Submitted:
16 December 2023
Posted:
18 December 2023
You are already at the latest version
Abstract
Keywords:
INTRODUCTION
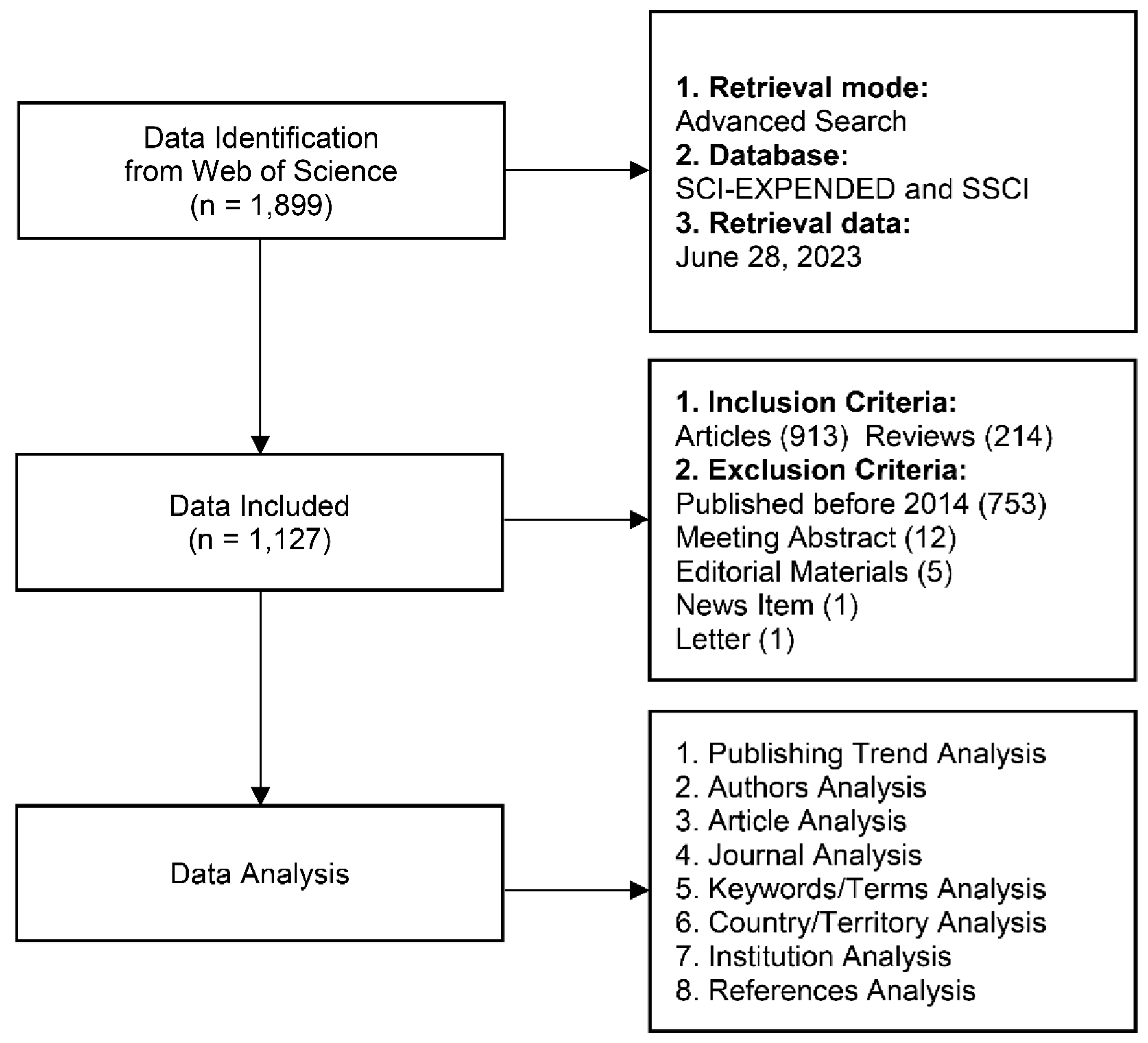
MATERIAL AND METHODS
Data Collection
Data Analysis
RESULTS
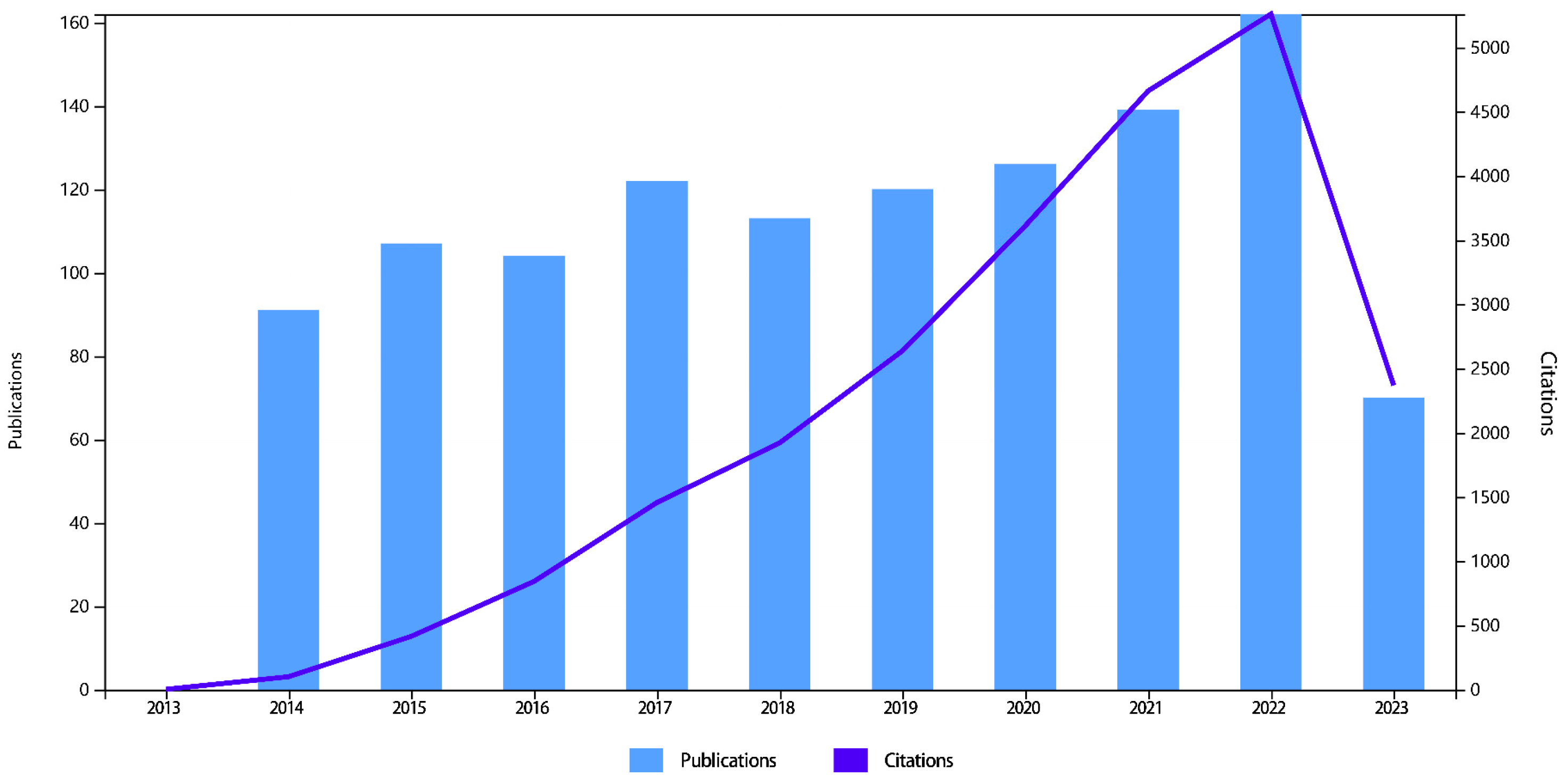
General Features of Publications
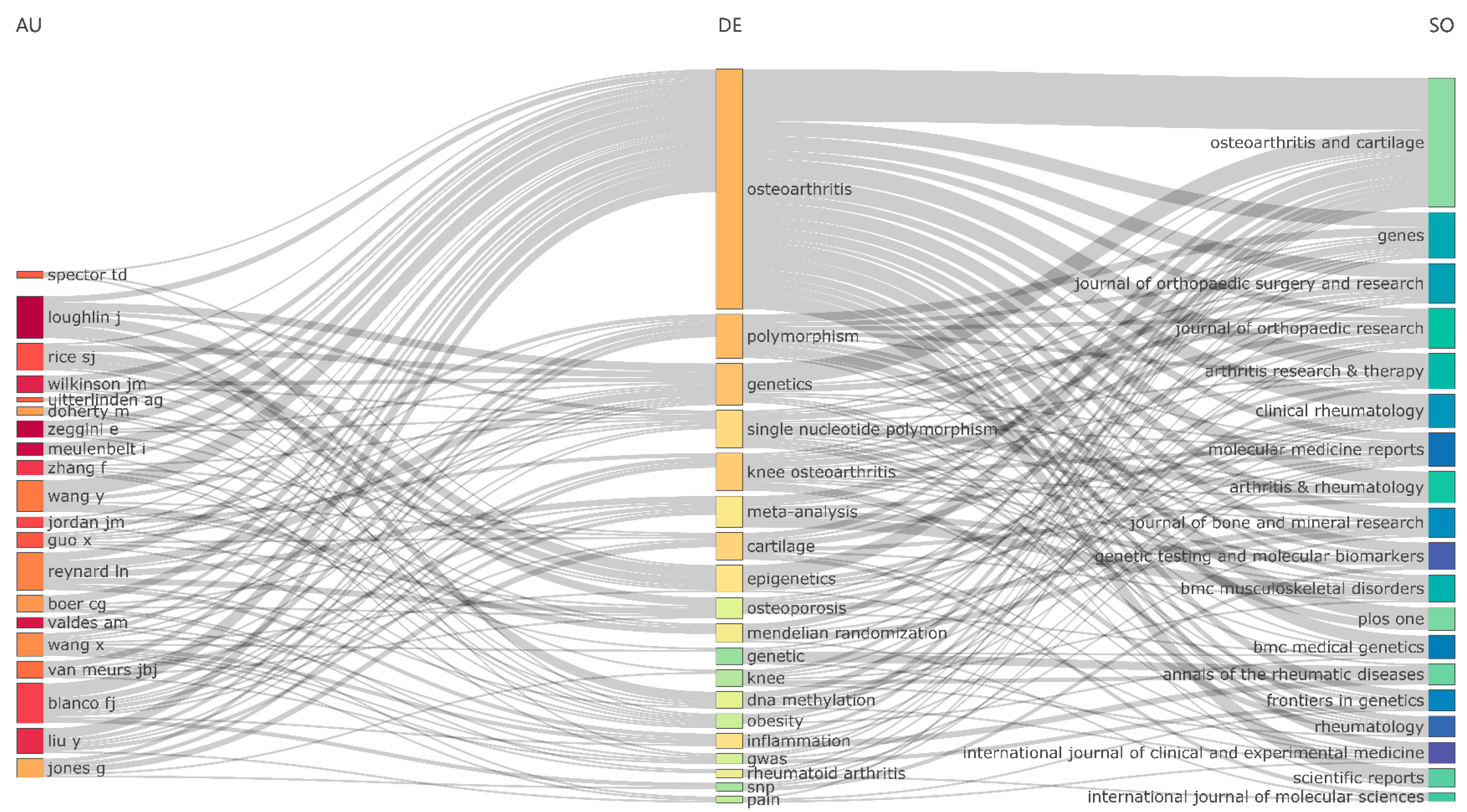
Author Analysis
Top Cited Papers
Sources Analysis
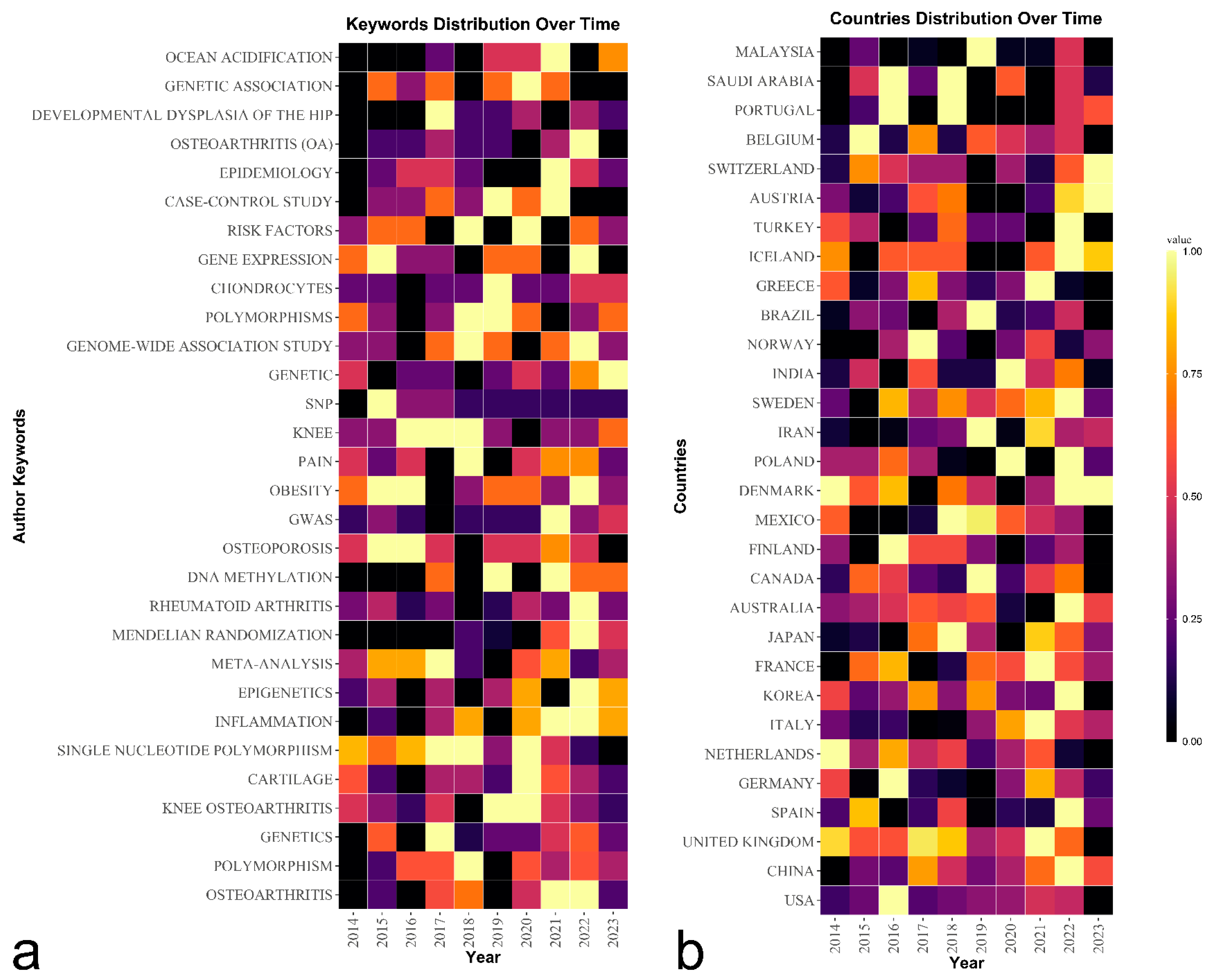
Trend Topics and Keywords Analysis
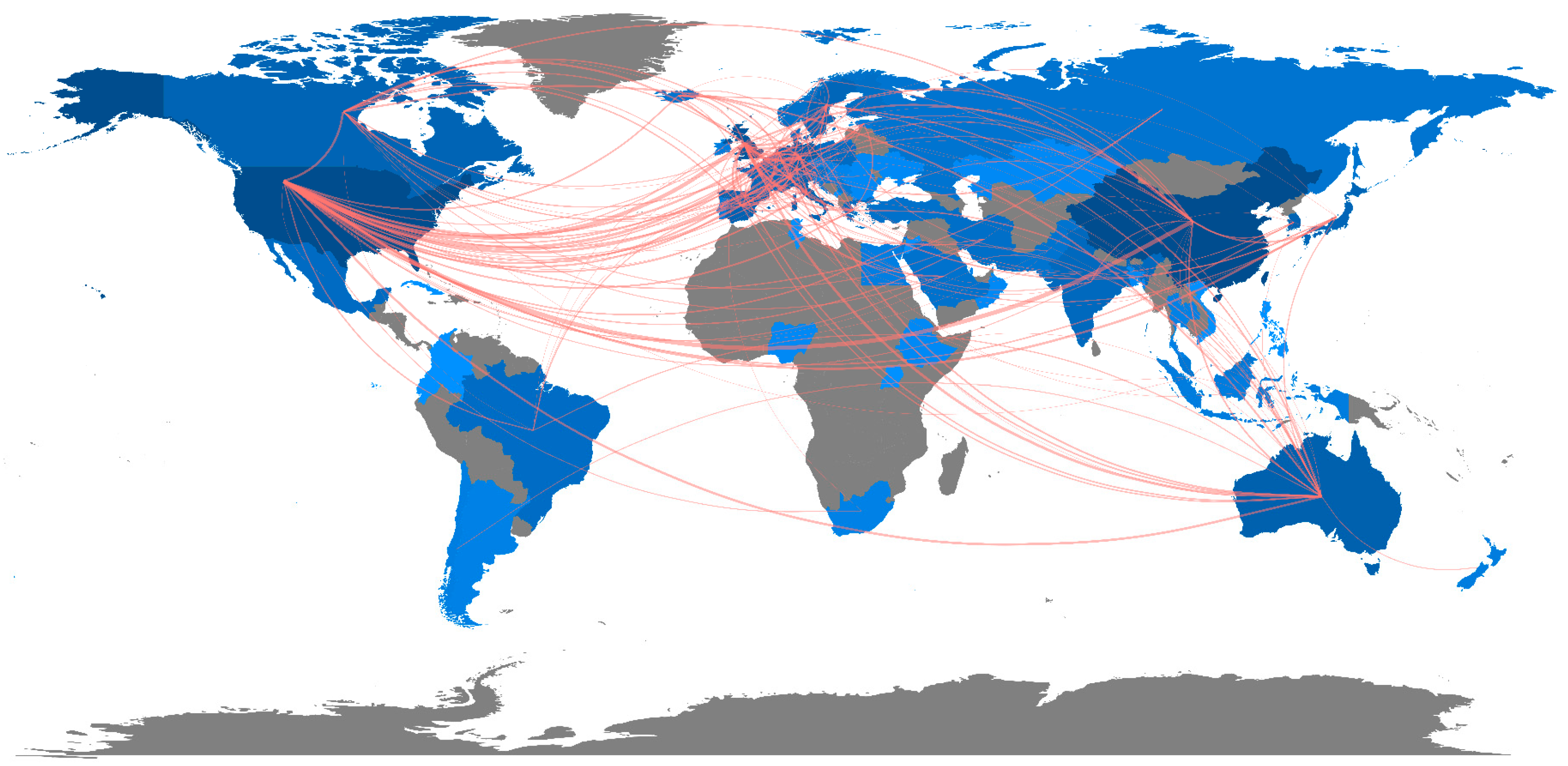
Country/Territory Analysis
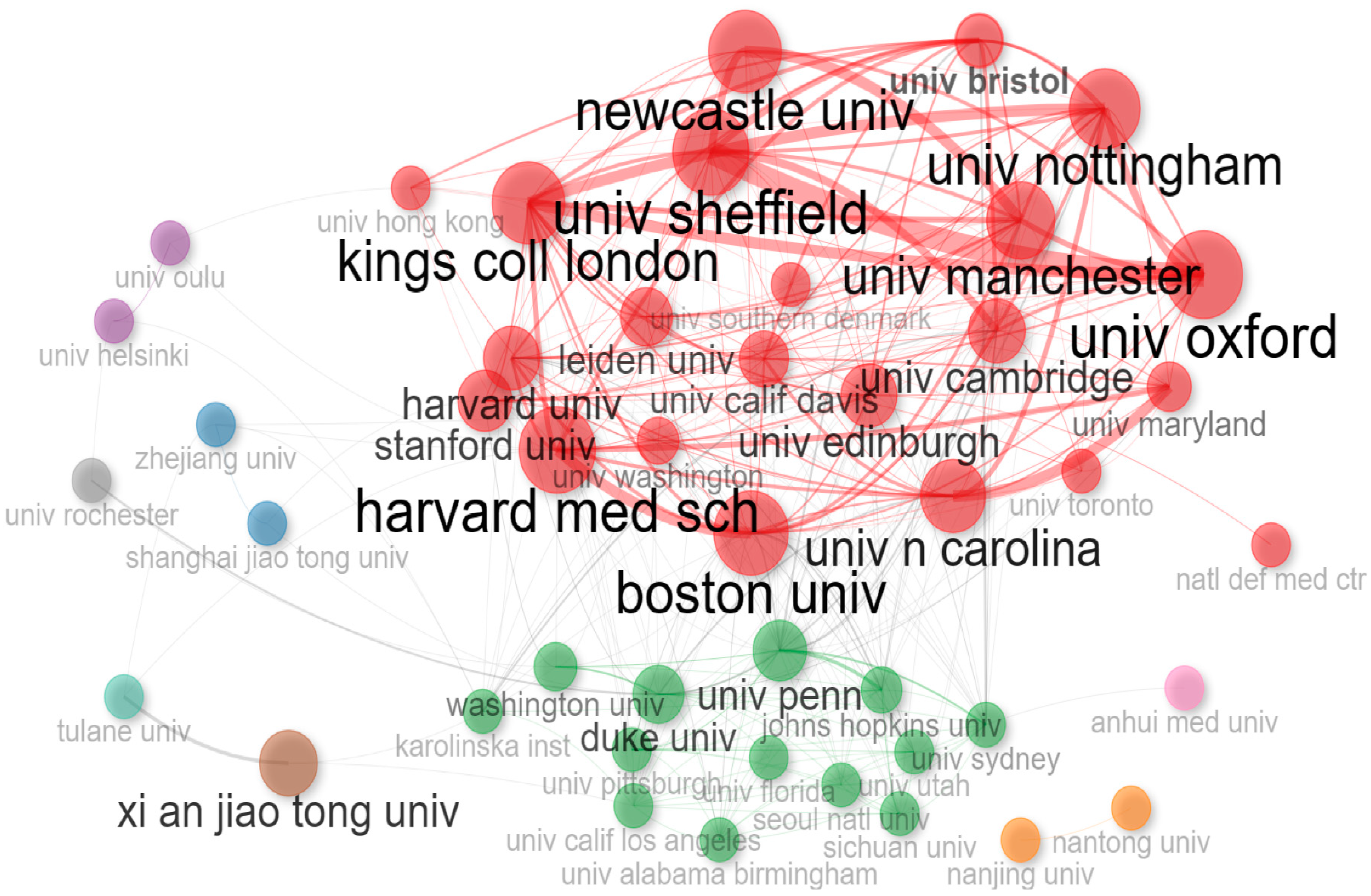
Institution Analysis
Reference Analysis
DISCUSSION
CONCLUSION
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- J. L. Allen, M. E. Cooke, T. Alliston, “ECM stiffness primes the TGF beta pathway to promote chondrocyte differentiation,” Molecular Biology of the Cell, vol. 23, no. 18, pp. 3731-3742, 2012. [CrossRef]
- V. Trachana, E. Ntoumou, L. Anastasopoulou, et al., “Studying microRNAs in osteoarthritis: Critical overview of different analytical approaches,” Mech Ageing Dev, vol. 171, pp. 15-23, 2018. [CrossRef]
- Y. Zhang, J. Jia, S. Yang, et al., “MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes,” Exp Mol Med, vol. 46, no. 2, pp. e79, 2014. [CrossRef]
- A. M. Carossino, R. Recenti, R. Carossino, et al., “Methodological models for in vitro amplification and maintenance of human articular chondrocytes from elderly patients,” Biogerontology, vol. 8, no. 5, pp. 483-498, 2007. [CrossRef]
- H. J. Kerkhof, R. J. Lories, I. Meulenbelt, et al., “A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22,” Arthritis Rheum, vol. 62, no. 2, pp. 499-510, 2010. [CrossRef]
- A. M. Valdes, T. D. Spector, “The contribution of genes to osteoarthritis,” Med Clin North Am, vol. 93, no. 1, pp. 45-66, x, 2009. [CrossRef]
- G. Aubourg, S. J. Rice, P. Bruce-Wootton, et al., “Genetics of osteoarthritis,” Osteoarthritis Cartilage, vol. 30, no. 5, pp. 636-649, 2022. [CrossRef]
- A. Buniello, J. A. L. MacArthur, M. Cerezo, et al., “The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019,” Nucleic Acids Res, vol. 47, no. D1, pp. D1005-d1012, 2019. [CrossRef]
- K. Chapman, A. M. Valdes, “Genetic factors in OA pathogenesis,” Bone, vol. 51, no. 2, pp. 258-264, 2012. [CrossRef]
- O. Ellegaard, J. A. Wallin, “The bibliometric analysis of scholarly production: How great is the impact?,” Scientometrics, vol. 105, no. 3, pp. 1809-1831, 2015. [CrossRef]
- V. Durieux, P. A. Gevenois, “Bibliometric indicators: quality measurements of scientific publication,” Radiology, vol. 255, no. 2, pp. 342-351, 2010. [CrossRef]
- P. Kokol, H. Blažun Vošner, J. Završnik, “Application of bibliometrics in medicine: a historical bibliometrics analysis,” Health Info Libr J, vol. 38, no. 2, pp. 125-138, 2021. [CrossRef]
- A. W. K. Yeung, “Comparison between Scopus, Web of Science, PubMed and publishers for mislabelled review papers,” Current Science, vol. 116, no. 11, pp. 1909-+, 2019. [CrossRef]
- Q. Wang, L. Waltman, “Large-scale analysis of the accuracy of the journal classification systems of Web of Science and Scopus,” Journal of Informetrics, vol. 10, no. 2, pp. 347-364, 2016. [CrossRef]
- M. Aria, C. Cuccurullo, “bibliometrix: An R-tool for comprehensive science mapping analysis,” Journal of Informetrics, vol. 11, no. 4, pp. 959-975, 2017. [CrossRef]
- M. Weinstock, “Bradford's Law,” Nature, vol. 233, no. 5319, pp. 434, 1971. [CrossRef]
- R. Whittemore, A. Chao, M. Jang, et al., “Methods for knowledge synthesis: An overview,” Heart & Lung, vol. 43, no. 5, pp. 453-461, 2014. [CrossRef]
- A. D. Theocharis, D. Manou, N. K. Karamanos, “The extracellular matrix as a multitasking player in disease,” Febs Journal, vol. 286, no. 15, pp. 2830-2869, 2019. [CrossRef]
- S. Glyn-Jones, A. J. R. Palmer, R. Agricola, et al., “Osteoarthritis,” Lancet, vol. 386, no. 9991, pp. 376-387, 2015. [CrossRef]
- E. R. Vina, C. K. Kwoh, “Epidemiology of osteoarthritis: literature update,” Current Opinion in Rheumatology, vol. 30, no. 2, pp. 160-167, 2018. [CrossRef]
- J. H. Kim, J. Jeon, M. Shin, et al., “Regulation of the Catabolic Cascade in Osteoarthritis by the Zinc-ZIP8-MTF1 Axis,” Cell, vol. 156, no. 4, pp. 730-743, 2014. [CrossRef]
- G. A. Bray, W. E. Heisel, A. Afshin, et al., “The Science of Obesity Management: An Endocrine Society Scientific Statement,” Endocr Rev, vol. 39, no. 2, pp. 79-132, 2018. [CrossRef]
- D. Haffner, F. Emma, D. M. Eastwood, et al., “Clinical practice recommendations for the diagnosis and management of X-linked hypophosphataemia,” Nature Reviews Nephrology, vol. 15, no. 7, pp. 435-455, 2019. [CrossRef]
- C. K. F. Chan, E. Y. Seo, J. Y. Chen, et al., “Identification and Specification of the Mouse Skeletal Stem Cell,” Cell, vol. 160, no. 1-2, pp. 285-298, 2015. [CrossRef]
- J. H. D. Bassett, G. R. Williams, “Role of Thyroid Hormones in Skeletal Development and Bone Maintenance,” Endocrine Reviews, vol. 37, no. 2, pp. 135-187, 2016. [CrossRef]
- F. Guilak, R. J. Nims, A. Dicks, et al., “Osteoarthritis as a disease of the cartilage pericellular matrix,” Matrix Biology, vol. 71-72, pp. 40-50, 2018. [CrossRef]
- X. Xu, L. W. Zheng, Q. Yuan, et al., “Transforming growth factor-beta in stem cells and tissue homeostasis,” Bone Research, vol. 6, 2018. [CrossRef]
- L. Sharma, “Osteoarthritis of the Knee,” N Engl J Med, vol. 384, no. 1, pp. 51-59, 2021.
- E. Birney, “Mendelian Randomization,” Cold Spring Harb Perspect Med, vol. 12, no. 4, 2022. [CrossRef]
- T. D. Arnold, K. L. Zang, A. Vallejo-Illarramendi, “Deletion of integrin-linked kinase from neural crest cells in mice results in aortic aneurysms and embryonic lethality,” Disease Models & Mechanisms, vol. 6, no. 5, pp. 1205-1212, 2013. [CrossRef]
- Y. H. Lee, “An overview of meta-analysis for clinicians,” Korean J Intern Med, vol. 33, no. 2, pp. 277-283, 2018. [CrossRef]
- E. Zeggini, K. Panoutsopoulou, L. Southam, et al., “Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study,” Lancet, vol. 380, no. 9844, pp. 815-823, 2012. [CrossRef]
- E. Zengini, K. Hatzikotoulas, I. Tachmazidou, et al., “Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis,” Nat Genet, vol. 50, no. 4, pp. 549-558, 2018. [CrossRef]
- J. H. Kellgren, J. S. Lawrence, “Radiological assessment of osteo-arthrosis,” Ann Rheum Dis, vol. 16, no. 4, pp. 494-502, 1957. [CrossRef]
- T. D. Spector, F. Cicuttini, J. Baker, et al., “Genetic influences on osteoarthritis in women: a twin study,” Bmj, vol. 312, no. 7036, pp. 940-943, 1996. [CrossRef]
- R. Altman, E. Asch, D. Bloch, et al., “Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association,” Arthritis Rheum, vol. 29, no. 8, pp. 1039-1049, 1986. [CrossRef]
- P. O. Seglen, “Citations and journal impact factors: questionable indicators of research quality,” Allergy, vol. 52, no. 11, pp. 1050-1056, 1997. [CrossRef]
- C. G. Boer, K. Hatzikotoulas, L. Southam, et al., “Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations,” Cell, vol. 184, no. 18, pp. 4784-4818.e4717, 2021. [CrossRef]
- X. B. Zhu, F. Chen, K. Lu, et al., “PPAR gamma preservation via promoter demethylation alleviates osteoarthritis in mice,” Annals of the Rheumatic Diseases, vol. 78, no. 10, pp. 1420-1429, 2019. [CrossRef]
- L. Bornmann, H. D. Daniel, “What do we know about the h index?,” Journal of the American Society for Information Science and Technology, vol. 58, no. 9, pp. 1381-1385, 2007. [CrossRef]
- X. W. Zhang, H. Lai, F. Zhang, et al., “Visualization and Analysis in the Field of Pan-Cancer Studies and Its Application in Breast Cancer Treatment,” Frontiers in Medicine, vol. 8, 2021. [CrossRef]
- Z. Abbaszadeh, S. Çeşmeli, Ç. Biray Avcı, “Crucial players in glycolysis: Cancer progress,” Gene, vol. 726, pp. 144158, 2020. [CrossRef]
- Y. Na, R. Bai, Z. Zhao, et al., “IL1R1 gene polymorphisms are associated with knee osteoarthritis risk in the Chinese Han population,” Oncotarget, vol. 8, no. 3, pp. 4228-4233, 2017. [CrossRef]
- H. Takahashi, M. Nakajima, K. Ozaki, et al., “Prediction model for knee osteoarthritis based on genetic and clinical information,” Arthritis Res Ther, vol. 12, no. 5, pp. R187, 2010. [CrossRef]
- V. B. Kraus, M. A. Karsdal, “Osteoarthritis: Current Molecular Biomarkers and the Way Forward,” Calcif Tissue Int, vol. 109, no. 3, pp. 329-338, 2021. [CrossRef]
- S. Graw, K. Chappell, C. L. Washam, et al., “Multi-omics data integration considerations and study design for biological systems and disease,” Mol Omics, vol. 17, no. 2, pp. 170-185, 2021. [CrossRef]
| SCR | Authors | N | AF | TC | CPS | Country |
|---|---|---|---|---|---|---|
| 1 | Loughlin J | 23 | 4.48 | 248 | 10.78 | United Kingdom |
| 2 | Zeggini E | 17 | 1.40 | 260 | 15.29 | United Kingdom |
| 3 | Meulenbelt I | 15 | 1.61 | 197 | 13.13 | Netherlands |
| 4 | Valdes AM | 15 | 1.73 | 199 | 13.27 | United Kingdom |
| 5 | Wilkinson JM | 15 | 1.84 | 232 | 15.47 | United States |
| 6 | Liu Y | 14 | 1.59 | 17 | 1.21 | China |
| 7 | Zhang F | 14 | 1.73 | 14 | 1.00 | China |
| 8 | Blanco FJ | 13 | 1.45 | 97 | 7.46 | Spain |
| 9 | Jordan JM | 13 | 1.06 | 93 | 7.15 | United States |
| 10 | Rice SJ | 13 | 2.19 | 91 | 7.00 | United Kingdom |
| SCR | Title | Year | TC | Journal | FA | CA | Country |
|---|---|---|---|---|---|---|---|
| 1 | Osteoarthritis | 2015 | 1604 | Lancet | Glyn-Jones, S. | Palmer, AJR | United Kingdom |
| 2 | Epidemiology of osteoarthritis: literature update | 2018 | 475 | Current Opinion in Rheumatology | Vina, ER | Vina, ER | United States |
| 3 | Identification and Specification of the Mouse Skeletal Stem Cell | 2015 | 430 | Cell | Chan, CKF | Chan, CKF | United Kingdom |
| 4 | The Science of Obesity Management: An Endocrine Society Scientific Statement | 2018 | 403 | Endocrine Reviews | Bray, GA | Bray, GA | United States |
| 5 | Role of Thyroid Hormones in Skeletal Development and Bone Maintenance | 2016 | 273 | Endocrine Reviews | Bassett, JHD | Williams, GR | United Kingdom |
| 6 | Regulation of the Catabolic Cascade in Osteoarthritis by the Zinc-ZIP8-MTF1 Axis | 2014 | 247 | Cell | Kim, JH | Chun, JS | South Korea |
| 7 | Osteoarthritis as a disease of the cartilage pericellular matrix | 2018 | 234 | Matrix Biology | Guilak, F | Guilak, F | United States |
| 8 | Clinical practice recommendations for the diagnosis and management of X-linked hypophosphataemia | 2018 | 225 | Nature Reviews Nephrology | Haffner, D | Haffner, D | Germany |
| 9 | Transforming growth factor-beta in stem cells and tissue homeostasis | 2019 | 221 | Bone Research | Xu, X | Zhou, XD | China |
| 10 | The extracellular matrix as a multitasking player in disease | 2019 | 202 | FEBS Journal | Theocharis, AD | Theocharis, AD | Greece |
| SCR | Journal | N | H-index | TC |
|---|---|---|---|---|
| 1 | Osteoarthritis and Cartilage | 39 | 17 | 933 |
| 2 | Annals of the Rheumatic Disease | 21 | 14 | 687 |
| 3 | Plos One | 33 | 14 | 439 |
| 4 | Arthritis & Rheumatology | 19 | 13 | 440 |
| 5 | International Journal of Molecular Sciences | 20 | 10 | 505 |
| 6 | Journal Orthopaedic Research | 17 | 10 | 320 |
| 7 | Journal of Bone and Mineral Research | 12 | 10 | 298 |
| 8 | Rheumatology | 10 | 8 | 186 |
| 9 | Arthritis Research & Therapy | 16 | 8 | 173 |
| 10 | Molecular Medicine Reports | 10 | 8 | 126 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





