profiling showed that 93% isolates of
C. albicans and
C. tropicalis were fluconazole susceptible, although
C. utilis,
C. parapsilosis, and
C. glabrata showed diminishing susceptibility rates (77%–85%).
C. auris had lowest susceptibility percentages to fluconazole [
38]. According to a recent study in India during 2020, 67% of candidemia detected from Corona virus disease patients were caused by
C. auris. Several other major outbreaks of candidemia also been reported from various parts of world like Columbia, South Africa, United Kingdom and United States [
39,
40,
41].
C. auris is resistant to fluconazole, amphotericin B, and echinocandins and therefore it is unique and are multi-drug resistant [
42].
Molecular characterization of FRCA ERG 11 gene were done in this study to detect mutations in the gene. Out of 17 isolates, only 12 (70.6%) isolates of FRCA had amplified a part of ERG 11 gene in this study. It may be postulated that five FRCA isolates may have mutations at the primers site and therefore they may not had amplified. Sequencing of these amplified products detected a single missense mutation G1309A (V437I) in one FRCA isolate and eight different silent mutations were observed. Here most of the FRCA isolates had an average of four silent mutations (observed in six isolates) and a single isolate had six silent mutations. Eight different silent mutations were observed among FRCA isolates - T1296C 10 (83.3%), C1203T 8 (66.7%), A1440G 8 (66.7%), C1302T 6 (50%), T1470C 6 (50%), T1140C 3 (25%), T1110C 2 (16.7%), and T1284C 1 (8.3%) in this study. Fluconazole resistance in
C. albicans was said to be due to mutation in ERG 11 gene in other similar studies but limited in number. Sardari et al studied 216 clinical
Candida isolates including 100 FRCA isolates and identified sixteen different nucleic acid substitutions. Among which only two showed amino acid substitutions - E266D and V488I [
25]. In an another study conducted by Paul et al in Chennai, all the fluconazole resistant
Candida isolates showed mutation in ERG 11 gene including two extensive mutations. They found many non-sense mutations and missense mutations like Y18D, D23Y, V28L, G36D, Y41N, F49L, R53P, M63R, E243D etc. and they concluded that these mutations are contributing to fluconazole resistance [
43]. Mane et al in India observed 6 amino acid substitutions in FRCA isolates including E116D, F145L, E226D, I437V, P406L and Q474H. But they concluded that azole resistance was primarily due to CDR1 mediated mechanisms [
44]. Xiang et al, found 17 aminoacid substitutions in ERG 11 gene including seven novel mutations such as K143Q, Y205E, A255V, E260V, N435V, G472R, and D502. They observed V437I (G1309A) mutation which was also identified in this study. But they observed this mutation in both fluconazole resistant and susceptible isolates, suggesting that there is no association of this mutation with fluconazole resistance [
45]. We observed less number of mutations in FRCA isolates. It may be due to less number of FRCA isolates studied and we could only amplify a part of ERG 11 gene - from 1067 to 1576 bp part of the 1587 bp of ERG 11 gene. Mutation may be present in other part of ERG 11 gene and five of FRCA isolates failed to amplify. Even though there were many limitations in this study, but we are having opinion that resistant in FRCA isolates are primarily due to some other mechanism other than mutations in ERG 11 gene as suggested by Mane et al [
44].
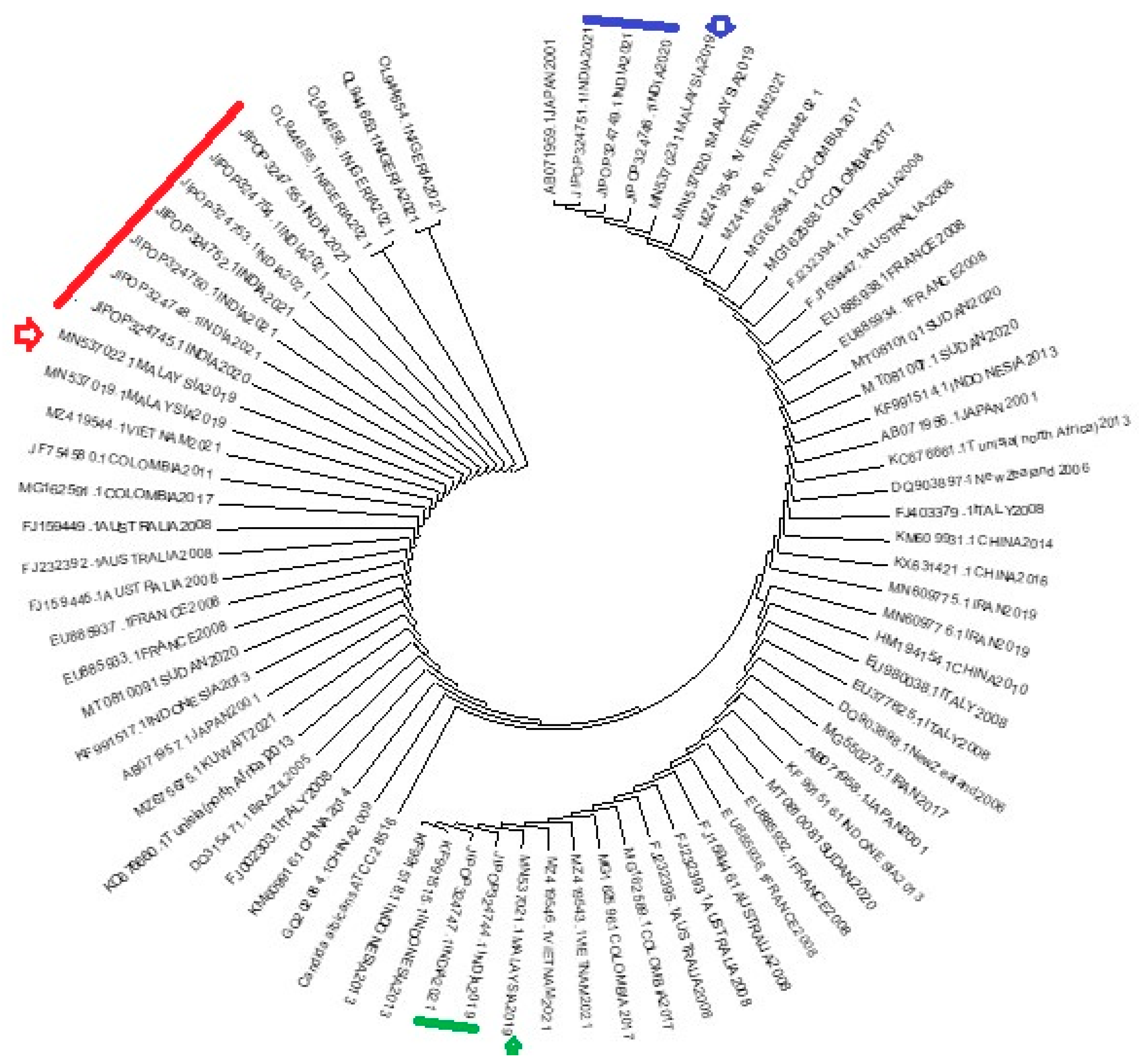
A phylogenetic tree was constructed combining 12 sequences of this study with a reference sequence of AF153844.1 ATCC 28516
C. albicans and 65 published sequences in GenBank from five different continents - Asia, Africa, Australia, Europe and South America. We observed that our isolates arranged in three clusters (
Figure 6). Interestingly, all twelve isolates and the three clusters showed relatedness to Malaysian 2019 isolates. Here all twelve isolates were recovered from 12 different patients, from different sites and at different time frame - from 2019 to 2021. We don’t have travel history of these 12 patients to Malayasia, as this is primarily a retrospective study. But it can be concluded that FRCA isolates circulating in our region might have ancestral source in Malayasia. Further study is required to identify the mode and source of transmission of FRCA isolates. It will help us to implement appropriate preventive measures.