Submitted:
01 December 2023
Posted:
04 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethical considerations
2.2. Study Site and Participants
2.3. Laboratory analysis of blood samples
2.4. Statistical analysis
Results
3.1. Study Participants
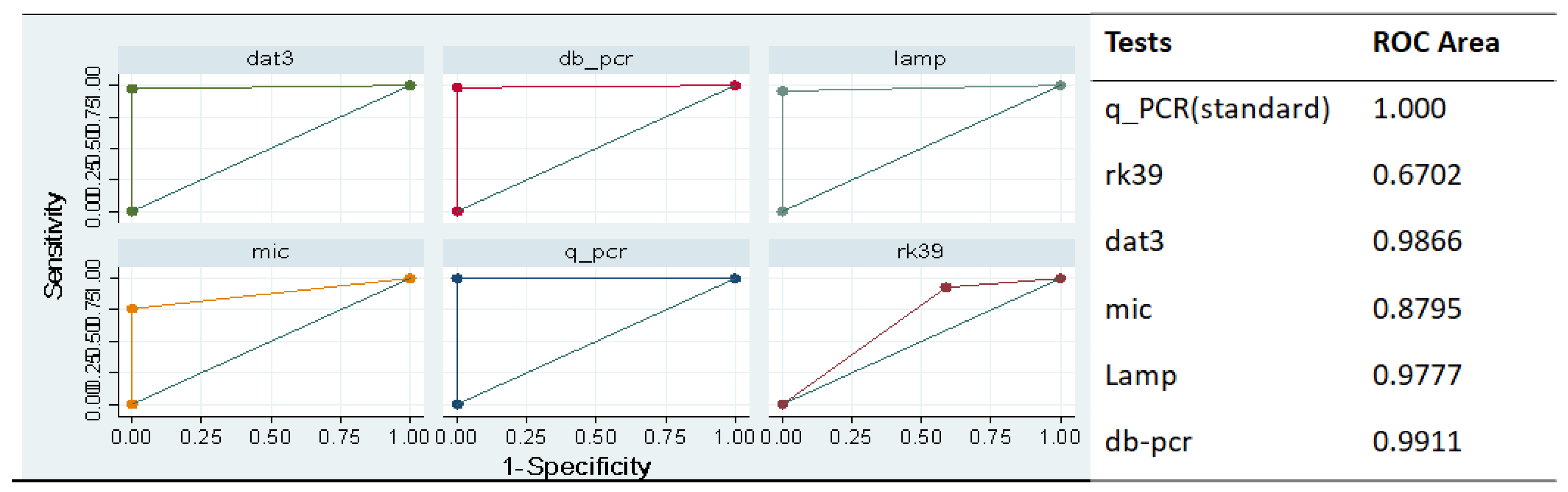
3.2. Performance of the tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kassa, M.; Abdellati, S.; Cnops, L.; Bremer Hinckel, B.C.; Yeshanew, A.; Hailemichael, W.; Vogt, F.; Adriaensen, W.; Mertens, P.; Diro, E. Diagnostic accuracy of direct agglutination test, rK39 ELISA and six rapid diagnostic tests among visceral leishmaniasis patients with and without HIV coinfection in Ethiopia. PLoS Negl. Trop. Dis. 2020, 14, e0008963. [Google Scholar] [CrossRef] [PubMed]
- Oryan, A.; Akbari, M. Worldwide risk factors in leishmaniasis. Asian Pac. J. Trop. Med. 2016, 9, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Challa¹, P. (2018). Amphotericin B and Fluconazole Susceptibility Profiles of Old World and New World Strains of Leishmania.
- Bi, K.; Chen, Y.; Kuang, S.Z.Y.; Wu, C.-H.J. Current visceral leishmaniasis research: a research review to inspire future study. BioMed research international. 2018. [Google Scholar] [CrossRef] [PubMed]
- De Ruiter, C.; Van Der Veer, C.; Leeflang, M.; Deborggraeve, S.; Lucas, C.; Adams, E. Molecular tools for diagnosis of visceral leishmaniasis: systematic review and meta-analysis of diagnostic test accuracy. J. Clin. Microbiol. 2014, 52, 3147–3155. [Google Scholar] [CrossRef] [PubMed]
- Diro, E.; Lynen, L.; Ritmeijer, K.; Boelaert, M.; Hailu, A.; van Griensven, J. Visceral leishmaniasis and HIV coinfection in East Africa. PLoS Negl. Trop. Dis. 2014, 8, e2869. [Google Scholar] [CrossRef] [PubMed]
- Ghodrati, M.; Spotin, A.; Hazratian, T.; Mahami-Oskouei, M.; Bordbar, A.; Ebrahimi, S.; Fallahi, S.; Parvizi, P. Diagnostic Accuracy of Loop-mediated Isothermal Amplification Assay as a Field Molecular Tool for Rapid Mass Screening of Old World Leishmania Infections in Sand Flies and In Vitro Culture. Iran J Parasitol. 2017, 12, 506–515. [Google Scholar] [PubMed]
- Lopes, E.; Sevá, A.d.P.; Ferreira, F.; Nunes, C.M.; Keid, L.B.; Hiramoto, R.; Ferreira, H.L.; Oliveira, T.M.F.d.S.; Bigotto, M.; Galvis-Ovallos, F. Serological and molecular diagnostic tests for canine visceral leishmaniasis in Brazilian endemic area: one out of five seronegative dogs are infected. Epidemiol. Infect. 2017, 145, 2436–2444. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, E.; Ali, M.S.; El-Hassan, A.; El-Toum, I.A.; Satti, M.; Ghalib, H.; Sondorp, E.; Winkler, A. Kala-azar in displaced people from southern Sudan: epidemiological, clinical and therapeutic findings. Trans. R. Soc. Trop. Med. Hyg. 1991, 85, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Cloots, K.; Marino, P.; Burza, S.; Gill, N.; Boelaert, M.; Hasker, E. Visceral Leishmaniasis-HIV coinfection as a predictor of increased Leishmania transmission at the village level in Bihar, India. Frontiers in Cellular and Infection Microbiology. 2021, 11, 604117. [Google Scholar] [CrossRef]
- Rijal, S.; Ostyn, B.; Uranw, S.; Rai, K.; Bhattarai, N.R.; Dorlo, T.P.; Beijnen, J.H.; Vanaerschot, M.; Decuypere, S.; Dhakal, S.S. Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin. Infect. Dis. 2013, 56, 1530–1538. [Google Scholar] [CrossRef]
- Ortalli, M.; Lorrai, D.; Gaibani, P.; Rossini, G.; Vocale, C.; Re, M.C.; Varani, S. Serodiagnosis of Visceral leishmaniasis in Northeastern Italy: Evaluation of seven serological tests. Microorganisms. 2020, 8, 1847. [Google Scholar] [CrossRef]
- Hagos, D.G.; Kiros, Y.K.; Abdulkader, M.; Arefaine, Z.G.; Nigus, E.; Schallig, H.H.; Wolday, D. Utility of the Loop-Mediated Isothermal Amplification Assay for the Diagnosis of Visceral Leishmaniasis from Blood Samples in Ethiopia. The American Journal of Tropical Medicine and Hygiene. 2021, 105, 1050. [Google Scholar] [CrossRef] [PubMed]
- Hagos, D.G.; Kebede, Y.; Abdulkader, M.; Nigus, E.; Gessesse Arefaine, Z.; Nega, G.; Schallig, H.D.; Wolday, D. Effect of rK39 testing in guiding treatment initiation and outcome in patients with visceral leishmaniasis in Ethiopia: A prospective cohort study. PLoS One. 2021, 16, e0253303. [Google Scholar] [CrossRef] [PubMed]
- da Silva, M.R.B.; Stewart, J.M.; Costa, C.H.N. Sensitivity of bone marrow aspirates in the diagnosis of visceral leishmaniasis. The American journal of tropical medicine and hygiene. 2005, 72, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Dayama, A.; Mehrotra, S.; Sundar, S. Diagnosis of visceral leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Osman, O.F.; Oskam, L.; Zijlstra, E.E.; Kroon, N.; Schoone, G.J.; Khalil, E.; El-Hassan, A.M.; Kager, P.A. Evaluation of PCR for diagnosis of visceral leishmaniasis. J. Clin. Microbiol. 1997, 35, 2454–2457. [Google Scholar] [CrossRef] [PubMed]
- ter Horst, R.; Tefera, T.; Assefa, G.; Ebrahim, A.Z.; Davidson, R.N.; Ritmeijer, K. Field evaluation of rK39 test and direct agglutination test for diagnosis of visceral leishmaniasis in a population with high prevalence of human immunodeficiency virus in Ethiopia. The American journal of tropical medicine and hygiene. 2009, 80, 929–934. [Google Scholar] [CrossRef] [PubMed]
- El-Moamly, A.; El-Sweify, M.; Hafeez, M. Performance of rK39 immunochromatography and freeze-dried direct agglutination tests in the diagnosis of imported visceral leishmaniasis. Parasitol. Res. 2012, 110, 349–354. [Google Scholar] [CrossRef]
- de Paiva Cavalcanti, M.; de Brito, M.E.F.; de Souza, W.V.; de Miranda Gomes, Y.; Abath, F.G. The development of a real-time PCR assay for the quantification of Leishmania infantum DNA in canine blood. The Veterinary Journal. 2009, 182, 356–358. [Google Scholar] [CrossRef]
- Opota, O.; Balmpouzis, Z.; Berutto, C.; Kaiser-Guignard, J.; Greub, G.; Aubert, J.D.; Prod'hom, G.; Manuel, O.; Jaton, K. Visceral leishmaniasis in a lung transplant recipient: usefulness of highly sensitive real-time polymerase chain reaction for preemptive diagnosis. Transpl. Infect. Dis. 2016, 18, 801–804. [Google Scholar] [CrossRef]
- Gedda, M.R.; Madhukar, P.; Shukla, A.; Mudavath, S.L.; Srivastava, O.N.; Singh, O.P.; Sundar, S. Nanodiagnostics in leishmaniasis: A new frontiers for early elimination. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 2021; 13, e1675. [Google Scholar]
- Mugasa, C.M.; Laurent, T.; Schoone, G.J.; Basiye, F.L.; Saad, A.A.; El Safi, S.; Kager, P.A.; Schallig, H.D. Simplified molecular detection of Leishmania parasites in various clinical samples from patients with leishmaniasis. Parasit Vectors. 2010, 3, 13. [Google Scholar] [CrossRef] [PubMed]
- Mens, P.; De Bes, H.; Sondo, P.; Laochan, N.; Keereecharoen, L.; Van Amerongen, A.; Flint, J.; Sak, J.; Proux, S.; Tinto, H. Direct blood PCR in combination with nucleic acid lateral flow immunoassay for detection of Plasmodium species in settings where malaria is endemic. J. Clin. Microbiol. 2012, 50, 3520–3525. [Google Scholar] [CrossRef] [PubMed]
- Mikita, K.; Maeda, T.; Yoshikawa, S.; Ono, T.; Miyahira, Y.; Kawana, A. The Direct Boil-LAMP method: a simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol. Int. 2014, 63, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.H.; Ding, D.; Wang, J.Y.; Steverding, D.; Wang, X.; Yang, Y.T.; Shi, F. Development of a LAMP assay for detection of Leishmania infantum infection in dogs using conjunctival swab samples. Parasit Vectors. 2015, 8, 370. [Google Scholar] [CrossRef]
- Notomi T; Mori Y; Tomita N; Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [CrossRef] [PubMed]
- Sriworarat, C.; Phumee, A.; Mungthin, M.; Leelayoova, S.; Siriyasatien, P. Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit Vectors. 2015, 8, 591. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Caceres, A.G.; Guerrero-Quincho, S.; Tineo-Villafuerte, E.; Rodriquez-Delfin, L.; Mimori, T.; Uezato, H.; Katakura, K.; Gomez, E.A.; Guevara, A.G.; et al. A rapid molecular diagnosis of cutaneous leishmaniasis by colorimetric malachite green-loop-mediated isothermal amplification (LAMP) combined with an FTA card as a direct sampling tool. Acta Trop. 2016, 153, 116–119. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, N.J.; Menting, S.; Wentink-Bonnema, E.M.; Broekhuizen-van Haaften, P.E.; Withycombe, E.; Schallig, H.D.; Mens, P.F. Laboratory evaluation of the miniature direct-on-blood PCR nucleic acid lateral flow immunoassay (mini-dbPCR-NALFIA), a simplified molecular diagnostic test for Plasmodium. Malar. J. 2023, 22, 98. [Google Scholar] [CrossRef] [PubMed]
- Erber, A.C.; Sandler, P.J.; de Avelar, D.M.; Swoboda, I.; Cota, G.; Walochnik, J. Diagnosis of visceral and cutaneous leishmaniasis using loop-mediated isothermal amplification (LAMP) protocols: a systematic review and meta-analysis. Parasites & vectors, 2022; 15, 1–16. [Google Scholar]
- Ruang-Areerate, T.; Sukphattanaudomchoke, C.; Thita, T.; Leelayoova, S.; Piyaraj, P.; Mungthin, M.; Suwannin, P.; Polpanich, D.; Tangchaikeeree, T.; Jangpatarapongsa, K. Development of loop-mediated isothermal amplification (LAMP) assay using SYBR safe and gold-nanoparticle probe for detection of Leishmania in HIV patients. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Sukphattanaudomchoke, C.; Siripattanapipong, S.; Thita, T.; Leelayoova, S.; Piyaraj, P.; Mungthin, M.; Ruang-Areerate, T. Simplified closed tube loop mediated isothermal amplification (LAMP) assay for visual diagnosis of Leishmania infection. Acta Trop. 2020, 212, 105651. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Kato, H.; Peters, N.C. Loop-mediated isothermal amplification (LAMP): An advanced molecular point-of-care technique for the detection of Leishmania infection. PLoS Negl. Trop. Dis. 2019, 13, e0007698. [Google Scholar] [CrossRef]
- Pecchia, S.; Da Lio, D. Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. J. Microbiol. Methods. 2018, 151, 118–128. [Google Scholar] [CrossRef]
- Harizanov, R.N.; Kaftandjiev Iskren, T. Interactions between parasite and host in human visceral leishmaniasis. J Cytol Tissue Biol. 2014, 1. [Google Scholar]
- Vaish, M.; Singh, O.P.; Chakravarty, J.; Sundar, S. rK39 antigen for the diagnosis of visceral leishmaniasis by using human saliva. The American journal of tropical medicine and hygiene. 2012, 86, 598. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, H.; Bhattacharya, S.K.; Verma, S.; Salotra, P. Serological and molecular analysis of Leishmania infection in healthy individuals from two districts of West Bengal, India, endemic for visceral leishmaniasis. The American Journal of Tropical Medicine and Hygiene. 2017, 96, 1448. [Google Scholar] [CrossRef]
- Adams, E.R.; Jacquet, D.; Schoone, G.; Gidwani, K.; Boelaert, M.; Cunningham, J. Leishmaniasis direct agglutination test: using pictorials as training materials to reduce inter-reader variability and improve accuracy. PLoS Negl. Trop. Dis. 2012, 6, e1946. [Google Scholar] [CrossRef]
- Ayelign, B.; Jemal, M.; Negash, M.; Genetu, M.; Wondmagegn, T.; Zeleke, A.J.; Worku, L.; Bayih, A.G.; Shumie, G.; Behaksra, S.W. Validation of in-house liquid direct agglutination test antigen: the potential diagnostic test in visceral Leishimaniasis endemic areas of Northwest Ethiopia. BMC Microbiol. 2020, 20, 1–7. [Google Scholar] [CrossRef]
- Roberts, T.; Keddie, S.H.; Rattanavong, S.; Gomez, S.R.; Bradley, J.; Keogh, R.H.; Bärenbold, O.; Falconer, J.; Mens, P.F.; Hopkins, H. Accuracy of the direct agglutination test for diagnosis of visceral leishmaniasis: a systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 782. [Google Scholar] [CrossRef] [PubMed]
- Federal Ministry of Health of Ethiopia (FMOH). Guideline for Diagnosis, Treatment and Prevention of Leishmaniasis in Ethiopia. 2013, 2nd edition, Addis Ababa, Ethiopia.
- Mukhtar, M.; Ali, S.S.; Boshara, S.A.; Albertini, A.; Monnerat, S.; Bessell, P.; Mori, Y.; Kubota, Y.; Ndung’u, J.M.; Cruz, I. Sensitive and less invasive confirmatory diagnosis of visceral leishmaniasis in Sudan using loop-mediated isothermal amplification (LAMP). PLoS Negl. Trop. Dis. 2018, 12, e0006264. [Google Scholar] [CrossRef]
- Hossain, F.; Picado, A.; Owen, S.I.; Ghosh, P.; Chowdhury, R.; Maruf, S.; Khan, M.A.A.; Rashid, M.U.; Nath, R.; Baker, J. Evaluation of Loopamp™ Leishmania Detection Kit and Leishmania Antigen ELISA for Post-Elimination Detection and Management of Visceral Leishmaniasis in Bangladesh. Frontiers in Cellular and Infection Microbiology. 2021, 11, 670759. [Google Scholar] [CrossRef]
- Georgiadou, S.P.; Stefos, A.; Spanakos, G.; Skrimpas, S.; Makaritsis, K.; Sipsas, N.V.; Dalekos, G.N. Current clinical, laboratory, and treatment outcome characteristics of visceral leishmaniasis: results from a seven-year retrospective study in Greece. Int. J. Infect. Dis. 2015, 34, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.; Rai, M. Laboratory diagnosis of visceral leishmaniasis. Clin. Vaccine Immunol. 2002, 9, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Boelaert, M.; Rijal, S.; Regmi, S.; Singh, R.; Karki, B.; Jacquet, D.; Chappuis, F.; Campino, L.; Desjeux, P.; Le Ray, D. A comparative study of the effectiveness of diagnostic tests for visceral leishmaniasis. Am. J. Trop. Med. Hyg. 2004, 70, 72. [Google Scholar] [CrossRef] [PubMed]
- Hagos, D.G.; Schallig, H.D.; Kiros, Y.K.; Abdulkadir, M.; Wolday, D. Performance of rapid rk39 tests for the diagnosis of visceral leishmaniasis in Ethiopia: a systematic review and meta-analysis. BMC Infect. Dis. 2021, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kiros, Y.K.; Regassa, B.F. The role of rk39 serologic test in the diagnosis of visceral leishmaniasis in a Tertiary Hospital, Northern Ethiopia. BMC Res. Notes. 2017, 10, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, T.; Boelaert, M.; Miles, M.A. Comparison of visceral leishmaniasis diagnostic antigens in African and Asian Leishmania donovani reveals extensive diversity and region-specific polymorphisms. PLoS Negl. Trop. Dis. 2013, 7, e2057. [Google Scholar] [CrossRef] [PubMed]
- Bezuneh, A.; Mukhtar, M.; Abdoun, A.; Teferi, T.; Takele, Y.; Diro, E.; Jemaneh, A.; Shiferaw, W.; Wondimu, H.; Bhatia, A. Comparison of point-of-care tests for the rapid diagnosis of visceral leishmaniasis in East African patients. The American journal of tropical medicine and hygiene. 2014, 91, 1109. [Google Scholar] [CrossRef] [PubMed]
- Boelaert, M.; El-Safi, S.; Hailu, A.; Mukhtar, M.; Rijal, S.; Sundar, S.; Wasunna, M.; Aseffa, A.; Mbui, J.; Menten, J. Diagnostic tests for kala-azar: a multi-centre study of the freeze-dried DAT, rK39 strip test and KAtex in East Africa and the Indian subcontinent. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Coulborn, R.M.; Gebrehiwot, T.G.; Schneider, M.; Gerstl, S.; Adera, C.; Herrero, M.; Porten, K.; den Boer, M.; Ritmeijer, K.; Alvar, J. Barriers to access to visceral leishmaniasis diagnosis and care among seasonal mobile workers in Western Tigray, Northern Ethiopia: A qualitative study. PLoS Negl. Trop. Dis. 2018, 12, e0006778. [Google Scholar] [CrossRef]
- Ready, P. Leishmaniasis emergence in Europe. Eurosurveillance. 2010, 15, 19505. [Google Scholar] [CrossRef]
- Chappuis, F.; Sundar, S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nature reviews microbiology. 2007, 5, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Cota, G.F.; De Sousa, M.R.; Demarqui, F.N.; Rabello, A. The diagnostic accuracy of serologic and molecular methods for detecting visceral leishmaniasis in HIV infected patients: meta-analysis. PLoS Negl. Trop. Dis. 2012, 6, e1665. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.; Ali, S.S.; Boshara, S.A.; Albertini, A.; Monnerat, S.; Bessell, P.; Mori, Y.; Kubota, Y.; Ndung'u, J.M.; Cruz, I. Sensitive and less invasive confirmatory diagnosis of visceral leishmaniasis in Sudan using loop-mediated isothermal amplification (LAMP). PLoS Negl. Trop. Dis. 2018, 12, e0006264. [Google Scholar] [CrossRef] [PubMed]
- Cota, G.F.; de Sousa, M.R.; de Freitas Nogueira, B.M.; Gomes, L.I.; Oliveira, E.; Assis, T.S.M.; de Mendonça, A.L.P.; Pinto, B.F.; Saliba, J.W.; Rabello, A. Comparison of parasitological, serological, and molecular tests for visceral leishmaniasis in HIV-infected patients: a cross-sectional delayed-type study. The American journal of tropical medicine and hygiene. 2013, 89, 570. [Google Scholar] [CrossRef] [PubMed]
- Melkamu, R.; Berhane, N.; Jacobs, B.K.; Mohammed, R.; Kassa, M.; Yeshanew, A.; Fikre, H.; Atnafu, S.; van Henten, S.; van Griensven, J. PCR for detection of Leishmania donovani from microscopically negative tissue smears of suspected patients in Gondar, Ethiopia. PLoS Negl. Trop. Dis. 2023, 17, e0011128. [Google Scholar] [CrossRef] [PubMed]
- Takele, Y.; Mulaw, T.; Adem, E.; Womersley, R.; Kaforou, M.; Franssen, S.U.; Taylor, G.P.; Müller, I.; Cotton, J.A.; Kropf, P. Recurrent visceral leishmaniasis relapses in HIV co-infected patients are characterised by less efficient immune responses and higher parasite load. iScience. 2022, 105867. [Google Scholar]
- Babiker, Z.O.; Davidson, R.; Mazinda, C.; Kipngetich, S.; Ritmeijer, K. Utility of lymph node aspiration in the diagnosis of visceral leishmaniasis in Sudan. The American journal of tropical medicine and hygiene. 2007, 76, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, K.R. How to Interpret Sensitivity and Specificity. Western Psychological Services. 2022, 2022. [Google Scholar]
- Waikar, S.S.; Betensky, R.A.; Emerson, S.C.; Bonventre, J.V. Imperfect gold standards for biomarker evaluation. Clinical Trials. 2013, 10, 696–700. [Google Scholar] [CrossRef]
- Narkhede, S. Understanding auc-roc curve. Towards Data Science. 2018, 26, 220–227. [Google Scholar]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | All VL cases (N=235) |
Q-PCR+ (n=144/235) |
rk39 RDT+ (n=158/235) |
DAT+ (n=141/235) |
Microscopic+ (n=85/135) |
LAMP+ (n=135/226) |
dbPCR-NALFIA+ (n=138/235) |
| Sex | |||||||
| Male | 220(93.6%) | 142(60.4%) | 155(66.0%) | 139(59.1%) | 84(62.2%) | 131(95.0%) | 13(58.3%) |
| Female | 15(6.450 | 2(0.85%) | 3(1.80%) | 2(0.85%) | 1(0.74%) | 4(1.77%) | 19).43%) |
| Signs & symptoms | |||||||
| Fever(>2wks) | 226 (96.2%), | 139(96.5)% | 148(93.7%) | 133(95.7%) | 83(98.8%) | 128(94.8%) | 132(95.7%) |
| Wt. Lost | 218 (92.8%) | 137(95.1%) | 139(87.9%) | 134(95.0%) | 79(92.9%) | 126(93.3%) | 130(94.2%) |
| Fatigue | 199 (84.6%). | 123985.4%) | 132(83.5) | 126(89.4%) | 82(96.5%) | 128(94.8%) | 131(94.9%) |
| Abdominal swelling | 112(77.8%) | 101(65.2%) | 108(76.6%) | 72(84.7%) | 103(76.3%) | 109(78.9%) | |
| Splenomegaly | 179 (76.2%), | 134(93.1%) | 118(74.7%) | 125(88.7%) | 77(90.6%) | 129(95.6%) | 111(80.4%) |
| Hepatomegaly | 73 (31.1%) | 64(44.4%) | 41(25.9%) | 48(34.0%) | 33(38.8%) | 42(31.1%) | 51(37.0%) |
| Laboratory findings | |||||||
| WBC(x109/L | 1.9(1.5-2.3) | 1.7(1.1-2.6) | 1.8(1.3-2.1) | 2.0(1.6-2.3) | 1.7(1.1-1.9) | 1.6(1.2-2.2) | 1.65(1.2-2.1) |
| Hemoglobin (mg/dL) | 7.6(6.3-11.3) | 7.8(6.3-12.0) | 7.9(6.6-10.1) | 8.1(6.8-12.0) | 7.2(6.1-9.4) | 7.8(6.5-10.2) | 7.4(5.9-9.9) |
| Platelet count (3109/L) | 172(71-352) | 167(165-345) | 177(154-243) | 171(129-271) | 169(116-237) | 174(124-299) | 169(153-253) |
| AST(U/L) | 175(66.8%) | 68(43-168) | 71(39-173) | 66(45-171) | 70(44-143) | 65(39-169) | 66(40-155) |
| ALT(U/L) | 41(23-65) | 42(21-63) | 41(27-67) | 39(28-65) | 38(26-59) | 42(27-61) | 39(24-67) |
| Alkaline phosphate (U/L) | 166(81-342) | 163(78-298) | 171(77-355) | 162(69-343) | 159(89-236) | 164(81-309) | 163(79-321) |
| Creatinine (mg/dL) | 0.7(0.48-0.9) | 0.69(0.46-0.87) | 0.7(0.5-0.97) | 0.69(0.5-0.87) | 0.68(0.5-0.87) | 0.73(0.5-1.01) | 0.68(0.47-0.96) |
| Tests | Sensitivity (95%CI) | Specificity (95%CI) | PPV (95%CI) | NPV (95%CI) |
| Rk39 RDT | 88.11% (84.68 - 91.55%) |
83.33% 79.38-87.29% |
79.25% (74.94-83.55%) | 90.66% 87.57-93.75% |
| DAT | 96.50% (94.55-98.46%) |
97.96% (96.45-99.46%) |
97.18% (95.42-98.94%) | 97.46% (95.79-99.14%) |
| Microscopy |
75.89% (68.68 - 83.11%) |
100.00% (100.0-100.0%) |
100.00% (100.0-100.0%) | 46.00% (37.59 - 54.41% |
| LAMP assay | 94.33% (91.84 - 96.81%) |
97.88% (96.34-99.43%) |
96.38% (94.37 - 98.39% | 95.88% (93.74 - 98.02%) |
| Mini-dbPCR-NALFIA | 95.80% (93.68 - 97.93%) |
98.99% (97.93 - 100.05%) |
98.56% (97.30 - 99.83%) |
97.03% (95.23 - 98.83%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





