1. Summary
Bacterial infections remain among the most serious problems in healthcare. The continuous grow of antimicrobial drug resistance in clinical pathogenic bacteria represents a serious threat to public health worldwide leading to a limited set, if any, of available treatment options [
1]. Making things worse, the antimicrobial resistance (AMR) has already outstepped hospitals and other healthcare institutions and become a significant matter in community settings [
2], so that AMR infections has become one of the top causes of death worldwide [
3].
The spread of AMR within particular bacterial species can be driven by several lineages usually called ‘global clones’ or ‘international clones of high risk’, which was shown to be the case for the most successful and widespread pathogens like
Klebsiella pneumoniae [
4] and
Acinetobacter baumannii [
5]. Thus, in order to perform the epidemiological surveillance and develop effective prevention measures against multidrug-resistant (MDR) bacteria it is essential to track the spread of such global clones and to check whether particular isolates belong to these lineages. Simply stated, you should know your enemy before you could fight it.
Isolate classification and assignment to a particular clone can be based on several characteristics revealed using molecular biology techniques, but currently the whole genome sequencing is increasingly used for this and many other purposes due to unprecedented amount of information it produces and its high cost-effectiveness [
6,
7].
Now, tens of thousands genomes of pathogenic bacteria are available in public databases, and this amount will continue to increase. The representation in genomic databases is interrelated with the level of concern raised by a particular pathogen and the incidence of infections caused by it.
A. baumannii is responsible for a significant share of nosocomial infections worldwide [
8], and the World Health Organization (WHO) listed its isolates resistant to antibiotics from carbapenem class as one of the most critical pathogens with the highest priority in new antibiotic development [
9].
Currently, more than 17,000 draft
A. baumannii genomes are available at NCBI (
https://www.ncbi.nlm.nih.gov/datasets/taxonomy/470/, accessed on 23 October 2023), but the data provided there usually lacks isolate typing information, which should be derived using various computational tools by users. Another commonly used database PubMLST [
10] contains epidemiologically related and typing information for more than 8,700 isolates, but the number of available genomes is about 3,300 (
https://pubmlst.org/organisms/acinetobacter-baumannii, accessed on 23 October 2023). However, as we described earlier, an assignment of a particular isolate to some international clone is not always straightforward even when whole genome data is available [
11], and, to the best of our knowledge, no public database currently presents such assignment for large isolate datasets.
Here we present the dataset containing typing information, including assignment to international clones, for the whole set of the A. baumannii isolates available at NCBI (accessed on 23 February 2023), which included 17,546 genomes. The information available contains multilocus sequence typing (MLST)-based types (STs), intrinsic blaOXA-51-like gene variants, capsular (KL) and oligosaccharide (OCL) types, core genome MLST (cgMLST) profiles, and the data regarding the presence of CRISPR-Cas systems in each of the isolates. The data regarding the presence of AMR genes providing resistance to various classes of antibiotics is also available.
We already used this dataset to study the representation of known international clones in Genbank [
11]. Deriving of all this information from the available genomes requires advanced level of bioinformatics expertise and significant computational resources. As it was noticed recently, the proper global genomic epidemiology investigations of
A. baumannii are very important for understanding global dissemination of important clones [
12]. We believe that the dataset will be useful for epidemiological studies of
A. baumannii, including facilitation of selection of the proper reference isolate sets for any types of genome-based investigations.
This precomputed data will be especially helpful for the researchers starting their investigations in the emerging field of genomic epidemiology concerning this important and dangerous pathogen.
The dataset is available for academic use under Creative Commons Attribution-Non Commercial-ShareAlike (CC BY-NC-SA) 4.0 International License. The updates are scheduled to be provided at least twice a year.
2. Data Description
2.1. Data Structure
The dataset contains three tables provided in various formats: xlsx, txt (tab-delimited) and pdf (summary table only). When using xlsx format in table processing software, users can create their own filters to select the subsets of interest, change or add sorting parameters, build graphs etc. Text format (txt) is intended for computational processing using bioinformatics tools, while pdf format is provided for summary file containing main isolate typing results to present them in human-readable form.
The tables, which will be described below in more details, include:
- summary table, which contains typing information for all isolates, such as MLST Pasteur ST, OXA-51 like variant, capsule synthesis loci (KL) and lipooligosaccharide outer core loci (OCL)-based types, assignments of an isolate to known international clones of high risk (IC1-IC9) and the possible presence and type of CRISPR-Cas system in the genome of an isolate
- AMR gene table containing the information on the presence of genes known to provide, when properly expressed, the AMR to various classes of antimicrobial drugs for particular isolate
- the table containing cgMLST profiles for all isolates, which can be used for extended comparison and clustering purposes
2.1.1. Typing summary table
The format and exemplary data for typing summary table is given in
Table 1.
First column contains the assembly code assigned by Genbank, which uniquely identifies a particular A. baumannii genome assembly.
‘IC’ stands for ‘international clone of high risk’ and shows the assignment of a particular isolate to know international clones. The procedure for such an assignment is described in the Methods section and our previous publication [
11]. If an isolate was not assigned to any IC, this column contained ‘NOIC’ designation.
Third column contains a sequence type (ST) defined as a combination of 7 loci (cpn60, fusA, gltA, pyrG, recA, rplB and rpoB genes) typing scheme known as ‘Pasteur MLST’ [
13]. Each variant of a particular locus is numbered, and the combination of 7 loci numbers constitutes a ST, which has its own number. For example, the combination cpn60_3, fusA_29, gltA_30, pyrG_1, recA_9, rplB_1 and rpoB_4 was defined as ST17. Loci variants and the definitions of corresponding STs can be found in PubMLST database (
https://pubmlst.org/organisms/acinetobacter-baumannii). ‘ND’ in this column means that ST was not determined due to either low sequencing quality or the presence of a novel MLST allele not uploaded to the databases yet.
Fourth column shows the variant of a gene encoding intrinsic OXA-51-like β-lactamase, which is possessed by all A. baumannii isolates and was sometimes used for typing purposes [
14]. ‘NOT_FOUND’ can appear in this column due to low sequencing quality or low similarity with known OXA-51-like variants.
KL- and OCL-type show the typing classes based on the corresponding sets of genes, respectively. Capsular polysaccharide is an essential factor determining bacterial virulence and its susceptibility to phages, which makes it useful epidemiological marker [
15]. Capsular polysaccharide gene cluster consists of about 30 genes, while OC locus includes only five. Each distinct gene cluster found between the flanking genes is assigned a unique number identifying the locus type, and these data can be found in public databases [
16].
Final column shows the presence of CRISPR-Cas system in the isolate. Clustered regularly interspaced short palindromic repeat (CRISPR) arrays and CRISPR-associated genes (cas) constitute bacterial adaptive immune systems and function as a variable genetic element. Each CRISPR-Cas locus includes a strain-specific array of so-called spacer sequences, which can be used for strain subtyping [
17]. CRISPR-Cas systems can be divided into six major types (I-VI) and several subtypes (A-I, K, U) based on a combination of evidence from phylogenetic, comparative genomic, and structural analysis [
18,
19]. ‘UNKN’ in this column denotes incomplete system, in which some genes are absent, while ‘NF’ means that CRISPR-Cas system was not revealed in a particular isolate genome.
2.1.2. AMR gene table
Another part of the dataset includes the information regarding the presence of various genes known to provide antimicrobial resistance in the investigated A. baumannii genomes. First column holds the assembly code, which is the same as in typing summary table, second column shows the number of AMR genes found in a particular isolate, while other columns exhibit the presence of a particular gene given in a first row and its sequence similarity level with the corresponding allele from the database. The absence of a gene is marked with a dot for better readability purposes.
An example is provided in
Table 2. Only a small part of available genes is presented in this example.
The presence of a particular gene by itself does not confirm the resistance to the corresponding antimicrobial drug since this gene, for example, might not be expressed [
20,
21]. However, this information is very useful for estimation of AMR potential within bacterial population.
2.1.3. cgMLST profiles
Third part of the dataset includes cgMLST profiles for all isolates. The cgMLST typing scheme is similar to MLST in that it enumerates gene variants and uses their combination to form a profile, but the difference is that cgMLST relies on all conservative genes within particular species. Such a scheme for A. baumannii was proposed in 2017 [
22] and included 2390 loci. The allele variants for these loci are available in regularly updated public database at cgmlst.org (
https://www.cgmlst.org/ncs/schema/schema/3956907).
cgMLST profiles can be used in cluster analysis of a set of isolates in order to estimate their genomic closeness and, possibly, obtain some evolutionary or epidemiologically valuable insights. The threshold of 3 different cgMLST loci was proposed to check whether two A. baumannii isolates belonged to a single strain [
23], but less stringent criterion can be used depending on specific investigation case.
In this dataset, cgMLST profiles are given in a table format. First column contains the same assembly identifier as in other dataset tables, and other columns show the numbers representing the variants of genes given in a header row. Some special characters can appear besides numbers, namely, N - indicates novel allele variant not present in the database; 0? - locus is missing in assembly (probably, due to low sequencing quality); "-" - allele is partially covered; “+” represents multiple possible alleles, in which case the most probable is shown.
2.2. General Statistics
Some general statistics based on the dataset is provided below. As it was noticed previously [
11,
24], Genbank set of genomes cannot be considered representative for the whole A. baumannii population since it is strongly biased towards multidrug-resistant or other clinically relevant isolates. At the same time, the statistics on representation of particular STs, ICs, AMR genes etc. can provide useful information for reference set selection and comparison purposes.
We will refer to any Genbank assembly record containing complete or partial genome as ‘‘isolate” for simplicity, although some different assemblies could represent the same isolate, or some record could contain only a part of the isolate genome.
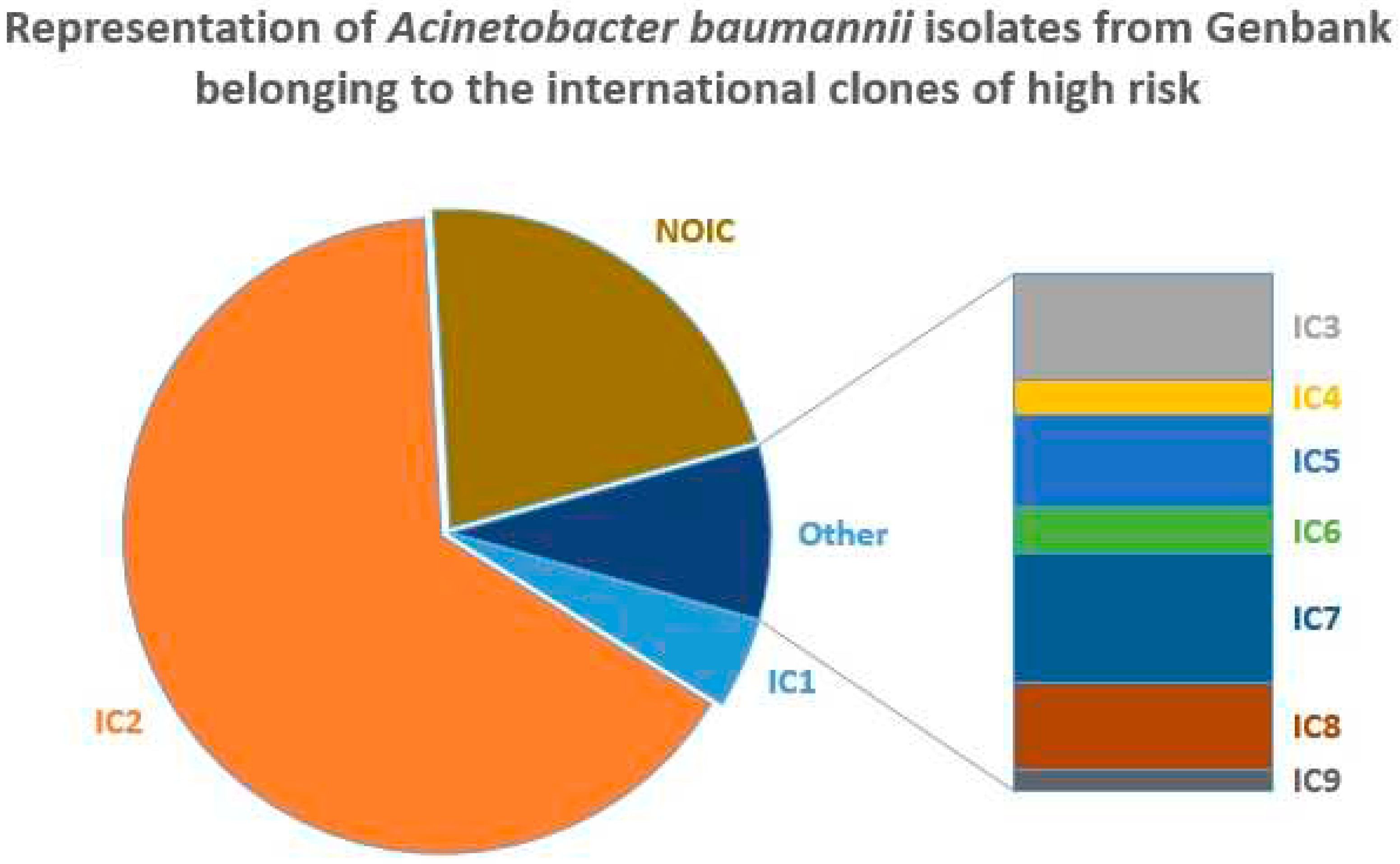
The distribution of ICs in Genbank is shown in
Figure 1.
The summary of top three dominating characteristics in each category is given in
Table 3.
In total, 78.5% of the isolates belonged to IC1-IC9, with IC2 accounting for 65% of all Genbank genomes and 83% of A. baumannii isolates belonging to ICs. The second largest IC - IC1 - was revealed in about 3.6%/4.5% of all isolates and isolates belonging to ICs, respectively. These results are typical since IC2 is known to be the most successful and widespread clone of A. baumannii being responsible for the majority of outbreaks worldwide [
25]. The dominating ST was, not surprisingly, ST2, which constituted the most part of IC2 and was revealed in 63.3% (11108) of cases. ST1 (3.59%) and ST499 (3.4%) also were in the top three. The number of distinct STs revealed was 482. However, 398 of them were presented by 10 or less isolates each, with 236 STs featured only a single isolate.
The dominating OXA-51-like variant was OXA-66 (about 60%) associated with IC2, followed by OXA-82 (also associated with IC2) and OXA-69 (IC1), each accounting for ten times less isolates than the former. In this case, the statistics is nearly full since the genes encoding OXA-51-like beta-lactamases were revealed in 98% of the isolates due to low similarity caused by bad sequencing quality or low coverage of particular genomic regions. The data on intrinsic beta-lactamases clearly correlates with IC and ST distribution, which was as expected [
11,
14]. Totally, 130 distinct OXA-51-like variants were revealed.
Top three KL types included KL2 (15.5% of the isolates), KL3 (11.6%) and KL22 (5.4%). KL2 was recently reported to be the most common type in A. baumannii and was associated with increased AMR [
26], which conforms with the previously noted bias of Genbank A. baumannii isolates towards more resistant ones. In total, 229 types were present.
The diversity of OCL types was lower with OCL1 accounting for 70% of the isolates. OCL3 and OCL6 were found in about 11% and 5%, respectively. In total, 22 types were revealed. These results corresponds to the previous study, in which OCL1 was the most common type among A. baumannii isolates belonging to ST1, ST2, ST3 and ST78, which together constituted about 71% of the isolates available in Genbank [
27].
The median number of AMR genes possessed by the isolates was 10, with the number ranging from 1 to 25. The number was never equal to zero since intrinsic blaOXA-51-like and blaADC genes were also included. In several cases, intrinsic genes were not revealed due to low similarity or insufficient genome region coverage. We revealed 1236 (7.0%) isolates possessing only intrinsic genes denoted above.
The most abundant gene except intrinsic ones were bla
OXA-23, aph(6)-Id and aph(3'')-Ib revealed in about 64% of the isolates each. First gene, encoding OXA-23 carbapenemase, was shown to be associated with IC2 [
28], and our study confirmed that it was found in about 95% of IC2 isolates. The latter two genes, usually being of plasmid origin [
29], encode aminoglycoside phosphotransferases providing AMR to streptomycin.
CRISPR-Cas systems were revealed in about 8% (1342) of the isolates. The dominating type in this case was I-F, with I-F1 subtype accounting for 67% and I-F2 – for about 20% of the CRISPR-Cas positive isolates, which corresponds to previous RefSeq analysis, in which it was found that most CRISPR-Cas systems in A. baumannii belonged to the types I-F1 and I-F2 [
30]. The systems were predominantly found in IC1 (39%), NOIC (29%) and IC7 (17%) groups, and were not revealed in IC2, which also conforms with previous investigations [
30]. The relatively low fraction of the isolates in Genbank containing apparently functional CRISPR-Cas system could also be attributed to overrepresentation of IC2, which usually does not possess such a system.
3. Methods
We retrieved 17,546 genomic sequences of
A. baumannii from Genbank (
https://www.ncbi.nlm.nih.gov/genbank/, accessed on 14 January 2023), for which the assembly level was defined as ‘Complete Genome’, ‘Chromosome’, or ‘Scaffold’.
Multilocus sequence typing (MLST) was performed using the PubMLST database (
https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_seqdef, accessed 14 February 2023) using Pasteur [
13] typing scheme. When we tried to get STs based on Oxford scheme [
31], the reliable assignment was obtained for only about 23% of the isolates due to known issue of
gdhB gene paralogy and technical artifacts [
32]. It was also shown that the Pasteur scheme is more appropriate for population biology and epidemiological studies of
A. baumannii than the Oxford one since it can be used for more precise isolate classification in clonal groups [
33]. For these reasons, the data for Oxford STs are not presented in our dataset.
The detection of capsule synthesis loci (KL) and lipooligosaccharide outer core loci (OCL) was made using Kaptive v. 2.0.3 [
34] with default parameters (last update of the databases on 13 February 2023).
The presence of CRISPR-Cas systems in the genomes analyzed was investigated using CRISPRCasFinder [
35] version 4.2.20 with the following parameters: ‘-fast -rcfowce -ccvRep -vicinity 1200 –cas -useProkka’. Recent classifications based on the multiparametric analysis reported type I loci with
cas3 as a signature gene and type I-F with fused
cas3 and
cas2 genes [
36,
37]. Types I-F1 and I-F2 both contain
cas1 and
cas2-3 gene, but the rest part of the loci is different and includes four genes
csy1 (
cas8f1),
csy2 (
cas5f1),
csy3 (
cas7f1),
csy4 (
cas6f) in I-F1 and three genes
cas5fv (
cas5f2),
cas6f, and
cas7fv (
cas7f2) in I-F2 [
30].
Additional data processing and output formatting was performed using computational pipeline developed earlier by us [
39,
40].
Assignment of the isolates to known international clones of high risk (IC) was based on Pasteur MLST ST and
blaOXA-51-like gene variant supported by cgMLST profile when needed, as described earlier by us [
11]. In order to make the classification reliable, we performed cluster analysis of STs using currently available data and compared it with the information derived from literature analysis of experimentally verified IC assignments.
4. User Notes
The regularly updated description of possible dataset applications to genomic epidemiology investigations, including detailed scripting commands for UNIX-based systems, will be provided on the dataset webpage in the file how_to_use.txt.
Funding
This work was supported by the Ministry of Science and Higher Education of the Russian Federation within the framework of a grant in the form of a subsidy for the creation and development of the «World-class Genomic Research Center for Ensuring Biological Safety and Technological Independence under the Federal Scientific and Technical Program for the Development of Genetic Technologies», agreement No. 075-15-2019-1666.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Salam:, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. , et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Shaidullina, E.R.; Schwabe, M.; Rohde, T.; Shapovalova, V.V.; Dyachkova, M.S.; Matsvay, A.D.; Savochkina, Y.A.; Shelenkov, A.A.; Mikhaylova, Y.V.; Sydow, K. , et al. Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395. Genome Med. 2023, 15, 9. [Google Scholar] [CrossRef]
- Zhang, X.; Li, F.; Awan, F.; Jiang, H.; Zeng, Z.; Lv, W. Molecular Epidemiology and Clone Transmission of Carbapenem-Resistant Acinetobacter baumannii in ICU Rooms. Front. Cell. Infect. Microbiol. 2021, 11, 633817. [Google Scholar] [CrossRef]
- Rafei, R.; Osman, M.; Dabboussi, F.; Hamze, M. Update on the epidemiological typing methods for Acinetobacter baumannii. Future Microbiol. 2019, 14, 1065–1080. [Google Scholar] [CrossRef]
- Atxaerandio-Landa, A.; Arrieta-Gisasola, A.; Laorden, L.; Bikandi, J.; Garaizar, J.; Martinez-Malaxetxebarria, I.; Martinez-Ballesteros, I. A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data. Microorganisms 2022, 10, 2364. [Google Scholar] [CrossRef]
- Castanheira, M.; Mendes, R.E.; Gales, A.C. Global Epidemiology and Mechanisms of Resistance of Acinetobacter baumannii-calcoaceticus Complex. Clin. Infect. Dis. 2023, 76, S166–S178. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. , et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Shelenkov, A.; Akimkin, V.; Mikhaylova, Y. International Clones of High Risk of Acinetobacter Baumannii-Definitions, History, Properties and Perspectives. Microorganisms 2023, 11, 2115. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Ramirez, S. Genomic epidemiology of Acinetobacter baumannii goes global. mBio 2023, 14, e0252023. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 2010, 5, e10034. [Google Scholar] [CrossRef] [PubMed]
- Pournaras, S.; Gogou, V.; Giannouli, M.; Dimitroulia, E.; Dafopoulou, K.; Tsakris, A.; Zarrilli, R. Single-locus-sequence-based typing of blaOXA-51-like genes for rapid assignment of Acinetobacter baumannii clinical isolates to international clonal lineages. J. Clin. Microbiol. 2014, 52, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Arbatsky, N.P.; Kasimova, A.A.; Shashkov, A.S.; Shneider, M.M.; Popova, A.V.; Shagin, D.A.; Shelenkov, A.A.; Mikhailova, Y.V.; Yanushevich, Y.G.; Azizov, I.S. , et al. Structure of the K128 capsular polysaccharide produced by Acinetobacter baumannii KZ-1093 from Kazakhstan. Carbohydr. Res. 2019, 485, 107814. [Google Scholar] [CrossRef] [PubMed]
- Cahill, S.M.; Hall, R.M.; Kenyon, J.J. An update to the database for Acinetobacter baumannii capsular polysaccharide locus typing extends the extensive and diverse repertoire of genes found at and outside the K locus. Microb. Genom. 2022, 8, 000878. [Google Scholar] [CrossRef]
- Tyumentseva, M.; Mikhaylova, Y.; Prelovskaya, A.; Tyumentsev, A.; Petrova, L.; Fomina, V.; Zamyatin, M.; Shelenkov, A.; Akimkin, V. Genomic and Phenotypic Analysis of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates Carrying Different Types of CRISPR/Cas Systems. Pathogens 2021, 10, 205. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Lipinski, D.; Zeyland, J. Evolution of CRISPR/Cas Systems for Precise Genome Editing. Int. J. Mol. Sci. 2023, 24, 14233. [Google Scholar] [CrossRef]
- Stasiak, M.; Mackiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef]
- Shelenkov, A.; Mikhaylova, Y.; Yanushevich, Y.; Samoilov, A.; Petrova, L.; Fomina, V.; Gusarov, V.; Zamyatin, M.; Shagin, D.; Akimkin, V. Molecular Typing, Characterization of Antimicrobial Resistance, Virulence Profiling and Analysis of Whole-Genome Sequence of Clinical Klebsiella pneumoniae Isolates. Antibiotics 2020, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Prior, K.; Harmsen, D.; Seifert, H. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS ONE 2017, 12, e0179228. [Google Scholar] [CrossRef] [PubMed]
- Schurch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef] [PubMed]
- Cain, A.K.; Hamidian, M. Portrait of a killer: Uncovering resistance mechanisms and global spread of Acinetobacter baumannii. PLoS Pathog. 2023, 19, e1011520. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Yang, C.J.; Chuang, Y.C.; Sheng, W.H.; Chen, Y.C.; Chang, S.C. Association of capsular polysaccharide locus 2 with prognosis of Acinetobacter baumannii bacteraemia. Emerg. Microbes Infect. 2022, 11, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Sorbello, B.M.; Cahill, S.M.; Kenyon, J.J. Identification of further variation at the lipooligosaccharide outer core locus in Acinetobacter baumannii genomes and extension of the OCL reference sequence database for Kaptive. Microb. Genom. 2023, 9, 001042. [Google Scholar] [CrossRef] [PubMed]
- Khuntayaporn, P.; Kanathum, P.; Houngsaitong, J.; Montakantikul, P.; Thirapanmethee, K.; Chomnawang, M.T. Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: a nationwide study. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 19. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Yadav, G.; Singh, R. In silico analysis reveals the co-existence of CRISPR-Cas type I-F1 and type I-F2 systems and its association with restricted phage invasion in Acinetobacter baumannii. Front. Microbiol. 2022, 13, 909886. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodriguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J.; Hall, R.M. Problems with the Oxford Multilocus Sequence Typing Scheme for Acinetobacter baumannii: Do Sequence Type 92 (ST92) and ST109 Exist? J. Clin. Microbiol. 2017, 55, 2287–2289. [Google Scholar] [CrossRef]
- Gaiarsa, S.; Batisti Biffignandi, G.; Esposito, E.P.; Castelli, M.; Jolley, K.A.; Brisse, S.; Sassera, D.; Zarrilli, R. Comparative Analysis of the Two Acinetobacter baumannii Multilocus Sequence Typing (MLST) Schemes. Front. Microbiol. 2019, 10, 930. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020, 6, e000339. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P. , et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Barth, Z.K.; Makarova, K.S.; Wolf, Y.I.; Brover, V.; Peters, J.E.; Koonin, E.V. Widespread CRISPR-derived RNA regulatory elements in CRISPR-Cas systems. Nucleic Acids Res. 2023, 51, 8150–8168. [Google Scholar] [CrossRef]
- Feijao, P.; Yao, H.T.; Fornika, D.; Gardy, J.; Hsiao, W.; Chauve, C.; Chindelevitch, L. MentaLiST—A fast MLST caller for large MLST schemes. Microb. Genom. 2018, 4, e000146. [Google Scholar] [CrossRef]
- Shelenkov, A.; Petrova, L.; Fomina, V.; Zamyatin, M.; Mikhaylova, Y.; Akimkin, V. Multidrug-Resistant Proteus mirabilis Strain with Cointegrate Plasmid. Microorganisms 2020, 8, 1775. [Google Scholar] [CrossRef]
- Shelenkov, A.; Petrova, L.; Zamyatin, M.; Mikhaylova, Y.; Akimkin, V. Diversity of International High-Risk Clones of Acinetobacter baumannii Revealed in a Russian Multidisciplinary Medical Center during 2017–2019. Antibiotics 2021, 10, 1009. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).